Abstract
The genome size, complexity, and ploidy of the dimorphic pathogenic fungus Histoplasma capsulatum was determined by using DNA renaturation kinetics, genomic reconstruction, and flow cytometry. Nuclear DNA was isolated from two strains, G186AS and Downs, and analyzed by renaturation kinetics and genomic reconstruction with three putative single-copy genes (calmodulin, α-tubulin, and β-tubulin). G186AS was found to have a genome of approximately 2.3 × 107 bp with less than 0.5% repetitive sequences. The Downs strain, however, was found to have a genome approximately 40% larger with more than 16 times more repetitive DNA. The Downs genome was determined to be 3.2 × 107 bp with approximately 8% repetitive DNA. To determine ploidy, the DNA mass per cell measured by flow cytometry was compared with the 1n genome estimate to yield a DNA index (DNA per cell/1n genome size). Strain G186AS was found to have a DNA index of 0.96, and Downs had a DNA index of 0.94, indicating that both strains are haploid. Genomic reconstruction and Southern blot data obtained with α- and β-tubulin probes indicated that some genetic duplication has occurred in the Downs strain, which may be aneuploid or partially diploid.
The dimorphic fungus Histoplasma capsulatum is the etiologic agent of histoplasmosis, a respiratory disease affecting an estimated 500,000 people per year in the United States (10). In the environment or in vitro at 25°C the organism grows in a multicellular mold form. In the infected host or in the laboratory at 37°C the fungus grows as a unicellular budding yeast. Because this mold-yeast conversion is reversible and easily accomplished in the laboratory by temperature shifts, H. capsulatum serves as an interesting model for cell differentiation in a lower eucaryote. More importantly, the mold-to-yeast conversion is required for pathogenesis (20).
Very little information regarding the genome of H. capsulatum is available. The nuclear guanine plus cytosine (G+C) content has been reported to be 45.4 to 49.8%, with an observed mean of 47.3% (2). Originally, isolates of H. capsulatum were separated into three classes based on restriction fragment length polymorphism (RFLP) analysis of the mitochondrial DNA (27). Subsequent studies with ribosomal DNA polymorphisms (24), yps-3 RFLP analysis (15), and arbitrary primer PCR (16) demonstrated that strains of H. capsulatum are actually quite diverse. Analysis of chromosome number by field-inversion gel electrophoresis and contour-clamped homogeneous electric field electrophoresis also showed major differences between strains. Steele et al. (25) showed that the Downs strain has at least seven chromosomes, G186B has at least four, and G217B has at least three. While these studies have yielded important information regarding the H. capsulatum genome, the fundamental features in terms of genome size, complexity, and ploidy have not been reported.
MATERIALS AND METHODS
Strains.
The H. capsulatum strain G186AS (a low-virulence derivative from G186B) was a kind gift from William Goldman (Washington University Medical School, St. Louis, Mo.). The H. capsulatum strains Downs (ATCC 38904) and G186B (ATCC 26030) and Escherichia coli K-12 (ATCC 23588) were purchased from the American Type Culture Collection (Manassas, Va.). Haploid and diploid strains of Saccharomyces cerevisiae were a kind gift from Joanne Tornow and George Santangelo (University of Southern Mississippi). Tetraploid strains of S. cerevisiae were obtained from the Yeast Genetic Stock Center (Berkeley, Calif.).
Media and growth conditions.
H. capsulatum yeast cells were grown to mid-log phase in GYE (2% glucose, 1% yeast extract; Difco Laboratories, Detroit, Mich.) at 37°C. S. cerevisiae cells were grown to mid-log phase in GYE broth at 30°C. E. coli was grown to mid-log phase in nutrient broth (Difco) at 37°C.
Single-copy genes.
Three genes previously reported to be present as single copy in H. capsulatum, calmodulin (6), α-tubulin, and β-tubulin (11), were selected for genomic reconstruction experiments. The H. capsulatum α-tubulin (GenBank M28358) and β-tubulin (GenBank L39132) clones were kindly provided by Grace Spatafora (Middlebury College, Middlebury, Vt.). The H. capsulatum calmodulin gene was isolated in our laboratory. Restriction fragments containing most of the amino acid-encoding sequences used as probes were as follows: CAM1 (GenBank AF072882), a 1-kb HindIII fragment containing exons 3 to 5; α-tub, a 900-bp BamHI/EcoRI fragment containing exons 5 and 6; and β-tub, a 1.1-kb BamHI fragment containing exons 5 to 7.
DNA extraction.
E. coli DNA was extracted and purified by standard molecular techniques (1). DNA was extracted from H. capsulatum G186AS by a modification of the technique of Worsham and Goldman (29). Briefly, yeast cells in mid-log phase were washed in phosphate-buffered saline (PBS; 4.3 mM Na2HPO4, 1.4 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl [pH 7.4]) and resuspended in spheroplasting buffer (1 M sorbitol, 100 mM sodium citrate, 60 mM disodium EDTA [pH 5.9]), and 120,000 U of β-glucuronidase (Sigma Chemical Co., St. Louis, Mo.) per ml was added. The cells were placed at 37°C with gentle shaking for 4 to 6 h. Cells were pelleted, washed three times with breakage buffer (0.9 M sorbitol, 10 mM EDTA, 0.1 M Tris-HCl [pH 7.5]), and lysed in lysis buffer (0.5 M Tris [pH 9.0], 20 mM NaCl, 0.2 M EDTA, 3% [wt/vol] sodium dodecyl sulfate [SDS]). The lysate was extracted with an equal volume of phenol saturated with 1 M Tris (pH 8.0) on a rotary shaker at 225 rpm for 2 h. The supernatant was mixed with an equal volume of Tris-saturated phenol:chloroform and shaken overnight at 25°C. The aqueous phase was extracted with chloroform, and the DNA was ethanol precipitated. The DNA pellet was washed with 70% ethanol and dissolved in 10 mM Tris-HCl (pH 7.5)–20 mM NaCl–1 mM EDTA. Because the Downs strain was recalcitrant to the β-glucuronidase procedure, cells were broken by physical means. Yeast cells were grown to mid-log phase, pelleted, washed with PBS, and resuspended in breakage buffer at 4°C. The cells were mechanically disrupted in a Bead Beater (Biospec Products, Bartlesville, Okla.) by shaking with 0.5-mm-diameter acid-washed glass beads for 3 min at 4°C. The homogenate was mixed with 40 ml of lysis buffer and an equal volume of Tris-saturated phenol and was gently shaken at 25°C for 35 min. The homogenate was centrifuged at 3,000 × g for 20 min at 25°C, and the aqueous-phase supernatant was extracted with an equal volume of Tris-saturated phenol:chloroform (1:1) followed by a chloroform extraction. The DNA was ethanol precipitated as described above. DNA from both H. capsulatum strains was purified by ultracentrifugation through successive cesium chloride-bisbenzimide gradients (12). The nuclear DNA band was carefully recovered from each gradient and repurified until no mitochondrial DNA was visible by fluorescence. This typically required three to five sequential CsCl gradients. Bisbenzimide was removed by several extractions with 20× SSC-saturated isopropanol (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate [pH 7.0]). Before analysis by reassociation kinetics, the DNA was ethanol precipitated twice to eliminate carryover of cesium chloride. Examination of the DNA by agarose gel electrophoresis showed high-molecular-weight DNA (greater than 23 kb) with no detectable rRNA or tRNA contamination, even in overloaded lanes.
DNA radiolabeling.
DNA was radiolabeled with [α-32P]dATP or α-35S-dATP by the random primer method with a Deca Prime labeling kit (Ambion, Austin, Tex.) according to the manufacturer’s directions.
Reassociation kinetics.
Purified DNA from E. coli and H. capsulatum was diluted to 0.1 mg per ml in sterile TE (10 mM Tris [pH 7.4], 1 mM EDTA) and sonicated to yield fragments between 200 and 2,000 bp in length. The DNA was ethanol precipitated and redissolved in 2× SSC–1 mM EDTA (pH 7.0) to a concentration of 0.25 μg/μl. The DNA was aliquoted in 10-μl amounts (2.5 μg), overlayed with sterile mineral oil, denatured by boiling, and placed at a reassociation temperature of 72°C. After reassociation to the desired EC0t (equivalent C0t corrected for nonstandard salt concentration [4]), the aliquots were quick frozen in a dry ice-ethanol bath. After thawing of the aliquots, single-stranded DNA (ssDNA) was digested with S1 nuclease (2 U per 100 ng of DNA) at 37°C for 45 min. The reaction was stopped with 20 μl of termination buffer (1 M Tris [pH 9.0]–0.1 M EDTA). The renatured DNA was quantitated by diluting a 50-μl aliquot of each reaction mixture to 1-ml volume with detection buffer (10 mM Tris [pH 7.4], 1 mM EDTA, 0.1 M NaCl, 0.1 μg [per ml] bisbenzimide) followed by fluorescence measurement (28) in a Hitachi F-2000 fluorescence spectrophotometer (with excitation at 356 nm and detection at 456 nm). Data were analyzed with a reassociation kinetics least-squares computer program kindly provided by William Pearson (21).
Minicot analysis.
Minicot curves are reassociation kinetic curves for an isolated fraction of DNA. A minicot uses the isolated fraction which has been radiolabeled as a tracer in a C0t curve with total unlabeled DNA. This proves especially useful in estimating the curve for a very small amount of repetitive DNA (4, 26). H. capsulatum DNA enriched for repetitive sequences was prepared by a modification of the method of Timberlake (26). A 5-μg aliquot of sonicated H. capsulatum DNA (0.25 μg/μl) in 1× SSC and 1 mM EDTA was overlayed with sterile mineral oil and denatured by boiling. The DNA was placed at a reassociation temperature of 52°C until reaching an EC0t of 1, after which the sample was quick frozen in a dry ice-ethanol bath. The ssDNA was digested with S1 nuclease as described above. After ethanol precipitation the renatured DNA was quantitated by bisbenzimide fluorescence as described above. This rapidly reassociating fraction was labeled with α-35S-dATP, dissolved in 1× SSC and 1 mM EDTA, and used as a tracer at 0.01% of the total DNA weight in a reassociation reaction of H. capsulatum nuclear DNA as described above. Denaturation, reassociation, and S1 nuclease treatment were the same as for the reassociation kinetic experiments described above. After S1 nuclease treatment, 200 μl was removed from under the oil, mixed with an ice-cold mixture of 0.2 M sodium pyrophosphate and 2 M HCl, and incubated on ice for 10 min. The precipitate was collected onto a Whatman GF/C glass microfiber filter. The filter was washed four times with 3 ml of a mixture of 0.1 M sodium pyrophosphate and 1 M HCl, washed one time with 3 ml of 95% ethanol, and dried overnight. The amount of radiolabeled double-stranded DNA (dsDNA) retained on the filter was determined by liquid scintillation counting.
Genomic reconstruction.
Genomic reconstruction experiments were done by a modification of the method of Francis et al. (8). DNA was quantitated by fluorescence spectrophotometry as described above. Serial twofold dilutions of H. capsulatum nuclear DNA were prepared to yield 400 to 12.5 ng in 200 μl of TE buffer. Herring sperm DNA was added as a carrier in increasing amounts so that the amount of total DNA (H. capsulatum DNA plus herring sperm DNA) was constant at 800 ng per 200 μl of aliquot. In a similar manner, DNA from single-copy H. capsulatum genes was prepared to yield 15 to 0.47 pg in 200 μl of TE buffer. Herring sperm DNA was added to yield 800 ng of total DNA per 200-μl aliquot. The final samples (200 μl) were mixed with 44 μl of a mixture of 2 M NaOH and 50 mM EDTA, boiled for 10 min, neutralized with an equal volume of 0.5 M ammonium acetate (pH 7.0), and vacuum filtered on a nitrocellulose membrane with a slot blot apparatus. The nitrocellulose filters were baked at 80°C for 2 h and prehybridized in a mixture of 0.5 M sodium phosphate (pH 7.0) and 5% (wt/vol) SDS for 1 h at 65°C. Approximately 20 ng of each single-copy gene was radiolabeled with 32P and used to probe the appropriate filter for 18 h at 65°C. Filters were washed two times for 15 min each time in 0.1× SSC–0.1% SDS at 65°C and exposed to X-ray film. Exposures were adjusted to be within the linear range of the film. Autoradiographic images were digitized with a Kohu high-resolution monochrome camera attached to a Macintosh Quadra computer, and hybridization intensities were quantitated by image analysis with the public domain NIH Image program (developed at the National Institutes of Health and available from the Internet by anonymous FTP from zippy.nimh.nih.gov or on floppy disk from the National Technical Information Service, Springfield, Va. [part no. PB95-500195GEI]).
Flow cytometry.
H. capsulatum yeast cells were pelleted by centrifugation at 800 × g for 10 min at 8°C, washed with sterile PBS, and fixed with 70% ethanol for 1 to 2 h at 25°C. The cells were sonicated gently to reduce clumping, filtered through a 20-μm-pore-size nylon mesh, washed with PBS, and resuspended in PBS to a concentration of 2 × 106 to 3 × 106 cells per ml. Thirty microliters of the cell suspension was treated with 150 μl of 1-mg/ml RNase A (Sigma Chemical Co.) at 37°C for 1 h. The cells were pelleted at 1,100 × g for 3 min and resuspended in 400 μl of 5-mg/ml pepsin (Sigma Chemical Co.). After 15 min at 25°C, the cells were pelleted (1,100 × g, 3 min) and resuspended in 1 ml of a modified Krishan hypotonic sodium citrate staining buffer containing 5-μg/ml propidium iodide, 4 mM sodium citrate, and 0.1% (vol/vol) Triton X-100 (5, 19). After a 1-h incubation at 25°C in the dark, the cells were pelleted and resuspended in 1 ml of PBS and analyzed in a Coulter EPICS-Profile II flow cytometer (Hialeah, Fla.) according to the manufacturer’s recommendations.
Southern blots.
Two-microgram aliquots of genomic DNA from H. capsulatum G186AS and Downs were digested to completion with several six-base-recognition restriction enzymes. The DNA was separated on a 0.7% agarose gel and downward blotted with 0.4 M NaOH onto a Hybond N+ positively charged nylon membrane (Amersham) according to the manufacturer’s directions. The blots were probed as described for the genomic reconstruction slot blots.
RESULTS
Reassociation kinetics.
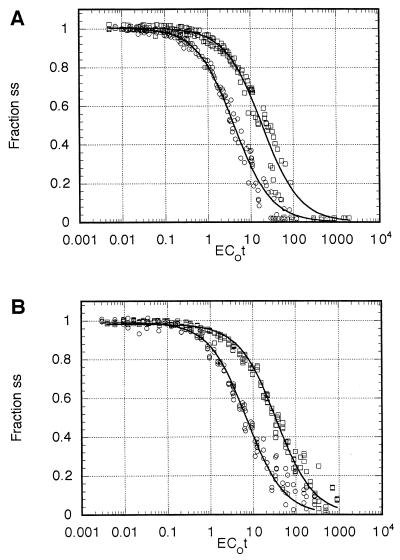
Three independent reassociation experiments, each with 30 EC0t values spanning the range from 0.01 to 1,000, were performed for G186AS and Downs DNA. Each experiment was done in tandem with E. coli DNA as a standard. The combined data from each H. capsulatum strain were analyzed by the nonlinear least-squares computer program of Pearson et al. (21). C0t1/2 values for H. capsulatum and E. coli were determined from the computer model best-fitted curve. Comparison of these values, using a genome size of 4.7 × 106 bp for E. coli (18), was used to estimate the genome size of H. capsulatum. The best-fitted curve for G186AS (root mean squares [RMS] of 0.046) was a single-component genome of 2.2 × 107 bp (Fig. 1A, Table 1), and the best-fitted curve for Downs (RMS of 0.028) was a two-component genome of 3.5 × 107 bp with 92% single-copy sequence and 8% moderately repetitive component (Fig. 1B, Table 1). The complexity of the repetitive component was 1.3 × 105 bp with a repetition frequency of 22.
FIG. 1.
Renaturation kinetics. Data shown represent the computer model best-fitted composite of three independent experiments for each curve for (A) G186AS DNA and (B) Downs DNA. Circles, E. coli DNA; squares, H. capsulatum DNA. ss, ssDNA.
TABLE 1.
Reassociation kinetics for nuclear DNA from H. capsulatum strains G186AS and Downsa
| Strain (component) | Fractionb | kc | C0t1/2d | kpuree | Copy no.f | Complexity (107 bp)g | Size (107 bp)h |
|---|---|---|---|---|---|---|---|
| G186AS | 1.0 | 0.0496 | 20.16 | 0.0478 | 1 | 2.18 | 2.18 |
| Downs (repetitive) | 0.083 | 0.587 | 1.703 | 7.042 | 22 | 0.0133 | 0.29 |
| Downs (single copy) | 0.917 | 0.265 | 37.74 | 0.0289 | 1 | 3.25 | 3.25 |
Combined data from three independent reassociation experiments.
Fraction of each component derived from computer modeling.
Reassociation rate expressed as M−1 sec−1.
Product of DNA concentration (nucleotide; moles) and time (s) at which the reassociation reached half completion.
Modified second-order rate constant for each component (C0t1/2pure = 1/kpure).
Copy number of repetitive fraction calculated by (C0t1/2pure/single-copy DNA fraction) divided by (C0t1/2pure/repetitive DNA fraction).
Total length of different DNA sequences present in each fraction.
Total size of each component (copy number × complexity).
Minicot analysis.
To increase the sensitivity of the analysis for repetitive DNA, renaturation experiments were performed in triplicate with the following modifications. The salt concentration was reduced to 1× SSC to slow the reassociation rate. A small amount (approximately 0.01% of the total DNA) of radiolabeled repetitive-enriched DNA was used as a tracer. Reassociation was quantitated by acid precipitation and scintillation counting, and the data were analyzed by computer modeling as described above.
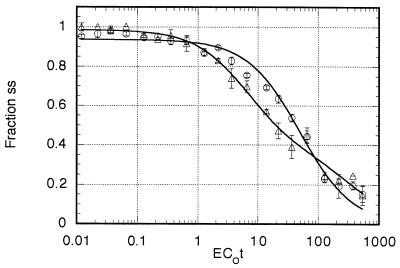
DNA from the Downs strain (Fig. 2) was best modeled as a two-component genome (RMS of 0.041) with the faster-reassociating component, representing 63.7% of the radiolabeled DNA, reassociating at a rate (k) of 0.18 M−1 sec−1. The remaining 36.3% reassociated at a rate of 0.007 M−1 sec−1. When enriching for the repetitive fraction, the amount of DNA renatured at an EC0t of 1, determined by S1 nuclease digestion of the ssDNA followed by fluorescence quantitation, was 1.9%. Therefore, the minicot repetitive fraction represents 1.21% of the total genome (0.019 × 0.637 × 100). However, at an EC0t of 1, only 15.2% of the repetitive component was renatured, as calculated (4) by the rate equation {100 × [1 − 1/1 + k EC0t)]} = {100 × [1 − 1/(1 + (0.18 × 1))]} = 15.2. Therefore, the actual representation of the repetitive component in the total genome was 8.0% [(0.0121/0.152) × 100]. This is in excellent agreement with the original renaturation analysis estimate of 8.3%. The copy number as determined from the ratio of the rate constants was 26, with a complexity of approximately 108,000 (3.5 × 107 × 0.08/26), in good agreement with the original determinations (Table 1).
FIG. 2.
Minicot renaturation kinetics. Data shown represent the computer model best-fitted curve. Circles, G186AS DNA; triangles, Downs DNA. Each error bar shows the standard deviation of triplicate measurements.
The best-fitted solution (RMS = 0.029) for G186AS was a two-component model. The faster-reassociating component represented 10.7% of the labeled DNA and reassociated at a rate (k) of 0.93 M−1 sec−1. The remaining 89.3% reassociated at a rate of 0.022 M−1 sec−1 and represented the single-copy component. The amount of renatured DNA at an EC0t of 1 in the tracer enrichment was 1.0%. Computer modeling showed that 10.7% of this dsDNA represented the repetitive component. Therefore, the minicot repetitive fraction represented 0.107% of the total genome (0.01 × 0.107 × 100 = 0.107%). However, at an EC0t of 1, 48.2% of the repetitive component was dsDNA {100 × [1 − 1/(1 + k EC0t)]} = {100 × [1 − 1/(1 + (0.93 × 1))]} = 48.2. Thus, the actual representation of the repetitive component in the genome was approximately 0.22% [(0.00107/0.482) × 100]. The copy number determined from the ratio of the rate constants was 42 with a complexity of approximately 1,000 (2.2 × 107 × 0.002/42). This small fraction of repetitive DNA is below the resolution limit of the original renaturation kinetic analysis (Table 1). Our confidence in the accuracy of the minicot method is about ±0.2%; therefore, our best estimate is that G186AS has less than 0.5% repetitive sequences.
Genomic reconstruction.
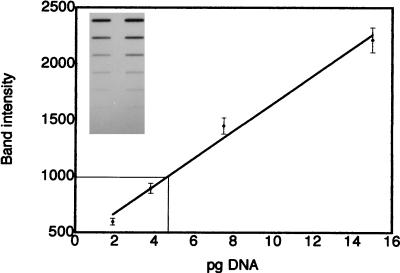
As an independent method of genome size estimation, genomic reconstruction experiments with three single-copy genes were performed. Carefully quantitated amounts of genomic DNA and cloned DNA from H. capsulatum single-copy genes were applied to a nitrocellulose membrane. The membrane was hybridized with the appropriate radiolabeled single-copy gene. Autoradiograms within the linear range of the film were digitized and analyzed by image analysis as shown in Fig. 3. A standard curve was calculated to correlate band intensity with amount (in picograms) of single-copy gene (Fig. 3). This standard curve was then used to quantitate the amount of the gene in several hundred nanograms of genomic DNA. With these data the amount of genomic DNA required to yield one copy of a single-copy gene (i.e., n or haploid genome size) was calculated. In the representative example shown in Fig. 3, 100 ng of genomic DNA was found to contain 4.7 pg of the 1-kb H. capsulatum calmodulin probe; thus, the calculated haploid genome size was 2.1 × 107 bp (0.0047 ng/100 ng = 1,000 bp/n). Results for each triplicate experiment are shown in Table 2. The genome size estimate for G186AS when calmodulin was used as the probe was 2.13 × 107 ± 0.09 × 107 bp, that when α-tubulin was used was 2.73 × 107 ± 0.51 × 107 bp, and that when β-tubulin was used was 2.35 × 107 ± 0.62 × 107 bp. Therefore, the average estimated genome size for G186AS based on nine independent genomic reconstruction analyses is 2.4 × 107 ± 0.3 × 107 bp.
FIG. 3.
Genomic reconstruction. Inset photo shows a representative autoradiograph of G186AS DNA and calmodulin probe. The lane on the left shows twofold serial dilutions of genomic DNA starting with 400 ng. The lane on the right shows twofold serial dilutions of H. capsulatum calmodulin probe starting with 15 pg. The nitrocellulose blot was probed with radiolabeled H. capsulatum calmodulin probe. The graph depicts a standard curve of relative band intensity plotted against the amount of calmodulin (inset, lane 2). Each bar shows the standard deviation of triplicate experiments. As an example, the thin line represents the intensity of the 100-ng genomic band and yields an estimate of 4.7 pg of calmodulin target per 100 ng of genomic DNA.
TABLE 2.
Genomic reconstructiona
| Strain | Single-copy probe
|
Overall avg (107 bp) | ||
|---|---|---|---|---|
| Calmodulin (107 bp) | α-Tubulin (107 bp) | β-Tubulin (107 bp) | ||
| G186AS | 2.13 ± 0.09 | 2.73 ± 0.51 | 2.35 ± 0.62 | 2.4 ± 0.3 |
| Downs | 2.99 ± 0.25 | 1.91 ± 0.29 | 2.04 ± 0.08 | 2.3 ± 0.59 |
Data shown are the averages of triplicate experiments ± standard deviation and represent the number of bp of nuclear DNA required to yield one copy of the single-copy gene probe.
The genome size estimate for Downs when calmodulin was used as the probe was 2.99 × 107 ± 0.25 × 107 bp, that when α-tubulin was used was 1.91 × 107 ± 0.29 × 107 bp, and that when β-tubulin was used was 2.04 × 107 ± 0.08 × 107 bp. Therefore, the overall estimated genome size for Downs, based on nine independent genomic reconstruction analyses, was 2.3 × 107 ± 0.6 × 107 bp. We were concerned that both the tubulin probes gave 1n genome estimates significantly lower than those given by the calmodulin probe, renaturation kinetics, or flow cytometry (see below). Additional replications with new preparations of DNA confirmed the precision of these measurements.
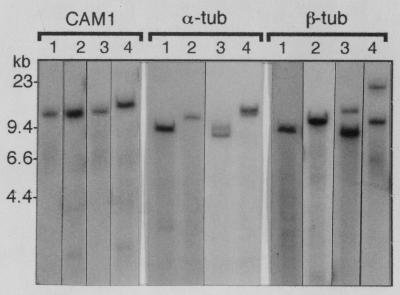
Since the published data indicating that α- and β-tubulin are single copy in H. capsulatum are based on strain G217B (11), the most simple explanation for the tubulin genomic reconstruction data in the Downs strain is that these genes are not present as single copies in Downs. Comparison of the kinetic haploid genome estimate with the α- and β-tubulin genomic reconstruction data is more consistent with two-copy genes (35 Mb/19 Mb = 1.8 for α-tub and 35 Mb/20 Mb = 1.75 for β-tub). To test this, we prepared Southern blots from genomic DNA cut to completion with several six-base-recognition restriction enzymes which either do not cut within the gene or cut only once within the gene but outside the probe sequence region. The blots were then probed with the same CAM1, α-tub, and β-tub probes. Figure 4 shows side-by-side comparisons of several representative lanes from G186AS and Downs blots. Complete digestion was confirmed in each blot by stripping the blot and reprobing with CAM1. Blots from both G186AS DNA and Downs DNA probed with CAM1 had a single band in each lane, as expected for a single-copy gene. In contrast, blots probed with α-tub and β-tub typically showed single bands in G186AS DNA and two bands in Downs DNA, consistent with a single-copy gene in G186AS and a two-copy gene in Downs. Based on this result we excluded the tubulin genomic reconstruction data from the calculations below for the Downs strain.
FIG. 4.
Southern blot of G186AS and Downs genomic DNA. Genomic DNA from strain G186AS and Downs was cut to completion with several six-base-recognition restriction enzymes, separated on a 0.7% agarose gel and transferred to a nylon membrane. The blot was probed with the radiolabeled fragments of CAM1, α-tub, and β-tub used for genomic reconstruction experiments. For CAM1, α-tub, and β-tub probes, lanes 1 and 2 contain G186AS DNA and lanes 3 and 4 contain Downs DNA. For CAM1, lanes 1 and 3, HpaI; lanes 2 and 4, SalI. For α-tub probe, lanes 1 and 3, EcoRI; lanes 2 and 4, HindIII. Although difficult to see on this composite figure, the band seen in lane 4 is a closely spaced doublet. For β-tub probe, lanes 1 and 3, HindIII; lanes 2 and 4, HpaI.
Flow cytometry.
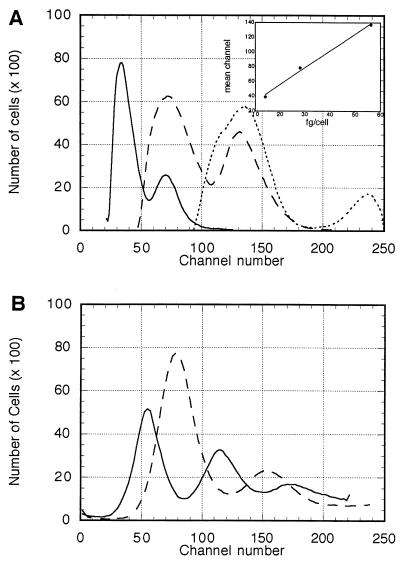
To determine the average mass of DNA per cell, H. capsulatum cells were analyzed by fluorescence flow cytometry. Channel number represents the intensity of fluorescence, which is directly proportional to the amount of DNA contained in each cell. The first peak in each graph represents the cells that are in the G0 and G1 phases of the cell cycle. The second peak, which appears at approximately twice the channel number of the G0-G1 peak, represents the cells that are in the G2 and M phases of the cell cycle. Haploid, diploid, and tetraploid strains of S. cerevisiae were analyzed by flow cytometry and used to construct a standard curve (r2 is >0.99). The theoretical amount of DNA in these S. cerevisiae strains was calculated from the recently completed sequence of the S. cereisiae genome (http://www-genome.stanford.edu). Based on a total sequence of 13,026,500 bp per haploid genome (12,057,500-bp unique sequence plus 969,000-bp repetitive sequence) and calculation of an average of 650 Da per bp of DNA, the haploid mass of DNA is 14.06 fg. The channel numbers of the G0-G1 peaks of 1n, 2n, and 4n strains of S. cerevisiae were plotted against the amount of DNA per cell that each strain contains (Fig. 5A). The channel mean of the G0-G1 peak of each Histoplasma strain (Fig. 5B) was then used to calculate the DNA content per Histoplasma cell. These data were then converted to base pairs using an average value of 650 Da/bp.
FIG. 5.
Flow cytometry results for (A) S. cerevisiae and (B) H. capsulatum G186AS (solid line) and Downs (dashed line). In panel A, the number of cells is plotted against channel number (fluorescence intensity) for S. cerevisiae. Solid line, 1n; dashed line, 2n; dotted line, 4n. Inset graph shows a typical standard curve relating channel number to theoretical nuclear DNA mass per cell.
Five independent experiments, each with a S. cerevisiae standard curve, were conducted. Examination of cell preparations by fluorescence microscopy showed strong nuclear staining with very low cellular background fluorescence in both S. cerevisiae and H. capsulatum (data not shown). The average amount of DNA per G186AS cell was 24 ± 1.8 fg or 2.2 × 107 ± 0.17 × 107 bp. The average amount of DNA per Downs cell was 32 ± 2.0 fg or 2.96 × 107 ± 0.2 × 107 bp.
Ploidy.
Ploidy can be determined by comparing haploid genome size with DNA content per cell to calculate the DNA index (DNA per cell per haploid genome size). A haploid organism should have a DNA index of approximately 1, a diploid organism should have an index of approximately 2, and so forth. To determine ploidy in H. capsulatum we compared the DNA content (flow cytometry data) with the average 1n genome estimate (reassociation kinetics and genomic reconstruction) to determine the DNA index.
The estimated genome size of G186AS as calculated by reassociation kinetic analysis was 22 Mb (Table 1), and that calculated by genomic reconstruction was 24 Mb (Table 2). The average size obtained by these two methods was 23 Mb. Analysis by flow cytometry estimated the amount of DNA per cell to be 22 Mb. The DNA index is 0.96 (22 Mb/23 Mb), indicating that H. capsulatum G186AS is haploid.
The 1n genome of Downs was estimated to be 35 Mb by reassociation kinetic analysis and 30 Mb by genomic reconstruction, for an average of approximately 32 Mb. Flow cytometry indicated a DNA content of 30 Mb; thus, the DNA index is 0.94 (30 Mb/32 Mb), indicating that the Downs strain is haploid.
Genome size.
Since both strains of H. capsulatum appear to be haploid we can directly include the flow cytometry 1n value in the overall genome size estimate. Our best estimate for the genome size derived from kinetic analysis (22 Mb), genomic reconstruction (24 Mb), and flow cytometry (22 Mb) in G186AS is 23 ± 1 Mb. We estimate the Downs genome size, based on the average by kinetic analysis (35 Mb), calmodulin genomic reconstruction (30 Mb), and flow cytometry (30 Mb), to be 32 ± 3 Mb.
DISCUSSION
Our goal in this work was to determine the basic features of the H. capsulatum genome regarding genome size, complexity, and ploidy. Because of the laborious nature of these experiments, most studies of this type have used only a single strain of the test organism. Considering the apparent diversity of H. capsulatum, we selected two strains for this analysis: the G186AS strain, because it is currently the most amenable to molecular manipulation, and the Downs strain, because a large body of dimorphism research has used this strain. These strains have no close association to each other aside from being clinical isolates. Downs was isolated in 1969 in Illinois (9), and the parent strain of G186AS (G186B) was isolated in Panama in 1967 (3).
Data from reassociation kinetic experiments with H. capsulatum G186AS DNA indicated a haploid genome size of 22 Mb (Fig. 1, Table 1) with less than 0.5% repetitive DNA (Fig. 2). This result is similar to that seen with Aspergillus nidulans DNA, which has a haploid genome size (based on kinetic analysis) of 2.6 × 107 bp and only slightly over 2% repetitive sequences (26). This similarity is not surprising since molecular phylogenetic analysis of several H. capsulatum genes isolated in our lab (6, 7, 23) indicate that the gene structure of H. capsulatum is remarkably similar to that of A. nidulans. The repetitive fraction in H. capsulatum most likely represents ribosomal DNA, as is the case for A. nidulans (26). Genomic reconstruction and flow cytometry gave average estimates of 24 Mb and 22 Mb, respectively, both of which closely accord with the renaturation kinetic estimates. The average estimate of haploid genome size by all three methods is 23 Mb. Data comparing haploid genome size and DNA content per cell clearly indicate that the organism is haploid (DNA index of 0.96).
The data from Downs DNA indicated that this strain has the larger genome and more repetitive DNA. Reassociation kinetic experiments indicated a haploid genome size of 35 Mb with 8.3% repetitive DNA (Fig. 1, Table 1). Minicot analysis (Fig. 2) indicated approximately 8% repetitive sequence, in close agreement with the primary reassociation data. The size derived from the calmodulin probe genomic reconstruction 1n estimate was 30 Mb. Flow cytometry gave an estimate of 30 Mb. The average estimate of haploid genome size by all three methods was 32 Mb. Data comparing haploid genome size and DNA content per cell indicate that Downs is also haploid (DNA index of 0.94).
The overall genome size estimates of G186AS (23 Mb) and Downs (32 Mb) were significantly larger than that of the ascomycetous yeast S. cerevisiae (13 Mb; see Materials and Methods) and similar to those of the filamentous ascomycetes such as A. nidulans (26 Mb [26]), Penicillium paxilli (23 Mb [13]), Podospora anserina (34 Mb [14]), and Neurospora crassa (43 to 45 Mb [22]). This result is consistent with our data indicating that H. capsulatum is more closely related to filamentous ascomycetes than to ascomycetous yeasts, as discussed above.
The difference in genome size in G186AS and Downs is certainly intriguing. Apparently Downs has a genome approximately 40% larger and with over 16 times more repetitive DNA than G186AS. What is this “extra” 9-Mb amount of genomic DNA in Downs? The repetitive DNA (approximately 2 to 3 Mb) is sufficient to account for only about one third of this amount. Steele et al. (25) showed that Downs apparently has more chromosomes (at least seven) than does G186B (at least four). A possible explanation is that chromosomal duplication has occurred in Downs. A single repeat of a few large regions of the chromosome(s) would be detected by neither renaturation kinetics nor genomic reconstruction unless a DNA probe within the duplicated region was fortuitously selected. Alternatively, it is possible that G186AS has lost 6 to 9 Mb of DNA and that the Downs genome more closely represents the “normal” H. capsulatum genome. It would be unlikely, however, that a haploid organism could lose such a large fraction of its genome with no apparent phenotypic effect. We have noted no major differences between the two strains in terms of their growth rates, nutritional requirements, or dimorphic transition during several years of work with these organisms. In addition, other empirical data indicate that Downs is a somewhat unusual strain, as discussed below.
The surprising data obtained with the tubulin probes in the Downs strain genomic reconstruction experiments also suggest some duplication of chromosomal DNA. The DNA index calculation indicates that Downs, like G186AS, is haploid, but the data obtained by genomic reconstruction and Southern blotting with the α- and β-tubulin probes indicate that these genes are present in two copies in Downs while G186AS (Table 2, Fig. 4) and G217B (11) have a single copy of each tubulin gene.
Since Downs is clearly not diploid it either (i) has extra copies of at least two chromosomes (since α- and β-tubulin probes hybridize to different bands on chromosome gels [25]) and is thus aneuploid or (ii) has intrachromosomal repeats in at least two chromosomes and is partially diploid. Since Downs has more chromosomes (25) and aneuploid strains are found in other filamentous fungi (17, 30), it is certainly tempting to favor the aneuploidy hypothesis. Either hypothesis is consistent with our data, however. We are currently conducting studies to test these hypotheses.
Our data support the concept that H. capsulatum strains are quite diverse in their genomic makeup. Studies that have examined more than one strain have shown that a number of fungi have a wide range of chromosome numbers and genome sizes in natural isolates (reviewed in reference 30). Fusarium oxysporum isolates, for example, have a genome size of 41 to 51 Mb with 11 to 14 chromosomes. We do not know if either of the two strains used in this study is more representative of H. capsulatum or if there is a single isolate that is most typical of the species. Some of our preliminary flow cytometry data (not shown) indicate that H. capsulatum G186B and G217B have the same DNA mass per cell as does G186AS. There are some data to indicate that Downs may be a somewhat unusual strain, however. Vincent et al. (27) grouped 23 H. capsulatum isolates into three classes based on mitochondrial RFLP analysis. Class 3 had 6 representatives, class 2 had 16 representatives, and Downs was the only member of class 1. More recently, Keath et al. examined 76 clinical and soil isolates and found most to belong to class 2 (15). Only four isolates, all from AIDS patients, were grouped in class 1 with the Downs strain, which is more heat sensitive and less virulent than most H. capsulatum isolates.
The work presented here provides data from two common laboratory stains of H. capsulatum. It would be interesting to characterize the genomes of several recent clinical and soil isolates to determine whether long-term laboratory culture has altered their chromosomal structure or if H. capsulatum is as variable in nature as lab strains apparently are. Nothing is known about chromosome number or genome size of H. capsulatum in wild isolates, and arguments can be made both for and against the survival value of a highly variable genome. As more strains are characterized, we will be able to determine the true genetic variability of this important human pathogen.
ACKNOWLEDGMENTS
We thank Elizabeth Keath for providing additional Downs strain DNA and George Santangelo for critical reading of the manuscript.
This work was supported in part by grants from the National Institutes of Health (AI31192) and the Mississippi Lung Association (RG-032-L).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1993. [Google Scholar]
- 2.Bawdon R E, Garrison R G, Fina L R. Deoxyribonucleic acid base composition of the yeastlike and mycelial phases of Histoplasma capsulatum and Blastomyces dermatitidis. J Bacteriol. 1972;111:593–596. doi: 10.1128/jb.111.2.593-596.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berliner M D. Primary subcultures of Histoplasma capsulatum: I. macro- and micromorphology of the mycelial phase. Sabouraudia. 1968;6:111–118. [PubMed] [Google Scholar]
- 4.Britten R J, Graham D E, Neufeld B R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- 5.Dressler L G, Seame R L C, Owens M A, Clark G M, McGuire W L. DNA flow cytometry and prognostic factors in 1331 frozen breast cancer specimens. Cancer. 1988;61:420–427. doi: 10.1002/1097-0142(19880201)61:3<420::aid-cncr2820610303>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 6.El-Rady J, Shearer G. Isolation and characterization of a calmodulin-encoding cDNA from the pathogenic fungus Histoplasma capsulatum. J Med Vet Mycol. 1996;34:163–169. doi: 10.1080/02681219680000271. [DOI] [PubMed] [Google Scholar]
- 7.El-Rady J, Shearer G. Cloning and analysis of an actin-encoding cDNA from the dimorphic pathogenic fungus Histoplasma capsulatum. J Med Vet Mycol. 1997;35:159–166. doi: 10.1080/02681219780001091. [DOI] [PubMed] [Google Scholar]
- 8.Francis D M, Hulbert S H, Michelmore R W. Genome size and complexity of the obligate fungal pathogen, Bremia lactucae. Exp Mycol. 1990;14:299–309. [Google Scholar]
- 9.Gass M, Kobayashi G S. Histoplasmosis. An illustrative case with unusual vaginal and joint involvement. Arch Dermatol. 1969;100:724–727. doi: 10.1001/archderm.100.6.724. [DOI] [PubMed] [Google Scholar]
- 10.Hammerman K J, Powell K E, Tosh F E. The incidence of hospitalized cases of systemic mycotic infections. Sabouraudia. 1974;12:33–45. doi: 10.1080/00362177485380061. [DOI] [PubMed] [Google Scholar]
- 11.Harris G S, Keath E J, Medoff J. Characterization of the alpha and beta tubulin genes in the dimorphic fungus Histoplasma capsulatum. J Gen Microbiol. 1989;135:1817–1832. doi: 10.1099/00221287-135-7-1817. [DOI] [PubMed] [Google Scholar]
- 12.Hudspeth M E S, Shumard D S, Tatti K M, Grossman L I. Rapid purification of yeast mitochondrial DNA in high yield. Biochim Biophys Acta. 1980;610:221–228. doi: 10.1016/0005-2787(80)90003-9. [DOI] [PubMed] [Google Scholar]
- 13.Itoh Y, Johnson R, Scott B. Integrative transformation of the mycotoxin-producing fungus Penicillium paxilli. Curr Genet. 1994;25:508–513. doi: 10.1007/BF00351670. [DOI] [PubMed] [Google Scholar]
- 14.Javerzat J P, Jacquier C, Barreau C. Assignment of linkage groups to the electrophoretically-separated chromosomes of the fungus Podospora anserina. Curr Genet. 1993;24:219–222. doi: 10.1007/BF00351795. [DOI] [PubMed] [Google Scholar]
- 15.Keath E J, Kobayashi G S, Medoff G. Typing of Histoplasma capsulatum by restriction fragment length polymorphisms in a nuclear gene. J Clin Microbiol. 1992;30:2104–2107. doi: 10.1128/jcm.30.8.2104-2107.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kersulyte D, Woods J P, Keath E J, Goldman W E, Berg D E. Diversity among clinical isolates of Histoplasma capsulatum detected by polymerase chain reaction with arbitrary primers. J Bacteriol. 1992;174:7075–7079. doi: 10.1128/jb.174.22.7075-7079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kistler H C, Benny U, Boehm W A, Katan T. Genetic duplication in Fusarium oxysporum. Curr Genet. 1995;28:173–176. doi: 10.1007/BF00315784. [DOI] [PubMed] [Google Scholar]
- 18.Kohara Y, Ariyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 19.Krishan A. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J Cell Biol. 1975;66:188–193. doi: 10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medoff G, Sacco M, Maresca B, Schlessinger D, Painter A, Kobayashi G S, Carratu L. Irreversible block of the mycelial to yeast phase transition of Histoplasma capsulatum. Science. 1986;231:476–479. doi: 10.1126/science.3001938. [DOI] [PubMed] [Google Scholar]
- 21.Pearson W R, Davidson E H, Britten R J. A program for least squares analysis of reassociation and hybridization data. Nucleic Acids Res. 1977;4:1727–1735. doi: 10.1093/nar/4.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radford A, Parish J H. The genome and genes of Neurospora crassa. Fungal Genet Biol. 1997;21:258–266. doi: 10.1006/fgbi.1997.0979. [DOI] [PubMed] [Google Scholar]
- 23.Shearer G. Cloning and analysis of cDNA encoding an elongation factor 1α from the dimorphic fungus Histoplasma capsulatum. Gene. 1995;161:119–123. doi: 10.1016/0378-1119(95)00269-c. [DOI] [PubMed] [Google Scholar]
- 24.Spitzer E D, Lasker B A, Travis S, Kobayashi G S, Medoff G. Use of mitochondrial and ribosomal DNA polymorphisms to classify clinical and soil isolates of Histoplasma capsulatum. Infect Immun. 1989;57:1409–1412. doi: 10.1128/iai.57.5.1409-1412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steele P E, Carle G F, Kobayashi G S, Medoff G. Electrophoretic analysis of Histoplasma capsulatum chromosomal DNA. Mol Cell Biol. 1989;9:983–987. doi: 10.1128/mcb.9.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timberlake W E. Low repetitive DNA content in Aspergillus nidulans. Science. 1978;202:973–975. doi: 10.1126/science.362530. [DOI] [PubMed] [Google Scholar]
- 27.Vincent R D, Goewert R, Goldman W E, Kobayashi G S, Lambowitz A M, Medoff G. Classification of Histoplasma capsulatum isolates by restriction fragment polymorphisms. J Bacteriol. 1986;165:813–818. doi: 10.1128/jb.165.3.813-818.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vytasek R. A sensitive fluorometric assay for the determination of DNA. Anal Biochem. 1982;120:243–248. doi: 10.1016/0003-2697(82)90342-6. [DOI] [PubMed] [Google Scholar]
- 29.Worsham P L, Goldman W E. Development of a genetic transformation system for Histoplasma capsulatum: complementation of uracil auxotrophy. Mol Gen Genet. 1990;221:358–362. doi: 10.1007/BF00259400. [DOI] [PubMed] [Google Scholar]
- 30.Zolan M E. Chromosome-length polymorphism in fungi. Microbiol Rev. 1995;59:686–698. doi: 10.1128/mr.59.4.686-698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]