Abstract
Background
Even with contemporary treatment strategies, more than 10% of HER2-positive early stage breast cancer patients may experience distant metastasis as first event during follow-up. Tools for predicting unique patterns of metastatic spread are needed to plan personalized surveillance. We evaluated how molecular heterogeneity affects the pattern of distant relapse in HER2-positive breast cancer.
Methods
A total of 677 HER2-positive stage I-III breast cancer patients from ShortHER trial, Cher-LOB trial, and 2 institutional cohorts were included. PAM50 molecular subtypes and research-based HER2DX scores were evaluated. The cumulative incidence of distant relapse as the first event (any site and site specific) was evaluated using competing risk analysis. Median follow-up was 8.4 years. Tests of statistical significance are 2-sided.
Results
Stage III and high HER2DX risk score identified patients at the highest risk of distant relapse as first event (10-year incidence 24.5% and 19.7%, respectively). Intrinsic molecular subtypes were associated with specific patterns of metastatic spread: compared with other subtypes, HER2-enriched tumors were more prone to develop brain metastases (10-year incidence 3.8% vs 0.6%, P = .005), basal-like tumors were associated with an increased risk of lung metastases (10-year incidence 11.1% vs 2.6%, P = .001), and luminal tumors developed more frequently bone-only metastases (10-year incidence 5.1% vs 2.0%, P = .042). When added to stage or HER2DX risk score in competing risk regression models, intrinsic subtype maintained an independent association with site-specific metastases.
Conclusions
The integration of intrinsic molecular subtypes with stage or HER2DX risk score predicts site-specific metastatic risk in HER2-positive breast cancer, with potential implications for personalized surveillance and clinical trials aimed at preventing site-specific recurrence.
Owing to the evolution of anti-HER2 therapies, the outcome of patients diagnosed with HER2-positive breast cancer has dramatically improved (1,2). However, even with the most modern treatment strategies, up to 11% of patients experience distant metastasis as the first event during follow-up (3,4). Moreover, the rate of brain metastases as the first site of relapse has been little affected by treatment improvements, still ranging between 2% and 6%, indicating a major unmet need for this patient population (3-8).
Currently, international guidelines do not recommend systemic radiological examinations during follow-up after treatment with curative intent for early breast cancer, because the anticipated detection of subclinical distant relapses has never been associated with improved subsequent outcomes (9,10). However, this evidence is derived from obsolete studies that do not account for more sophisticated diagnostic procedures, modern effective therapies, and breast cancer classification into biological subtypes (11-14). Indeed, the increased availability of effective multidisciplinary personalized strategies for advanced disease is putting into question the robustness of this approach, leading to the hypothesis that individually tailored intensive follow-up might be appropriate, at least for patients at a higher risk of relapse (15). Two of the main challenges limiting the implementation of personalized follow-up are the lack of valid tools to identify patients at increased risk of distant relapse and to predict specific patterns of distant metastatic spread.
It is known since more than a decade that in unselected breast cancer patients, breast cancer subtypes as defined by routine pathology influence the risk, timing, and pattern of metastatic spread (16-18). In HER2-positive breast cancer, several reports have highlighted differences in the pattern of relapse according to hormone receptor status (19,20). However, clinically defined HER2-positive breast cancer is highly heterogeneous from a molecular perspective, encompassing all 5 main intrinsic breast cancer subtypes, as evaluated by the PAM50 gene expression–based assay (21). Although the distribution of intrinsic subtypes varies according to the hormone receptor status, molecular heterogeneity can still be recognized in hormone receptor–positive and hormone receptor–negative subgroups and has relevant implications in terms of prognosis and treatment sensitivity (21,22). Intrinsic subtyping is not the only tool used to dissect the biological heterogeneity of HER2-positive diseases. Recently, the HER2DX genomic test was developed, providing 2 independent scores to predict both long-term prognosis and likelihood of pathological complete response (pCR) in HER2-positive early breast cancer (23). To the best of our knowledge, the impact of molecular heterogeneity on the pattern of metastatic spread has never been explored in clinically defined HER2-positive breast cancer patients.
We hypothesized that the integration of molecular profiles could improve our ability to select patients with HER2-positive breast cancer at increased risk of developing distant relapse at specific sites, to support individualized monitoring strategies during follow-up.
Methods
Patients
A total of 677 patients with available gene expression data and diagnosed with early HER2-positive breast cancer, all treated with surgery, chemotherapy, and anti-HER2 therapy with curative intent, were included in this study. Patients were derived from the following cohorts: ShortHER trial (n = 437), Cher-LOB trial (n = 84), hospital clinic (Barcelona, Spain), institutional cohort (n = 117), and Padova University, Veneto Institute of Oncology (Padova, Italy) institutional cohort (n = 39).
The study design, procedures, and median follow-up of the ShortHER (24) and Cher-LOB (25) trials, as well as the main features and median follow-up of the 2 institutional cohorts, are detailed in the Supplementary Methods (available online). The median follow-up of the entire study cohort was 8.4 years (95% confidence interval [CI] = 8.2 to 8.6 years).
The protocols of the 2 clinical trials and the 2 observational institutional studies were approved by competent ethical committees. Written informed consent was obtained from all the patients. Further details regarding the patient population are reported in Supplementary Methods (available online).
Intrinsic subtyping and HER2DX score
Gene expression analyses were performed on RNA extracted from tumor samples. Intrinsic molecular subtyping was performed using the previously reported PAM50 subtype predictor (26,27) and categorized as luminal A, luminal B, HER2-enriched, basal-like, and normal-like (normal). Further details are reported in Supplementary Methods (available online).
The HER2DX genomic test provides 2 independent scores, which predict long-term survival (risk score) and the likelihood of pCR (pCR score). Both HER2DX risk scores and pCR scores range from 0 to 100, defining 2 HER2DX risk groups (low = 0-50 and high = 50-100) and 3 HER2DX pCR groups (low = 0-33.3, medium = 33.3-66.7, and high = 66.7-100). This classifier represents a supervised learning algorithm that is based on the integration of clinical features (ie, tumor size and nodal status) with 4 gene signatures (23). To assess HER2DX scores (risk score and pCR score), the expression of 185 breast cancer–related genes and 5 housekeeping genes was measured using the same technology as previously reported (23). The 4 HER2DX gene signatures included a 14-gene immunoglobulin signature tracking immune infiltration (CD27, CD79A, HLA-C, IGJ, IGKC, IGL, IGLV3-25, IL2RG, CXCL8, LAX1, NTN3, PIM2, POU2AF1, and TNFRSF17), a 4-gene tumor cell proliferation signature (EXO1, ASPM, NEK2, and KIF23), a 5-gene luminal differentiation signature (BCL2, DNAJC12, AGR3, AFF3, and ESR1), and a 4-gene HER2 amplicon signature (ERBB2, GRB7, STARD3, and TCAP). Gene expression for each sample was independently normalized to the geometric mean of the 5 housekeeping genes. The HER2DX risk score was calculated based on the immunoglobulin, luminal, and proliferation signatures. The HER2DX pCR score was calculated based on HER2, IgG, luminal, and proliferation signatures. The 2 HER2DX scores were reported according to pre-established and above-specified cutoff values (23). All gene expression analyses were done blinded from clinical data.
Statistical analysis
Patient and tumor characteristics were compared across breast cancer subtypes and HER2DX scores using the χ2 test (categorical variables) and Mann–Whitney or Kruskal–Wallis test (continuous variables).
The cumulative incidence of distant metastases as the first event (at any site and at specific sites) was evaluated using a competing risk methodology [Fine and Gray method (28,29)]. Univariate and multivariate competing risk regression models were used to calculate the subdistribution hazard ratios (HRs) and their 95% confidence intervals (CIs) (28,29). Details regarding the competing risk analyses are reported in Supplementary Methods (available online). We then evaluated the association of the site of relapse with molecular and clinicopathological variables in patients who had a distant relapse as the first event using the χ2 test.
All P values were 2-sided with the level of statistical significance set at a P value less than .05. No formal sample size calculation was performed because the sample population was based on the number of cases with available molecular data suitable for the present analysis. Statistical analyses were conducted using IBM SPSS version 28.0.1.0 and R project software version 4.2.2 (30).
Ethical considerations
The study was approved by the ethics committee of the participating centers, and all relevant ethical regulations were complied with. Tumor samples were collected after approval from the respective institutional review board and in accordance with the Declaration of Helsinki. Informed written consent was obtained from each participant who was alive at the time of study entry.
Results
Patients’ characteristics
The distribution of PAM50 intrinsic subtypes across all 677 patients was as follows: HER2 enriched, 51.3%; luminal A, 19.2%; normal-like, 11.7%; luminal B, 11.1%; and basal-like, 6.8%.
The HER2DX scores were available for 640 patients. The HER2DX risk score showed that 51.2% of tumor samples were categorized as high risk and 48.8% as low risk. The HER2DX pCR score was high in 37.7%, medium in 31.7%, and low in 30.6% of the patients.
There was no statistically significant difference in the distribution of HER2DX risk scores between the different intrinsic subtypes, highlighting that the 2 classifications describe nonsuperimposable information (Supplementary Figure 1, A, available online). The distribution of HER2DX pCR scores across intrinsic subtypes is shown in Supplementary Materials (Supplementary Results, available online) and Supplementary Figure 1, B (available online).
The main clinicopathological features of the patients according to tumor molecular categories are detailed in Table 1. As expected, the intrinsic subtype distribution was statistically significantly different according to the histologic grade and hormone receptor status. The median age was younger in patients with normal-like tumors, which, accordingly, were more frequently premenopausal. Because the luminal and HER2 signatures are the main drivers of the HER2DX pCR score, we found a statistically significant association between this score and hormone receptor status and histologic grade.
Table 1.
Main clinicopathological patients’ characteristics according to intrinsic subtype by PAM50, HER2DX risk score, and HER2DX pCR scorea
| Intrinsic subtype |
HER2DX risk score |
HER2DX pCR score |
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | Total, No. (%) (n = 677) | Luminal A, No. (%) (n = 130) | Luminal B, No. (%) (n = 75) | HER enriched, No. (%) (n = 347) | Basal, No. (%) (n = 46) | Normal, No. (%) (n = 79) | P | Low, No. (%) (n = 312) | High, No. (%) (n = 328) | P | Low, No. (%) (n = 196) | Medium, No. (%) (n = 203) | High, No. (%) (n = 241) | P | ||||||||||||
| Age, median (range), y | 55 (25-83) | 57 (36-77) | 58 (27-76) | 54 (28-83) | 54 (31-83) | 49 (25-78) | .004 | 57 (25-83) | 54 (31-80) | .117 | 57 (34-80) | 53 (27-78) | 56 (25-83) | .054 | ||||||||||||
| Menopausal status | Pre | 213 | 37% | 37 | 35% | 14 | 23% | 117 | 37% | 10 | 32% | 35 | 56% | .004 | 94 | 34% | 110 | 40% | .200 | 58 | 35% | 74 | 43% | 72 | 33% | .131 |
| Post | 360 | 63% | 70 | 65% | 46 | 77% | 196 | 63% | 21 | 68% | 27 | 44% | 180 | 66% | 168 | 60% | 107 | 65% | 98 | 57% | 143 | 67% | ||||
| 7th AJCC stage | I | 179 | 27% | 46 | 35% | 15 | 20% | 84 | 24% | 9 | 20% | 25 | 32% | .152 | 163 | 52% | 15 | 5% | <.001 | 43 | 22% | 40 | 20% | 95 | 39% | <.001 |
| II | 384 | 57% | 65 | 50% | 50 | 67% | 198 | 57% | 30 | 65% | 41 | 52% | 143 | 46% | 217 | 66% | 118 | 60% | 127 | 63% | 115 | 48% | ||||
| III | 112 | 17% | 19 | 15% | 10 | 13% | 63 | 18% | 7 | 15% | 13 | 16% | 6 | 2% | 96 | 29% | 35 | 18% | 36 | 18% | 31 | 13% | ||||
| Histologic grade | 1 | 10 | 2% | 6 | 5% | 1 | 1% | 1 | 0% | 1 | 2% | 1 | 1% | <.001 | 4 | 1% | 6 | 2% | .170 | 5 | 3% | 3 | 2% | 2 | 1% | <.001 |
| 2 | 186 | 30% | 62 | 50% | 22 | 32% | 77 | 24% | 4 | 10% | 21 | 30% | 100 | 33% | 82 | 27% | 79 | 43% | 61 | 31% | 42 | 18% | ||||
| 3 | 423 | 68% | 57 | 46% | 45 | 66% | 237 | 75% | 36 | 88% | 48 | 69% | 196 | 65% | 221 | 72% | 98 | 54% | 131 | 67% | 188 | 81% | ||||
| Hormone receptors | Negative | 205 | 30% | 4 | 3% | 3 | 4% | 137 | 39% | 31 | 67% | 30 | 38% | <.001 | 97 | 31% | 94 | 29% | .502 | 7 | 4% | 38 | 19% | 146 | 61% | <.001 |
| Positive | 472 | 70% | 126 | 97% | 72 | 96% | 210 | 61% | 15 | 33% | 49 | 62% | 215 | 69% | 234 | 71% | 189 | 96% | 165 | 81% | 95 | 39% | ||||
| Neoadjuvant therapy | No | 437 | 65% | 87 | 67% | 43 | 57% | 232 | 67% | 27 | 59% | 48 | 61% | .403 | 216 | 69% | 217 | 66% | .406 | 119 | 61% | 136 | 67% | 178 | 74% | .014 |
| Yes | 240 | 35% | 43 | 33% | 32 | 43% | 115 | 33% | 19 | 41% | 31 | 39% | 96 | 31% | 111 | 34% | 77 | 39% | 67 | 33% | 63 | 26% | ||||
| Anthracycline | Yes | 649 | 96% | 127 | 98% | 73 | 97% | 328 | 95% | 44 | 96% | 77 | 97% | .464 | 299 | 96% | 319 | 97% | .323 | 191 | 97% | 198 | 98% | 229 | 95% | .250 |
| Taxane | Yes | 675 | 100% | 130 | 100% | 74 | 99% | 347 | 100% | 46 | 100% | 78 | 99% | .146 | 311 | 100% | 328 | 100% | .305 | 196 | 100% | 202 | 100% | 241 | 100% | .340 |
| Trastuzumab | Yes | 648 | 96% | 124 | 95% | 71 | 95% | 336 | 97% | 43 | 93% | 74 | 94% | .617 | 299 | 96% | 312 | 95% | .665 | 188 | 96% | 191 | 94% | 232 | 96% | .512 |
| Lapatinib | Yes | 62 | 9% | 14 | 11% | 11 | 15% | 19 | 5% | 7 | 15% | 11 | 14% | .011 | 29 | 9% | 33 | 10% | .743 | 19 | 10% | 25 | 12% | 18 | 7% | .228 |
| Pertuzumab | Yes | 20 | 3% | 6 | 5% | 3 | 4% | 8 | 2% | 1 | 2% | 2 | 3% | .700 | 7 | 2% | 7 | 2% | .925 | 11 | 6% | 3 | 1% | 0 | 0% | <.001 |
| Trastuzumab-emtansine | Yes | 7 | 1% | 0 | 0% | 0 | 0% | 4 | 1% | 0 | 0% | 3 | 4% | .073 | 1 | 0% | 4 | 1% | .197 | 4 | 2% | 0 | 0% | 1 | 0% | .049 |
| Cohort | ShortHER | 437 | 65% | 87 | 67% | 43 | 57% | 232 | 67% | 27 | 59% | 48 | 61% | <.001 | 216 | 69% | 217 | 66% | .457 | 119 | 61% | 136 | 67% | 178 | 74% | .058 |
| Cher-LOB | 84 | 12% | 21 | 16% | 14 | 19% | 22 | 6% | 12 | 26% | 15 | 19% | 38 | 12% | 46 | 14% | 29 | 15% | 29 | 14% | 26 | 11% | ||||
| Barcelona | 117 | 17% | 20 | 15% | 15 | 20% | 67 | 19% | 5 | 11% | 10 | 13% | 44 | 14% | 42 | 13% | 37 | 19% | 27 | 13% | 22 | 9% | ||||
| Padova | 39 | 6% | 2 | 2% | 3 | 4% | 26 | 7% | 2 | 4% | 6 | 8% | 14 | 4% | 23 | 7% | 11 | 6% | 11 | 5% | 15 | 6% | ||||
7th AJCC = American Joint Committee on Cancer 7th edition; pCR = pathological complete response.
Cumulative incidence of distant relapse in the whole population
In the entire study cohort, the cumulative incidence rates of distant relapse as the first event were 5.6% at 3 years, 8.6% at 5 years, and 13.0% at 10 years, highlighting a persistent risk beyond 5 years from diagnosis, as shown in Table 2. The cumulative incidence of brain metastases as the first site of relapse was approximately 2%, and all events occurred in the first 5 years of follow-up. The most frequently affected sites were the bone and liver.
Table 2.
Cumulative incidence rates at 3, 5, and 10 years for any distant relapse as first event and site-specific distant relapse as first event according to main clinicopathological characteristics and molecular features
| Site-specific distant relapse |
||||||||
|---|---|---|---|---|---|---|---|---|
| Clinicopathological characteristics | Years | Any distant relapse | Brain | Brain only | Lung | Bone | Bone only | Liver |
| All | 3 y | 5.6% | 1.8% | 1.0% | 1.0% | 2.2% | 0.9% | 2.2% |
| 5 y | 8.6% | 2.2% | 1.2% | 2.2% | 3.9% | 1.5% | 3.1% | |
| 10 y | 13.0% | 2.2% | 1.2% | 3.2% | 6.2% | 3.0% | 5.1% | |
| Hormone receptor positive | 3 y | 4.9% | 1.3% | 0.8% | 1.1% | 2.1% | 1.1% | 2.1% |
| 5 y | 7.7% | 1.7% | 1.1% | 2.2% | 4.3% | 1.9% | 3.0% | |
| 10 y | 13.1% | 1.7% | 1.1% | 2.9% | 7.2% | 3.8% | 5.7% | |
| Hormone receptor negative | 3 y | 7.3% | 2.9% | 1.5% | 1.0% | 2.5% | 0.5% | 2.4% |
| 5 y | 10.8% | 3.4% | 1.5% | 2.4% | 2.9% | 0.5% | 3.4% | |
| 10 y | 13.1% | 3.4% | 1.5% | 3.7% | 4.1% | 1.0% | 4.1% | |
| Gray P value | .535 | .164 | .656 | .682 | .209 | .080 | .781 | |
| Histologic grade 1-2 | 3 y | 3.1% | 2.0% | 0.5% | 0.0% | 1.0% | 0.5% | 1.0% |
| 5 y | 5.6% | 2.6% | 0.5% | 2.1% | 2.6% | 0.5% | 2.6% | |
| 10 y | 13.3% | 2.6% | 0.5% | 2.7% | 6.3% | 3.7% | 5.9% | |
| Histologic grade 3 | 3 y | 7.1% | 1.9% | 1.4% | 1.7% | 3.1% | 1.2% | 2.6% |
| 5 y | 9.5% | 2.4% | 1.7% | 2.1% | 4.3% | 1.9% | 3.3% | |
| 10 y | 12.1% | 2.4% | 1.7% | 3.0% | 5.7% | 2.8% | 4.1% | |
| Gray P value | .505 | .897 | .241 | .854 | .947 | .979 | .883 | |
| Stage I | 3 y | 1.1% | 0.6% | 0.6% | 0.0% | 0.6% | 0.0% | 0.1% |
| 5 y | 1.7% | 0.6% | 0.6% | 0.0% | 0.6% | 0.0% | 1.1% | |
| 10 y | 7.9% | 0.6% | 0.6% | 0.1% | 0.6% | 0.0% | 5.3% | |
| Stage II | 3 y | 5.2% | 0.8% | 0.3% | 0.8% | 2.1% | 1.0% | 2.9% |
| 5 y | 8.4% | 1.5% | 0.3% | 2.6% | 4.2% | 1.6% | 3.7% | |
| 10 y | 12.7% | 1.5% | 0.3% | 4.0% | 7.5% | 3.3% | 5.6% | |
| Stage III | 3 y | 14.3% | 7.1% | 4.5% | 3.6% | 5.4% | 1.8% | 2.7% |
| 5 y | 20.6% | 8.9% | 5.4% | 4.5% | 8.1% | 3.6% | 4.4% | |
| 10 y | 24.5% | 8.9% | 5.4% | 4.5% | 11.1% | 6.6% | 5.4% | |
| Gray P value | <.001 | <.001 | <.001 | .076 | .001 | .015 | .156 | |
| HER2DX risk score low | 3 y | 2.9% | 0.6% | 0.3% | 0.6% | 1.0% | 0.0% | 1.3% |
| 5 y | 3.2% | 0.6% | 0.3% | 0.6% | 1.0% | 0.0% | 1.6% | |
| 10 y | 5.3% | 0.6% | 0.3% | 1.1% | 2.3% | 1.0% | 2.0% | |
| HER2DX risk score high | 3 y | 8.2% | 2.7% | 1.5% | 1.5% | 3.4% | 1.8% | 3.1% |
| 5 y | 13.2% | 3.7% | 1.8% | 3.7% | 6.1% | 2.8% | 4.6% | |
| 10 y | 19.7% | 3.7% | 1.8% | 4.8% | 9.3% | 4.8% | 8.0% | |
| Gray P value | <.001 | .009 | .067 | .006 | <.001 | .003 | .008 | |
| Luminal A | 3 y | 1.5% | 0.0% | 0.0% | 0.8% | 1.5% | 0.1% | 0.0% |
| 5 y | 3.1% | 0.0% | 0.0% | 0.8% | 3.1% | 2.3% | 0.0% | |
| 10 y | 7.9% | 0.0% | 0.0% | 1.9% | 8.7% | 5.3% | 2.7% | |
| Luminal B | 3 y | 6.7% | 0.0% | 0.0% | 0.0% | 2.7% | 2.7% | 2.7% |
| 5 y | 10.7% | 0.0% | 0.0% | 1.4% | 6.7% | 2.7% | 5.4% | |
| 10 y | 14.8% | 0.0% | 0.0% | 3.3% | 8.9% | 4.8% | 5.4% | |
| HER2 enriched | 3 y | 7.2% | 3.2% | 2.0% | 1.2% | 2.6% | 0.9% | 2.9% |
| 5 y | 10.5% | 3.8% | 2.3% | 2.6% | 3.8% | 1.2% | 4.1% | |
| 10 y | 14.7% | 3.8% | 2.3% | 3.3% | 4.4% | 1.5% | 6.6% | |
| Basal | 3 y | 6.5% | 0.0% | 0.0% | 4.3% | 2.2% | 0.0% | 2.2% |
| 5 y | 10.9% | 0.0% | 0.0% | 8.7% | 4.3% | 0.0% | 2.2% | |
| 10 y | 15.5% | 0.0% | 0.0% | 11.1% | 6.6% | 0.0% | 4.4% | |
| Normal | 3 y | 3.8% | 1.3% | 0.0% | 0.0% | 1.3% | 0.0% | 2.5% |
| 5 y | 6.3% | 2.5% | 0.0% | 0.0% | 2.5% | 1.3% | 2.5% | |
| 10 y | 10.4% | 2.5% | 0.0% | 0.0% | 6.6% | 5.4% | 2.5% | |
| Gray P value | .179 | .049 | .102 | .010 | .679 | .187 | .383 | |
Cumulative incidence of distant relapse by clinicopathological characteristics
Among the classic clinicopathological characteristics, only stage statistically significantly affected the incidence of distant relapse as the first event (Table 2). Stage statistically significantly affected the cumulative incidence rate of any distant relapse as well as metastases in the brain, brain only, and bone (Table 2). The results of the statistically significant univariate competing risk regression models are shown in Table 3, and the relative incidence curves are shown in Supplementary Figure 2 (available online).
Table 3.
Univariate and multivariate competing risk regression models for any distant relapse as first event and site-specific distant relapse as first event by stage, intrinsic subtypes, and HER2DX risk score
| Univariate competing risk regression models: stagea |
|||
|---|---|---|---|
| Endpoint | Variables | Subdistribution HR (95% CI) | P |
| Any distant | Stage II vs stage I | 3.07 (1.39 to 6.8) | .0057 |
| Stage III vs stage I | 6.73 (2.92 to 15.5) | <.001 | |
| Brain | Stage II vs stage I | 1.87 (0.21 to 16.8) | .580 |
| Stage III vs stage I | 16.62 (2.11 to 130.8) | .0076 | |
| Brain only | Stage II vs stage I | 0.47 (0.03 to 7.44) | .590 |
| Stage III vs stage I | 9.76 (1.71 to 81.29) | .035 | |
| Bone | Stage II vs stage I | 12.1 (1.64 to 89.7) | .015 |
| Stage III vs stage I | 18.5 (2.39 to 144.1) | .005 | |
|
| |||
|
Univariate competing risk regression models: intrinsic subtype
|
|||
| Endpoint | Variables | Subdistribution HR (95% CI) | P |
| Any distant | Luminal A vs others | 0.46 (0.22 to 0.95) | .035 |
| Brain | HER2 enriched vs others | 6.33 (1.44 to 27.9) | .015 |
| Lung | Basal vs others | 4.74 (1.73 to 13.00) | .002 |
| Bone only | Luminal A and luminal B vs others | 2.6 (1.01 to 6.72) | .048 |
|
| |||
|
Univariate competing risk regression models: HER2DX risk score
|
|||
| Endpoint | Variables | Subdistribution HR (95% CI) | P |
| Any distant | HER2DX risk score high vs low | 3.77 (2.13 to 6.66) | <.001 |
| Brain | HER2DX risk score high vs low | 5.78 (1.29 to 25.8) | .022 |
| Lung | HER2DX risk score high vs low | 4.79 (1.39 to 16.6) | .013 |
| Bone | HER2DX risk score high vs low | 4.41 (1.83 to 10.7) | <.001 |
| Bone only | HER2DX risk score high vs low | 6.78 (1.55 to 29.6) | .011 |
| Liver | HER2DX risk score high vs low | 3.22 (1.29 to 8.03) | .012 |
|
| |||
|
Multivariate competing risk regression models: stage and intrinsic subtypeb
|
|||
| Endpointc | Variables | Subdistribution HR (95% CI) | P |
| Any distant | Stage II vs stage I | 2.93 (1.32 to 6.53) | .009 |
| Stage III vs stage I | 6.45 (2.79 to 14.93) | <.001 | |
| Luminal A vs others | 0.50 (0.24 to 1.04) | .064 | |
| Brain | Stage II vs stage I | 1.75 (0.20 to 15.7) | .620 |
| Stage III vs stage I | 14.87 (1.86 to 118.9) | .011 | |
| HER2 enriched vs others | 5.73 (1.30 to 25.3) | .021 | |
| Lung | Stage II vs stage I | 6.31 (0.84 to 47.3) | .073 |
| Stage III vs stage I | 8.03 (0.94 to 68.4) | .057 | |
| Basal vs others | 4.49 (1.65 to 12.2) | .003 | |
|
Multivariate competing risk regression models: HER2DX and intrinsic subtyped
|
|||
| Endpoint | Variables | Subdistribution HR (95% CI) | P |
| Any distant | HER2DX risk score high vs low | 3.71 (2.09 to 6.56) | <.001 |
| Luminal A vs others | 0.43 (0.20 to 0.94) | .033 | |
| Brain | HER2DX risk score high vs low | 5.13 (1.17 to 22.5) | .030 |
| HER2 enriched vs others | 5.32 (1.23 to 23.1) | .026 | |
| Lung | HER2DX risk score high vs low | 5.04 (1.48 to 17.2) | .010 |
| Basal vs others | 6.00 (2.19 to 16.4) | <.001 | |
| Bone only | HER2DX risk score high vs low | 6.90 (2.19 to 16.4) | .010 |
| Luminal A and luminal B vs others | 2.94 (1.10 to 7.85) | .032 | |
Only statistically significant and evaluable models showed. CI = confidence interval; HR = hazard ratio.
Models for those events for which stage and intrinsic subtype showed a statistically significant association in univariate analysis.
Regression model not evaluable for bone-only events.
Models for those events for which HER2DX risk score and intrinsic subtype showed a statistically significant association in univariate analysis.
Compared with patients with stage I disease, those with stage II and stage III had a statistically significantly increased risk of developing distant metastatic disease as the first event, statistically significantly increased risk of developing bone lesions, and numerically higher risk of lung metastases (Tables 2 and 3; Supplementary Figure 2, available online). The cumulative incidence of brain metastases at 5 and 10 years in stage III patients was as high as 8.9% (both at 5 and 10 years) as compared with 0.6% for stage I (both at 5 and 10 years) and 1.5% for stage II (both at 5 and 10 years) (Tables 2 and 3; Supplementary Figure 2, available online).
Cumulative incidence of distant relapse by intrinsic subtype and correction by stage
The intrinsic subtype was statistically significantly associated with the risk of any distant relapse as the first event, as well as metastases to the brain, lung, and bone only. The cumulative incidence rates are reported in Table 2, and the relevant cumulative incidence curves are shown in Figure 1. The results of the statistically significant competing risk models are presented in Table 3.
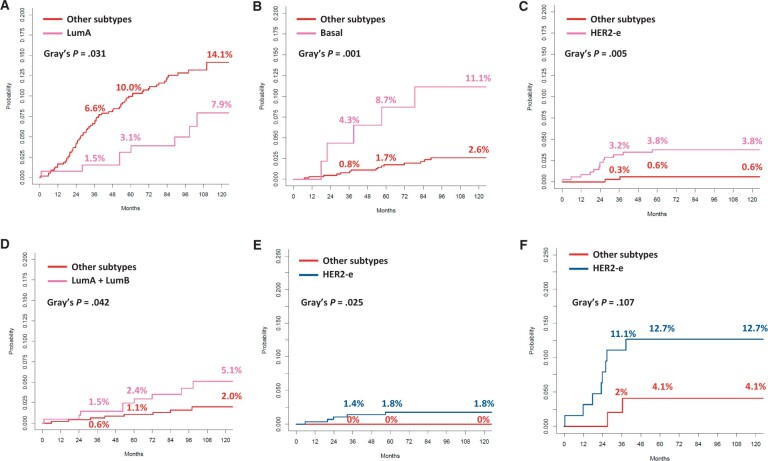
Figure 1.
Cumulative incidence curves according to PAM50 intrinsic molecular subtype for the following distant relapses as first event: any distant metastasis (A), lung metastasis (B), brain metastasis (C), bone-only metastasis (D), brain metastasis in patients with stage I-II disease at diagnosis (E), and brain metastasis in patients with stage III disease at diagnosis (F). HER2-e = HER2 enriched; LumA = luminal A; LumB = luminal B.
Patients with luminal A tumors experienced the lowest cumulative incidence of distant relapse across all intrinsic subtypes, with no substantial numerical differences among luminal B, HER2-enriched, basal-like, and normal subtypes (Table 2, Figure 1). However, in multivariate regression analysis including intrinsic subtype and stage, luminal A was not statistically significantly associated with a lower risk of distant relapse as the first event (subdistribution HR = 0.50, 95% CI = 0.24 to 1.04), whereas stage maintained a statistically significant association (Table 3).
Regarding site-specific metastases, HER2-enriched, basal-like, and luminal tumors were more prone as compared with other subtypes to generate brain, lung, and bone-only metastases as first event, respectively (Table 2, Figure 1). Compared with other patients, those with a HER2-enriched tumor had a statistically significantly higher risk of developing brain metastasis as the first event (subdistribution HR = 6.33, 95% CI = 1.44 to 27.9; P = .015; Table 2 and 3, Figure 1), with isolated brain metastases without extracranial disease occurring only in HER2-enriched patients (Table 2). The risk of lung metastasis as the first event was more than 4.5-fold higher in patients with basal-like tumors than in those with other tumors (Table 2 and 3, Figure 1). Finally, patients with luminal A and luminal B tumors had a statistically significant 2.6-fold increased risk of developing bone-only disease compared with other subtypes (Table 2 and 3, Figure 1). When correcting these regression models by stage, the intrinsic subtype maintained a statistically significant association with site-specific metastasis. Both HER2-enriched subtype and stage III were independently associated with an increased risk of brain metastases (Table 3). To show the independence of these 2 factors in determining the risk of brain metastases, we reported in Figure 1 cumulative incidence curves by intrinsic subtypes in stage I-II and stage III groups separately. Notably, the incidence at 5 years of brain metastasis in patients with HER2-enriched and stage III tumors was 12.7%. In the multivariate regression model for lung metastases, including intrinsic subtype and stage, only the basal-like subtype was an independent risk factor (Table 3).
Cumulative incidence of distant relapse by HER2DX and correction by intrinsic subtype
As expected, the HER2DX risk score was associated with the risk of any distant relapse: 10-year cumulative incidence of 19.7% vs 5.3% for high vs low risk score (P < .001; Tables 2 and 3; Supplementary Figure 3, available online). Moreover, we found a statistically significant association of HER2DX risk score with all site-specific metastases (with the exception of brain only), supporting an increased generic risk of developing distant metastases in patients with a high-risk score, without a specific preferential site. Table 2 shows the cumulative incidence rates and Gray P value, and cumulative incidence curves are shown as Supplementary Figure 3 (available online), and competing risk regression models are shown in Table 3.
Because intrinsic subtypes and the HER2DX risk score provide nonsumperimposable information and because we already showed that intrinsic subtype refined some site-specific regression models beyond stage, we conducted bivariate analyses integrating intrinsic subtype and HER2DX risk score for any distant, brain, lung, and bone-only metastases.
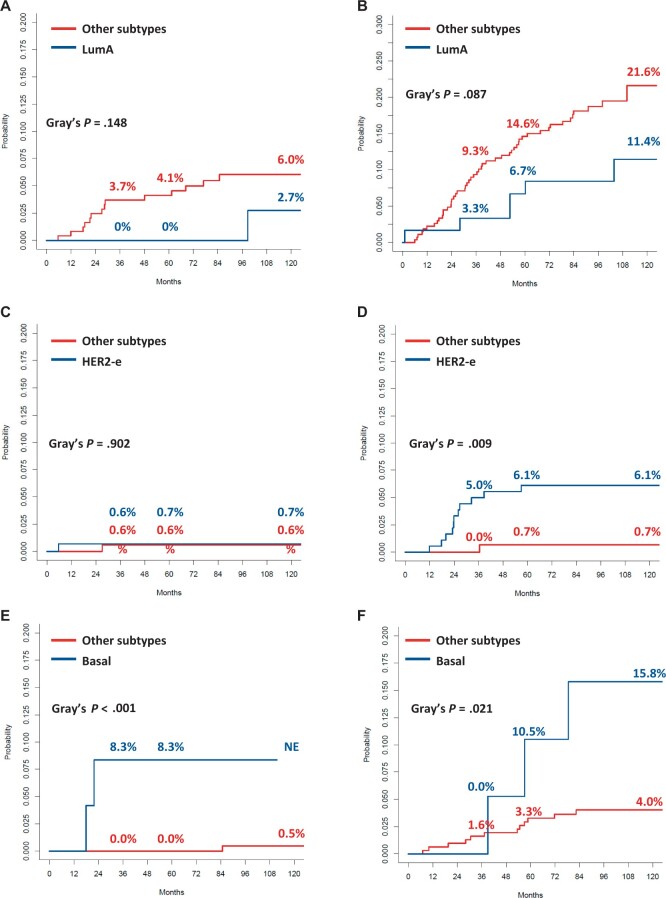
The results show that intrinsic subtype and HER2DX risk score were independently associated with the event of interest in each model, as detailed in Table 3. Cumulative incidence curves for any distant, brain, lung, and bone-only metastases according to intrinsic subtype in the HER2DX high-risk score and low-risk score are shown in Figure 2. Luminal A tumors, compared with other subtypes, showed lower cumulative incidence rates of any distant metastasis in HER2DX low-risk and HER2DX high-risk groups, without reaching statistical significance. The cumulative incidence of brain metastases was extremely low irrespective of intrinsic subtype in the HER2DX low-risk group, whereas in the HER2DX high-risk score group, there was a statistically significantly higher incidence for HER2-enriched cases than for other subtypes. For lung metastases, the cumulative incidence was statistically significantly higher for basal-like vs other subtypes in both HER2DX risk score groups. Finally, patients with a luminal A and luminal B tumor showed at least a numerically increased cumulative incidence of bone-only metastases in both the low HER2DX risk score group and in the high HER2DX risk score group.
Figure 2.
Cumulative incidence curves for distant relapse as first event according to intrinsic molecular subtype in HER2DX risk score high and low groups: any distant relapse in low (A) and high (B) HER2DX risk score; brain metastasis in low (C) and high (D) HER2DX risk score; lung metastasis in low (E) and high (F) HER2DX risk score. HER2-e = HER2 enriched; LumA = luminal A; LumB = luminal B.
Next, we evaluated the impact of HER2DX pCR score on the pattern of distant relapse. The results are presented in Supplementary Materials (Supplementary Results, available online) and Supplementary Tables 1 and 2 (available online).
Frequency of site-specific metastases in patients with a distant relapse as first event
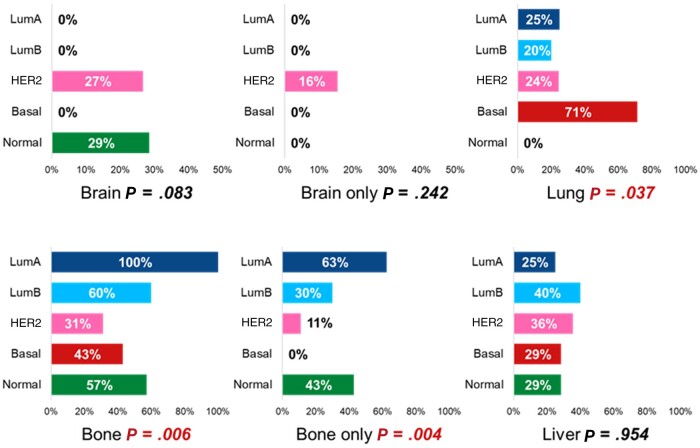
Finally, we analyzed the pattern of metastatic spread according to classic clinicopathological and molecular features among 77 patients who experienced distant relapse as the first event (Figure 3; Supplementary Table 3, available online). The overall distribution of metastatic sites was as follows: brain, 18%; brain only, 9%; lung, 26%; bone, 45%; bone only, 21%; and liver, 34%.
Figure 3.
Frequency of site-specific metastasis by intrinsic subtypes among patients who developed distant relapse as first event. Statistically significant P values are coloured in Red. LumA = luminal A; LumB = luminal B.
The frequency of brain metastases was higher in patients with stage III disease (stage I vs II vs III: 14.3% vs 9.1% vs 34.6%; P = .027). Isolated first distant relapse in the bone was more frequently observed in patients with hormone receptor–positive breast cancer than in those with hormone receptor–negative breast cancer (27.5% vs 7.7%; P = .043). The frequency of patients with lung, bone, and bone-only metastases was statistically significantly different according to intrinsic subtype (Figure 3). Lung metastases occurred more frequently in patients with basal-like tumors, whereas bone and bone-only disease occurred more frequently in patients with luminal A tumors. Brain-only relapses were exclusively observed in patients with HER2-enriched disease. HER2DX risk score was not associated with any site-specific distant relapse.
Discussion
This study represents the first evaluation of the pattern of metastatic spread in patients diagnosed with early HER2-positive breast cancer based on molecular features, such as intrinsic subtype and HER2DX. Our work led to 2 main results. First, we identified a higher disease stage at diagnosis and high HER2DX risk score, which integrates information on stage, as the main determinant of an increased general risk of metastatic disease as the first event, without being associated with a preferential site-specific pattern of metastatic spread. Second, our data revealed that gene expression tools reflecting intrinsic tumor molecular features, such as PAM50-based intrinsic subtype and HER2DX pCR score, tracking luminal, and HER2 signatures, were associated with unique patterns of preferential metastatic sites. In particular, the intrinsic molecular subtype could refine the site-specific prediction of the first metastatic event when integrated with the disease stage or HER2DX risk score. This result was further reinforced by the association between the PAM50 intrinsic subtype and the frequency of site-specific metastases in patients who developed a distant relapse as the first event.
The first set of results may help identify patients at increased risk of metastatic disease, which should be prioritized for intensive follow-up approaches. According to the stage and HER2DX risk score, patients at the highest risk had a 10-year cumulative incidence rate of distant relapse of 24.5% (stage III) and 19.7% (high HER2DX risk score). These results were not unexpected: stage is a well-known strong prognostic factor for metastatic risk in early HER2-positive breast cancer patients, and HER2DX risk score has been developed as a prognostic tool to predict distant disease-free survival (31). The intrinsic molecular subtype was also associated with an increased incidence of distant relapse at any site, and the integration of this factor further refined the prediction beyond the HER2DX risk score in regression models. In particular, patients with a low HER2DX risk score and a luminal A tumor experienced an extremely low incidence of metastasis as the first event at 10 years, identifying a subset of patients for whom intensive follow-up strategies are likely to be inappropriate. A second fundamental question to be answered when planning intensive follow-up is, how long? In this study, taking advantage of a long median follow-up period, we reported a persistent risk of metastatic disease beyond 5 years from diagnosis across all subgroups defined by clinicopathological characteristics or molecular features. Although there was no overall difference in the cumulative incidence of metastasis as the first event between patients with hormone receptor–positive and hormone receptor–negative tumors across the entire follow-up period, the absolute difference between the incidence rates at 5 and 10 years was larger in the case of hormone receptor–positive tumors than in patients with hormone receptor–negative disease. This observation is in line with other studies showing that the continuous risk of relapse is more evident in patients with hormone receptor–positive disease but not exclusively (19,20).
Once patients at an increased risk of distant relapse suitable for intensified surveillance have been identified, the next step is to decide which type of radiological examination is considered the most cost-effective and appropriate in a personalized approach. We demonstrated that PAM50-based intrinsic molecular subtype classification can discriminate between patients with distinct patterns of metastatic spread. The propensity of cancer cells to colonize specific distant organs arises from a complex and largely unknown interaction between seeds and soil (32). Our clinical observations indicate the impact of the intrinsic molecular biology of the primary tumor as a seed in the metastatic disease process. Several genomic-based signatures have been proposed to predict site-specific metastatic spread, although none have been developed specifically for HER2-positive breast cancer, and none has been definitively clinically validated to date (32-36).
Brain metastases represent a major clinical challenge for patients with HER2-positive breast cancer (37,38). Indeed, the incidence of brain metastases has been unaffected by advances in therapies for early disease (3-5,7,8). In our cohort, the incidence of brain relapse as the first event was 2.2% at 5 years, which is in line with data from the HERA and APHINITY trials (4,7). Recently, access to new anti-HER2 drugs with remarkable intracranial activity and the availability of sensitive diagnostic tools have attracted interest for brain metastasis prevention and early diagnosis. International guidelines have opened up the possibility of performing radiological brain screening for asymptomatic patients with HER2-positive stage IV disease, whereas they still recommend against surveillance during follow-up for patients with stage I-III disease (39,40). In this study, among all breast cancer subtypes, HER2 enriched clearly emerged as specifically associated with the development of brain metastases as the first event. In addition, brain relapse in the absence of other disease sites occurred exclusively in patients with a HER2-enriched tumor. A specific tropism of HER2-driven cancer cells for the central nervous system has been previously described, supported by studies describing the acquisition of alterations in the HER2 pathway and/or intrinsic subtype switching to HER2 enriched as frequent phenomena in brain metastases (41,42). In the context of their association with an increased general risk of distant relapse, the disease stage and HER2DX risk score modulated the risk of brain metastases. A higher disease burden at diagnosis is already known as a classic tumor-associated factor that modifies the risk of brain metastases (7,43), as confirmed by our data, and showed in our analysis a stronger association with brain relapse compared with the HER2DX risk score. When the disease stage was combined with the intrinsic subtype, both resulted in independent predictors of brain metastases, and the group of patients with a HER2-enriched tumor presenting with stage III at diagnosis emerged as the one with the highest absolute cumulative incidence of such an event. This combined biomarker could drive the implementation of personalized central nervous system screening and could be used as stratification factor and/or enrichment criteria in ongoing and future trials looking at brain metastasis screening/prevention (NCT04030507, NCT03881605, NCT05130840). Finally, in our large dataset with a long follow-up, brain relapses as the first event occurred in the first 5 years, which is potentially useful information in the planning of dedicated strategies.
The intrinsic subtype was also associated with the risk of lung metastases beyond the disease stage or HER2DX risk score, with basal-like tumors showing a major propensity to colonize this distant organ. Interestingly, previous preclinical and translational studies have linked breast cancer tropism for the lung with cancer stem cell properties, epithelial-to-mesenchymal transition, and Wnt pathway signaling, all of which are typically enriched in basal-like breast cancer (36,44). This information could be helpful in guiding individualized surveillance and may support clinical reasoning in the differential diagnosis of lung nodules detected during follow-up.
Finally, luminal tumors show a preferential predisposition for bone-only metastases. It has been previously shown that in HER2-positive breast cancer patients who develop distant recurrence, bone metastases are more common in hormone receptor–positive tumors than in hormone receptor–negative tumors (19). However, hormone receptor–positive status is a largely suboptimal surrogate for the luminal subtype (21,22). In our study, among patients who developed distant metastasis as the first event, we found an association between bone-only disease and HER2-positive/hormone receptor–positive breast cancer as well as a higher frequency of bone metastases in patients with luminal tumors, confirmed at the cumulative incidence analyses, which, on the other hand, did not confirm the same association for hormone receptor status. This suggested that intrinsic subtyping information may be more efficient in guiding bone-directed imaging during follow-up. Moreover, HER2-positive breast cancer patients with luminal tumors may be ideal candidates for bone-modifying agents in the adjuvant setting (45).
Our study has limitations. Surveillance protocols were not homogeneous across population cohorts. However, this potential diagnostic bias is unlikely to be unbalanced among different molecular categories. Moreover, some screening procedures, such as central nervous system or bone imaging in the absence of symptoms, were consistently not recommended for all cohorts. The main caveat is that the patients included in our study did not receive the most updated therapeutic strategies that are currently available for HER2-positive early breast cancer. This is the price to pay for a clinical cohort with a long-term follow-up. Notably, we do not expect results on brain metastases to be affected by this issue. It should also be acknowledged as an intrinsic bias that the ShortHER trial represented the training dataset for HER2DX risk score as a prognostic tool. Finally, although we provided possible explanations driving the observation of specific metastatic patterns across different molecular subtypes, they should be interpreted as speculative.
The main strengths include the unique design, being the first study to explore the pattern of distant metastases in HER2-positive disease according to molecular features, a large cohort of patients profiled by PAM50 and HER2DX with complete available information on the type and site of the first relapse, the long median follow-up, high treatment homogeneity, and the statistical approach taking advantage of the competing risk methodology.
In conclusion, we demonstrated that the integration of stage with molecular characterization can identify distinct patterns of metastatic spread in patients with early HER2-positive disease. This approach may have implications in planning individualized follow-up strategies, in the design and conduct of clinical trials testing, intensive surveillance, or aiming to prevent distant recurrences at specific sites.
Supplementary Material
Acknowledgements
This work was supported by Agenzia Italiana del Farmaco (AIFA, grant FARM62MC97), Italian Association for Cancer Research (AIRC, project MFAG 2014-15938; to V Guarneri), funding from the University of Padova, Department of Surgery, Oncology and Gastroenterology DOR 2019 (to VG, MVD, PFC), DOR 2020 (to VG, MVD), DOR 2021 (to VG, MVD), Fondazione AIRC under 5 per mille 2019 (ID. 22759 program—group leader VG).
The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Contributor Information
Maria Vittoria Dieci, Department of Surgery, Oncology and Gastroenterology, University of Padova, Padova, Italy; Oncology 2, Veneto Institute of Oncology Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS), Padova, Italy.
PierFranco Conte, Veneto Oncology Network, Padova, Italy.
Giancarlo Bisagni, Department of Oncology and Advanced Technologies, Oncology Unit, Azienda Unità Sanitaria Locale-Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS), Reggio Emilia, Italy.
Stefania Bartolini, Nervous System Medical Oncology Department, IRCCS Istituto delle Scienze Neurologiche di Bologna, Bologna, Italy.
Antonio Frassoldati, Clinical Oncology, Department of Translational Medicine and for Romagna, S. Anna University Hospital, Ferrara, Italy.
Daniele Generali, Department of Medicine, Surgery and Health Sciences, University of Trieste, Trieste, Italy; Multidisciplinary Unit of Breast Pathology and Translational Research, Cremona Hospital, Cremona, Italy.
Federico Piacentini, Department of Medical and Surgical Sciences for Children and Adults, University Hospital of Modena, Modena, Italy.
Gaia Griguolo, Department of Surgery, Oncology and Gastroenterology, University of Padova, Padova, Italy; Oncology 2, Veneto Institute of Oncology Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS), Padova, Italy.
Enrico Tagliafico, Center for Genome Research, University of Modena and Reggio Emilia, Modena, Italy; Department of Laboratory Medicine and Pathology, Diagnostic Hematology and Clinical Genomics Unit, Modena University Hospital, Modena, Italy.
Fara Brasó Maristany, Translational Genomics and Targeted Therapies in Solid Tumors, August Pi i Sunyer Biomedical Research Institute, Barcelona, Spain.
Nuria Chic, Translational Genomics and Targeted Therapies in Solid Tumors, August Pi i Sunyer Biomedical Research Institute, Barcelona, Spain; Department of Medical Oncology, Hospital Clinic of Barcelona, Spain.
Laia Paré, Reveal Genomics, Barcelona, Spain.
Federica Miglietta, Department of Surgery, Oncology and Gastroenterology, University of Padova, Padova, Italy; Oncology 2, Veneto Institute of Oncology Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS), Padova, Italy.
Roberto Vicini, Department of Medical and Surgical Sciences for Children and Adults, University Hospital of Modena, Modena, Italy.
Roberto D’Amico, Department of Medical and Surgical Sciences for Children and Adults, University Hospital of Modena, Modena, Italy.
Sara Balduzzi, Department of Medical and Surgical Sciences for Children and Adults, University Hospital of Modena, Modena, Italy.
Aleix Prat, Translational Genomics and Targeted Therapies in Solid Tumors, August Pi i Sunyer Biomedical Research Institute, Barcelona, Spain; Department of Medical Oncology, Hospital Clinic of Barcelona, Spain; Reveal Genomics, Barcelona, Spain; Department of Medicine, University of Barcelona, Barcelona, Spain.
Valentina Guarneri, Department of Surgery, Oncology and Gastroenterology, University of Padova, Padova, Italy; Oncology 2, Veneto Institute of Oncology Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS), Padova, Italy.
Data availability
The datasets that support the findings of this study are not publicly available in order to protect patient privacy. The data will be available on reasonable request from the corresponding author: MVD, mariavittoria.dieci@unipd.it.
Author Contributions
Maria Vittoria Dieci, MD (Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Resources; Validation; Visualization; Writing—original draft; Writing—review & editing), Pierfranco Conte, MD (Conceptualization; Funding acquisition; Investigation; Writing—review & editing), Giancarlo Bisagni, MD (Investigation; Writing—review & editing), Stefania Bartolini, MD (Investigation; Writing—review & editing), Antonio Frassoldati, MD (Investigation; Writing—review & editing), Daniele Generali, MD (Investigation; Writing—review & editing), Federico Piacentini, MD (Investigation; Writing—review & editing), Gaia Griguolo, MD (Formal analysis; Investigation; Writing—review & editing), Enrico Tagliafico, PhD (Investigation; Writing—review & editing), Fara Brasó Maristany, PhD (Investigation; Writing—review & editing), Nuria Chic, MD (Investigation; Writing—review & editing), Laia Paré, PhD (Investigation; Writing—review & editing), Federica Miglietta, MD (Investigation; Writing—review & editing), Roberto Vicini, MSc (Investigation; Writing—review & editing), Roberto D’Amico, PhD (Investigation; Writing—review & editing), Sara Balduzzi, PhD (Investigation; Writing—review & editing), Aleix Prat, MD, PhD (Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Resources; Supervision; Validation, Visualization; Writing—original draft; Writing—review & editing), and Valentina Guarneri, MD (Conceptualization; Funding acquisition; Investigation; Methodology; Project administration; Resources; Supervision; Validation; Writing –original draft; Writing—review & editing).
Conflicts of interest
MVD reports personal fees from EliLilly, Exact Sciences, Novartis, Pfizer, Seagen, Gilead, MSD, AstraZeneca, Daiichi Sankyo, and Roche outside of the submitted work. PFC reports personal fees from Novartis, EliLilly, AstraZeneca, Tesaro, Daiichi-Sankyo, Gilead, Reveal Genomics, BMS, and Roche outside the submitted work. MVD is also listed as an inventor of patent applications for the HER2DX assay. PFC reports personal fees for advisory board from Daiichi Sankyo, Lilly, Reveal Genomics, Gilead Science, personal fees for Speakers’ bureau from Roche, Genetech, Novartis, Astrazeneca, Lilly, BMS, institutional research funding from Merck KGaA, PFC is also listed as an inventor on patent applications for the HER2DX assay. AF reports personal fees from Roche, Novartis, EliLilly, AstraZeneca, Daiichi Sankyo, Seagen, and Gilead, outside the submitted work. GG reports personal fees from Eli Lilly, Novartis, and Gilead. FB-M has the HER2DX patent application filed. FM reports personal fees from Roche, Novartis, Gilead, and Pfizer. VG reports personal fees from EliLilly, Exact Sciences, Novartis, Pfizer, Gilead, MSD, Amgen, Sanofi, Merck Serono, and Eisai outside the submitted work. VG is listed as an inventor of patent applications for the HER2DX assay. AP reports advisory and consulting fees from Roche, Pfizer, Novartis, Amgen, BMS, Puma, Oncolytics Biotech, MSD, Guardant Health, Peptomyc, and Lilly; lecture fees from Roche, Pfizer, Novartis, Amgen, BMS, Nanostring Technologies, and Daiichi Sankyo; institutional financial interests from Boehringer, Novartis, Roche, Nanostring, Sysmex Europa GmbH, Medica Scientia Innovation Research, SL, Celgene, Astellas, and Pfizer; stockholder and consultant of Reveal Genomics, SL. AP is also listed as an inventor of patent applications for the HER2DX assay. LP is listed as an inventor of patent PCT/EP2021/070788. The other authors declare no conflicts of interest.
References
- 1. Miglietta F, Dieci MV, Griguolo G, Guarneri V.. Neoadjuvant approach as a platform for treatment personalization: focus on HER2-positive and triple-negative breast cancer. Cancer Treat Rev. 2021;98:102222. doi: 10.1016/j.ctrv.2021.102222 [DOI] [PubMed] [Google Scholar]
- 2. Guarneri V, Barbieri E, Dieci MV, Piacentini F, Conte P.. Anti-HER2 neoadjuvant and adjuvant therapies in HER2 positive breast cancer. Cancer Treat Rev. 2010;36(suppl 3):S62-6. doi: 10.1016/S0305-7372(10)70022-0 [DOI] [PubMed] [Google Scholar]
- 3. von MG, Huang CS, Mano MS, et al. ; KATHERINE Investigators. Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N Engl J Med. 2019;380(7):617-628. doi: 10.1056/NEJMoa1814017 [DOI] [PubMed] [Google Scholar]
- 4. Loibl S, Jassem J, Sonnenblick A, et al. VP6-2022: Adjuvant pertuzumab and trastuzumab in patients with early HER-2 positive breast cancer in APHINITY: 8.4 years’ follow-up. Ann Oncol. 2022;33:986-987. doi: 10.1016/j.annonc.2022.06.009 [DOI] [Google Scholar]
- 5. Stavrou E, Winer EP, Lin NU.. How we treat HER2-positive brain metastases. ESMO Open. 2021;6(5):100256. doi: 10.1016/j.esmoop.2021.100256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pestalozzi BC, Zahrieh D, Price KN, et al. ; International Breast Cancer Study Group (IBCSG). Identifying breast cancer patients at risk for Central Nervous System (CNS) metastases in trials of the International Breast Cancer Study Group (IBCSG). Ann Oncol. 2006;17(6):935-944. doi: 10.1093/annonc/mdl064 [DOI] [PubMed] [Google Scholar]
- 7. Pestalozzi BC, Holmes E, de Azambuja E, et al. CNS relapses in patients with HER2-positive early breast cancer who have and have not received adjuvant trastuzumab: a retrospective substudy of the HERA trial (BIG 1-01). Lancet Oncol. 2013;14(3):244-248. doi: 10.1016/S1470-2045(13)70017-2 [DOI] [PubMed] [Google Scholar]
- 8. Ferraro E, Singh J, Patil S, et al. Incidence of brain metastases in patients with early HER2-positive breast cancer receiving neoadjuvant chemotherapy with trastuzumab and pertuzumab. NPJ Breast Cancer. 2022;8(1):37. doi: 10.1038/s41523-022-00380-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cardoso F, Kyriakides S, Ohno S, et al. ESMO Guidelines Committee. Electronic address: clinicalguidelines@esmo.org. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2019;30(8):1194-1220. doi: 10.1093/annonc/mdz173 [DOI] [PubMed] [Google Scholar]
- 10. Galjart B, Höppener DJ, Aerts JGJV, Bangma CH, Verhoef C, Grünhagen DJ.. Follow-up strategy and survival for five common cancers: a meta-analysis. Eur J Cancer. 2022;174:185-199. doi: 10.1016/j.ejca.2022.07.025 [DOI] [PubMed] [Google Scholar]
- 11. Bornhak S, Heidemann E, Herschlein HJ, et al. Symptom-oriented follow-up of early breast cancer is not inferior to conventional control. Results of a prospective multicentre study. Onkologie. 2007;30(8-9):443-449. doi: 10.1159/000105257 [DOI] [PubMed] [Google Scholar]
- 12. Turco MRD, Palli D, Cariddi A, Ciatto S, Pacini P, Distante V.. Intensive diagnostic follow-up after treatment of primary breast cancer. A randomized trial. National Research Council Project on Breast Cancer follow-up. JAMA. 1994;271(20):1593-1597. doi: 10.1001/jama.271.20.1593 [DOI] [PubMed] [Google Scholar]
- 13. Palli D, Russo A, Saieva C, et al. Intensive vs clinical follow-up after treatment of primary breast cancer: 10-year update of a randomized trial. National Research Council Project on Breast Cancer Follow-up. JAMA. 1999;281(17):1586. doi: 10.1001/jama.281.17.1586 [DOI] [PubMed] [Google Scholar]
- 14. Ghezzi P, Magnanini S, Rinaldini M, et al. Impact of follow-up testing on survival and health-related quality of life in breast cancer patients. A multicenter randomized controlled trial. The GIVIO Investigators. JAMA. 1994;271(20):1587-1592. doi: 10.1001/jama.1994.03510440047031 [DOI] [PubMed] [Google Scholar]
- 15. Rose FD, Meduri B, Santis MCD, et al. Rethinking breast cancer follow-up based on individual risk and recurrence management. Cancer Treat Rev. 2022;109:102434. doi: 10.1016/j.ctrv.2022.102434 [DOI] [PubMed] [Google Scholar]
- 16. Kennecke H, Yerushalmi R, Woods R, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28(20):3271-3277. doi: 10.1200/JClinOncol.2009.25.9820 [DOI] [PubMed] [Google Scholar]
- 17. Ribelles N, Perez-Villa L, Jerez JM, et al. Pattern of recurrence of early breast cancer is different according to intrinsic subtype and proliferation index. Breast Cancer Res. 2013;15(5):R98. doi: 10.1186/bcr3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gerratana L, Fanotto V, Bonotto M, et al. Pattern of metastasis and outcome in patients with breast cancer. Clin Exp Metastasis. 2015;32(2):125-133. doi: 10.1007/s10585-015-9697-2 [DOI] [PubMed] [Google Scholar]
- 19. Lambertini M, Campbell C, Gelber RD, et al. Dissecting the effect of hormone receptor status in patients with HER2-positive early breast cancer: exploratory analysis from the ALTTO (BIG 2-06) randomized clinical trial. Breast Cancer Res Treat. 2019;177(1):103-114. doi: 10.1007/s10549-019-05284-y [DOI] [PubMed] [Google Scholar]
- 20. Chumsri S, Li Z, Serie DJ, et al. Incidence of late relapses in patients with HER2-positive breast cancer receiving adjuvant trastuzumab: combined analysis of NCCTG N9831 (alliance) and NRG oncology/NSABP B-31. J Clin Oncol. 2019;37(35):3425-3435. doi: 10.1200/JClinOncol.19.00443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schettini F, Prat A.. Dissecting the biological heterogeneity of HER2-positive breast cancer. Breast. 2021;59:339-350. doi: 10.1016/j.breast.2021.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dieci MV, Miglietta F, Griguolo G, Guarneri V.. Biomarkers for HER2-positive metastatic breast cancer: beyond hormone receptors. Cancer Treat Rev. 2020;88:102064. doi: 10.1016/j.ctrv.2020.102064 [DOI] [PubMed] [Google Scholar]
- 23. Prat A, Guarneri V, Pascual T, et al. Development and validation of the new HER2DX assay for predicting pathological response and survival outcome in early-stage HER2-positive breast cancer. EBioMedicine. 2022;75:103801. doi: 10.1016/j.ebiom.2021.103801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Conte P, Frassoldati A, Bisagni G, et al. Nine weeks versus 1 year adjuvant trastuzumab in combination with chemotherapy: final results of the phase III randomized Short-HER study‡. Ann Oncol. 2018;29(12):2328-2333. doi: 10.1093/annonc/mdy414 [DOI] [PubMed] [Google Scholar]
- 25. Guarneri V, Generali DG, Frassoldati A, et al. Double-blind, placebo-controlled, multicenter, randomized, Phase IIB neoadjuvant study of letrozole-lapatinib in postmenopausal hormone receptor–positive, human epidermal growth factor receptor 2–negative, operable breast cancer. J Clin Oncol. 2014;32(10):1050-1057. doi: 10.1200/JClinOncol.2013.51.4737 [DOI] [PubMed] [Google Scholar]
- 26. Parker JS, Mullins M, Cheang MCU, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160-1167. doi: 10.1200/JClinOncol.2008.18.1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guarneri V, Dieci MV, Bisagni G, et al. PIK3CA mutation in the ShortHER randomized adjuvant trial for patients with early HER2+ breast cancer: association with prognosis and integration with PAM50 subtype. Clin Cancer Res. 2020;26(22):5843-5851. doi: 10.1158/1078-0432.CCR-20-1731 [DOI] [PubMed] [Google Scholar]
- 28. Fine JP, Gray RJ.. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94(446):496-509. doi: 10.1080/01621459.1999.10474144 [DOI] [Google Scholar]
- 29. Gray RJ. A class of $K$-sample tests for comparing the cumulative incidence of a competing risk. Ann. Statist. 1988;16(3):doi: 10.1214/aos/1176350951 [DOI] [Google Scholar]
- 30. The R Foundation. R: The R Project for Statistical Computing.https://www.r-project.org/. Accessed September 23, 2023.
- 31. Prat A, Guarneri V, Pascual T, et al. Development and validation of the new HER2DX assay for predicting pathological response and survival outcome in early-stage HER2-positive breast cancer. EBioMedicine. 2022;75:103801. doi: 10.1016/j.ebiom.2021.103801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gao Y, Bado I, Wang H, Zhang W, Rosen JM, Zhang XHF.. Metastasis organotropism: redefining the congenial soil. Dev Cell. 2019;49(3):375-391. doi: 10.1016/j.devcel.2019.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518-524. doi: 10.1038/nature03799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bos PD, Zhang XHF, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005-1009. doi: 10.1038/nature08021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klein A, Olendrowitz C, Schmutzler R, et al. Identification of brain- and bone-specific breast cancer metastasis genes. Cancer Lett. 2009;276(2):212-220. doi: 10.1016/j.canlet.2008.11.017 [DOI] [PubMed] [Google Scholar]
- 36. DiMeo TA, Anderson K, Phadke P, et al. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009;69(13):5364-5373. doi: 10.1158/0008-5472.CAN-08-4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Darlix A, Griguolo G, Thezenas S, et al. Hormone receptors status: a strong determinant of the kinetics of brain metastases occurrence compared with HER2 status in breast cancer. J Neurooncol. 2018;138(2):369-382. doi: 10.1007/s11060-018-2805-9 [DOI] [PubMed] [Google Scholar]
- 38. Darlix A, Louvel G, Fraisse J, et al. Impact of breast cancer molecular subtypes on the incidence, kinetics and prognosis of central nervous system metastases in a large multicentre real-life cohort. Br J Cancer. 2019;121(12):991-1000. doi: 10.1038/s41416-019-0619-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rhun EL, Guckenberger M, Smits M, et al. ; EANO Executive Board and ESMO Guidelines Committee. Electronic address: clinicalguidelines@esmo.org. EANO-ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up of patients with brain metastasis from solid tumours. Ann Oncol. 2021;32(11):1332-1347. doi: 10.1016/j.annonc.2021.07.016 [DOI] [PubMed] [Google Scholar]
- 40. Ramakrishna N, Anders CK, Lin NU, et al. Management of advanced human epidermal growth factor receptor 2-positive breast cancer and brain metastases: ASCO guideline update. J Clin Oncol. 2022;40(23):2636-2655. doi:10.1200/J Clin Oncol.22.00520 [DOI] [PubMed] [Google Scholar]
- 41. Priedigkeit N, Hartmaier RJ, Chen Y, et al. Intrinsic subtype switching and acquired ERBB2/HER2 amplifications and mutations in breast cancer brain metastases. JAMA Oncol. 2017;3(5):666-671. doi: 10.1001/jamaoncol.2016.5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Varešlija D, Priedigkeit N, Fagan A, et al. Transcriptome characterization of matched primary breast and brain metastatic tumors to detect novel actionable targets. J Natl Cancer Inst. 2019;111(4):388-398. doi: 10.1093/jnci/djy110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Laakmann E, Witzel I, Fasching PA, et al. Development of central nervous system metastases as a first site of metastatic disease in breast cancer patients treated in the neoadjuvant trials GeparQuinto and GeparSixto. Breast Cancer Res. 2019;21(1):60. doi: 10.1186/s13058-019-1144-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jin L, Han B, Siegel E, Cui Y, Giuliano A, Cui X.. Breast cancer lung metastasis: molecular biology and therapeutic implications. Cancer Biol Ther. 2018;19(10):858-868. doi: 10.1080/15384047.2018.1456599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Group (EBCTCG) EBCTC. Adjuvant bisphosphonate treatment in early breast cancer: meta-analyses of individual patient data from randomised trials. Lancet (Lond Engl). 2015;386(10001):1353-1361. doi: 10.1016/S0140-6736(15)60908-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets that support the findings of this study are not publicly available in order to protect patient privacy. The data will be available on reasonable request from the corresponding author: MVD, mariavittoria.dieci@unipd.it.