Abstract
Novel screening and diagnostic tests based on artificial intelligence (AI) image recognition algorithms are proliferating. Some initial reports claim outstanding accuracy followed by disappointing lack of confirmation, including our own early work on cervical screening. This is a presentation of lessons learned, organized as a conceptual step-by-step approach to bridge the gap between the creation of an AI algorithm and clinical efficacy. The first fundamental principle is specifying rigorously what the algorithm is designed to identify and what the test is intended to measure (eg, screening, diagnostic, or prognostic). Second, designing the AI algorithm to minimize the most clinically important errors. For example, many equivocal cervical images cannot yet be labeled because the borderline between cases and controls is blurred. To avoid a misclassified case-control dichotomy, we have isolated the equivocal cases and formally included an intermediate, indeterminate class (severity order of classes: case>indeterminate>control). The third principle is evaluating AI algorithms like any other test, using clinical epidemiologic criteria. Repeatability of the algorithm at the borderline, for indeterminate images, has proven extremely informative. Distinguishing between internal and external validation is also essential. Linking the AI algorithm results to clinical risk estimation is the fourth principle. Absolute risk (not relative) is the critical metric for translating a test result into clinical use. Finally, generating risk-based guidelines for clinical use that match local resources and priorities is the last principle in our approach. We are particularly interested in applications to lower-resource settings to address health disparities. We note that similar principles apply to other domains of AI-based image analysis for medical diagnostic testing.
Recent advances in artificial intelligence (AI)–based image recognition technology have led to many potential AI-based clinical tests. However, numerous initially promising applications have not proven ultimately to be useful, including our group’s early success in AI-based analysis of cervical images for cervical screening (1-3). Several years of interdisciplinary research and redesign have subsequently revealed some important lessons. We have developed a step-by-step approach for design and evaluation of AI (deep learning) algorithms that fit into a risk-based screening strategy, based on the principle of “equal management of individuals at equal risk” (4). This strategy can be tailored to individual settings including low-resource ones, taking into consideration their unique risk tolerance and treatment capacity.
We are focusing on visual images, but the same principles can also be applied to microscopic images (5). Although our focus in this paper centers around cervical cancer screening, other work on radiologic diagnostics (6,7) and retinal analyses (8-10) indicates that the approach presented here has the potential for application in other fields as well.
Background: importance of cervical cancer prevention and designing a screening program in low-resource settings
For those not familiar with cervical screening and diagnosis, we briefly provide the background necessary to understand this example. Persistent cervical infection with carcinogenic genotypes of human papillomavirus (HPV) is the main cause of cervical cancer, and the causal pathway from sequential states of infection to precancer to invasion is well understood (11). There are prevention and treatment strategies, nevertheless, every year more than 300 000 women lose their lives because of this disease. Almost 90% of these losses occur in resource-limited countries. To address this worsening inequity, there is a critical need for streamlined vaccination efforts and the development of cost-effective and easily accessible, accurate, screening methods. Regarding screening, diagnosing and treating cervical precancer is proven to prevent invasive cancer. Currently, the World Health Organization recommends testing for high-risk HPV types as the primary screening method, ideally from a self-collected cervical–vaginal swab sample to permit more rapid population screening (12). HPV is common and usually benign at the early phases of the infection. Therefore, in high-resource settings, an additional triage test is used to further stratify the risk of cervical precancer or cancer (will be denoted as precancer/cancer) among HPV-positive individuals (ie, screen-triage-treat strategy) thus reducing overtreatment. In lower-resource settings as well, HPV screening is recommended if practical. A screen-triage-treat strategy may be preferable to treating all HPV-positive individuals when HPV prevalence is very high, treatment availability is limited, and/or there is a desire to balance test sensitivity with minimal unnecessary treatment. Progress in creating affordable HPV testing is detailed elsewhere (13,14). Currently, there is no triage test adequate for resource-limited settings that is rapid, simple, and cost-effective and does not require laboratory and pathology capabilities.
AI-assisted visual evaluation could help triage HPV-positive individuals. Magnified and highly illuminated visual evaluation of the cervix by clinicians using colposcopy is considered the standard of care in most high-resource settings, primarily to rule out cancer and to determine which areas of the cervix to biopsy. In resource-limited settings, a simpler unmagnified visual inspection with acetic acid is frequently used for triage to assess the need for treatment and eligibility for a simple thermal ablation procedure, rather than excision. However, visual assessment exhibits notable limitations in terms of intra- and interrater repeatability, even when magnified and performed by experts (12). Nonetheless, if standardized and made more reliable with the assistance of an AI algorithm, visual evaluation of the cervix could help fulfill the need for a rapid and inexpensive triage test.
We have successfully developed a deep learning–based method (a revised version of automated visual evaluation) to help recognize the precancerous changes among HPV-positive individuals who require treatment to prevent invasive disease (15). A large-scale validation initiative is underway. The main lessons learned to date are detailed below.
Developing an AI-based visual classification that fits into a risk-based screening program
Step 1: Defining the purpose of an AI-based medical test like automated visual evaluation
Medical tests, whether for screening, diagnosis, or prognosis, are used to measure where an individual stands along the pathway from healthy to increasingly severe disease and/or death. Screening tests find abnormalities within the generally healthy population. Among cases (case is defined as an outcome of a particular disease or any transitional state on the pathway to a specific disease, in our example we define cases as cervical precancers with a high chance of progressing into cancer or worse situation like cervical cancer) with abnormalities, triage and diagnostic tests further determine who has abnormalities that represent true disease. Among cases with disease, prognostic tests predict the outcome. For visual tests, the creation of an AI algorithm comes down to labeling many images to “train” the algorithm as to who are the cases and who are the controls (control is defined as being disease-free or being at the risk of transitioning into a case status on the causal pathway of a specific disease) at that point in the causal pathway. Careful consideration of case definition, for a given test, is sometimes overlooked in the search for sufficient “big data” sets. But larger numbers of cases do not overcome faulty labeling in producing statistical power for training an AI algorithm. For example, some AI algorithms for evaluation of cervical images have been trained to identify cases defined by any abnormal cytology, any grade of histologic cervical intraepithelial neoplasia, or visual abnormalities defined by gynecologist reviewers. The considerable error and imprecision in all of these subjective definitions guarantee that the AI algorithm will incorporate misclassification (ie, if the labeling of the training set has error, it will be reflected in the test performance). The optimal target of cervical screening programs overall, to permit effective treatment to prevent cancer, would be precancers that are reasonably likely to invade if untreated. No widely available means to define precancers perfectly are available, but a reasonable pragmatic definition is to label as cases those individuals with histologic diagnosis of cervical intraepithelial neoplasia grade 3 (which subsumes carcinoma in situ) who also are known to be positive for 1 of the 13 known carcinogenic genotypes of HPV. As to the question of “who are the controls,” identification of precancer cases among all screened individuals implies the controls for training would be those without precancer, chosen to represent the broad variety of appearances of the healthy or, at most, HPV-infected cervix.
Two additional considerations of the purpose of an AI algorithm in clinical testing refer to what AI cannot overcome, as novel as the method can seem. First, the value of a test (ie, the average risk stratification obtained that justifies using a test), whether AI-based or not, depends on how common the case outcome is. The reader is referred for a more complete discussion on the topic that the same test of a given accuracy will provide less risk stratification when the outcome it is predicting is rare (16-18).
Finally, it is possible that an AI visual algorithm might someday identify (ie, discover for the first time) features predicting case status that are currently completely unknown to us. But, for cervical screening so far, this has not been evident. The performance of automated visual evaluation as any other visual method that evaluates the cervix depends on the adequate visualization of the transformation zone bounded by the squamous columnar junction (a ring of metaplastic epithelium at greatly increased risk for the development of cervical cancer). Thus, to assure adequate performance, cervical image analysis performs best at a specific age range (ie, approximately 25-45) at which the squamous columnar junction is fully visible, which defines our target population for triage with automated visual evaluation [for more details about our study, please refer to de Sanjose et al. (19)].
Step 2: Building the automated visual evaluation model incorporating clinical epidemiologic principles
Our early attempts to classify cervical images were vulnerable in retrospect to several weaknesses in algorithm development (3). Those included lack of repeatability across the replicate images captured from the same individual at the same screening visit, a fraction of precancer/cancer patients labeled as normal, and near-complete failure of what initially seemed to be an accurate algorithm when applied to a dataset from a different source (ie, images captured with a different camera type). To address these issues, we conducted a series of experiments to determine the best performing automated visual evaluation algorithm with differing deep-learning architectures, loss functions, balancing strategies, dropout methods, and different ground truth levels (20). We evaluated these models according to repeatability (percentage of agreement in repeat evaluations) and true classification (percentage of overall true classifications and percentages of misclassified precancer/cancers as normal and vice versa) (20).
The most important of the lessons learned through this systematic approach was to choose a multiclass ordinal classification (positive, indeterminate, negative) rather than binary (positive, negative). A large group of equivocal cervical images are neither clear cases nor clear controls that arise from difficult or unfamiliar clinical presentations and look-alike lesions. We realized the need to recognize an indeterminate class while assessing automated visual evaluation repeatability on replicate images of indeterminate cases. In our early work, classification of replicates flip-flopped profoundly between case and control. For uncertain cervical appearance that would ideally be classified repeatedly as indeterminate, one replicate might be classified as definitely normal, another image captured at the same visit from the same patient might be classified as high-probability precancer/cancer (repeatability in Supplementary Table 1, available online). To reduce severe misclassification and lack of repeatability in automated visual evaluation screening tests, we formalized the indeterminate category to act as a buffer zone. This category consists of images in between clearly normal and clearly precancer/cancer. By having these 3 ordinal classes, we were able to increase the distance between the 2 extreme classes (ie, normal and precancer/cancer), force equivocal changes into the indeterminate class, and have more homogeneous and accurate prediction of those images classified as precancer/cancer or normal.
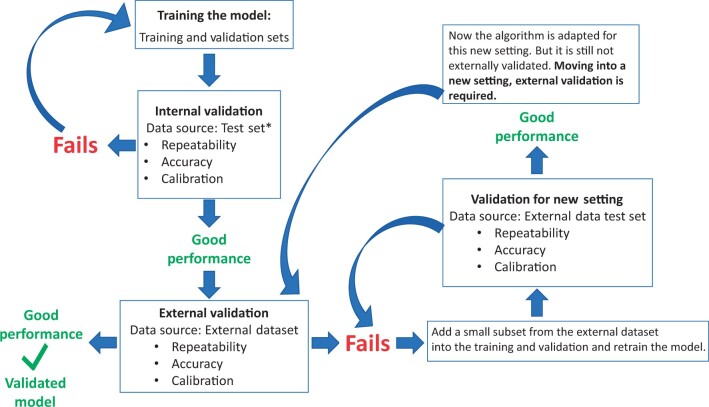
The other fundamental lesson involved external validation and avoidance of overfitting. Deep-learning algorithms are created by extracting innumerable features of the image, estimating multiple parameters (ie, weights and bias terms), and back-and-forth iterations of the training process until the algorithm’s ability to mimic the labels taught to it are maximized on a held-back test set without obvious overfitting. The error term between the truth labels given it, and the predictions, is iteratively minimized. When the fit is good, the algorithm is considered to be well calibrated. Overfitting is common despite procedures meant to prevent it (21,22). The resultant area under the curve (AUC) of the receiver operating characteristic that crosses sensitivity and (1 − specificity) to summarize accuracy can be unrealistically high. The algorithms can lack real-world generalizability and cannot meet our aim of developing a screening test that can reliably be used in many different settings. Therefore, external validation (eg, different settings, different camera types) is necessary to evaluate whether the model generalizes to the target population (Figure 1).
Figure 1.
Model validation steps and portability of the algorithm. *Test set is part of the dataset where training and validation sets belong as well. For this reason, validation by using this test set is called internal validation. In other words, checking model performance through this test set can only prove validation of the model on the dataset it is trained on.
Incorporating a method called Monte-Carlo dropout [and “ensemble learning” not covered here (23-25)] increased repeatability and reduced overfitting in our algorithm (22). This method involves running dozens of replicates of the original deep-learning algorithm. At each iteration, some nodes in the neural network are masked at random, assuring that the algorithm is not learning itself into an unrealistic perfect state of prediction by weighting the vagaries of the features peculiar to that dataset. The final result of the algorithm, when Monte-Carlo dropout is used, is the average of these purposely varied iterations, each with its own idiosyncrasies. The average is robust. Another notable enhancement that mitigated overfitting in our algorithm was the amalgamation of multiple distinct datasets (20) for training purposes. This strategic integration of diverse image collections, encompassing different camera types, populations, and settings, not only aided in diminishing model overfitting but also helped accumulate a sufficient number of representative images across all stages of the disease’s natural history. Of note, however, if a new image type is still out of the varied training distribution, the algorithm may still fail to perform well when applied to the new set.
Step 3: Evaluation of model performance
AI produces results that arise from a hidden, or latent, combination of weighting predictive features and their interactions. The hidden process and apparently exceptional accuracy can make AI algorithms seem distinct from usual assays. However, the data of all clinical tests regardless of the method must eventually pass standard clinical epidemiologic evaluation of the output. The key criteria we require in an AI-based medical test include the usual ones, in order of sufficiency (as they are all ultimately necessary); 1) repeatability, which is required for 2) accuracy, which is required in turn for 3) risk stratification (Supplementary Table 1, available online).
Repeatability is the first and foremost component to be satisfied no matter how novel and intricate the test is, whether it is molecular biological or clinical or AI (see Supplementary Table 1, available online, for how to test for repeatability). If the test is repeatable, then we can move on to assess the accuracy of the test. The test accuracy is the ability of the test to correctly classify cases (in our case, cervical precancer/cancer) and control individuals. For rare diseases, AUC might not be a good measure by itself for accuracy; therefore, we also tested the percentages of severely misclassified precancer/cancers called normal. Finally, the results of the test should separate patients at high absolute risk (high positive predictive value) from those at low absolute risk (low 1 minus negative predictive value), so we assess the average amount of risk stratification that the test provides as a final component.
The assessment of repeatability, accuracy, and predictive values is the goal of validation of algorithm performance. To begin the validation of an AI algorithm, the labeled dataset (for which the truth of case/control status is known) is typically partitioned into 3 randomly divided subsets, that is, for training, validation (not to be confused with the process of validation of the finalized algorithm described below), and testing. Labels for training and validation sets are openly provided to train the algorithm. The validation set is used to check on the growing fit of the algorithm (eg, adjusting hyperparameters), a technical step beyond this discussion. It is obvious that the model be validated using the test set only (not the subsets used during algorithm construction). Less obvious is that the 3 sets are identical in terms of features except for random variation. It is therefore too easy to show good predictive ability in the test set that is so like the training set. AUC values near perfection are often achieved in this first, internal validation (with the test set), through the countless iterations of try, assess, try again used to build the AI latent model. If internal validation fails, the model must be retrained, as depicted in Figure 1. But too often it does not fail although it proves to have been overfitted on internal data and nonportable to external datasets. Following satisfactory internal validation with respect to accuracy, repeatability, and risk stratification (for definition and evaluation of each term, refer to Supplementary Table 1, available online), it is important to make the same assessments for an external dataset (external validation, Figure 1) that is realistically representative of the target population in which the test will be used.
A study showed that only 6% of the studies, evaluating medical images through AI algorithms for diagnostic purposes, published in 2018 had external validation (26). Recently, guidelines have been created to standardize reporting AI project results in medical imaging (27-30). External validation plays a crucial role in evaluating the generalizability of a model, which means being applicable to datasets that the model was not trained on, but rather datasets from populations of intended use. Examples of factors that make a test set external include geographic diversity (including data from various countries or regions), methodological variations (including images captured using different devices, as illustrated in our example), and a wide spectrum of population characteristics (such as age or data from immunosuppressed populations that are HIV positive) (31). In an early proof-of-concept analysis, we failed initially (because of small numbers of labeled image collections) to run an external validation test on the algorithm (1-3). The internal validation was nearly perfect, but the algorithm failed to classify the first external dataset we tried to classify.
After developing a new and optimized algorithm (20), we proceeded to perform an external validation of the approach using a screening cohort from Zambia (15) with images captured by a camera (Samsung Galaxy J8) not included in our training set.
We were able to retrain the algorithm to work on the Zambian camera type. In other words, when first applied, the new algorithm again failed to distinguish cases from controls. The image collection device remains a major barrier to the application of AI-based image recognition, and to date, complete portability between camera types is not possible. To make our algorithm work well on new camera choices required retraining with expanded training and validation sets that include a small subset from the new (external) dataset (thus making the validation mainly but not completely external). Our experiments continue to gauge the ideal size of this small subset, but approximately 40-80 individuals’ data for each ground truth class proved sufficient; this number might vary (15). Thus, on the basis of our experience from our studies, we have learned that changing image capture device is a powerful factor in AI-based image analysis. However, change in geography and disease spectrum (ie, HIV positive in our example) does not affect the algorithm performance in our experience so far (15).
Figure 1 shows how the model is validated for the new setting after retraining. It is worth reiterating that, at this stage (with current supervised learning techniques), algorithm validation on an external dataset cannot be accomplished without limited retraining of the algorithm, and this predicament is a common challenge encountered by many deep-learning algorithms (32,33). In our automated visual evaluation project, we have chosen to use a dedicated camera device designed specifically for cervical screening to minimize the need for retraining.
Step 4: Estimating absolute risks from AI algorithm results
After validating the performance of the AI algorithm, the next step is to translate the results to a form that leads to management decisions; we estimate absolute risks of having or developing precancer/cancer. The observed risk of precancer/cancer is calculated for all possible screening test results generated by our screening strategy with HPV genotyping and automated visual evaluation tests. Although the common view of AI algorithms is that they recognize an object (like a face), estimating the probability of disease status based on disease-related features in images, it is important to recognize that the algorithm actually learns common features from images with the same ground truth values. Consequently, the algorithm outputs classification probabilities that reflect the algorithm’s prediction on each image belonging to each ground truth class (ie, simply classification probability). Therefore, one needs to be careful about the interpretation of the probabilities created by the algorithm. These probabilities should not be evaluated as the probability of having a certain disease. To estimate the risk of having a disease status (in our example, having precancer/cancer), a risk model is necessary (Supplementary Table 1, available online).
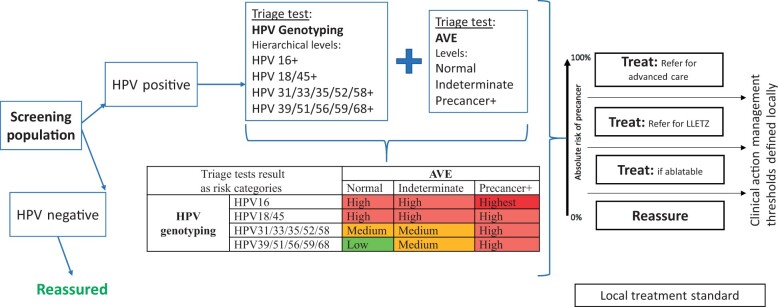
We are building a statistical model to estimate disease risk. It can be built either using the AI-based test result alone or adding other covariates to obtain more precise estimates. In our example, we are combining HPV genotype status with the automated visual evaluation test result to obtain precancer/cancer risk of patients (Figure 2). According to our screening strategy defined above, HPV-positive individuals will be assigned to 1 of 4 hierarchical HPV genotype groups (HPV 16 positive, else HPV 18 or 45 positive, else HPV 31 and/or 33 and/or 35 and/or 52 and/or 58 positive, else HPV 39 and/or 51 and/or 56 and/or 59 and/or 68 positive), and their cervix image will be classified as 1 of 3 categories (normal, indeterminate, precancer/cancer) by automated visual evaluation. Combining (crossing) HPV genotype and automated visual evaluation class, each individual will be allocated to 1 of 12 risk categories, as outlined in Figure 2. Through our initial risk estimation analyses, we have been able to directly assess the risk of precancer/cancer occurrence for each of these categories (15). Although the precise rank order (and absolute risk) of these 12 categories may vary across different regions, subsequently consolidating them into 4 groups, as illustrated in Figure 2 (highest, high, medium, low) permits consistency of the order across various settings.
Figure 2.
Intended screening program with HPV genotyping and automated visual evaluation screening tests. AVE = automated visual evaluation; HPV = human papillomavirus; LLETZ = large loop excision of the transformation zone; Precancer+ = cervical precancer or cancer.
Step 5: Creating risk-based management guidelines
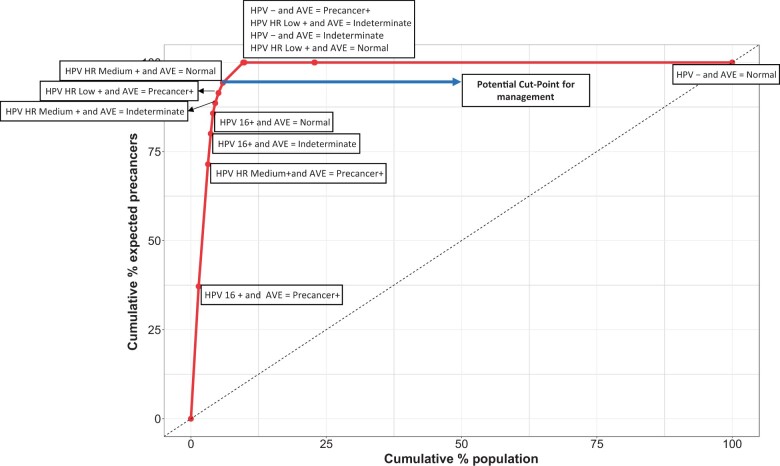
The absolute risks in the risk model can be organized using a concentration curve (see Supplementary Methods, available online) to display the risk distribution from highest to lowest yield of cases. This curve can be used by local decision makers in formulating their own risk-based management guidelines tailored to their specific setting. We illustrate the use of a concentration curve on a hypothetical population in Figure 3. This approach is a visual representation that aids in understanding the risk distribution within the population and supports the development of context-specific guidelines. Specifically, a concentration curve shows the percentage of the population that needs to be managed to eliminate a certain percentage of precancers or cancers expected in that population (see Supplementary Methods, available online). This is a helpful tool for local experts to define cutoff points (also called clinical action management thresholds) for available management options in that setting (34,35). The right side of Figure 2 demonstrates an anticipated local management and treatment standard, which is based on the principle of “equal management of individuals at equal risk” (4). Below a specified risk, individuals will be reassured, and the rest will be either treated locally, if eligible for ablation, or referred for excision or advanced care as appropriate. In Figure 3, the long arrow arrow at the top points to a cutoff point for treatment in this example setting. If this threshold is chosen, 5.9% of the screening population will be treated resulting in eliminating 94% of the expected precancer or cancer cases in this population. This could be a reasonable option in settings with few planned screens per lifetime, for which treatment is favored over expectant management for most patients. The remaining 41% of HPV-positive patients (4.1% of the total screening population) have low-risk combinations of HPV genotype and automated visual evaluation results and are therefore less likely to benefit from treatment. Note that approximately 90% of the population has an HPV-negative result and would therefore not undergo triage testing.
Figure 3.
Concentration curve for the estimation of the expected percent of precancer or cancer cases in relation to the percentage of the population that would require treatment. This is a hypothetical example of a concentration curve. This curve visualizes the entire population, ranked from the highest to the lowest predicted risk, on the x-axis and presents the expected precancers or cancers on the y-axis. The first point on this curve (which is labeled as HPV 16+ and automated visual evaluation test result is precancer/cancer, denoted AVE = Precancer+ in the figure) represents the highest risk group in this population, which refers to 1.4% of the total screening population. If only this group is referred for treatment, it will result in treating 1.4% of the total screening population, in return eliminating approximately 37% of the expected precancer/cancer cases from this population. Each data point on the curve corresponds to a screening result, arranged in decreasing severity order from the lower left corner to the upper right corner. In this example, a favorable cut point for treatment is highlighted by the long arrow at the top of the figure. Choosing this potential cut point means, every individual in these risk categories falling in and below this point emphasized with this arrow as a cut point for management will be treated (they correspond to 5.9% of the total screening population) to eliminate approximately 94% of the expected precancer/cancer cases within the population. We believe the concentration curve will serve as a valuable tool for local experts and decision makers in formulating their individualized risk-based guidelines. AVE = automated visual evaluation; Precancer+ = precancer or worse situation like cancer; HPV- = human papillomavirus negative for cervical cancer high-risk types; HPV HR Medium + = HPV high-risk medium types (any of 18, 45, 31, 33, 35, 52, or 58) positive; HPV HR Low + = HPV high-risk low types (any of 39, 51, 56, 59, or 68) positive. The hierarchical order of the HPV genotype groups is HPV 16 positive, else HPV HR Medium positive, else HPV HR Low positive, else HPV negative.
Our objective is for each setting to establish its own risk-based management guidelines, taking into account its specific HPV genotyping and disease prevalence, risk tolerance level, and available treatment and screening capacities. This entails defining the optimal thresholds for clinical action management, as depicted in Figure 2, and aligning them with the local treatment standards (36). By tailoring the management guidelines to the unique characteristics of each setting, we aim to optimize the allocation of resources and enhance the effectiveness of clinical interventions.
AI provides a novel approach to pattern recognition and classification. However, as a clinical test, the output still follows general epidemiologic and biostatistical principles. This entails ensuring the repeatability of results when testing under the same conditions, obtaining accurate results for both disease-positive and disease-negative cases, establishing a buffer zone to address unavoidably equivocal diagnoses, and most importantly, being able to help clinicians make informed clinical management decisions that ultimately provide information that is helpful to those being tested. Trust in AI-assisted clinical testing depends on overcoming understandable skepticism and proving stable worth.
Supplementary Material
Acknowledgements
The funder did not play a role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Contributor Information
Didem Egemen, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Rockville, MD, USA.
Rebecca B Perkins, Department of Obstetrics and Gynecology, Boston Medical Center/Boston University School of Medicine, Boston, MA, USA.
Li C Cheung, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Rockville, MD, USA.
Brian Befano, Information Management Services Inc, Calverton, MD, USA; Department of Epidemiology, School of Public Health, University of Washington, Seattle, WA, USA.
Ana Cecilia Rodriguez, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Rockville, MD, USA.
Kanan Desai, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Rockville, MD, USA.
Andreanne Lemay, Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Boston, MA, USA.
Syed Rakin Ahmed, Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Boston, MA, USA; Harvard Graduate Program in Biophysics, Harvard Medical School, Harvard University, Cambridge, MA, USA; Massachusetts Institute of Technology, Cambridge, MA, USA; Geisel School of Medicine at Dartmouth, Dartmouth College, Hanover, NH, USA.
Sameer Antani, National Library of Medicine, National Institutes of Health, Bethesda, MD, USA.
Jose Jeronimo, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Rockville, MD, USA.
Nicolas Wentzensen, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Rockville, MD, USA.
Jayashree Kalpathy-Cramer, Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Boston, MA, USA.
Silvia De Sanjose, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Rockville, MD, USA; ISGlobal, Barcelona, Spain.
Mark Schiffman, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Rockville, MD, USA.
Data availability
No new data were generated or analyzed for this commentary.
Author contributions
Didem Egemen, PhD (Conceptualization; Formal analysis; Methodology; Writing—original draft), Rebecca B Perkins, MD, MSc (Conceptualization; Methodology; Writing—original draft; Writing—review & editing), Li C Cheung, PhD (Conceptualization; Methodology; Writing—review & editing), Brian Befano, MPH (Conceptualization; Methodology; Writing—review & editing), Ana Cecilia Rodriguez, MD, MPH (Conceptualization; Methodology; Writing—review & editing), Kanan Desai, MD, MPH (Conceptualization; Methodology; Writing—review & editing), Andreanne Lemay, MS (Conceptualization; Methodology; Writing—review & editing), Syed Rakin Ahmed, BSc (Conceptualization; Methodology; Writing—review & editing), Sameer Antani, PhD (Conceptualization; Methodology; Writing—review & editing), Jose Jeronimo, MD, MPH (Conceptualization; Methodology; Writing—review & editing), Nicolas Wentzensen, MD, PhD, MS (Conceptualization; Methodology; Writing—review & editing), Jayashree Kalpathy-Cramer, PhD (Conceptualization; Methodology; Writing—review & editing), Silvia de Sanjose, MD, PhD (Conceptualization; Methodology; Writing—review & editing), and Mark Schiffman, MD, MPH (Conceptualization; Methodology; Writing—original draft).
Funding
These analyses have been supported by the Intramural Research Program of the National Institutes of Health.
Conflicts of interest
The authors declare that they have no conflicts of interest.
References
- 1. Hu L, Bell D, Antani S, et al. An observational study of deep learning and automated evaluation of cervical images for cancer screening. J Natl Cancer Inst. 2019;111(9):923-932. doi: 10.1093/jnci/djy225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xue Z, Novetsky AP, Einstein MH, et al. A demonstration of automated visual evaluation of cervical images taken with a smartphone camera. Int J Cancer. 2020;147(9):2416-2423. doi: 10.1002/ijc.33029 [DOI] [PubMed] [Google Scholar]
- 3. Desai KT, Befano B, Xue Z, et al. The development of “automated visual evaluation” for cervical cancer screening: the promise and challenges in adapting deep-learning for clinical testing. Int J Cancer. 2022;150(5):741-752. doi: 10.1002/ijc.33879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Katki HA, Kinney WK, Fetterman B, et al. Cervical cancer risk for women undergoing concurrent testing for human papillomavirus and cervical cytology: a population-based study in routine clinical practice. Lancet Oncol. 2011;12(7):663-672. doi: 10.1016/S1470-2045(11)70145-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wentzensen N, Lahrmann B, Clarke MA, et al. Accuracy and efficiency of deep-learning-based automation of dual stain cytology in cervical cancer screening. J Natl Cancer Inst 2021;113(1):72-79. doi: 10.1093/JNCI/DJAA066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li MD, Arun NT, Gidwani M, et al. Automated assessment and tracking of COVID-19 pulmonary disease severity on chest radiographs using convolutional siamese neural networks MGH and BWH center for clinical data science. Radiol Artif Intell 2020;2(4):e200079. doi: 10.1148/ryai.2020200079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bridge CP, Best TD, Wrobel MM, et al. A fully automated deep learning pipeline for multi- vertebral level quantification and characterization of muscle and adipose tissue on chest CT scans. Radiol Artif Intell 2022;4(1):e210080. doi: 10.1148/ryai.210080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen JS, Coyner AS, Ostmo S, et al. Deep learning for the diagnosis of stage in retinopathy of prematurity: accuracy and generalizability across populations and cameras. Ophthalmol Retina. 2021;5(10):1027-1035. doi: 10.1016/j.oret.2020.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alryalat SA, Singh P, Kalpathy-Cramer J, Kahook MY.. Artificial intelligence and glaucoma: going back to basics. Clin Ophthalmol. 2023;17:1525-1530. doi: 10.2147/OPTH.S410905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. deCampos-Stairiker MA, Coyner AS, Gupta A, et al. Epidemiologic evaluation of retinopathy of prematurity severity in a large telemedicine program in india using artificial intelligence. Ophthalmology. 2023;130(8):837-843. doi: 10.1016/j.ophtha.2023.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S.. Human papillomavirus and cervical cancer. Lancet. 2007;370(9590):890-907. doi: 10.1016/S0140-6736(07)61416-0 [DOI] [PubMed] [Google Scholar]
- 12. Bouvard V, Wentzensen N, Mackie A,. et al. The IARC perspective on cervical cancer screening. N Engl J Med. 2021;385(20):1908-1918. doi: 10.1056/NEJMsr2030640 [DOI] [PubMed] [Google Scholar]
- 13. Desai KT, Adepiti CA, Schiffman M, et al. Redesign of a rapid, low-cost HPV typing assay to support risk-based cervical screening and management. Int J Cancer 2022;151(7):1142-1149., doi: 10.1002/ijc.34151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Inturrisi F, De Sanjosé S, Desai KT, et al. A rapid HPV typing assay to support global cervical cancer screening and risk-based management: a cross-sectional study. Int J Cancer. 2023;10.1002/ijc.34698. doi: 10.1002/ijc.34698 [DOI] [PubMed] [Google Scholar]
- 15. Parham G, Egemen D, Befano B, et al. Validation in Zambia of a cervical screening strategy including HPV genotyping and artificial intelligence (AI)-based automated visual evaluation. Infect Agents Cancer 2023;18(61). doi: 10.1186/s13027-023-00536-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Katki HA, Schiffman M.. A novel metric that quantifies risk stratification for evaluating diagnostic tests: the example of evaluating cervical-cancer screening tests across populations. Prev Med 2018;110:100-105. doi: 10.1016/j.ypmed.2018.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Katki HA. Quantifying risk stratification provided by diagnostic tests and risk predictions: comparison to AUC and decision curve analysis. Stat Med 2019;38(16):2943-2955. doi: 10.1002/sim.8163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wentzensen N, Wacholder S.. From differences in means between cases and controls to risk stratification: a business plan for biomarker development. Cancer Discov. 2013;3(2):148-157. doi: 10.1158/2159-8290.CD-12-0196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Sanjose S, Perkins R, Campos N, et al. Design of the HPV-Automated Visual Evaluation (PAVE) study: validating a novel cervical screening strategy. medRxiv [Preprint]. 2023. doi: 10.1101/2023.08.30.23294826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ahmed RS, Befano B, Lemay A, et al. Reproducible and clinically translatable Deep Neural Networks for cervical screening. medRxiv [Preprint]. 2022. doi: 10.1101/2022.12.17.22282984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gidwani M, Chang K, Patel JB, et al. Inconsistent partitioning and unproductive feature associations yield idealized radiomic models. Radiology. 2023;307(1):e220715. doi: 10.1148/radiol.220715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lemay A, Hoebel K, Bridge CP, et al. Improving the repeatability of deep learning models with Monte Carlo dropout. NPJ Digit Med. 2022;5(1):174. doi: 10.1038/s41746-022-00709-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pan I, Thodberg HH, Halabi SS, Kalpathy-Cramer J, Larson DB.. Improving automated pediatric bone age estimation using ensembles of models from the 2017 RSNA machine learning challenge. Radiol Artif Intell. 2019;1(6):e190053. doi: 10.1148/ryai.2019190053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kurc T, Bakas S, Ren X, et al. Segmentation and classification in digital pathology for glioma research: challenges and deep learning approaches. Front Neurosci. 2020;14:27. doi: 10.3389/fnins.2020.00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Halabi SS, Prevedello LM, Kalpathy-Cramer J, et al. The rSNA pediatric bone age machine learning challenge. Radiology. 2019;290(2):498-503. doi: 10.1148/radiol.2018180736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim DW, Jang HY, Kim KW, Shin Y, Park SH.. Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J Radiol. 2019;20(3):405-410. doi: 10.3348/kjr.2019.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klontzas ME, Gatti AA, Tejani AS, Kahn CE.. AI Reporting Guidelines: How to Select the Best One for Your Research. Radiol Artif Intell. 2023;5(3):e230055. doi: 10.1148/ryai.230055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mongan J, Moy L, Kahn CE.. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): a guide for authors and reviewers. Radiol Artif Intell. 2020;2(2):e200029. doi: 10.1148/ryai.2020200029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu X, Cruz Rivera S, Moher D, et al. ; SPIRIT-AI and CONSORT-AI Working Group. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat Med. 2020;26(9):1364-1374. doi: 10.1038/s41591-020-1034-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lekadir K, Osuala R, Gallin C, et al. FUTURE-AI: Guiding principles and consensus recommendations for trustworthy artificial intelligence in medical imaging.CoRR. 2021;abs/2109.09658.
- 31. Justice AC, Covinsky KE, Berlin JA, Pittsburgh F, Francisco S.. Assessing the generalizability of prognostic information. Ann Intern Med. 1999;130(6):515-524. doi: 10.7326/0003-4819-130-6-199903160-00016 [DOI] [PubMed] [Google Scholar]
- 32. Chang K, Beers AL, Brink L, et al. Multi-institutional assessment and crowdsourcing evaluation of deep learning for automated classification of breast density. J Am Coll Radiol. 2020;17(12):1653-1662. doi: 10.1016/j.jacr.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Van Calster B, Steyerberg EW, Wynants L, van Smeden M.. There is no such thing as a validated prediction model. BMC Med 2023;21(1):70. doi: 10.1186/s12916-023-02779-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perkins RB, Guido RS, Castle PE, et al. 2019 ASCCP Risk-Based Management Consensus Guidelines Committee. 2019 ASCCP risk-based management consensus guidelines for abnormal cervical cancer screening tests and cancer precursors. J Low Genit Tract Dis. 2020;24(2):102-131. doi: 10.1097/LGT.0000000000000525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Egemen D, Cheung LC, Chen X, et al. Risk estimates supporting the 2019 ASCCP risk-based management consensus guidelines. J Low Genit Tract Dis. 2020;24(2):132-143. doi: 10.1097/LGT.0000000000000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perkins RB, Smith DL, Jeronimo J, et al. Use of risk-based cervical screening programs in resource-limited settings. Cancer Epidemiol 2023;84:102369. doi: 10.1016/j.canep.2023.102369 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were generated or analyzed for this commentary.