Abstract
Background
Neuroblastoma is an embryonal cancer of the developing sympathetic nervous system. The genetic contribution of rare pathogenic or likely pathogenic germline variants in patients without a family history remains unclear.
Methods
Germline DNA sequencing was performed on 786 neuroblastoma patients. The frequency of rare cancer predisposition gene pathogenic or likely pathogenic variants in patients was compared with 2 cancer-free control cohorts. Matched tumor DNA sequencing was evaluated for second hits, and germline DNA array data from 5585 neuroblastoma patients and 23 505 cancer-free control children were analyzed to identify rare germline copy number variants. Patients with germline pathogenic or likely pathogenic variants were compared with those without to test for association with clinical characteristics, tumor features, and survival.
Results
We observed 116 pathogenic or likely pathogenic variants involving 13.9% (109 of 786) of neuroblastoma patients, representing a statistically significant excess burden compared with cancer-free participants (odds ratio [OR] = 1.60, 95% confidence interval [CI] = 1.27 to 2.00). BARD1 harbored the most statistically significant enrichment of pathogenic or likely pathogenic variants (OR = 32.30, 95% CI = 6.44 to 310.35). Rare germline copy number variants disrupting BARD1 were identified in patients but absent in cancer-free participants (OR = 29.47, 95% CI = 1.52 to 570.70). Patients harboring a germline pathogenic or likely pathogenic variant had a worse overall survival compared with those without (P = 8.6 x 10−3).
Conclusions
BARD1 is an important neuroblastoma predisposition gene harboring both common and rare germline pathogenic or likely pathogenic variations. The presence of any germline pathogenic or likely pathogenic variant in a cancer predisposition gene was independently predictive of worse overall survival. As centers move toward paired tumor-normal sequencing at diagnosis, efforts should be made to centralize data and provide an infrastructure to support cooperative longitudinal prospective studies of germline pathogenic variation.
Neuroblastoma is an embryonal malignancy of early childhood that arises from developing postganglionic sympathetic neurons and accounts for 12% of all childhood cancer-related deaths (1). Patients are classified into low, intermediate, and high risk based on a series of clinical and tumor biological features, and this risk group is used for treatment stratification purposes (1). Despite aggressive multimodal therapy, nearly 50% of high-risk neuroblastoma patients diagnosed at older than 18 months of age eventually succumb to their disease. A subset of these tumors harbor somatic MYCN amplification and/or an activating somatic ALK mutation or gene amplification (1). However, sequencing studies of neuroblastoma tumors have revealed a low overall somatic mutation rate and few recurrently mutated genes (2-4). The young median age at diagnosis and standardized incidence ratio of siblings of children with neuroblastoma of approximately 9.7 (5) are consistent with an underlying genetic etiology.
The genetic basis of neuroblastoma predisposition has come into focus over the past decade. Familial neuroblastoma, which accounts for 1%-2% of cases, arises primarily from pathogenic germline variants in ALK (6), with rarer neurocristopathy syndrome cases explained by germline pathogenic variants in PHOX2B (7,8). However, the vast majority of neuroblastomas appear to arise sporadically, without a family history. Genome-wide association studies (GWAS) have identified common variants associated with sporadic neuroblastoma at more than a dozen loci. These genetic associations have implicated multiple candidate genes including CASC15, NBAT1, BARD1, LMO1, DUSP12, DDX4, IL31RA, HSD17B12, HACE1, LIN28B, TP53, RSRC1, MLF1, CPZ, MMP20, KIF15, and NBPF23 (9-17). Several susceptibility genes identified by GWAS not only influence disease initiation but also drive tumor aggressiveness and/or maintenance of the malignant phenotype (11,13,15,18-21). A rare 16p11.2 microdeletion syndrome has also been associated with neuroblastoma (22). Finally, recent sequencing efforts have reported rare pathogenic germline variants in multiple cancer predisposition genes (2,23-33); however, the prevalence and clinical significance of these and other rare variants in neuroblastoma remain unclear and require evaluation in larger patient cohorts with detailed phenotypic data.
Here, we analyzed germline whole genome sequencing, whole exome sequencing, and targeted capture sequencing data from 786 children diagnosed with neuroblastoma and profiled through the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative. Our aims were to 1) determine the prevalence, spectrum, and pathogenicity of rare germline variants in known cancer predisposition genes; 2) test for enrichment of rare variants in children with neuroblastoma compared with cancer-free control populations; and 3) evaluate clinical features and outcomes in neuroblastoma patients with and without germline pathogenic or likely pathogenic variants in cancer predisposition genes to identify translational opportunities.
Methods
Detailed methods are provided in Supplementary Methods (available online). Briefly, the study cohort consisted of 786 neuroblastoma patients accrued through the Children’s Oncology Group (COG) ANBL00B1 biology study, unselected for family history (see Table 1; Supplementary Table 1, available online). Germline DNA and matched diagnostic tumor DNA and RNA were sequenced through the TARGET initiative. The original set of tumor-normal pairs were sequenced with Complete Genomics whole genome sequencing (n = 134) and/or Illumina whole exome sequencing (n = 222), as previously described (2,34). A total of 59 samples were sequenced by whole genome sequencing, and whole exome sequencing provided internal validation. We have previously reported a small number of germline variants based on the whole exome sequencing cohort (2); however, an in-depth study of pathogenic germline variation in these children was not performed at that time. Germline DNA from an independent neuroblastoma cohort (n = 489) was sequenced using Illumina custom capture panels, including a germline panel newly designed for this study (n = 166 genes; Supplementary Table 2, available online). Cancer-free control data were obtained from the Penn Medicine BioBank (PMBB; n = 6295) (35,36) and the Genome Aggregation Database (gnomAD v2.1) without cancer (n = 15 708). Ancestry for neuroblastoma and PMBB participants was inferred through principal component analysis using matched germline DNA array data. Germline variants were called using Genome Analysis Toolkit (GATK) best practices (whole exome sequencing and custom capture data) or the Complete Genomics pipeline (v2) with custom filtering (37) (whole genome sequencing data) then annotated with SnpEff (38) (v4.3t) and ANNOVAR (39). For all cohorts (neuroblastoma, PMBB, and gnomAD), rare germline variants (<0.1% across each population in public control databases) in 166 cancer predisposition genes were then assessed for pathogenicity with a clinically focused pipeline incorporating ClinVar (40) evidence and a modified implementation of InterVar (41), a tool that seeks to automate pathogenicity classification based on guidelines from the American College of Medical Genetics and Genomics and the Association of Molecular Pathology (42) (Supplementary Methods, Supplementary Figure 1, available online). A subset of germline variants were validated through Sanger sequencing. Patients harboring a germline pathogenic or likely pathogenic variant in a cancer predisposition gene were further assessed using matched tumor sequencing data when available. Fisher’s exact test was used to compare the enrichment of pathogenic or likely pathogenic variants in neuroblastoma patients with 2 cancer-free control cohorts and across clinical and biological subsets of neuroblastoma. Pathogenic or likely pathogenic variant enrichment was also compared at gene and pathway levels, using a Bonferroni correction for multiple testing. Kaplan–Meier analyses of event-free and overall survival were performed to compare outcomes of patients with and without germline pathogenic or likely pathogenic variants. A multivariate Cox proportional hazards regression model was used to assess if the presence of a cancer predisposition gene pathogenic or likely pathogenic variant was independently predictive of survival.
Table 1.
Clinical and tumor biological characteristics for 786 neuroblastoma patientsa
| Characteristics | Neuroblastoma sequencing cohort No. (%) |
|---|---|
| Age | |
| Younger than 18 months | 242 (30.8%) |
| 18 months and older | 544 (69.2%) |
| Sex | |
| Female | 339 (43.1%) |
| Male | 447 (56.9%) |
| COG risk | |
| Low | 103 (13.2%) |
| Intermediate | 119 (15.1%) |
| High | 564 (71.8%) |
| INSS stage | |
| Stage 1 | 38 (4.8%) |
| Stage 2 | 60 (7.7%) |
| Stage 3 | 93 (11.8%) |
| Stage 4 | 546 (69.5%) |
| Stage 4S | 49 (6.2%) |
| MYCN status | |
| Not amplified | 552 (71.1%) |
| Amplified | 224 (28.9%) |
| Not available | 10b |
| Histology | |
| Favorable | 211 (28.8%) |
| Unfavorable | 522 (71.2%) |
| Not available | 53b |
| Ploidy | |
| Hyperdiploid | 473 (61.7%) |
| Diploid | 293 (38.3%) |
| Not available | 20b |
COG = Children’s Oncology Group; INSS = International Neuroblastoma Staging System.
Not included in % calculation
Results
Neuroblastoma patient characteristics
A total of 786 neuroblastoma patients were included in the study (Table 1; Supplementary Table 2, available online). Overall, 564 (71.8%) patients were classified as high-risk based on the COG risk stratification system, and 546 of these patients had stage IV disease according to the International Neuroblastoma Staging System (INSS) criteria. Cases profiled by whole genome sequencing or whole exome sequencing were intentionally enriched for high-risk disease, consistent with the overall goals of the TARGET initiative. In contrast, cases that underwent targeted capture (custom capture) sequencing were representative of the general neuroblastoma risk group profile. A total of 769 patients had available matched germline DNA array data and were evaluated for ancestry by principal component analysis. As expected, the majority (66.8%) of cases were inferred to be of European ancestry (Supplementary Figure 2, available online).
Frequency of pathogenic or likely pathogenic variants in known cancer predisposition genes
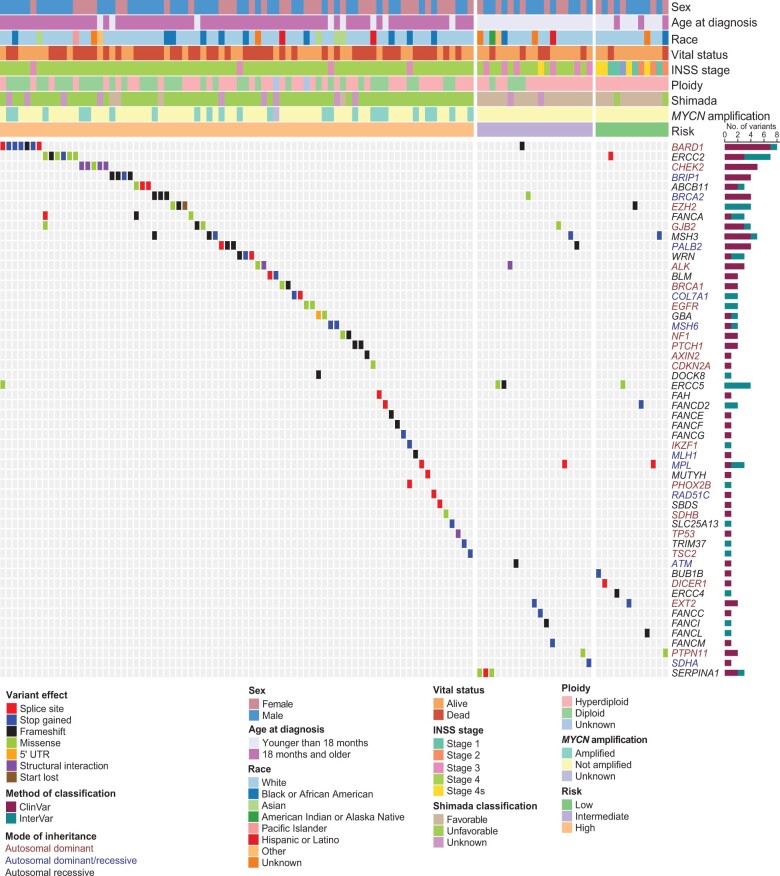
We observed 116 pathogenic or likely pathogenic variants involving 54 of the 166 cancer predisposition genes studied (Figure 1; Supplementary Table 3, available online). Of these variants, 73 were classified as pathogenic or likely pathogenic on the basis of ClinVar evidence, and 43 variants were assigned pathogenic or likely pathogenic on the basis of our revised InterVar assessment. Overall, pathogenic or likely pathogenic variants were detected in 109 of 786 (13.9%) neuroblastoma patients. Classic familial neuroblastoma germline variants were observed in 0.4% (3 of 786) of cases. These included 2 patients with ALK (p.R1275Q) activating variants and a single patient with a PHOX2B splice variant (NM_003924: exon3: c.430-2A>G). An additional ALK variant (p.I1250T) was predicted to be likely pathogenic; however, this variant was not found to be activating in a previous study (43). Six cases harbored more than 1 pathogenic or likely pathogenic variant in a known cancer predisposition gene (Supplementary Table 4, available online). A total of 27 genes harbored pathogenic or likely pathogenic variants in 2 or more cases, with variants in the BARD1 gene being most frequent (8 of 786 cases or 1.0% overall; Supplementary Figure 3, Supplementary Table 3, available online). Select variants were validated by Sanger sequencing (Supplementary Table 5, available online). Notably, of the 9 neuroblastoma genes identified by GWAS and included in this study, only BARD1 and TP53 harbored rare pathogenic or likely pathogenic coding variants. This suggests that causal variants at other GWAS loci may be in the noncoding genome or tied to common variants not considered here.
Figure 1.
Germline pathogenic or likely pathogenic variants in cancer predisposition genes across 786 neuroblastoma patients. Oncoprint of known cancer predisposition genes harboring rare germline variants classified as pathogenic or likely pathogenic. Patients without pathogenic or likely pathogenic variants in these genes are not shown. Patients are ordered by Children’s Oncology Group risk group and annotated with clinical and tumor biologic features. Genes are color-coded according to mode of inheritance, when known. Bar chart to the right indicates the number of variants detected for each gene and whether pathogenicity was determined based on ClinVar or our modified InterVar automated assessment. All variants were manually reviewed for quality and evidence of pathogenicity. INSS = International Neuroblastoma Staging System; UTR = Untranslated Region.
Matched tumor DNA sequencing reveals pathogenic or likely pathogenic variants are retained and second hits are rare
We examined somatic alterations affecting genes with germline pathogenic or likely pathogenic variants using matched published neuroblastoma tumor whole genome sequencing and whole exome sequencing variant calls (34) and our analysis of available custom capture sequencing from TARGET. Nearly all (95%, 73 of 77) germline pathogenic or likely pathogenic variants were detected in matched tumor DNA, when available (Supplementary Table 3, available online). Pathogenic or likely pathogenic variants detected in tumor DNA had an average variant allele fraction of 0.46. In contrast, variants not detected in the matched tumor (n = 4) exhibited a lower variant allele fraction (range = 0.25-0.31), suggesting these variants may be mosaic. No second hit single nucleotide variations (SNVs) or indels were observed in the tumor DNA of patients harboring a germline pathogenic or likely pathogenic variant in a cancer predisposition gene. We detected 1 focal somatic deletion within EZH2 observed in the tumor from patient PASEGA, who also harbored a pathogenic germline EZH2 variant (p.T536fs). Phase could not be determined, but this somatic event was confirmed by Sanger sequencing of the tumor DNA (Supplementary Figure 4, available online).
Germline pathogenic or likely pathogenic variants in cancer predisposition genes are enriched in neuroblastoma patients
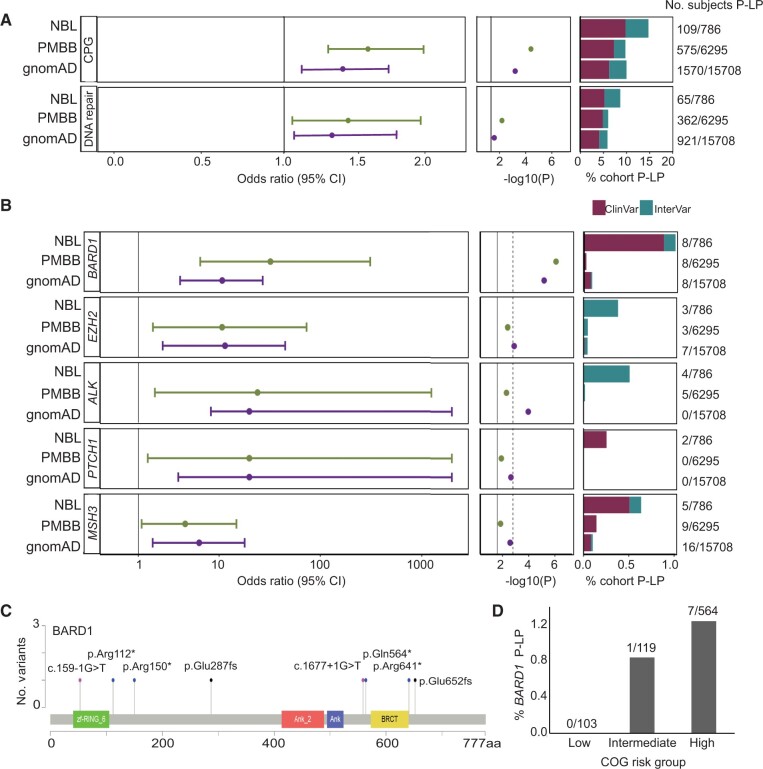
To assess whether children diagnosed with neuroblastoma harbor an excess of rare pathogenic germline variation in the 166 cancer predisposition genes considered in this study, we compared the pathogenic or likely pathogenic burden in neuroblastoma patients to 2 independent control cohorts (Figure 2, A, top panel). First, we applied our full analytic pipeline (alignment, variant calling, quality control, and pathogenicity assessment) to participants sequenced through the PMBB and without a history of cancer or benign tumors (n = 6295; Supplementary Figure 5, available online). Germline pathogenic or likely pathogenic variants were statistically significantly enriched in neuroblastoma patients compared with PMBB (PPMBB = 5.14 x 10−5; odds ratio [OR] = 1.60, 95% confidence interval [CI] = 1.27 to 2.00). To assess reproducibility of this result, identical filtering and pathogenicity assessment was applied to rare variants in gnomAD excluding cancer samples (n = 15 708 individuals). This confirmed the excess burden of pathogenic or likely pathogenic variants in neuroblastoma (PgnomAD = 1.82 x 10−3; OR = 1.41, 95% CI = 1.34 to 1.74).
Figure 2.
Neuroblastoma patients harbor an excess burden of rare pathogenic or likely pathogenic (P-LP) variants in cancer predisposition genes. A) Overall excess burden of P-LP variants (single nucleotide variations and indels) in neuroblastoma (NBL) vs Penn Medicine BioBank (PMBB) and the Genome Aggregation Database (gnomAD) v2.1 controls is shown for cancer predisposition genes and the subset of genes studied involved in DNA repair. B) Gene-based rare variant burden test results comparing the number of neuroblastoma subjects with P-LP variants to those detected in gnomAD v2.1 and PMBB. C) Lollipop figure depicting 8 germline P-LP variants in BARD1. D) Rare P-LP variants in BARD1 are observed predominantly in patients diagnosed with high-risk neuroblastoma. CI = confidence interval; COG = Children’s Oncology Group; CPG = cancer predisposition gene.
Germline pathogenic or likely pathogenic variants in BARD1 are enriched in neuroblastoma patients
Next, we performed gene-based rare variant burden testing comparing neuroblastoma patients with PMBB and gnomAD control cohorts (Supplementary Tables 6 and 7, available online). Five genes (BARD1, EZH2, ALK, PTCH1, and MSH3) exhibited statistically significant enrichment (P < .05), in both control cohort comparisons (Figure 2, B). Pathogenic variants in BARD1 and EZH2 were validated by Sanger sequencing in neuroblastoma patients when DNA was available (Supplementary Figures 3 and 4, available online). ALK and BARD1 remained statistically significant after Bonferroni adjustment for multiple testing in at least 1 control comparison. ALK is the main major familial neuroblastoma predisposition gene (6). Only 1 likely pathogenic variant in ALK was detected in PMBB (PPMBB = 5.00 x 10−3; OR = 24.09, 95% CI = 1.93 to 1255.78). This variant (p.A1168T) was classified likely pathogenic based on InterVar and was not reported in ClinVar. No pathogenic or likely pathogenic ALK variants were observed in gnomAD v2.1 whole genome controls (PgnomAD = 1.08 x 10−4; OR = 140.3, 95% CI = 7.24 to 2719.0).
BARD1 is the only gene that passed a Bonferroni adjustment in both control comparisons (Figure 2, B). Common variation at the BARD1 locus is known to be associated with high-risk neuroblastoma from our prior GWAS (10). We and others have also reported germline BARD1 rare variants in neuroblastoma patients (2,44,45). However, to date, the number of patients analyzed has been limited. Here, we observed rare pathogenic or likely pathogenic variants in BARD1 in 8 of 786 (1.0%) neuroblastoma patients, all predicted to be loss-of-function (Figure 2, C; Supplementary Table 8, available online). Moreover, all but 1 variant was observed in the high-risk subset (Figure 2, D). Only 2 of 6295 (0.03%) control participants in PMBB harbored a pathogenic or likely pathogenic germline variant in BARD1 (PPMBB = 8.18 x 10−7; OR = 32.30, 95% CI = 6.44 to 310.35). Similarly, only 15 of 15 708 (0.09%) control participants in gnomAD harbored a pathogenic or likely pathogenic germline variant in BARD1 (PgnomAD = 6.64 x 10−6; OR = 10.75, 95% CI = 3.93 to 27.13). Enrichment of pathogenic or likely pathogenic variants in BARD1 remained statistically significant when we restricted the analysis to neuroblastoma patients and cancer-free control particpants of European ancestry, considering PMBB (PPMBB = 2.70 x 10−5; OR = 21.36, 95% CI = 4.55 to 131.80) and gnomAD (PgnomAD = 1.55 x 10−4; OR = 10.13, 95% CI = 3.75 to 28.74.86), suggesting this result is not likely due to population stratification.
Germline pathogenic or likely pathogenic variants in DNA repair genes are enriched in neuroblastoma patients
BARD1 is known to bind BRCA1 and influence DNA repair (46). We hypothesized that neuroblastoma patients harbor an excess burden of pathogenic or likely pathogenic variants in DNA repair pathway genes overall. To explore this hypothesis, we interrogated genes in our 166-gene panel that intersected published DNA repair genes (47). A total of 48 DNA repair genes were assayed in the full study cohort and included in the analysis (Supplementary Table 2, available online). We observed 68 pathogenic or likely pathogenic variants in 27 distinct DNA repair genes, affecting 64 of 786 (8.1%) neuroblastoma patients (Figure 2, A, bottom panel). In contrast, only 362 of 6295 (5.8%) PMBB participants harbored a pathogenic or likely pathogenic variant (PPMBB = 0.011; OR = 1.45, 95% CI = 1.08 to 1.92). Similarly, only 959 of 15 708 (6.1%) gnomAD participants harbored a pathogenic or likely pathogenic variant in a DNA repair gene (PgnomAD = 0.028; OR = 1.36, 95% CI = 1.03 to 1.77).
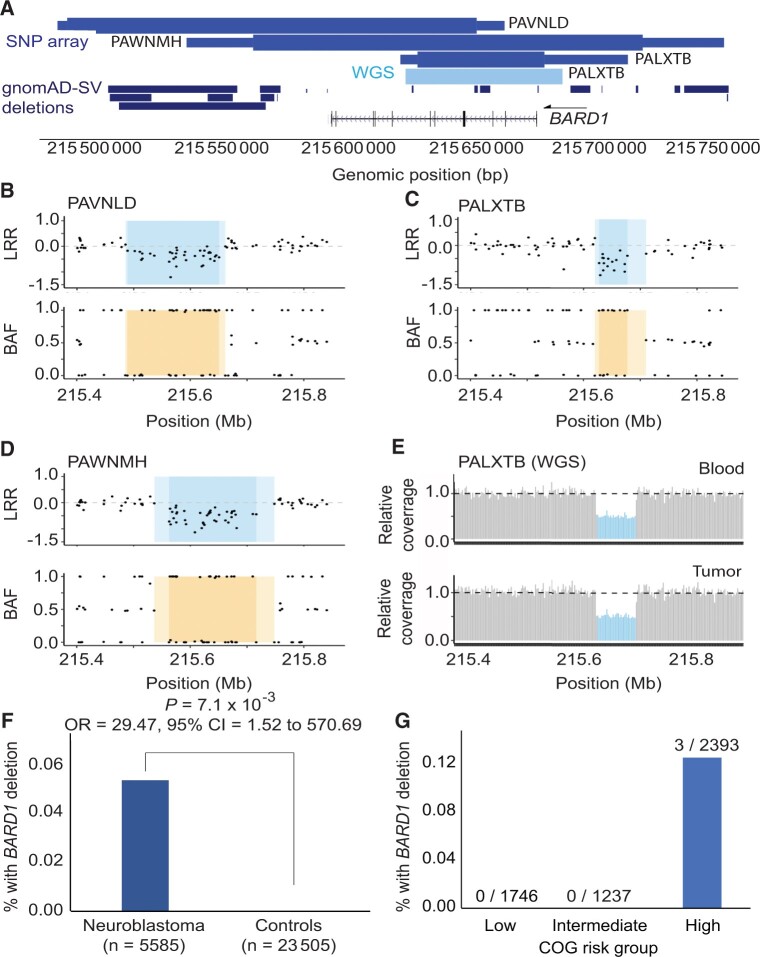
Germline copy number variants disrupting BARD1 are enriched in neuroblastoma
Given the enrichment of BARD1 rare pathogenic or likely pathogenic SNVs and indels in this study and the previous association of BARD1 common variants with neuroblastoma through GWAS, we next sought to determine if rare germline copy number variants at BARD1 also associate with neuroblastoma. We analyzed copy number variants in germline DNA array data from 5585 neuroblastoma patients and 23 505 cancer-free control children genotyped in our neuroblastoma GWAS efforts (22). We detected 3 focal germline deletions fully or partially encompassing BARD1 in neuroblastoma (0.05%; Figure 3, A-E; Supplementary Table 9, available online). No copy number variants affecting BARD1 were observed in 23 505 chip-matched cancer-free GWAS participants, and no protein coding deletions affecting BARD1 were observed in 10 847 individuals in the gnomAD v2.1 structural variant dataset (48) (Figure 3, A). The rare germline deletions at BARD1 were statistically significantly associated with neuroblastoma (P = 7.08 x 10−3; OR = 29.47, 95% CI = 1.52 to 570.70; Figure 3, F) and were detected only in high-risk neuroblastoma patients (Figure 3, G). Collectively, these data suggest that BARD1 alterations are an important genetic determinant of neuroblastoma, including common germline variants and rare germline SNVs, indels, and structural variants.
Figure 3.
Rare germline copy number variants disrupting BARD1 in neuroblastoma patients. A) BARD1 deletions were identified in 3 of 5585 neuroblastoma patients through copy number analysis of a large germline single nucleotide polymorphism (SNP) array dataset (medium blue, top track). No deletions were observed in 23 505 array-matched cancer-free control participants. The thick and thin bars represent minimum and maximum deletion coordinates, respectively. One deletion was validated and fine-mapped by whole genome sequencing (WGS; light blue, middle track). No BARD1 protein coding deletions were observed in 10 847 individuals in the gnomAD v2.1 structural variant dataset (dark blue, bottom track). B-D) The 3 array-based copy number variant calls are shown in log R ratio (LRR) and B allele frequency (BAF) plots. Darker shading indicates the minimum deleted region, whereas lighter shading indicates the maximum region. E) WGS validation for patient PALXTB is shown as relative sequencing coverage for matched blood and tumor samples. F) Rare BARD1 deletion copy number variants are enriched in neuroblastoma compared to cancer-free participants. G) Deletions disrupting BARD1 were observed exclusively in patients diagnosed with high-risk subset of neuroblastoma. gnomAD = Genome Aggregation Database.
Neuroblastoma patients harboring germline pathogenic or likely pathogenic variants in cancer predisposition genes have worse overall survival
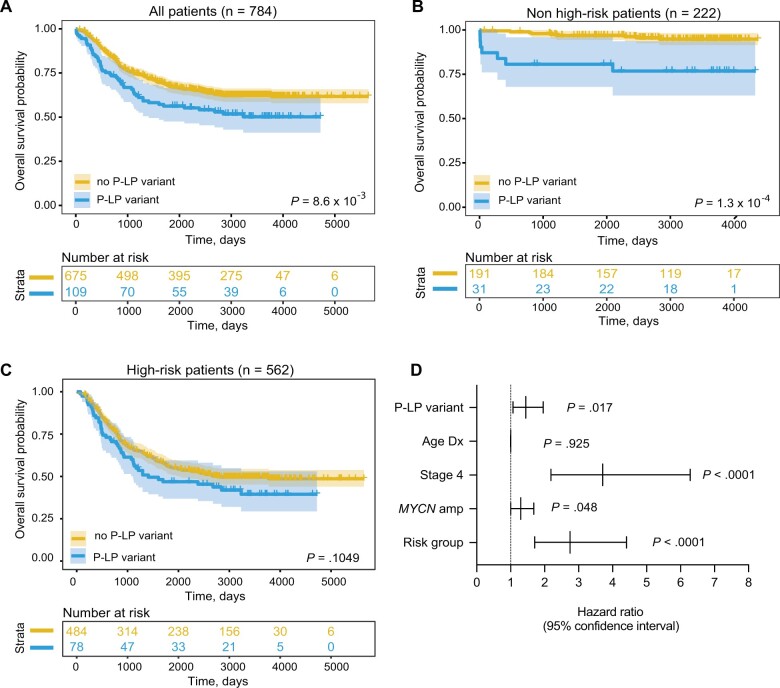
Finally, we investigated whether rare pathogenic or likely pathogenic variants (SNVs and indels) in cancer predisposition genes were associated with specific clinical and tumor biological characteristics and patient survival. A nominally statistically significant enrichment of pathogenic or likely pathogenic variants was observed in patients with tumors harboring loss of heterozygosity of chromosome 11q (P = .012); however, no association with age at diagnosis, stage, MYCN amplification status, COG risk group, or other characteristics was detected (Supplementary Table 10, available online). We repeated this analysis using the custom capture data only; however, the results were similar (Supplementary Table 11, available online). We next evaluated overall survival probability based on the presence or absence of germline pathogenic or likely pathogenic variants in cancer predisposition genes. We observed that patients with a germline pathogenic or likely pathogenic variant have worse overall survival compared with subjects without a germline pathogenic or likely pathogenic variant in a cancer predisposition gene (log-rank test P = 8.6 x 10−3; Figure 4, A). Furthermore, if restricted to only low- and intermediate-risk patients, overall survival remains worse for patients with a germline pathogenic or likely pathogenic variant (log-rank test: P = 1.3 x 10−4; Figure 4, B). A similar trend was observed when restricted to high-risk only, though this did not reach statistical significance (log-rank test P = .1049; Figure 4, C). Finally, a multivariate Cox proportional hazards regression model revealed the presence of a germline pathogenic or likely pathogenic variant was independently predictive of overall survival when considering age and diagnosis, INSS stage, MYCN amplification status, and COG risk group (Figure 4, D; P = .017; hazard ratio = 1.44, 95% CI = 1.07 to 1.96; Supplementary Table 12, available online). Taken together, these data demonstrate that the presence of a germline pathogenic or likely pathogenic variant in a cancer predisposition gene is associated with worse survival, independent of risk group.
Figure 4.
Neuroblastoma patients harboring germline pathogenic or likely pathogenic (P-LP) variants in cancer predisposition genes have worse overall survival. Kaplan–Meier plots of overall survival probability in neuroblastoma patients with and without P-LP variants cancer predisposition genes. A) All patients. B) Restricted to low- and intermediate-risk (non-high-risk) groups. C) Restricted to high-risk group. Statistical significance in panels A-C assessed by log-rank test (P < .05). D) Forest plot of hazard ratios from Cox proportional hazards model. amp = amplified; Dx = diagnosis.
Discussion
Neuroblastoma is a cancer of the developing sympathetic nervous system with an established genetic basis. Patients who present with a family history of the disease most commonly harbor rare pathogenic variants in ALK (6) or PHOX2B (7,8). In contrast, GWAS studies have identified common variation associated with sporadic neuroblastoma implicating more than a dozen susceptibility genes (49), including BARD1 (10). Germline sequencing studies have reported rare pathogenic variation in cancer predisposition genes, including APC, AXIN2, BARD1, BRCA1, BRCA2, CHEK2, LZTR1, PALB2, PINK1, SDHB, SMARCA4, and TP53 (50). Neuroblastoma has also been reported in several childhood-onset tumor-predisposition syndromes (50). However, a study of germline pathogenic variants in a clinically annotated cohort of patients large enough to investigate excess burden of pathogenic variation and clinical features associated with such variation has not been previously reported. Thus, in this study, we sought to define the prevalence, spectrum, and clinical significance of rare pathogenic germline variants in cancer predisposition genes in neuroblastoma.
To accomplish these goals, we analyzed germline DNA sequencing from 786 neuroblastoma patients with detailed clinical covariate and outcomes data. Using a conservative, clinically focused pipeline to classify pathogenicity of rare variants in cancer predisposition genes, we observed pathogenic or likely pathogenic germline variants in a substantial (13.9%) portion of neuroblastoma patients studied. This percentage is slightly higher but in-line with recent pan-childhood cancer germline studies (23,24,29,32,33,44). Two genes (ALK and BARD1) showed enrichment of pathogenic or likely pathogenic variants in neuroblastoma patients compared with independent cancer-free control cohorts after adjusting for multiple testing. Neuroblastoma patients carrying a germline pathogenic or likely pathogenic variant had worse overall survival compared with those without pathogenic or likely pathogenic variants, independent of age at diagnosis, INSS stage, MYCN amplification, and COG risk group.
The greatest number of pathogenic germline variants were observed in BARD1, BRCA2, ERCC2, CHEK2, and MSH3. Notably, BARD1, BRCA2, CHEK2, and MSH3 variants were primarily classified as pathogenic or likely pathogenic based on ClinVar annotation, suggesting that these variants have previously been observed in patients in a clinical lab. All 5 genes are involved in DNA repair, and indeed we observed an overall enrichment of pathogenic or likely pathogenic variants in DNA repair genes considered here. Although this finding requires validation using a full repertoire of DNA repair genes, the result suggests that neuroblastoma is another cancer initiated by germline defects in DNA repair. The current study was large enough to demonstrate a statistically significant enrichment of rare BARD1 pathogenic or likely pathogenic variants (SNVs and copy number variants) in neuroblastoma, adding to the common variants in BARD1 identified by GWAS and previously implicated in disease pathogenesis. In addition to BARD1, we observed 1.7% (13 of 786) of children in our cohort with a pathogenic or likely pathogenic variant in BRCA1, BRCA2, or a mismatch repair gene, which is similar to that observed (approximately 1.2%) in a large (n = 3975) meta-analysis of childhood cancer studies (51). Notably, 2 of these patients harbored multiple pathogenic or likely pathogenic variants in these genes, including 1 patient with a pathogenic or likely pathogenic variant in both BRCA1 and BRCA2 and the other patient with pathogenic or likely pathogenic variants in BRCA2 and MSH3.
In a parallel study, we used multiple lines of evidence to demonstrate the impact of BARD1 germline pathogenic or likely pathogenic variants identified here on DNA repair processes in neuroblastoma (52). Briefly, a subset of the rare BARD1 variants identified in the current study were introduced as monoallelic knock-ins in neuroblastoma cell models via CRISPR-Cas9 genome editing. These heterozygous variants induced BARD1 haploinsufficiency, DNA repair deficiency, ineffective RAD51 foci formation at DNA double-strand break sites, and enhanced sensitivity to cisplatin and poly-adenosine diphosphate ribose polymerase inhibition. Taken together, these data further implicate BARD1 and defective DNA repair as important driving factors in neuroblastoma tumorigenesis that may have important therapeutic implications.
Evidence for bi-allelic inactivation and/or loss of heterozygosity in neuroblastoma patients with pathogenic or likely pathogenic germline variants was observed in only 1 tumor and involved EZH2. There are several possible explanations for the low rate of bi-allelic inactivation in neuroblastoma. First, haploinsufficiency may be sufficient to tumorigenesis, as seen in our companion BARD1 functional studies (52). Alternatively, other inactivation mechanisms may be present but not detected by our approach (eg, epigenetic and noncoding alterations). Finally, there have been a limited number of patients with germline pathogenic or likely pathogenic variants and matched tumor data evaluated to date. Functional studies, such as those presented for BARD1 (52), and large cohort analyses incorporating the full spectrum of potential inactivation mechanisms are needed to resolve these important questions.
Universal germline and somatic genomic testing for adults with cancer has been advocated for using scientific and moral arguments (53). Similar arguments may also apply to patients diagnosed with neuroblastoma for multiple reasons: 1) from a prognostic standpoint, identification of a pathogenic or likely pathogenic variant predicts worse overall survival, independent of clinical risk stratification; 2) identification of pathogenic or likely pathogenic variants in some genes (eg, BARD1) may suggest eligibility for specific therapies, especially at time of relapse; 3) cascade testing of adult family members may guide gene-specific surveillance and therapies (eg, in BARD1, BRCA1, BRCA2, CHEK2, Lynch, PALB2, and TP53); 4) cascade testing of children for select genes (eg, TP53, PTPN11, and DICER1) may guide surveillance and therapy; and 5) identification of pathogenic variation in genes associated with specific syndromes (eg, PTCH1 in Gorlin syndrome, EZH2 in Weaver syndrome) may guide prognosis and clinical management. However, as the field is adopting paired germline-tumor DNA sequencing at the time of diagnosis for neuroblastoma and other pediatric cancer patients, there remains uncertainty on how to act on findings. This is particularly true for variants identified with low penetrance or lack of functional data or in clinical situations where access to high-quality genetic counseling is limited or there are challenges in performing effective surveillance (51). Genetic counseling should become an integral part of every patient’s multidisciplinary cancer care planning, something that has already been implemented in many large academic centers. Moreover, centralization of these data, through the Childhood Cancer Data Initiative or similar efforts, will facilitate longitudinal studies of patient survival and provide a resource for prioritizing functional and mechanistic studies to identify specific actionable insights to improve outcomes.
There are some limitations to this study. Patients studied were enrolled in the North American neuroblastoma biology study, ANBL00B1. These patients are predominantly of European ancestry and may not be representative of other geographic locations and ancestries. Second, we attempted to control for this in our burden testing; however, the use of different sequencing methods (whole genome sequencing, whole exome sequencing, custom capture) may affect variant detection at some genetic loci. Third, noncoding variants, epigenetic alterations, and an exhaustive set of structural variants were not analyzed here because of the different sequencing methods used. Finally, because of the lack of parental DNA, we cannot say whether the pathogenic or likely pathogenic variants identified in this study are inherited or acquired de novo. Patients in this study also did not include family history, multifocal tumor status, or secondary malignancy data. Additional large studies from diverse populations, including parental DNA and expanded annotations, are needed to replicate and extend our results. Recent sequencing studies, such as those supported by the Gabriella Miller Kids First research program, will be key in addressing many of these questions.
In conclusion, this study of 786 neuroblastoma patients found that 13.9% harbor rare germline pathogenic variants in 1 or more cancer predisposition genes. Rare pathogenic variants (SNVs and copy number variants) in BARD1 and other DNA repair genes were statistically significantly enriched in neuroblastoma compared with cancer-free controls. The presence of 1 or more germline pathogenic or likely pathogenic variants in a cancer predisposition gene was independently associated with worse overall survival. These data may be used to inform decision making regarding genetic testing and potential therapeutic options for children diagnosed with neuroblastoma.
Supplementary Material
Acknowledgements
This study involved collaboration with Penn Medicine BioBank and Regeneron Genetics Center. A complete list of individuals and their contributions is included in the Supplementary Information (available online). This work used resources of the NIH High Performance Computing Biowulf cluster and the Children’s Hospital of Philadelphia High Performance Compute cluster. The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; or the writing of the manuscript and decision to submit it for publication.
Contributor Information
Jung Kim, Clinical Genetics Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Rockville, MD, USA.
Zalman Vaksman, Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Laura E Egolf, Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Cell and Molecular Biology Graduate Group, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Rebecca Kaufman, Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
J Perry Evans, Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Karina L Conkrite, Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Arnavaz Danesh, Princess Margaret Cancer Centre, University Health Network, Toronto, Ontario, ON, Canada.
Gonzalo Lopez, Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Michael P Randall, Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Maiah H Dent, Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Lance M Farra, Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Neil L Menghani, Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Malwina Dymek, Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Heena Desai, Division of Hematology/Oncology, Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA; Abramson Family Cancer Research Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Ryan Hausler, Division of Hematology/Oncology, Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA; Abramson Family Cancer Research Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Belynda Hicks, Cancer Genome Research Laboratory, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Rockville, MD, USA.
Jaime Guidry Auvil, Office of Cancer Genomics, National Cancer Institute, Bethesda, MD, USA.
Daniela S Gerhard, Office of Cancer Genomics, National Cancer Institute, Bethesda, MD, USA.
Hakon Hakonarson, Center for Applied Genomics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Kara N Maxwell, Division of Hematology/Oncology, Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA; Abramson Family Cancer Research Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Kristina A Cole, Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Trevor J Pugh, Princess Margaret Cancer Centre, University Health Network, Toronto, Ontario, ON, Canada; Department of Medical Biophysics, University of Toronto, Toronto, ON, Canada.
Kristopher R Bosse, Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Abramson Family Cancer Research Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA; Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Javed Khan, Oncogenomics Section, Genetics Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA.
Jun S Wei, Oncogenomics Section, Genetics Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA.
John M Maris, Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Abramson Family Cancer Research Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA; Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Douglas R Stewart, Clinical Genetics Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Rockville, MD, USA.
Sharon J Diskin, Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Abramson Family Cancer Research Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA; Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Data availability
All neuroblastoma sequencing data analyzed in this study are available through the database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap/) under study-id phs000218 and accession number phs000467.
Author contributions
Jung Kim, PhD (Formal analysis; Investigation; Methodology; Visualization; Writing—original draft; Writing—review & editing), John M. Maris, MD (Data curation; Investigation; Writing—review & editing), Jun S. Wei, PhD (Investigation), Javed Khan, MD (Investigation), Kristopher R. Bosse, MD (Investigation), Trevor J. Pugh, PhD (Data curation; Supervision), Kristina A. Cole, MD, PhD (Investigation), Kara N. Maxwell, MD (Data curation; Investigation), Hakon Hakonarson, MD, PhD (Data curation), Daniela S. Gerhard, PhD (Data curation), Jaime Guidry Auvil, PhD (Data curation), Belynda Hicks, MS (Data curation), Ryan Hausler, MS (Data curation; Investigation), Heena Desai, MS (Data curation; Investigation), Malwina Dymek, BS (Investigation; Validation), Neil L. Menghani, MS (Investigation; Validation), Lance M. Farra, BS (Investigation; Validation), Maiah H. Dent, MS (Investigation; Validation), Michael P. Randall, MD (Investigation; Validation), Gonzalo Lopez, PhD (Data curation; Formal analysis; Investigation), Arnavaz Danesh, MS (Data curation; Formal analysis; Investigation), Karina L. Conkrite, BA (Investigation; Validation), J. Perry Evans, PhD (Formal analysis; Writing—review & editing), Rebecca Kaufman, MS (Data curation; Formal analysis; Investigation; Methodology; Validation; Writing—review & editing), Laura E. Egolf, PhD (Data curation; Formal analysis; Investigation; Methodology; Visualization; Writing—review & editing), Zalman Vaksman, PhD (Data curation; Formal analysis; Investigation; Methodology; Software; Validation; Visualization; Writing—original draft; Writing—review & editing), Douglas R. Stewart, MD (Funding acquisition; Investigation; Supervision; Writing—original draft; Writing—review & editing), and Sharon J Diskin, PhD (Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Resources; Software; Supervision; Validation; Visualization; Writing—original draft; Writing—review & editing).
Funding
This work was supported by a supplement to the Children’s Oncology Group Chair’s grant CA098543 and with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E to SJD and Complete Genomics. This work was also supported by National Institutes of Health grants R01CA204974 (SJD), R01CA237562 (SJD), R03CA230366 (SJD), R35CA220500 (M.M), K08CA230223 (KRB), K08CA215312 (KNM), a Howard Hughes Medical Institute Medical Fellows grant (MPR), Alex’s Lemonade Stand Foundation (KRB), the EVAN Foundation (KRB), Burroughs Wellcome Fund—1017184 (KNM), Basser Center for BRCA (KNM), the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and Center for Cancer Research of the National Cancer Institute, Bethesda, MD, and the Cancer Genome Research Laboratory, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Rockville, MD. KRB is a Damon Runyon Physician-Scientist supported (in part) by the Damon Runyon Cancer Research Foundation (PST-07-16).
Conflicts of interest
The authors have no disclosures.
References
- 1. Matthay KK, Maris JM, Schleiermacher G, et al. Neuroblastoma. Nat Rev Dis Primers. 2016;2:16078. doi: 10.1038/nrdp.2016.78 [DOI] [PubMed] [Google Scholar]
- 2. Pugh TJ, Morozova O, Attiyeh EF, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45(3):279-284. doi: 10.1038/ng.2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sausen M, Leary RJ, Jones S, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45(1):12-17. doi: 10.1038/ng.2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Molenaar JJ, Koster J, Zwijnenburg DA, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483(7391):589-593. doi: 10.1038/nature10910 [DOI] [PubMed] [Google Scholar]
- 5. Friedman DL, Kadan-Lottick NS, Whitton J, et al. Increased risk of cancer among siblings of long-term childhood cancer survivors: a report from the childhood cancer survivor study. Cancer Epidemiol Biomarkers Prev. 2005;14(8):1922-1927. doi: 10.1158/1055-9965.EPI-05-0066 [DOI] [PubMed] [Google Scholar]
- 6. Mossé YP, Laudenslager M, Longo L, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455(7215):930-935. doi: 10.1038/nature07261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mosse YP, Laudenslager M, Khazi D, et al. Germline PHOX2B mutation in hereditary neuroblastoma. Am J Hum Genet. 2004;75(4):727-730. doi: 10.1086/424530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trochet D, Bourdeaut F, Janoueix-Lerosey I, et al. Germline mutations of the paired-like homeobox 2B (PHOX2B) gene in neuroblastoma. Am J Hum Genet. 2004;74(4):761-764. doi: 10.1086/383253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maris JM, Mosse YP, Bradfield JP, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. New Engl J Med. 2008;358(24):2585-2593. doi: 10.1056/NEJMoa0708698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Capasso M, Devoto M, Hou C, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41(6):718-723. doi: 10.1038/ng.374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang K, Diskin SJ, Zhang H, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469(7329):216-220. doi: 10.1038/nature09609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nguyen LB, Diskin SJ, Capasso M, et al. Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility Loci. PLoS Genet. 2011;7(3):e1002026. doi: 10.1371/journal.pgen.1002026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diskin SJ, Capasso M, Schnepp RW, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet. 2012;44(10):1126-1130. doi: 10.1038/ng.2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diskin SJ, Capasso M, Diamond M, et al. Rare variants in TP53 and susceptibility to neuroblastoma. J Natl Cancer Inst. 2014;106(4):dju047. doi: 10.1093/jnci/dju047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McDaniel LD, Conkrite KL, Chang X, et al. Common variants upstream of MLF1 at 3q25 and within CPZ at 4p16 associated with neuroblastoma. PLoS Genet. 2017;13(5):e1006787. doi: 10.1371/journal.pgen.1006787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hungate EA, Applebaum MA, Skol AD, et al. Evaluation of genetic predisposition for MYCN-amplified neuroblastoma. J Natl Cancer Inst. 2017;109(10):djx093. doi: 10.1093/jnci/djx093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Diskin SJ, Hou C, Glessner JT, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459(7249):987-991. doi: 10.1038/nature08035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bosse KR, Diskin SJ, Cole KA, et al. Common variation at BARD1 results in the expression of an oncogenic isoform that influences neuroblastoma susceptibility and oncogenicity. Cancer Res. 2012;72(8):2068-2078. doi: 10.1158/0008-5472.CAN-11-3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oldridge DA, Wood AC, Weichert-Leahey N, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;528(7582):418-421. doi: 10.1038/nature15540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schnepp RW, Khurana P, Attiyeh EF, et al. A LIN28B-RAN-AURKA signaling network promotes neuroblastoma tumorigenesis. Cancer Cell. 2015;28(5):599-609. doi: 10.1016/j.ccell.2015.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Russell MR, Penikis A, Oldridge DA, et al. CASC15-S is a tumor suppressor lncRNA at the 6p22 neuroblastoma susceptibility locus. Cancer Res. 2015;75(15):3155-3166. doi: 10.1158/0008-5472.CAN-14-3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Egolf LE, Vaksman Z, Lopez G, et al. Germline 16p11.2 microdeletion predisposes to neuroblastoma. Am J Hum Genet. 2019;105(3):658-668. doi: 10.1016/j.ajhg.2019.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang J, Walsh MF, Wu G, et al. Germline mutations in predisposition genes in pediatric cancer. New Engl J Med. 2015;373(24):2336-2346. doi: 10.1056/NEJMoa1508054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parsons DW, Roy A, Yang Y, et al. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol. 2016;2(5):616-624., doi: 10.1001/jamaoncol.2015.5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gröbner SN, Worst BC, Weischenfeldt J, et al. ; ICGC PedBrain-Seq Project The landscape of genomic alterations across childhood cancers. Nature. 2018;555(7696):321-327. doi: 10.1038/nature25480 [DOI] [PubMed] [Google Scholar]
- 26. Wang Z, Wilson CL, Easton J, et al. Genetic risk for subsequent neoplasms among long-term survivors of childhood cancer. J Clin Oncol. 2018;36(20):2078-2087. doi:10.1200/JClin Oncol.2018.77.8589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fransson S, Martinez-Monleon A, Johansson M, et al. Whole-genome sequencing of recurrent neuroblastoma reveals somatic mutations that affect key players in cancer progression and telomere maintenance. Sci Rep. 2020;10(1):22432. doi: 10.1038/s41598-020-78370-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Akhavanfard S, Padmanabhan R, Yehia L, Cheng F, Eng C.. Comprehensive germline genomic profiles of children, adolescents and young adults with solid tumors. Nat Commun. 2020;11(1):2206. doi: 10.1038/s41467-020-16067-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fiala EM, Jayakumaran G, Mauguen A, et al. Prospective pan-cancer germline testing using MSK-IMPACT informs clinical translation in 751 patients with pediatric solid tumors. Nat Cancer. 2021;2:357-365. doi: 10.1038/s43018-021-00172-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim J, Gianferante M, Karyadi DM, et al. Frequency of pathogenic germline variants in cancer-susceptibility genes in the Childhood Cancer Survivor Study. JNCI Cancer Spectr. 2021;5(2):pkab007. doi: 10.1093/jncics/pkab007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wagener R, Taeubner J, Walter C, et al. Comprehensive germline-genomic and clinical profiling in 160 unselected children and adolescents with cancer. Eur J Hum Genet. 2021;29(8):1301-1311. doi: 10.1038/s41431-021-00878-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wong M, Mayoh C, Lau LMS, et al. Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat Med. 2020;26(11):1742-1753. doi: 10.1038/s41591-020-1072-4 [DOI] [PubMed] [Google Scholar]
- 33. Oberg JA, Glade Bender JL, Sulis ML, et al. Implementation of next generation sequencing into pediatric hematology-oncology practice: moving beyond actionable alterations. Genome Med. 2016;8(1):133. doi: 10.1186/s13073-016-0389-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brady SW, Liu Y, Ma X, et al. Pan-neuroblastoma analysis reveals age- and signature-associated driver alterations. Nat Commun. 2020;11(1):5183. doi: 10.1038/s41467-020-18987-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park J, Levin MG, Haggerty CM, et al. ; Regeneron Genetics Center A genome-first approach to aggregating rare genetic variants in LMNA for association with electronic health record phenotypes. Genet Med. 2020;22(1):102-111. doi: 10.1038/s41436-019-0625-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang L, Desai H, Verma SS, et al. ; Regeneron Genetics Center Performance of polygenic risk scores for cancer prediction in a racially diverse academic biobank. Genet Med. 2022;24(3):601-609. doi: 10.1016/j.gim.2021.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brohl AS, Patidar R, Turner CE, et al. Frequent inactivating germline mutations in DNA repair genes in patients with Ewing sarcoma. Genet Med. 2017;19(8):955-958. doi: 10.1038/gim.2016.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cingolani P, Platts A, Wang LL, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80-92. doi: 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang K, Li M, Hakonarson H.. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Landrum MJ, Lee JM, Benson M, et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862-868. doi: 10.1093/nar/gkv1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Q, Wang K.. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet. 2017;100(2):267-280. doi: 10.1016/j.ajhg.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bresler SC, Weiser DA, Huwe PJ, et al. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell. 2014;26(5):682-694. doi: 10.1016/j.ccell.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mody RJ, Wu Y-M, Lonigro RJ, et al. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA. 2015;314(9):913-925. doi: 10.1001/jama.2015.10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lasorsa VA, Formicola D, Pignataro P, et al. Exome and deep sequencing of clinically aggressive neuroblastoma reveal somatic mutations that affect key pathways involved in cancer progression. Oncotarget. 2016;7(16):21840-21852. doi: 10.18632/oncotarget.8187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tarsounas M, Sung P.. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat Rev Mol Cell Biol. 2020;21(5):284-299. doi: 10.1038/s41580-020-0218-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Knijnenburg TA, Wang L, Zimmermann MT, et al. ; Cancer Genome Atlas Research Network. Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell Rep. 2018;23(1):239-254 e236. doi: 10.1016/j.celrep.2018.03.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Collins RL, Brand H, Karczewski KJ, et al. ; Genome Aggregation Database Consortium. A structural variation reference for medical and population genetics. Nature. 2020;581(7809):444-451. doi: 10.1038/s41586-020-2287-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ritenour LE, Randall MP, Bosse KR, Diskin SJ.. Genetic susceptibility to neuroblastoma: Current knowledge and future directions. Cell Tissue Res. 2018;372(2):287-307. doi: 10.1007/s00441-018-2820-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Capasso M, Montella A, Tirelli M, et al. Genetic predisposition to solid pediatric cancers. Front Oncol. 2020;10:590033. doi: 10.3389/fonc.2020.590033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kratz CP, Smirnov D, Autry R, et al. Heterozygous BRCA1/2 and mismatch repair gene pathogenic variants in children and adolescents with cancer. J Natl Cancer Inst. 2022;114(11):1523-1532. doi: 10.1093/jnci/djac151 [DOI] [PubMed] [Google Scholar]
- 52. Randall MP, Egolf LE, Vaksman Z, et al. BARD1 germline variants induce haploinsufficiency and DNA repair defects in neuroblastoma. J Natl Cancer Inst. 2024;116(1):149-159. [DOI] [PMC free article] [PubMed]
- 53. Subbiah V, Kurzrock R.. Universal germline and tumor genomic testing needed to win the war against cancer: genomics is the diagnosis. J Clin Oncol. 2023;41(17):3100-3103. doi:10.1200/JClin Oncol.22.02833 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All neuroblastoma sequencing data analyzed in this study are available through the database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap/) under study-id phs000218 and accession number phs000467.