Abstract
Background
High-risk neuroblastoma is a complex genetic disease that is lethal in more than 50% of patients despite intense multimodal therapy. Through genome-wide association studies (GWAS) and next-generation sequencing, we have identified common single nucleotide polymorphisms and rare, pathogenic or likely pathogenic germline loss-of-function variants in BARD1 enriched in neuroblastoma patients. The functional implications of these findings remain poorly understood.
Methods
We correlated BARD1 genotype with expression in normal tissues and neuroblastomas, along with the burden of DNA damage in tumors. To validate the functional consequences of germline pathogenic or likely pathogenic BARD1 variants, we used CRISPR-Cas9 to generate isogenic neuroblastoma (IMR-5) and control (RPE1) cellular models harboring heterozygous BARD1 loss-of-function variants (R112*, R150*, E287fs, and Q564*) and quantified genomic instability in these cells via next-generation sequencing and with functional assays measuring the efficiency of DNA repair.
Results
Both common and rare neuroblastoma-associated BARD1 germline variants were associated with lower levels of BARD1 mRNA and an increased burden of DNA damage. Using isogenic heterozygous BARD1 loss-of-function variant cellular models, we functionally validated this association with inefficient DNA repair. BARD1 loss-of-function variant isogenic cells exhibited reduced efficiency in repairing Cas9-induced DNA damage, ineffective RAD51 focus formation at DNA double-strand break sites, and enhanced sensitivity to cisplatin and poly (ADP-ribose) polymerase (PARP) inhibition both in vitro and in vivo.
Conclusions
Taken together, we demonstrate that germline BARD1 variants disrupt DNA repair fidelity. This is a fundamental molecular mechanism contributing to neuroblastoma initiation that may have important therapeutic implications.
High-risk neuroblastoma remains a substantial clinical challenge, with mortality exceeding 50% despite intensive multimodal chemoradiotherapy and immune-based treatment regimens (1). In recent years, the genetic basis of neuroblastoma has come into focus. Germline variants in ALK are the predominant cause of familial neuroblastoma, accounting for 1%-2% of overall patient cases (2), mutations in PHOX2B cause neuroblastoma in the context of a global neurocristopathy that can be familial (3,4), and multiple genomic loci have been implicated in predisposition to the more common sporadic neuroblastoma through a large genome-wide association study (GWAS) (5,6). One of the most statistically significant neuroblastoma-associated genomic regions in this GWAS is a complex linkage disequilibrium block centered on the BRCA1-associated RING domain 1 (BARD1) gene (7). The BARD1 protein is a well-documented heterodimerization partner of BRCA1 (8,9), a critical interaction required for BRCA1 stability, ubiquitin ligase activity, and other critical cellular functions including repair of DNA double-strand breaks by homologous recombination (10-14). Common single nucleotide polymorphisms (SNPs) at the BARD1 locus are associated with neuroblastoma across multiple ethnicities and are enriched in high-risk patients (5,7,15,16). Additionally, BARD1 risk variants correlate with increased expression of an oncogenically activated BARD1 isoform and reduced expression of full-length BARD1 (17-19).
More recently, rare coding variants with larger predicted effect sizes were also found to be enriched in the germline of sporadic neuroblastoma patients, including multiple putative loss-of-function variants in the BARD1 gene (20-22). Notably, BARD1 coding variants are also enriched in the germline of patients with several other malignancies (23-29), suggesting a potential shared mechanism of tumor predisposition across multiple cancers. Large germline sequencing studies in pediatric and adult cancers have successfully described the landscape of cancer-associated germline variation across many malignancies (30-32), but the precise functional implications of these germline variants on tumor development remain largely undefined. To address this need, here, we evaluated the functional impact of both common and rare germline variation at the BARD1 locus in neuroblastoma. Specifically, we aimed to determine whether BARD1 variants perturb DNA repair efficiency in neuroblastoma cells.
Methods
Detailed methods are provided in the Supplementary Methods (available online). BARD1 isogenic cellular models were generated via CRISPR-Cas9 where Cas9 enzyme, guide RNAs, and single-stranded mutated donor oligonucleotides were introduced into neuroblastoma cells via electroporation. Genotypes were confirmed by Sanger sequencing. Quantitative real-time polymerase chain reaction results were derived via the 2–ΔΔCt method. Whole genome sequencing was performed on isogenic and control cellular models. Quantification of DNA structural variants, copy number, indels, single-nucleotide variants, and double-strand breaks was performed as previously described (33-37). Immunofluorescence images were obtained using a Leica fluorescence microscope and standard staining protocols. RAD51 foci were quantified using Focinator v2.0 software (38). The Clover-LMNA assay was performed according to published methods (39). Cytotoxicity studies were performed via serial dilution of drugs and vehicle controls, and cell viability was measured via CellTiter-Glo assays. In vivo studies were performed as previously described (40) using murine xenografts generated from BARD1 isogenic cells.
Quantification and statistical analysis
Differences between groups were presented as the mean (SD) as noted in the figure legends. All statistical analyses were 2-sided and done with GraphPad Prism. P values less than .05 were considered statistically significant.
Results
Neuroblastoma-associated BARD1 common germline variation correlates with increased somatic DNA double-strand breaks
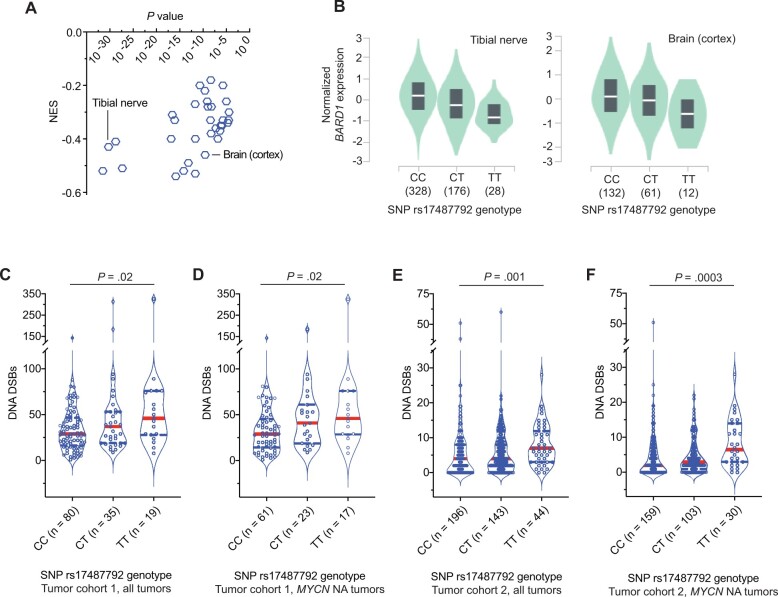
We previously reported that common SNPs at the BARD1 locus are associated with neuroblastoma predisposition across multiple ethnicities (7,15,16,18) and that a subset of these SNPs are correlated with the expression of an oncogenically activated BARD1 isoform (17). However, we suspect that there are additional mechanisms involving BARD1 that contribute to neuroblastoma predisposition. In view of BARD1’s critical role in stabilizing BRCA1 and facilitating accurate DNA repair (11,41,42), we first tested the hypothesis that common BARD1 variation is associated with reduced BARD1 expression that results in a genome-wide DNA repair deficiency. We selected the most statistically signficant, directly genotyped common risk allele (T) at SNP rs17487792 from our GWAS (5) for further analysis. We first queried the multitissue expression quantitative trait loci data from the Genotype-Tissue Expression portal (https://gtexportal.org/) for the risk allele of this SNP and found that the T risk allele was associated with a reduction in BARD1 expression across 33 normal tissues, notably including multiple nervous system tissues (Figure 1, A and B; P = 1 x 10−5 - 3 x 10−32). Next, to determine if there was any association between the rs17487792 T risk allele and genomic instability in neuroblastoma tumors, we used 2 large sets of primary neuroblastoma tumors subjected to either paired tumor-normal whole-genome sequencing (n = 134 tumors; cohort 1) or SNP genotyping (n = 383 tumors; cohort 2). Using methods previously described (33,34), we quantified the tumor burden of DNA double-strand breaks and correlated these data with rs17487792 genotype in these 2 tumor datasets. Tumors arising in children harboring a germline homozygous risk allele genotype (T/T) at SNP rs17487792 had an increased burden of double-strand breaks when compared with the homozygous nonrisk allele genotype at this SNP (C/C; Figure 1, C-F). This correlation between SNP rs17487792 genotype and quantity of DNA double-strand breaks was more pronounced in patients harboring tumors without MYCN amplification, an association that was also found when limiting these analyses to only high-risk neuroblastomas (Supplementary Figure 1, A-F, available online). Taken together, these findings suggest that deficiencies in DNA repair associated with decreased BARD1 expression are an additional mechanism by which common variants at the BARD1 locus contribute to neuroblastoma predisposition.
Figure 1.
Common BARD1 germline risk variants correlate with decreased BARD1 expression and genome-wide deficiencies in DNA repair. A) Plot of the normalized effect size (NES) of the single nucleotide polymorphism (SNP) rs17487792 expression quantitative trait loci vs P value for 33 human tissues derived from the Genotype-Tissue Expression (GTEx) project. B) Plots of BARD1 expression vs SNP rs17487792 genotype in tibial nerve (left) and brain cortex (right) derived from the GTEx project. C-F) Violin plots depicting the number of DNA double-strand breaks (DSBs) in neuroblastoma tumors from patients with different germline SNP rs17487792 genotypes. Panels C and D depict quantity of DNA DSBs in cohort 1 among all tumors or among only tumors without MYCN amplification, respectively. Panels E and F depict quantity of DNA DSBs in cohort 2 among all tumors or among only tumors without MYCN amplification, respectively. Red solid line denotes median, and blue dotted lines denote quartiles. MYCN NA = MYCN non-amplified; C = cytosine; T = thymine.
Generation of isogenic neuroblastoma cellular models harboring neuroblastoma-associated BARD1 germline loss-of-function variants (BARD1+/mut)
In a parallel study of 786 patients with neuroblastoma, we observed that BARD1 was the most altered cancer predisposition gene in the germline of neuroblastoma patients (22). We identified rare pathogenic or likely pathogenic nonsense, frameshift, or splice site variants in BARD1 in the germline DNA in 8 (1%) of the 786 patients (22). These variants were distributed throughout the BARD1 coding sequence and displayed no evidence of somatic loss of heterozygosity (LOH) at the BARD1 locus in available matched tumor DNA (Supplementary Table 1, available online). Further, many of these variants are enriched in the germline DNA of adults with other malignancies (Supplementary Table 1, available online).
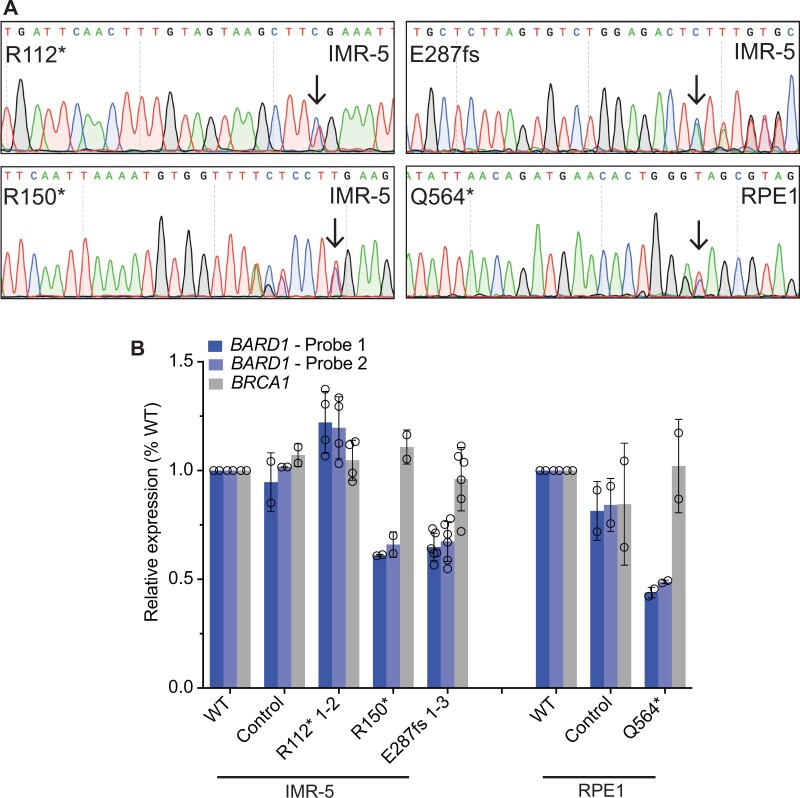
We next aimed to study the functional implications of these germline pathogenic and likely pathogenic loss-of-function BARD1 variants, focusing on DNA repair mechanisms of the BARD1-BRCA1 heterodimer (11,41,42). We introduced a subset of these BARD1 variants as monoallelic knock-ins via CRISPR-Cas9 genome editing (Supplementary Tables 1 and 2, available online) utilizing 2 complementary cell lines to study their functional impact: IMR-5 (a MYCN-amplified, TP53 wild-type neuroblastoma cell line; hereafter designated as IMR-5 BARD1+/mut) and hTERT RPE1 (an immortalized cell line of neural crest origin; hereafter designated as RPE1 BARD1+/mut) (43). We chose to focus on the identified pathogenic and likely pathogenic BARD1 loss-of-function nonsense and frameshift variants rather than the BARD1 splice site alterations or the common noncoding SNP variations identified via GWAS, hypothesizing that these loss-of-function variants may result in the most reproducible phenotypes. We successfully engineered 4 of these variants (BARD1R112*, R150*, E287fs, and Q564*) across these 2 cellular models (Supplementary Table 1, available online). Heterozygosity for the appropriate BARD1 variant in the cells to be used for functional assays was confirmed by Sanger sequencing (Figure 2, A). The most likely exonic off-target CRISPR sites with cutting frequency determination scores of at least 0.04 (n = 2-3 loci per mutation) as determined by the CRISPOR tool (44) were also sequenced to ensure no aberrant Cas9 editing had occurred (Supplementary Table 3, available online). Clones with no evidence of editing at either BARD1 allele, which had undergone similar single-cell selection pressure, were chosen at random to use as controls for subsequent sequencing and functional studies.
Figure 2.
Neuroblastoma cells heterozygous for disease-associated BARD1 loss-of-function variants (BARD1+/mut) have reduced BARD1 expression. A) Representative chromatograms from IMR-5 and RPE1 BARD1+/mut isogenic cell lines. Black arrows indicate CRISPR-introduced BARD1 heterozygous variants. Other variants reflect synonymous protospacer adjacent motif (PAM) changes (R150*, Q564*) or frameshift-induced nucleotide alterations (E287fs). B) BARD1 and BRCA1 expression in IMR-5 BARD1+/mut (left) and RPE1 BARD1+/mut (right) cells and nontargeted clonal control cells. BARD1 expression measured via 2 unique TaqMan probes. Data in panel B is represented as means (SD) of 2 biological replicates of each unique cell line, including multiple cell lines with identical BARD1 variants (n = 2 IMR-5 BARD1+/R112*; n = 1 for IMR-5 BARD1+/R150*; and n = 3 for IMR-5 BARD1+/E287fs cell lines). WT = wild-type.
Cells heterozygous for BARD1 loss-of-function variants have reduced BARD1 expression
We first sought to characterize BARD1 expression in these IMR-5 and RPE1 BARD1+/mut cellular models, given that similar BRCA1 monoallelic variants induce functionally relevant BRCA1 haploinsufficiency (45,46) and considering our findings that neuroblastoma-associated common BARD1 variation is associated with decreased BARD1 expression. Via real-time polymerase chain reaction using 2 unique BARD1 TaqMan probes, we found that 3 of 4 BARD1+/mut isogenic cellular models exhibited a substantial reduction in BARD1 expression compared to wild-type parental cells or nontargeted clonal control cells (33%-56% reduction in BARD1 mRNA in IMR-5/RPE1 BARD1+/mut cells; Figure 2, B). However, IMR-5 BARD1+/R112* cells had BARD1 mRNA expression comparable to wild-type cells and nontargeted clonal control cells (Figure 2, B). The BARD1R112* variant is unique among the prioritized BARD1 nonsense and frameshift variants studied in that it has a nearby downstream putative start codon (M145) distal to the aberrant stop codon, potentially allowing for resumption of translation and avoidance of nonsense-mediated decay, a phenomenon that has been previously described (47). We also quantified BRCA1 mRNA as a control and found no difference in BRCA1 mRNA expression between wild-type, clonal control cells, and BARD1+/mut isogenic cells (Figure 2, B). Thus, most of the prioritized neuroblastoma associated BARD1 loss-of-function germline variants result in reduced BARD1 expression.
BARD1 +/mut cells exhibit widespread genomic instability
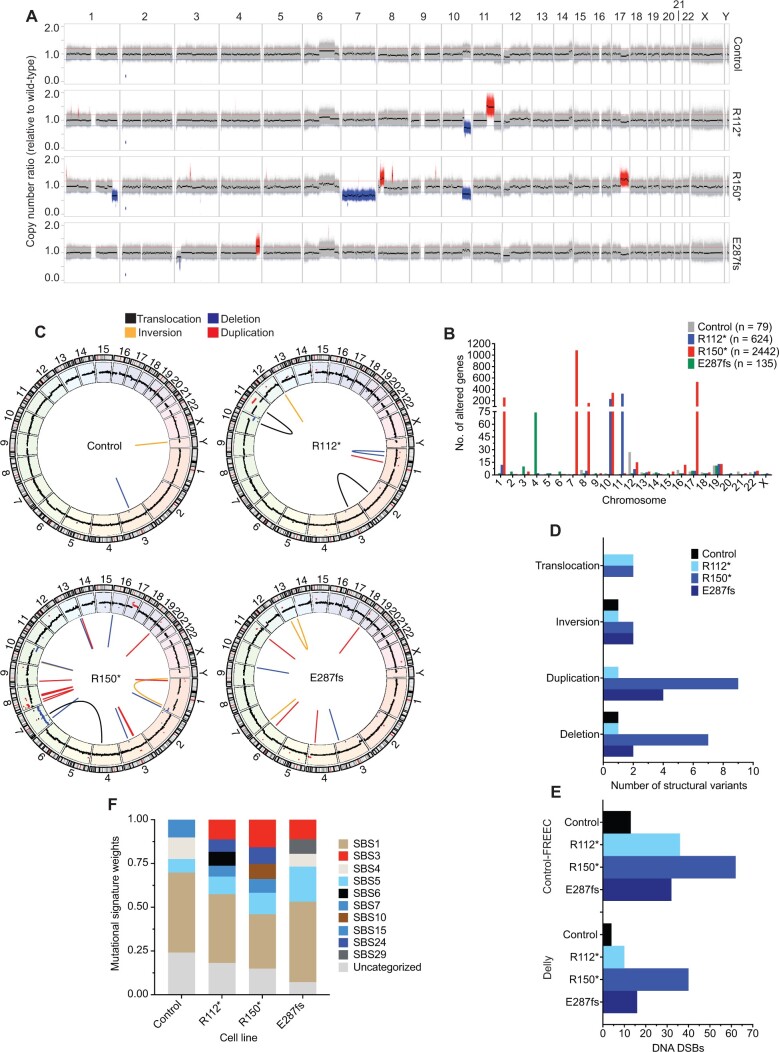
The BRCA1-BARD1 heterodimer is essential for maintaining genomic integrity, in part via the homology-directed repair of DNA double-strand breaks, and substantial homology-directed repair defects have been observed in cellular models with heterozygous loss-of-function BRCA1 mutants (10,45,48). Considering these data, along with the correlation of neuroblastoma-associated common variation with an increased number of DNA double-strand breaks, we first quantified the genome-wide impact of this BARD1 haploinsufficiency on DNA repair in isogenic IMR-5 BARD1+/mut cells. After 20 passages in cell culture, genomic DNA from 3 representative IMR-5 BARD1+/mut clones, a nontargeted control clone, and wild-type parental cells were examined via whole-genome sequencing. We identified genomic aberrations that each clonal cell line acquired relative to the wild-type parental cell line by applying Control-FREEC (35) and Delly (36) as complementary algorithms for copy number and structural variant analysis, respectively. Striking genomic instability was observed uniquely in the IMR-5 BARD1+/mut isogenic cell lines, including large-scale copy number alterations (Figure 3, A; Supplementary Figure 2, A, available online) altering a substantial number of genes (Figure 3, B), along with an increase in structural variants (Figure 3, C and D; Supplementary Figure 2, B and C, available online). We also quantified DNA double-strand breaks (33,34) based on Control-FREEC and Delly calls, and all 3 IMR-5 BARD1+/mut isogenic cell lines harbored increased double-strand breaks compared with the nontargeted control clone (Figure 3, E). These results were consistent using stringent (Figure 3, C-E; Supplementary Figure 2, B and C, available online) and more relaxed structural variant filtering parameters (Supplementary Figure 3, A-E, available online). Parallel analyses with MuTect (37) also revealed an increase in single-nucleotide variants and indels in IMR-5 BARD1+/mut clones vs the control clone, with variants in BARD1+/mut clones exhibiting higher allele frequencies (Supplementary Figure 4, A-C, available online). Finally, analysis of mutational signatures in the nontargeted control and isogenic IMR-5 BARD1+/mut cell lines revealed enhanced exposure of SBS3 (defective homologous recombination DNA repair; BRCA1 and 2 mutation) in the isogenic cell lines, along with other DNA repair deficiency signatures (SBS6, defective DNA mismatch repair; SBS10, POLE mutation) uniquely found in a subset of the isogenic IMR-5 BARD1+/mut cell lines but not the nontargeted control cells (Figure 3, F; Supplementary Figure 4, D, available online).
Figure 3.
Neuroblastoma IMR-5 BARD1+/mut cell lines exhibit widespread genomic instability. A) Copy number ratio across the genome for the IMR-5 nontargeted control clone and BARD1+/mut isogenic clones relative to wild-type parental IMR-5 cells. Large acquired genomic losses are indicated in blue (fold change < 0.8) and gains in red (fold change > 1.2). B) Quantification of the number of genes found within copy number variant-altered genomic regions in the IMR-5 BARD1+/mut and control isogenic cell lines shown in panel A. Total number of altered genes for each isogenic cell line is shown in the legend. C) Circos plots depicting structural variants identified in nontargeted control and BARD1+/mut IMR-5 cells. Copy number segments from panel A are shown in the outer circle for reference. D) Counts of structural variants in nontargeted control and BARD1+/mut IMR-5 cells. E) Counts of DNA double-strand breaks in nontargeted control and BARD1+/mut IMR-5 cells, quantified from the Control-FREEC copy number data (top) and Delly structural variant data (bottom). F) Plot of mutational signature weights in nontargeted control and BARD1+/mut IMR-5 cells using COSMIC mutational signatures v2. DSB = double-strand break.
BARD1 +/mut cells are deficient in DNA repair and are more sensitive to cisplatin and poly (ADP-ribose) polymerase (PARP) inhibition
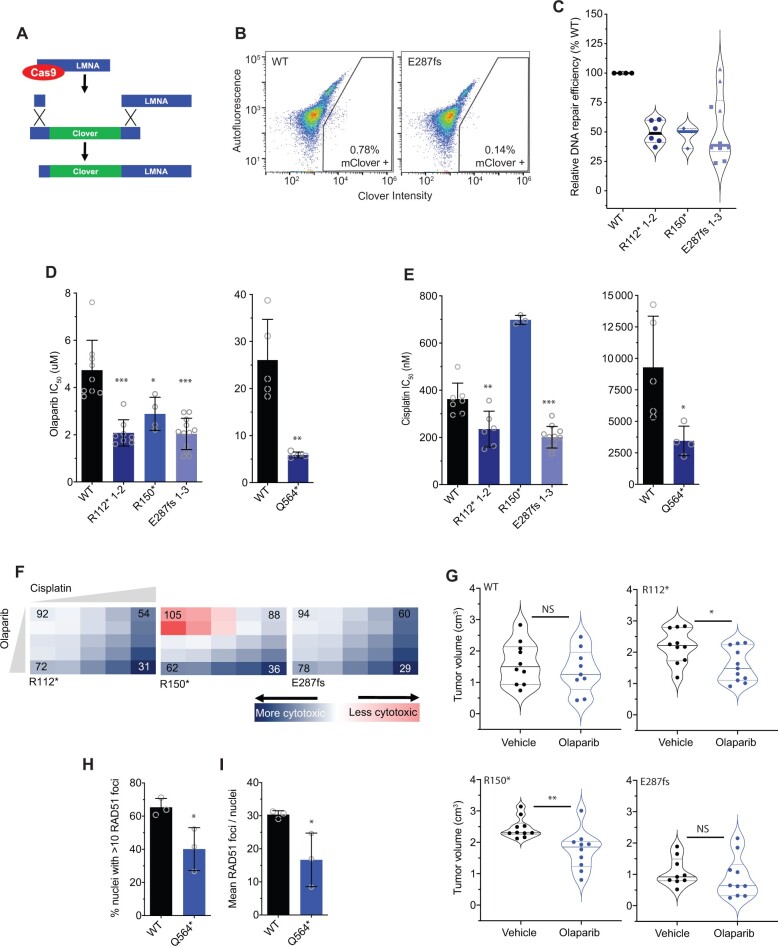
We next used several complementary functional studies to evaluate the ability of BARD1+/mut isogenic cells to perform efficient DNA repair. First, we used a CRISPR-based in vitro assay incorporating a fluorescent endogenous Clover tag (Figure 4, A) to directly quantify homology-directed repair efficiency in the BARD1+/mut isogenic models via flow cytometry (39). We found that after Cas9-induced DNA cutting, IMR-5 BARD1+/mut isogenic cellular models consistently integrated the DNA repair template with the Clover tag at approximately 50% the efficiency of wild-type IMR-5 cells (relative mean Clover-positive BARD1+/mut isogenic cells 46%-53% of wild-type IMR-5 cells; Figure 4, B and C). Notably, the relative reduction in homology-directed repair capacity was consistent among the different isogenic cell lines, except 1 of the 3 IMR-5+/E287fs isogenic cell clones, which had similar levels of Clover tag integration as wild-type IMR-5 cells (Figure 4, C, blue triangles).
Figure 4.
BARD1 +/mut cells are deficient in DNA repair and sensitive to poly (ADP-ribose) polymerase (PARP) inhibition. A) Schematic of the Clover-LMNA homology-directed repair assay (39). B) Representative flow cytometry plots of IMR-5 wild-type (WT) and BARD1+/E287fs cells cotransfected with pX330-LMNA1 guide RNA and pCR2.1 Clover-LMNA repair template plasmids with gating strategy for Clover-positive cells indicated. C) Violin plots showing relative DNA damage repair efficiency across IMR-5 BARD1+/mut cells as quantified with the Clover-LMNA homology-directed repair assay shown in panels A and B. D) Olaparib IC50 values in IMR-5 WT and BARD1+/mut cell lines (left) and in RPE1 WT and BARD1+/mut cell lines (right). E) Cisplatin IC50 values in IMR-5 WT and BARD1+/mut cell lines (left) and in RPE1 WT and BARD1+/mut cell lines (right). F) Relative cytotoxicity of combined olaparib and cisplatin in IMR-5 BARD1+/mut cell lines compared with WT. Each square represents a single dose combination. Blue squares represent drug combinations at which greater cytotoxicity is observed in BARD1+/mut cells; red squares represent drug combinations at which greater cytotoxicity is observed in WT cells. Numbers in corner cells represent the percent of isogenic cells alive compared with WT cells alive at equivalent doses. G) Violin plots of tumor volumes after 2 weeks of olaparib treatment in BARD1+/mut vs WT IMR-5 cell line xenografts. (n = 9-11 mice per cohort). Tumor volumes for olaparib-treated IMR-5 BARD1+/E287fs xenografts measured on day 13. Solid lines denote medians, and dotted lines denote quartiles. H) Proportion of RPE1 WT and BARD1+/Q564* cell nuclei with more than 10 RAD51 foci after treatment with cisplatin. I) Mean RAD51 foci per RPE1 WT and BARD1+/Q564* cell nucleus after treatment with cisplatin. Data in panel C are means (SD) of 3-10 biological replicates of each isogenic cell line, including multiple cell lines with identical BARD1 variants (n = 2 IMR-5 BARD1+/R112*; n = 1 for IMR-5 BARD1+/R150*; and n = 3 for IMR-5 BARD1+/E287fs cell lines). Data in panels D and E are means (SD) of 3-12 biological replicates of each isogenic cell line, including multiple cell lines with identical BARD1 variants (n = 2 IMR-5 BARD1+/R112*; n = 1 for IMR-5 BARD1+/R150*; and n = 3 for IMR-5 BARD1+/E287fs cell lines). Data in panels H and I represent means (SD) of 3 biological replicates for each cell line with a total of 1262 nuclei (range = 245-718 nuclei per replicate) analyzed for RPE1 WT cells and a total of 1596 nuclei (range = 300-905 nuclei per replicate) for RPE1 BARD1+/Q564*cells. *P < .05, **P < .01, ***P < .0001. LMNA = lamin A/C; NS = not statistically significant as measured by t test.
We next investigated whether BARD1+/mut cells displayed increased vulnerability to PARP inhibition, another well-validated marker of DNA damage repair deficiency (49). We treated both wild-type and isogenic IMR-5 BARD1+/mut cells with the PARP inhibitor olaparib (0.1-100 μM) and assessed cytotoxicity after 4 days (Figure 4, D). IMR-5 BARD1+/mut cells exhibited enhanced sensitivity to olaparib compared with wild-type cells (mean IMR-5+/mut IC50 2.1-2.9 μM vs 4.7 μM for wild-type IMR-5 cells; P < .05; Figure 4, D, left), as did RPE1 BARD1+/Q564* cells (mean IC50 5.9 μM vs 26.0 μM for wild-type RPE1 cells; P < .01; Figure 4, D, right). Similarly, except for IMR-5 BARD1+/R150*, BARD1+/mut cells also displayed increased sensitivity to the DNA intercalating agent cisplatin (mean IC50 201-235 nM for IMR-5 BARD1+/mut cells vs 362 nM for wild-type IMR-5 cells [P < .01; Figure 4, E, left] and 3450 nM for RPE1 BARD1+/mut cells vs 9282 nM for wild-type RPE1 cells [P < .05; Figure 4, E, right]). To further validate these data, we designed a cytotoxicity assay incorporating olaparib and cisplatin at escalating doses to evaluate whether the combination was also more potent in IMR-5 BARD1+/mut cells than wild-type IMR-5 cells. Compared with wild-type IMR-5 cells, BARD1+/R112* and BARD1+/E287fs IMR-5 cells were more sensitive to these drugs at every dose combination tested, and BARD1+/R150* cells were more sensitive at most doses (Figure 4, F). Next, to confirm the sensitivity of BARD1+/mut cells to olaparib, we treated cohorts of mice xenografted with each isogenic line with 20 mg/kg of olaparib daily for 28 days (50). Olaparib-treated BARD1+/R112* and BARD1+/R150* isogenic xenografts had a statistically significant reduction in tumor growth vs paired vehicle-treated, and wild-type IMR-5 and the much slower-growing IMR-5 BARD1+/E287fs xenografts did not show any olaparib-induced tumor growth delay (Figure 4, G; Supplementary Figure 5, A-D, available online). Mice harboring the BARD1+/R150* xenografts also had a longer progression-free survival compared with paired vehicle treated mice (P < .01; Supplementary Figure 5, E-H, available online).
Finally, we used the RPE1 isogenic model to evaluate the ability of BARD1+/mut cells to form RAD51 foci after treatment with cisplatin (51). The larger nucleus of RPE1 cells facilitated increased resolution of distinct RAD51 foci by immunofluorescence. Twenty-four hours after treatment with cisplatin, fewer RPE1 BARD1+/Q564* cells exhibited more than 10 RAD51 foci per nucleus (mean of 40.1% vs 65.3% for wild-type RPE1 cells; P < .05; Figure 4, H; Supplementary Figure 5, I, available online), and nuclei from RPE1 BARD1+/Q564* cells had fewer average RAD51 foci per nucleus overall (mean of 17 vs 30 for RPE1 wild-type cells; P < .05; Figure 4, I;Supplementary Figure 5, I, available online). Taken together, these findings suggest that neuroblastoma-associated BARD1 loss-of-function variants markedly impair DNA repair processes.
Discussion
Neuroblastoma, like all human cancers, is a genetic disease. Common germline alleles at several genomic loci (eg, BARD1, LMO1, CASC15) contribute to sporadic neuroblastoma predisposition, and rare heterozygous variants in neurodevelopmental genes (PHOX2B and ALK) underlie familial neuroblastoma (2-4,20). More recently, next-generation sequencing efforts focused on germline DNA from neuroblastoma patients have also identified enrichment of potentially pathogenic variants in cancer predisposition genes; among these, the most frequently altered gene is BARD1 (22). Although substantial progress has been made in defining the landscape of rare germline variation in cancer predisposition genes across pediatric and adult cancers (30-32), less effort has concentrated on elucidating how these disease-associated genetic variants influence cancer development at the molecular level, especially in pediatric malignancies. Thus, the functional validation of neuroblastoma-associated BARD1 germline variants described here not only further enhances our understanding of the contribution of the BARD1 locus to neuroblastoma predisposition but also represents a critical attempt to define the functional and potential clinical relevance of cancer-associated germline variation.
Given that BARD1 is one of the most statistically significant and replicated neuroblastoma-associated GWAS loci, we first looked for genome-wide evidence of a DNA damage repair defect in tumors from patients harboring a common BARD1 risk haplotype. We quantified DNA double-strand breaks in 2 large sets of neuroblastoma tumors and observed a correlation between germline SNP genotype and somatic DNA damage, which was most robust in tumors without MYCN amplification. Given MYCN’s central role in response to DNA damage (52,53), this suggests a potential compensatory mechanism for DNA repair in neuroblastoma cells with high levels of MYCN. However, given the low effect sizes of these neuroblastoma-associated common variants, we chose to primarily focus on functional validation of a subset of the recently identified loss-of-function BARD1 germline variants with much larger predicted effect sizes. Our findings support a model in which BARD1 loss-of-function variants induce BARD1 haploinsufficiency leading to genomic instability from deficient DNA damage repair. This model may be immediately relevant to other predicted loss-of-function variants in DNA repair–related genes that are enriched in the germline of children (and adults) with multiple tumor histotypes (30,32,54).
In further considering this haploinsufficiency model for heterozygous BARD1 loss-of-function variants in neuroblastoma tumorigenesis, it is important to note that the traditional model for BRCA1- and BRCA2-associated familial malignancies involves early LOH of the wild-type allele (55). Further, although emerging evidence suggests that a subset of breast and ovarian tumors from individuals with BRCA1 or BRCA2 germline variants lack BRCA1 and 2 locus-specific LOH, these heterozygous tumors exhibit homologous recombination deficiency scores similar to non-BRCA1 and 2 mutated tumors (56). In contrast, a single hit to a homologous recombination pathway gene may be sufficient to induce homologous recombination deficiency in neuroblastoma and other pediatric cancers as observed in a recent pan-pediatric cancer study (57) that identified 8 patients with monoallelic germline variants in homologous recombination pathway genes such as BRCA2. Although none of the matched tumors in this study carried a second hit to induce locus-specific LOH, they exhibited mutational signatures consistent with homologous recombination deficiency. Additionally, considering that homozygous loss of either BARD1 or BRCA1 results in embryonic lethality in murine models (58), LOH may not be well tolerated as an early event in pediatric cancers that commonly initiate during embryogenesis.
Interestingly, although the prioritized BARD1 pathogenic and likely pathogenic variants led to overall comparable defects in DNA repair, there were some notable differences in how each isogenic cell line performed in a subset of these functional assays. Considering the number of genomic alterations we observed by sequencing these BARD1 loss-of-function mutant isogenic cells, it is possible that acquired genetic changes uniquely endowed individual models with other tumor-enhancing properties. For example, although the isogenic IMR-5 BARD1+/R150* model showed increased sensitivity to PARP inhibition like other BARD1+/mut isogenic cell lines, these cells appeared to have uniquely acquired resistance to cisplatin. The IMR-5 BARD1+/R150* model harbored the largest number of altered genes within acquired copy number variants, with the observed copy number changes in at least 5 of these genes being previously implicated in platinum resistance (59). These data potentially explain the acquired cisplatin resistance in this BARD1 isogenic cell line. Additionally, the kinetics of in vivo growth were not identical across this panel of isogenic cells with BARD1+/R150* and BARD1+/R112* cellular models having faster in vivo growth, again suggesting the possible acquisition of additional genetic lesions that facilitated cell growth in these cell lines.
Although this study focused specifically on germline loss-of-function variants, other heterozygous germline and somatic variations in BARD1 have been identified in neuroblastoma (20,22). These variants may induce similar defects in DNA repair efficiency and genomic stability. The functional validation approach taken here can be extended to these and other potentially pathogenic germline variants recently identified across several childhood and adult malignancies, especially those that are predicted to disrupt DNA repair pathways. However, although we focus on utilizing established neuroblastoma and neural control cell lines as models to study the functional impact of BARD1 variants, it will also be important to study the function of pathogenic and likely pathogenic germline variants more directly in the development of neuroblastoma. Although the neuroblastoma cell of origin has ultimately remained elusive, patient-derived induced pluripotent stem cells and transgenic animal models may serve as appropriate systems to address these questions in preclinical studies (60-63). Although, considering the concerted effort and resources required for these experimental approaches, they may only be suitable for recurrent pathogenic and likely pathogenic germline variants that are found across several tumor histotypes. Another potential limitation of this study is the reliance on single cell cloning approaches to generate a homogenous population of cells harboring BARD1 germline pathogenic and likely pathogenic variants because of inefficiency of knocking in these variants using current CRISPR technology. Thus, we aimed to generate multiple paired clones for these studies controlling for any genomic differences in the parent cells. However, as the efficiency of this technology increases, similar studies can be performed on bulk cell populations that best recapitulate intratumoral heterogeneity.
As germline sequencing of pediatric cancer patients becomes more widely adopted into clinical practice, the role of germline variants in treatment stratification, normal tissue susceptibility to cancer therapies, and implications for genetic counseling must all be rigorously examined. For example, here, we suggest tumors harboring BARD1 loss-of-function variants may be more sensitive to DNA damaging agents, findings that may have important implications for selection of treatment regimens especially in the relapse setting where several different chemotherapeutic agents may be available to patients. Additionally, the presence of cancer-associated gene variants within normal tissues may result in substantial morbidities from cytotoxic therapies. This phenomenon has been noted in cancer predisposition syndromes such as Li-Fraumeni in which genotoxic treatments can precipitate severe acute toxicity and contribute to an elevated risk of second malignancy (64,65). Of note, during the preparation of this manuscript, a patient presented to our oncology clinic with a rare composite neuroblastoma-pheochromocytoma tumor that was ultimately found to have a BARD1R150* germline variant. This patient was treated with our standard high-risk neuroblastoma regimen, including conditioning with carboplatin, etoposide, and melphalan followed by autologous hematopoietic stem cell transplantation (66), which precipitated severe acute kidney injury progressing rapidly to renal failure and death. Although the contribution of this patient’s germline BARD1 variant to this fatal complication cannot be known, this case raises the possibility that the BARD1R150* variant caused increased renal sensitivity to the carboplatin-containing transplant conditioning regimen. A prior report from our institution focused on the toxicities in 44 patients who received the same carboplatin, etoposide, and melphalan conditioning places this patient’s complication in context (67). Approximately one-third of these patients developed modest increases in serum creatinine, with 2 requiring brief courses of dialysis, but none experienced irreversible or fatal kidney injury. Unfortunately, germline sequencing data were not available for these patients. However, clearly the influence of germline variation in DNA repair–related genes, or other genes critical in normal tissue homeostasis, on the development of normal tissue toxicity during multimodal cancer therapy merits further investigation.
Additionally, the identification of pathogenic variants in the germline of children with cancer has important genetic counseling implications as a majority of these are likely inherited in an autosomal manner similar to recent findings in pediatric sarcomas (68). Although these moderate penetrance pathogenic variants are unlikely to be sufficient to cause malignancy alone, they do substantially increase the risk of developing cancer, which has important repercussions for family members who may also harbor an identical germline genotype. Furthermore, it is now clear that pathogenic germline variants classically associated with adult cancer predisposition syndromes (eg, BRCA1 and 2) also contribute to cancer risk in children and adolescents (69), further emphasizing the importance for cascade testing. Finally, although at some larger academic medical centers it is common practice to sequence both tumor and germline tissues at diagnosis, these data support adopting this parallel sequencing practice for all pediatric cancer patients. Future studies will be essential to illuminate additional functional implications of cancer-predisposing germline genetic variants in tumorigenesis and how best to expand their utility in oncology clinical practice.
Supplementary Material
Acknowledgements
The funders did not play a role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Contributor Information
Michael P Randall, Division of Oncology and Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Laura E Egolf, Division of Oncology and Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Cell and Molecular Biology Graduate Group, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
Zalman Vaksman, Division of Oncology and Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Current affiliation: New York Genome Center, New York, NY.
Minu Samanta, Division of Oncology and Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Matthew Tsang, Division of Oncology and Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
David Groff, Division of Oncology and Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
J Perry Evans, Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Current affiliation: Genomics and Data Sciences, Spark Therapeutics, Philadelphia, PA.
Jo Lynne Rokita, Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Center for Data-Driven Discovery in Biomedicine, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Division of Neurosurgery, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Mehdi Layeghifard, Genetics and Genome Biology, The Hospital for Sick Children, Toronto, ON, Canada.
Adam Shlien, Genetics and Genome Biology, The Hospital for Sick Children, Toronto, ON, Canada; Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, ON, Canada; Department of Pediatric Laboratory Medicine, The Hospital for Sick Children, Toronto, ON, Canada.
John M Maris, Division of Oncology and Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Cell and Molecular Biology Graduate Group, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA; Department of Pediatrics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
Sharon J Diskin, Division of Oncology and Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Cell and Molecular Biology Graduate Group, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA; Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Pediatrics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
Kristopher R Bosse, Division of Oncology and Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Cell and Molecular Biology Graduate Group, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA; Department of Pediatrics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
Data availability
The data underlying this article will be available in NCBI’s database of Genotypes and Phenotypes (dbGaP) with a unique identifier upon publication.
Author contributions
Michael P Randall, MD (Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Validation; Visualization; Writing—original draft; Writing—review & editing), Laura E Egolf, PhD (Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Software; Validation; Visualization; Writing—original draft; Writing—review & editing), Zalman Vaksman, PhD (Data curation; Formal analysis; Investigation; Methodology; Validation; Visualization), Minu Samanta, MD (Data curation; Investigation), Matthew Tsang, MS (Data curation; Investigation; Methodology), David Groff, MES (Data curation; Investigation; Methodology), J. Perry Evans, PhD (Data curation; Formal analysis; Investigation; Methodology), Jo Lynne Rokita, PhD (Data curation; Formal analysis; Investigation; Methodology; Software; Writing—review & editing), Mehdi Layeghifard, PhD (Formal analysis; Investigation; Methodology; Software), Adam Shlien, PhD (Funding acquisition; Investigation; Methodology; Software), John M Maris, MD (Conceptualization; Funding acquisition; Project administration; Writing—original draft; Writing—review & editing), Sharon J Diskin, PhD (Conceptualization; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Writing—original draft; Writing—review & editing), and Kristopher R Bosse, MD (Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Resources; Supervision; Validation; Visualization; Writing—original draft; Writing—review & editing).
Funding
This work was supported by the Howard Hughes Medical Institute (Medical Fellows grant to MPR); the National Institutes of Health (grant number T32 GM008216 to LEE); Alex’s Lemonade Stand (a Young Investigator Award to KRB); the EVAN Foundation (to KRB); the Giulio D’Angio Endowed Chair (to JMM); the National Cancer Institute at the National Institutes of Health (grant numbers K08 CA230223 to KRB, R35 CA220500 to JMM, R01 CA204974 to SJD, and R01 CA237562 to SJD); the Ontario Ministry of Research and Innovation (an Early Researcher Award to AS); the Canada Research Chair in Childhood Cancer Genomics (to AS); the V Foundation (to AS); and the St. Baldrick’s Foundation (a Robert J. Arceci Innovation Award to AS). KRB is a Damon Runyon Physician-Scientist supported (in part) by the Damon Runyon Cancer Research Foundation (grant number PST-07-16).
Conflicts of interest
JMM and KRB hold patents for the discovery and development of immunotherapies for cancer, including patents related to glypican-2 (GPC2)-directed immunotherapies. KRB and JMM receive research funding from Tmunity for research on GPC2-directed immunotherapies and JMM and KRB receive royalties from Tmunity for licensing of GPC2-related intellectual property. No other authors report any conflict of interest.
References
- 1. Matthay KK, Maris JM, Schleiermacher G, et al. Neuroblastoma. Nat Rev Dis Primers. 2016;2:16078. doi: 10.1038/nrdp.2016.78 [DOI] [PubMed] [Google Scholar]
- 2. Mosse YP, Laudenslager M, Longo L, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455(7215):930-935. doi: 10.1038/nature07261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trochet D, Bourdeaut F, Janoueix-Lerosey I, et al. Germline mutations of the paired-like homeobox 2B (PHOX2B) Gene in Neuroblastoma. Am J Hum Genet. 2004;74(4):761-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mosse YP, Laudenslager M, Khazi D, et al. Germline PHOX2B Mutation in Hereditary Neuroblastoma. Am J Hum Genet. 2004;75(4):727-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McDaniel LD, Conkrite KL, Chang X, et al. Common variants upstream of MLF1 at 3q25 and within CPZ at 4p16 associated with neuroblastoma. PLoS Genet. 2017;13(5):e1006787. doi: 10.1371/journal.pgen.1006787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maris JM, Mosse YP, Bradfield JP, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358(24):2585-2593. doi: 10.1056/NEJMoa0708698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Capasso M, Devoto M, Hou C, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41(6):718-723. doi: 10.1038/ng.374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baer R, Ludwig T.. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr Opin Genet Dev. 2002;12(1):86-91. doi: 10.1016/s0959-437x(01)00269-6 [DOI] [PubMed] [Google Scholar]
- 9. Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE.. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol. 2001;8(10):833-837. doi: 10.1038/nsb1001-833 [DOI] [PubMed] [Google Scholar]
- 10. Zhao W, Steinfeld JB, Liang F, et al. BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature. 2017;550(7676):360-365. doi: 10.1038/nature24060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simons AM, Horwitz AA, Starita LM, et al. BRCA1 DNA-binding activity is stimulated by BARD1. Cancer Res. 2006;66(4):2012-2018. doi: 10.1158/0008-5472.CAN-05-3296 [DOI] [PubMed] [Google Scholar]
- 12. Fabbro M, Savage K, Hobson K, et al. BRCA1-BARD1 complexes are required for p53Ser-15 phosphorylation and a G1/S arrest following ionizing radiation-induced DNA damage. J Biol Chem. 2004;279(30):31251-31258. doi: 10.1074/jbc.M405372200 [DOI] [PubMed] [Google Scholar]
- 13. Joukov V, Groen AC, Prokhorova T, et al. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell. 2006;127(3):539-552. doi: 10.1016/j.cell.2006.08.053 [DOI] [PubMed] [Google Scholar]
- 14. Starita LM, Horwitz AA, Keogh MC, Ishioka C, Parvin JD, Chiba N.. BRCA1/BARD1 ubiquitinate phosphorylated RNA polymerase II. J Biol Chem. 2005;280(26):24498-24505. doi: 10.1074/jbc.M414020200 [DOI] [PubMed] [Google Scholar]
- 15. Capasso M, Diskin SJ, Totaro F, et al. Replication of GWAS-identified neuroblastoma risk loci strengthens the role of BARD1 and affirms the cumulative effect of genetic variations on disease susceptibility. Carcinogenesis. 2013;34(3):605-611. doi: 10.1093/carcin/bgs380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Latorre V, Diskin SJ, Diamond MA, et al. Replication of neuroblastoma SNP association at the BARD1 locus in African-Americans. Cancer Epidemiol Biomarkers Prev. 2012;21(4):658-663. doi: 10.1158/1055-9965.EPI-11-0830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bosse KR, Diskin SJ, Cole KA, et al. Common variation at BARD1 results in the expression of an oncogenic isoform that influences neuroblastoma susceptibility and oncogenicity. Cancer Res. 2012;72(8):2068-2078. doi: 10.1158/0008-5472.CAN-11-3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cimmino F, Avitabile M, Diskin SJ, et al. Fine mapping of 2q35 high-risk neuroblastoma locus reveals independent functional risk variants and suggests full-length BARD1 as tumor-suppressor. Int J Cancer. 2018;143(11):2828-2837. doi: 10.1002/ijc.31822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cimmino F, Avitabile M, Lasorsa VA, et al. Functional characterization of full-length BARD1 strengthens its role as a tumor suppressor in neuroblastoma. J Cancer. 2020;11(6):1495-1504. doi: 10.7150/jca.36164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pugh TJ, Morozova O, Attiyeh EF, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45(3):279-284. doi: 10.1038/ng.2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lasorsa VA, Formicola D, Pignataro P, et al. Exome and deep sequencing of clinically aggressive neuroblastoma reveal somatic mutations that affect key pathways involved in cancer progression. Oncotarget. 2016;7(16):21840-21852. doi: 10.18632/oncotarget.8187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim J, Vaksman Z, Egolf LE, et al. Germline pathogenic variants in neuroblastoma patients are enriched in BARD1 and predict worse survival. J Natl Cancer Inst. 2024;116(1):149-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weber-Lassalle N, Borde J, Weber-Lassalle K, et al. Germline loss-of-function variants in the BARD1 gene are associated with early-onset familial breast cancer but not ovarian cancer. Breast Cancer Res. 2019;21(1):55. doi: 10.1186/s13058-019-1137-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Venier RE, Maurer LM, Kessler EM, et al. A germline BARD1 mutation in a patient with Ewing Sarcoma: implications for familial testing and counseling. Pediatr Blood Cancer. 2019;66(9):e27824. doi: 10.1002/pbc.27824 [DOI] [PubMed] [Google Scholar]
- 25. DeLeonardis K, Sedgwick K, Voznesensky O, et al. Challenges in interpreting germline mutations in BARD1 and ATM in breast and ovarian cancer patients. Breast J. 2017;23(4):461-464. doi: 10.1111/tbj.12764 [DOI] [PubMed] [Google Scholar]
- 26. De Brakeleer S, De Greve J, Desmedt C, et al. Frequent incidence of BARD1-truncating mutations in germline DNA from triple-negative breast cancer patients. Clin Genet. 2016;89(3):336-340. doi: 10.1111/cge.12620 [DOI] [PubMed] [Google Scholar]
- 27. Ramus SJ, Song H, Dicks E, et al. ; Ovarian Cancer Association Consortium. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst. 2015;107(11):djv214. doi: 10.1093/jnci/djv214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schulz E, Valentin A, Ulz P, et al. Germline mutations in the DNA damage response genes BRCA1, BRCA2, BARD1 and TP53 in patients with therapy related myeloid neoplasms. J Med Genet. 2012;49(7):422-428. doi: 10.1136/jmedgenet-2011-100674 [DOI] [PubMed] [Google Scholar]
- 29. Ghimenti C, Sensi E, Presciuttini S, et al. Germline mutations of the BRCA1-associated ring domain (BARD1) gene in breast and breast/ovarian families negative for BRCA1 and BRCA2 alterations. Genes Chromosomes Cancer. 2002;33(3):235-242. doi: 10.1002/gcc.1223 [DOI] [PubMed] [Google Scholar]
- 30. Grobner SN, Worst BC, Weischenfeldt J, et al. ; ICGC MMML-Seq Project. The landscape of genomic alterations across childhood cancers. Nature. 2018;555(7696):321-327. doi: 10.1038/nature25480 [DOI] [PubMed] [Google Scholar]
- 31. Huang KL, Mashl RJ, Wu Y, et al. ; Cancer Genome Atlas Research Network. Pathogenic germline variants in 10,389 adult cancers. Cell. 2018;173(2):355-370.e14. doi: 10.1016/j.cell.2018.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang J, Walsh MF, Wu G, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373(24):2336-2346. doi: 10.1056/NEJMoa1508054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lopez G, Conkrite KL, Doepner M, et al. Somatic structural variation targets neurodevelopmental genes and identifies SHANK2 as a tumor suppressor in neuroblastoma. Genome Res. 2020;30(9):1228-1242. doi: 10.1101/gr.252106.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lopez G, Egolf LE, Giorgi FM, Diskin SJ, Margolin AA.. svpluscnv: analysis and visualization of complex structural variation data. Bioinformatics. 2021;37(13):1912-1914. doi: 10.1093/bioinformatics/btaa878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boeva V, Popova T, Bleakley K, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28(3):423-425. doi: 10.1093/bioinformatics/btr670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO.. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28(18):i333-i339. doi: 10.1093/bioinformatics/bts378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213-219. doi: 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oeck S, Malewicz NM, Hurst S, Al-Refae K, Krysztofiak A, Jendrossek V.. The focinator v2-0 - graphical interface, four channels, colocalization analysis and cell phase identification. Radiat Res. 2017;188(1):114-120. doi: 10.1667/RR14746.1 [DOI] [PubMed] [Google Scholar]
- 39. Pinder J, Salsman J, Dellaire G.. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015;43(19):9379-9392. doi: 10.1093/nar/gkv993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bosse KR, Raman P, Zhu Z, et al. Identification of GPC2 as an oncoprotein and candidate immunotherapeutic target in high-risk neuroblastoma. Cancer Cell. 2017;32(3):295-309.e12. doi: 10.1016/j.ccell.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Laufer M, Nandula SV, Modi AP, et al. Structural requirements for the BARD1 tumor suppressor in chromosomal stability and homology-directed DNA repair. J Biol Chem. 2007;282(47):34325-34333. doi: 10.1074/jbc.M705198200 [DOI] [PubMed] [Google Scholar]
- 42. Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM.. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006;20(1):34-46. doi: 10.1101/gad.1381306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rambhatla L, Chiu CP, Glickman RD, Rowe-Rendleman C.. In vitro differentiation capacity of telomerase immortalized human RPE cells. Invest Ophthalmol Vis Sci. 2002;43(5):1622-1630. [PubMed] [Google Scholar]
- 44. Haeussler M, Schonig K, Eckert H, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17(1):148. doi: 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Konishi H, Mohseni M, Tamaki A, et al. Mutation of a single allele of the cancer susceptibility gene BRCA1 leads to genomic instability in human breast epithelial cells. Proc Natl Acad Sci USA. 2011;108(43):17773-17778. doi: 10.1073/pnas.1110969108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pathania S, Bade S, Le Guillou M, et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat Commun. 2014;5:5496. doi: 10.1038/ncomms6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Neu-Yilik G, Amthor B, Gehring NH, et al. Mechanism of escape from nonsense-mediated mRNA decay of human beta-globin transcripts with nonsense mutations in the first exon. RNA. 2011;17(5):843-854. doi: 10.1261/rna.2401811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pathania S, Nguyen J, Hill SJ, et al. BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol Cell. 2011;44(2):235-251. doi: 10.1016/j.molcel.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917-921. doi: 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- 50. Takagi M, Yoshida M, Nemoto Y, et al. Loss of DNA damage response in neuroblastoma and utility of a PARP inhibitor. J Natl Cancer Inst. 2017;109(11):djx062. doi: 10.1093/jnci/djx062 [DOI] [PubMed] [Google Scholar]
- 51. Raderschall E, Golub EI, Haaf T.. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci USA. 1999;96(5):1921-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Newman EA, Chukkapalli S, Bashllari D, et al. Alternative NHEJ pathway proteins as components of MYCN oncogenic activity in human neural crest stem cell differentiation: Implications for neuroblastoma initiation. Cell Death Dis. 2017;8(12):3208. doi: 10.1038/s41419-017-0004-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang W, Liu B, Wu W, et al. Targeting the MYCN-PARP-DNA damage response pathway in neuroendocrine prostate cancer. Clin Cancer Res. 2018;24(3):696-707. doi: 10.1158/1078-0432.CCR-17-1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ma X, Liu Y, Liu Y, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature. 2018;555(7696):371-376. doi: 10.1038/nature25795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smith SA, Easton DF, Evans DG, Ponder BA.. Allele losses in the region 17q12-21 in familial breast and ovarian cancer involve the wild-type chromosome. Nat Genet. 1992;2(2):128-131. doi: 10.1038/ng1092-128 [DOI] [PubMed] [Google Scholar]
- 56. Maxwell KN, Wubbenhorst B, Wenz BM, et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat Commun. 2017;8(1):319. doi: 10.1038/s41467-017-00388-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wong M, Mayoh C, Lau LMS, et al. Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat Med. 2020;26(11):1742-1753. doi: 10.1038/s41591-020-1072-4 [DOI] [PubMed] [Google Scholar]
- 58. McCarthy EE, Celebi JT, Baer R, Ludwig T.. Loss of Bard1, the heterodimeric partner of the Brca1 tumor suppressor, results in early embryonic lethality and chromosomal instability. Mol Cell Biol. 2003;23(14):5056-5063. doi: 10.1128/mcb.23.14.5056-5063.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huang D, Savage SR, Calinawan AP, et al. A highly annotated database of genes associated with platinum resistance in cancer. Oncogene. 2021;40(46):6395-6405. doi: 10.1038/s41388-021-02055-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ozgencil M, Barwell J, Tischkowitz M, et al. Assessing BRCA1 activity in DNA damage repair using human induced pluripotent stem cells as an approach to assist classification of BRCA1 variants of uncertain significance. PLoS One. 2021;16(12):e0260852. doi: 10.1371/journal.pone.0260852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Terada Y, Jo N, Arakawa Y, et al. Human pluripotent stem cell-derived tumor model uncovers the embryonic stem cell signature as a key driver in atypical teratoid/rhabdoid tumor. Cell Rep. 2019;26(10):2608-2621.e6. doi: 10.1016/j.celrep.2019.02.009 [DOI] [PubMed] [Google Scholar]
- 62. Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM.. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16(11):2985-2995. doi: 10.1093/emboj/16.11.2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weichert-Leahey N, Shi H, Tao T, et al. Genetic predisposition to neuroblastoma results from a regulatory polymorphism that promotes the adrenergic cell state. J Clin Invest. 2023;133(10):e166919. doi: 10.1172/JCI166919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nutting C, Camplejohn RS, Gilchrist R, et al. A patient with 17 primary tumours and a germ line mutation in TP53: tumour induction by adjuvant therapy? Clin Oncol (R Coll Radiol). 2000;12(5):300-304. [DOI] [PubMed] [Google Scholar]
- 65. Limacher JM, Frebourg T, Natarajan-Ame S, Bergerat JP.. Two metachronous tumors in the radiotherapy fields of a patient with Li-Fraumeni syndrome. Int J Cancer. 2001;96(4):238-242. [DOI] [PubMed] [Google Scholar]
- 66. Ladenstein R, Potschger U, Pearson ADJ, et al. ; SIOP Europe Neuroblastoma Group (SIOPEN). Busulfan and melphalan versus carboplatin, etoposide, and melphalan as high-dose chemotherapy for high-risk neuroblastoma (HR-NBL1/SIOPEN): an international, randomised, multi-arm, open-label, phase 3 trial. Lancet Oncol. 2017;18(4):500-514. doi: 10.1016/S1470-2045(17)30070-0 [DOI] [PubMed] [Google Scholar]
- 67. Desai AV, Heneghan MB, Li Y, et al. Toxicities of busulfan/melphalan versus carboplatin/etoposide/melphalan for high-dose chemotherapy with stem cell rescue for high-risk neuroblastoma. Bone Marrow Transplant. 2016;51(9):1204-1210. doi: 10.1038/bmt.2016.84 [DOI] [PubMed] [Google Scholar]
- 68. Gillani R, Camp SY, Han S, et al. Germline predisposition to pediatric Ewing sarcoma is characterized by inherited pathogenic variants in DNA damage repair genes. Am J Hum Genet. 2022;109(6):1026-1037. doi: 10.1016/j.ajhg.2022.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kratz CP, Smirnov D, Autry R, et al. Heterozygous BRCA1/2 and mismatch repair gene pathogenic variants in children and adolescents with cancer. J Natl Cancer Inst. 2022;114(11):1523-1532. doi: 10.1093/jnci/djac151 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be available in NCBI’s database of Genotypes and Phenotypes (dbGaP) with a unique identifier upon publication.