Abstract
Background: Deficiencies in DNA damage repair responses promote chemotherapy sensitivity of tumor cells. The Nibrin homolog encoding gene Nijmegen Breakage Syndrome 1 (NBS1) is a crucial component of the MRE11-RAD50-NBN complex (MRN complex) and is involved in the response to DNA double-strand breaks (DSBs) repair that has emerged as an attractive strategy to overcome tumor drug resistance, but the functional relationship between NBS1 regulated DNA damage repair and cell cycle checkpoints has not been fully elucidated. Methods: In this study, lentivirus-mediated RNAi was used to construct NBS1-downregulated cells. Flow cytometry, qPCR, and immunohistochemistry were used to explore the regulatory relationship between NBS1 and CyclinB in vivo and in vitro. Results: Our findings suggest that NBS1 deficiency leads to defective homologous recombination repair. Inhibition of NBS1 expression activates CHK1 and CyclinB signaling pathways leading to cell cycle arrest and sensitizes ovarian cancer cells to Olaparib treatment in vitro and in vivo. NBS1-deficient ovarian cancer cells tend to maintain sensitivity to chemotherapeutic drugs through activation of cell cycle checkpoints. Conclusions: NBS1 may be a potential therapeutic target for epithelial ovarian cancer as it plays a role in the regulation of the DNA damage response and cell cycle checkpoints. Suppression of NBS1 upregulates CyclinB to induce Olaparib sensitivity in ovarian cancer.
Keywords: Nijmegen Breakage Syndrome 1, CyclinB, Olaparib, homologous recombination, epithelial ovarian cancer
Introduction
Maintaining the integrity of the human genome, the so-called DNA damage response is crucial in preventing tumor development and drug resistance. DNA double-strand breaks (DSBs) are primarily repaired through 2 pathways: homologous recombination (HR) repair and nonhomologous end joining (NHEJ).1,2 NHEJ is faster than HR and mainly occurs in the G1 phase. Nevertheless, there is recent evidence that NHEJ functions throughout the cell cycle. 3 The MRE11-RAD50-NBN (MRN) complex, which is responsible for initial and sustained responses to DSBs, is a central conductor in the cellular response to DNA damage and the maintenance of chromosome integrity. 4 NBS1, a protein required for the MRN complex to exert DNA damage signal transduction, plays a critical role in HR repair regulation. 5 Although the NBS1 protein itself does not possess DNA-binding and kinase activities, NBS1 deficiency significantly suppressed HR repair efficiency following mitomycin-C intervention by approximately 100-fold compared with wild-type cells, indicating its essential role in HR repair regulations.6,7 Furthermore, cell cycle checkpoints must be coordinated with DNA repair to prevent cell growth until the completion of DNA repair. DSB repair involves at least 2 cell cycle checkpoints at the G1 and G2 phases coordinating the DNA repair process. NBS1 has been suggested to play a critical role in regulating the S phase and has been associated with G1 and G2 checkpoints8–11; however, the functional relationship between NBS1-regulated DNA damage repair and cell cycle checkpoints has not been fully elucidated.
CyclinB, a critical protein in cell cycle regulation, targets G2/M-related proteins that regulate ionizing radiation (IR) responses to control HR repair in cancers. 12 The IR response is a critical component of the DNA damage response, which plays a vital role in the maintenance of genome integrity. It involves a complex network of signaling pathways and cellular processes that are activated in response to DNA damage, including cell cycle checkpoints, chromatin remodeling, and DNA repair. The sensitivity of tumor cells to radiation and chemotherapy is influenced by the cell cycle stage, with cells being most sensitive to radiation in the G2/M phase, less sensitive in the G1 phase, and least sensitive in the late S phase. Elucidating the spatiotemporal regulation mechanisms that coordinate the DNA damage response through functional interplay cell cycle regulation protein is crucial for the development of effective therapeutic strategies for cancer treatment.
Poly(ADP-ribose) polymerase (PARP) inhibitors, particularly, Olaparib, are indicated for the treatment of advanced BRCA-mutated ovarian cancer to overcome chemotherapy drug resistance, improve the treatment effect, and prolong the survival time of patients.13,14 Olaparib exhibits potent antitumor effects in BRCA-mutant ovarian cancer.15,16 Clinical models show that PARP affects T-cell activation in a dose-dependent manner, leading to enhanced apoptosis of cancer cells. 17 It has also been found to play a role in BRCA wild-type ovarian cancer,18,19 but its underlying mechanism of action remains largely unknown. This study aims to investigate the regulatory mechanism of NBS1 in regulating ovarian cancer sensitivity to chemotherapy and its association with cell cycle checkpoints.
Materials and Methods
Cell Lines and Cell Cultures
The human cell lines A2780, HEY-A8, SKOV3, and Hosepic were procured from the Chinese Academy of Sciences Committee in Shanghai, China. The cells were maintained in RPMI-1640 medium containing 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. HO8910 and HEK 293 T cells were cultured in high glucose Dulbecco's Modified Eagle Medium (DMEM, Gibco, USA). The cells were grown at 37 °C in a humidified environment with 5% CO2.
Establishment of NBS1 Downregulated Cell Lines
A lentivirus expressing a specific RNAi interference sequence targeting NBS1 was designed and cloned into a GV248-puro (Addgene, MA, USA) vector at the AgeI and EcoRI sites to generate recombinant vectors GV248-puro-NBS1 (KD1 and KD2) and GV248-puro-control (CON) (Table 1). Lentiviruses were packaged by transfecting 293 T cells with the described lentivirus recombinant vectors, the packaging plasmid psPAX2, and the envelope vector pMD2.G (Addgene, MA, USA) using Lipofectamine 3000 transfection reagent (Invitrogen, CA, USA). The medium containing lentiviral particles was collected after 48 hours. Ovarian cancer cells were transfected with 1 × 106 IFU/mL lentivirus in 8 μg/mL Polybrene (Sigma-Aldrich, MO, USA) for 24 hours. Stably transfected cells were selected by treating with 1 μg/mL puromycin (Sigma-Aldrich, MO, USA) for 7 days. Western blot analysis was used to verify RNAi knockdown efficiency.
Table 1.
Sequences of Primers and Targets.
| Primers/targets | Sequences |
|---|---|
| shNBS1-CON | 5′-TTCTCCGAACGTGTCACGT-3′ |
| shNBS1-KD1 | 5′-TCGAAAGAATACAGAACTA-3′ |
| shNBS1-KD2 | 5′-AAGCAGATACATGGGATTT-3′ |
| NBS1 qRT-PCR forward | 5′-CACAACCTGCTACACCCTC-3′ |
| NBS1 qRT-PCR reverse | 5′-ACATTGACATCTTCCTCCT-3′ |
| β-actin qRT-PCR forward | 5′-AAGGTGACAGCAGTCGGTT-3′ |
| β-actin qRT-PCR reverse | 5′-TGTGTGGACTTGGGAGAGG-3′ |
| CyclinB qRT-PCR forward | 5′-TCTGGATAATGGTGAATGGACA-3′ |
| CyclinB qRT-PCR reverse | 5′’-CGATGTGGCATACTTGTTCTTG-3′ |
Abbreviations: NBS1, Nijmegen Breakage Syndrome 1; qRT-PCR, quantitative reverse transcription polymerase chain reaction.
Western Blot
Total protein was collected from the cells. Cell lysates were separated by SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Millipore, USA). After blocking with 10% nonfat milk (Solarbio, Beijing, China), the PVDF membranes were probed with primary antibodies, including NBS1 (1:1000 dilution, Abcam, MA, USA), β-actin (1:3000 dilution, Abcam, MA, USA), pH2AX (1:400 dilution, CST, MA, USA), Cyclin B (1:3000 dilution, Abcam, MA, USA), CHK1 (1:200 dilution, Santa Cruz, TX, USA), CDC25C (1:200 dilution, Santa Cruz, TX, USA), RAD51(1:3000 dilution, Abcam, MA, USA), Bax (1:3000 dilution, Abcam, MA, USA), Bcl-2 (1:3000 dilution, Abcam, MA, USA), Mre11 (1:1000 dilution, CST, MA, USA), MUS81 (1:200 dilution, Santa Cruz, TX, USA), pNBS1 (1:1000 dilution, CST, MA, USA), CDK1 (1:1000 dilution, Abcam, MA, USA), and p-CDK1 (1:1000 dilution, Abcam, MA, USA).
Quantitative RT-PCR
RNA was extracted from the NBS1 down-regulated cell lines using TRIzol® reagent (Thermo Fisher Scientific, Shanghai, China) and converted into cDNA using the PrimeScript® RT reagent Kit (TaKaRa, Shiga, Japan) as per the manufacturer's protocol. RT-PCR was carried out using a 10 μL reaction solution comprising 10 ng cDNA, 0.1 mmol/L primer (Table 1), and 5 mL 2 × SYBR Premix Ex Taq (TaKaRa, Shiga, Japan). PCR amplification was performed in a Mastercycler® ep realplex (Quantstudio dx, Thermo Fisher Scientific) with initial denaturation at 95 °C for 5 minutes, followed by 40 cycles of denaturation at 95 °C for 15 seconds and annealing/extension at 65 °C for 40 seconds. The transcriptional levels of NBS1 and CyclinB were normalized to β-actin as a reference gene.
Measurement of HR Efficiency
HR efficiency was detected using a qPCR-based HR assay kit (Norgen Biotek Corp, Thorold, Canada). The dl-1 and dl-2 plasmids were co-transfected into CON, KD1, and KD2 cells, and total cellular DNA was isolated 48 hours later using the Blood & Cell Culture DNA Mini Kit (QIAGEN, Germany). qPCR was performed using a set of universal primers and assay primers, and a standard curve was made with a positive control plasmid. Based on the standard DNA content curve, the difference in the number of cycles that reaches the inflection point of the amplification curve generated using HR-specific primers and the number of cycles that reaches the inflection point of the amplification curve with universal primers is converted into the difference in DNA content, and the differences in HR copy number between each group are compared.
Cells Intervention With UV Irradiation
Cells were seeded at a density of 3 × 105 cells per well in six-well plates. Ultraviolet (UV) irradiation (dose rate: 20 mJ/40 mJ/80 mJ, wavelength 365 nm, field: 20 × 20 cm) was used to irradiate A2780 and SKOV3 cells, while a well without UV irradiation was used as a control. Subsequently, the cells were cultured for 24 hours, collected, and lysed for total protein extraction using immunoprecipitation (IP) lysis buffer (Beyotime Biotechnology, Shanghai, China). Western blotting was used to measure the expression of NBS1, CyclinB, and other protein molecules, with pH2AX serving as an indicator of double-strand break repair.
Cell Cycle Assay
For the cell cycle assay, cells were seeded at a density of 4 × 105 cells per well in six-well plates. Then, 1 × 105 cells were collected and washed twice with ice-cold phosphate-buffered saline (PBS, Gibco, USA). The cells were transduced and fixed in 70% alcohol overnight at 4 °C. After that, they were incubated with 500 mL of propidium iodide (PI, BD PharmingenTM, USA) for 15 minutes in the dark. The distribution of cells in different cell cycle phases was analyzed by flow cytometer (Caliburn, BD, USA), and the experiment was performed in triplicates.
TUNEL Apoptosis Detection
Cells were grown in six-well plates, then washed with PBS twice and fixed with 4% paraformaldehyde for 30 minutes at room temperature. After washing again with PBS, they were incubated with PBS containing 0.3% Triton X-100 for 5 minutes at room temperature. TUNEL detection solution (Beyotime, China) was prepared using a ratio of TdT enzyme: fluorescent labeling solution: TUNEL detection solution = 1:9:10. Next, 100 μL of TUNEL detection solution was added to the sample and incubated at 37 °C for 60 minutes in the dark. After washing with PBS thrice, the slides were mounted with an anti-fluorescence quenching mounting solution (Beyotime, China) and visualized under a fluorescence microscope (Olympus, Japan). The Cy3 dye was excited at 550 nm, and the red fluorescence was emitted at 570 nm.
In Vivo Experiments
Female BALB/c nude mice aged 5 to 8 weeks and weighing ∼ 18 to 20 g were inoculated with 2 × 106 CON, KD1, or KD2 A2780 cells into the right flank. Tumor size and weight were measured daily, and when the calculated tumor volume reached ∼ 0.8 to 1.0 cm3, mice were randomized into Olaparib (n = 5/group) or control (n = 5/group) groups. Dissolve Olaparib in a solution containing 10% DMSO, 40% PEG300, 5% Tween 80%, and 45% ddH2O. Olaparib group was administered with 50 mg/kg Olaparib, i.p., once daily for 7 days, while the control group was administered with equal volume of normal saline (0.2 mL/mice). Tumor tissue was collected upon the completion of treatment for subsequent experiments. Mice with tissue damage and cachexia were excluded from the group during data collection.
Immunohistochemistry
The monoclonal antibody NBS1 (1:200 dilution, Abcam, MA, USA) and CyclinB (1:200 dilution, Abcam, MA, USA) are diluted with PBS at 1:50 were used to perform immunohistochemistry (IHC). Briefly, each 4-μm-thick paraffin-embedded section was dewaxed and hydrated, followed by endogenous peroxidase inhibition with methanol containing 0.3% H2O2. After antigen retrieval and cooling, the sections were blocked with 1% BSA and incubated overnight at 4 °C with primary antibody. The following day, the sections were incubated with HRP-conjugated secondary antibody (Shanghai Long Island Biotech, Shanghai, China) for 1 hour at room temperature, followed by diaminobenzidine treatment and hematoxylin counterstaining. Slides were visualized and photographed under light microscopy at 400 × (Leica, China, SP5).
Statistical Analysis
Data are presented as the mean ± SD of 3 independent experiments, and SPSS 20.0 software was used for statistical analyses. The differences between groups were examined by analysis of variance (ANOVA) and Student's t-tests. A P value < 0.05 was considered statistically significant.
Results
NBS1 is Overexpressed in Ovarian Cancer Tissues and Cell Lines
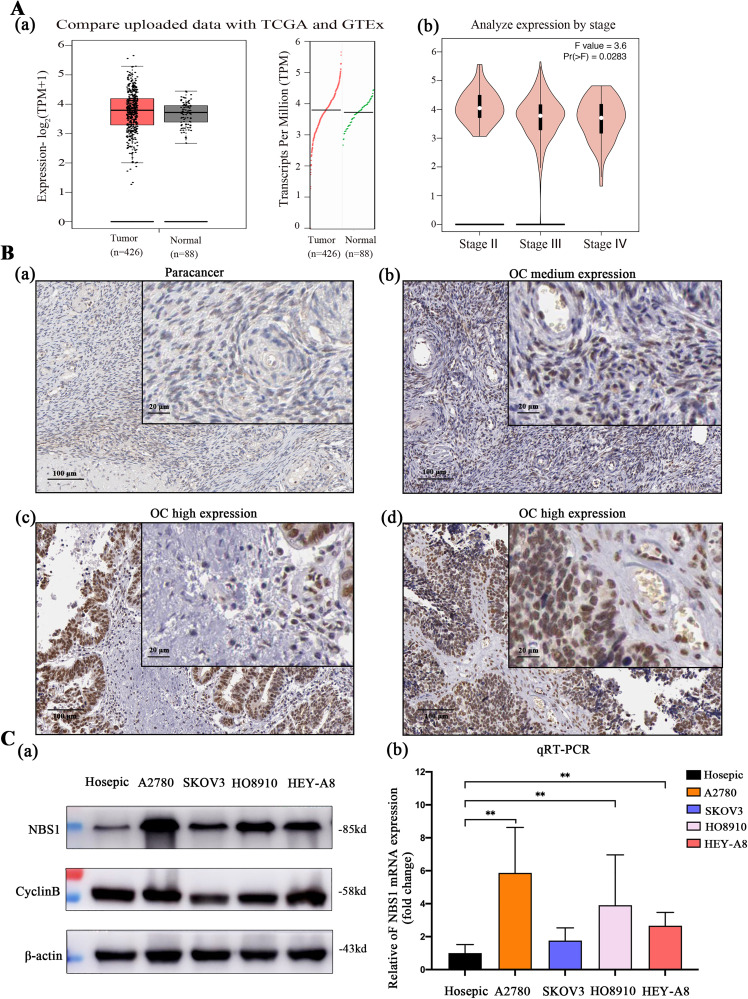
To investigate the expression of NBS1 in ovarian cancer, we conducted a comparative analysis of the TCGA and GTEx databases. The results indicated that the copy number of NBS1 in ovarian cancer (n = 456) was significantly higher than that in the control group (n = 55), as shown in Figure 1A(a). Furthermore, we studied the expression of NBS1 in different stages of ovarian cancer using the GEPIA database and found that the expression of the NBS1 gene was significantly higher in the early stage of ovarian cancer (P = .0283), as depicted in Figure 1A. Additionally, we compared HPA tissue banks and discovered that NBS1 was highly expressed in ovarian cancer tissues as compared to normal ovarian tissues (P < .05), as illustrated in Figure 1B and Table 2. Finally, we validated these results in ovarian cancer cell lines A2780, SKOV3, HO8910, and HEY-A8 by conducting western blot and RT-PCR analysis, which demonstrated that the expression level of NBS1 was significantly higher in these cell lines as compared to human normal ovarian Hosepic cells, as shown in Figure 1C. These findings are significant because they provide insights into the expression of NBS1 in ovarian cancer, which could potentially lead to the development of more effective treatments and drug resistance prevention strategies.
Figure 1.
(A) The expression of NBS1 in ovarian cancer and its relationship with the prognosis of ovarian cancer were analyzed through the database (http://www.cbioportal.org), (B) the HPA database (https://www.proteinatlas.org) analyzes the protein expression level of NBS1 in ovarian cancer tissues, and (C) protein and mRNA expression levels of NBS1 and its related protein CyclinB in ovarian cancer cell lines.
Abbreviations: NBS1, Nijmegen Breakage Syndrome 1; HPA, Human Protein Atlas.
Table 2.
Expression Levels of Nijmegen Breakage Syndrome 1 (NBS1) in Ovarian Cancer and Para-Carcinoma Tissues.
| Tumor | Normal | P | |
|---|---|---|---|
| High | 9 | 0 | |
| Medium | 0 | 3 | .002 |
| Low | 1 | 3 |
Down-Regulated NSB1 Inhibits HR Repair Pathway-Related Proteins and Promotes CyclinB Release
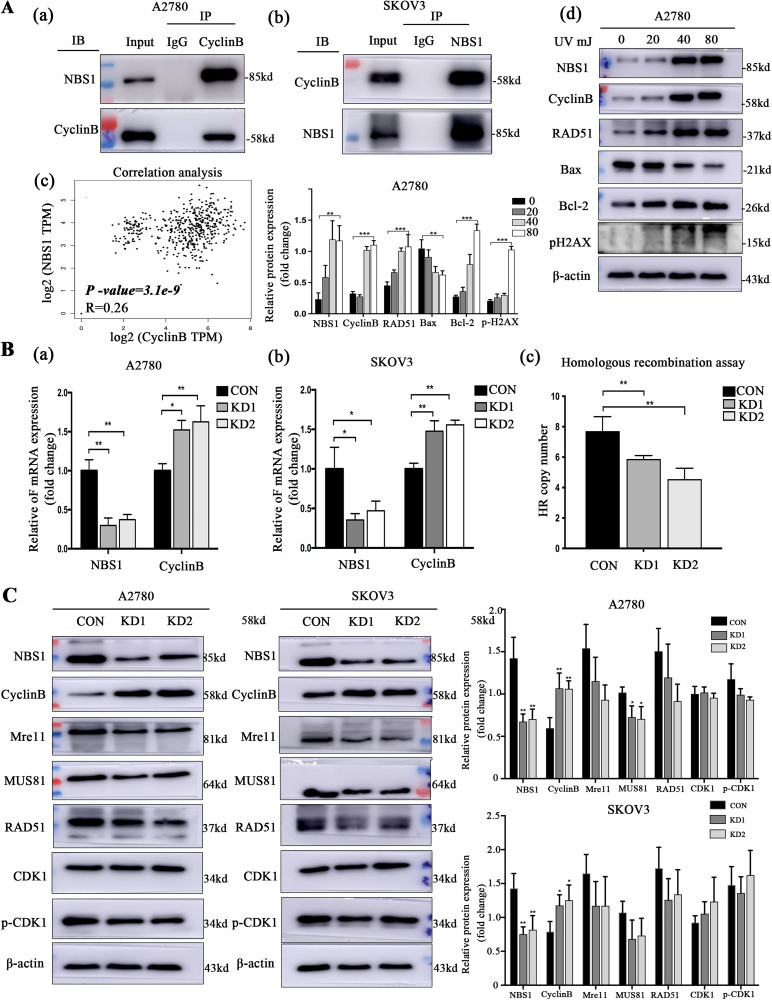
NBS1 is a crucial regulator of the HR repair process and cell cycle checkpoints, both of which play a critical role in the DNA damage response of tumors. CyclinB, an effector molecule, regulates the cell cycle, and its active state influences the repair process while also providing feedback regulation of the DNA damage response. Through co-immunoprecipitation experiments, we found that NBS1 and CyclinB co-expressed in A2780 cells, suggesting a protein interaction between the 2 (Figure 2A). Further gene correlation analysis using the TCGA database revealed a correlation between NBS1 and CyclinB (P < .05, R = 0.26, Figure 2A).To study the response to DNA damage, we established a model of DNA damage using UV irradiation in the A2780 cell lines. Western blot experiments revealed that both NBS1 and CyclinB were involved in the repair process, with their expression increasing with the dose of damage (Figure 2A). To investigate the mechanism by which NBS1 regulates CyclinB, we employed lentivirus-mediated RNA interference to down-regulate NBS1 in A2780 and SKOV3 cells. RT-PCR analysis revealed that the expression of NBS1 was significantly reduced in the KD1 and KD2 cell groups, while the expression of CyclinB was increased (Figure 2B). In order to study the relationship between NSB1 and HR activity, an HR assay kit was used to quantitatively detect HR products by qPCR, and it was observed that the HR efficiency of KD1 and KD2 cells was significantly lower than that of CON cells (Figure 2B). Western blot analysis of HR repair pathway proteins demonstrated that the expression of HR repair pathway-related proteins was suppressed in cells transfected with recombinant KD1/2 plasmid compared to those transfected with CON plasmid, and the CDK1 protein in the G2/M phase of the cell cycle checkpoints-related pathway was not phosphorylated. These results indicate that the inhibition of NSB1 expression hinders the repair of the HR repair pathway while enhancing CyclinB expression (Figure 2C).
Figure 2.
(A) NBS1 and CyclinB have participated in the HR repair pathway, (B) there is a regulatory relationship between NBS1 and CyclinB expression, and (C) the effect of NBS1 on the regulation of cell cycle pathway proteins.
Abbreviations: NBS1, Nijmegen Breakage Syndrome 1; HR, homologous recombination.
Downregulation of NSB1 Enhanced Olaparib Sensitivity in EOC Cells
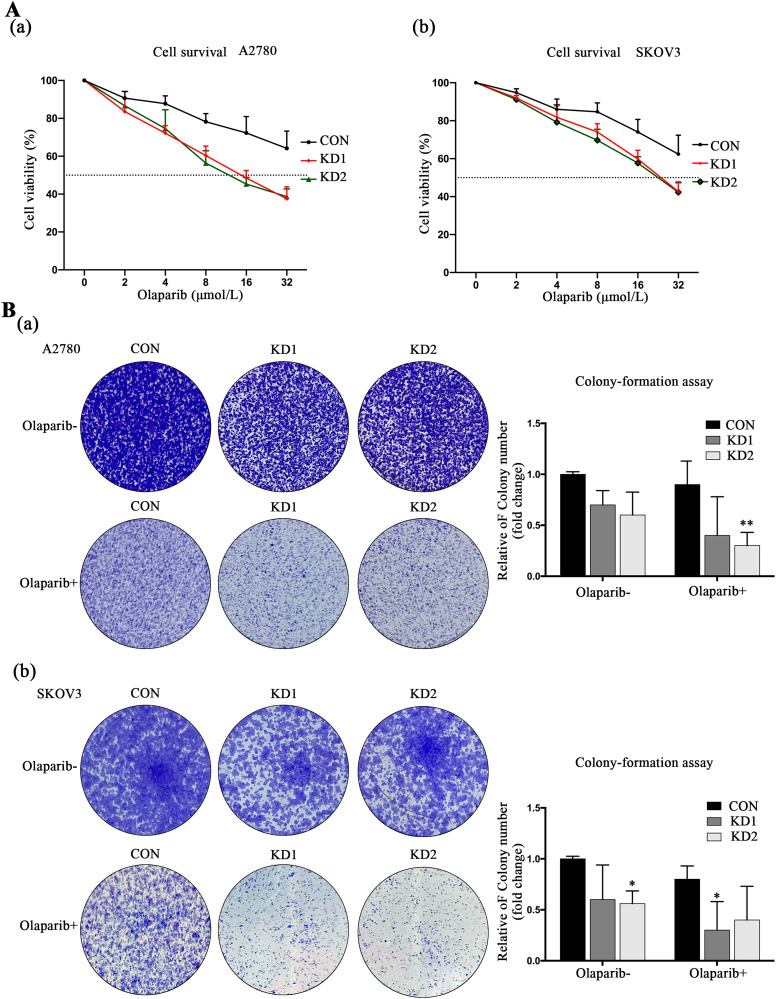
Our study has shown that Olaparib is effective in ovarian tumor cells with HR repair pathway defects, thereby increasing sensitivity to PARP inhibitors. CCK8 experiments revealed that Olaparib inhibited the growth of ovarian cancer cells in a dose-dependent manner, with a significantly lower IC50 value for KD1 and KD2 cells (A2780: 16.34 and 15.63 μmol/L, respectively; SKOV3: 20.38 and 20.45 μmol/L, respectively) compared to control cells (A2780: > 32 μmol/L; SKOV3: > 32 μmol/L) (Figure 3A). Furthermore, colony formation assays demonstrated a significant reduction in the number of colonies in KD1/2 cells treated with Olaparib compared to the CON group (Figure 3B). These findings suggest that downregulating NBS1 can sensitize ovarian tumor cells with HR repair pathway defects to Olaparib.
Figure 3.
(A) Olaparib inhibits the growth of cell lines by downregulating Nijmegen Breakage Syndrome 1 (NBS1) and (B) downregulation of NBS1 increases the drug sensitivity to Olaparib.
Activation of CyclinB in NBS1-Deficient Ovarian Cancer Promotes the Synergistic Inhibitory Effect of Olaparib
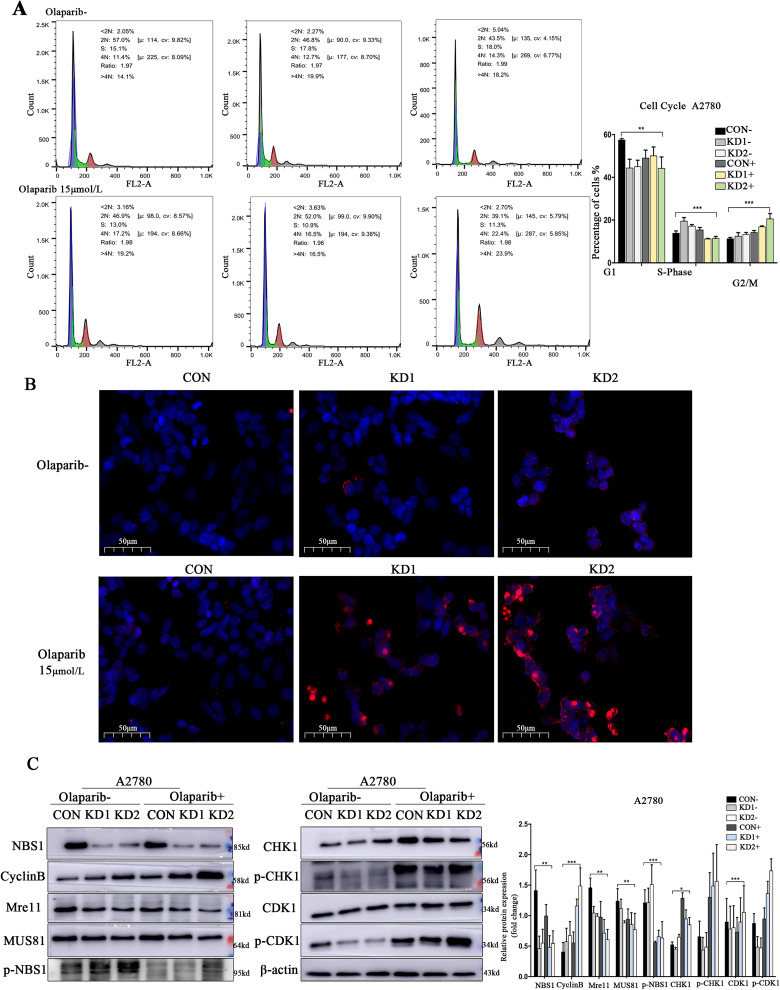
To investigate the regulatory mechanism of NBS1 sensitivity to Olaparib, flow cytometry was performed to detect the cell cycle. The results showed that after down-regulation of NBS1 expression, ovarian cancer cells were arrested in the S phase and G2/M phase. Olaparib treatment accelerated the cell cycle and caused significant G2/M phase arrest (Figure 4A). Additionally, the TUNEL assay revealed that Olaparib treatment significantly increased apoptosis in ovarian cancer cells with down-regulated NBS1 expression (Figure 4B). Western blot analysis was also conducted to assess protein expression levels after Olaparib treatment. The results indicated that in the absence of HR repair, CHK1 was phosphorylated, activating the G2/M cell cycle checkpoints and increasing CyclinB expression, leading to the release of p-CDK1, and thus inhibiting proliferation (Figure 4C). These findings suggest that Olaparib can produce a synergistic inhibitory effect on ovarian cancer cells with down-regulated NBS1 expression, and activate the G2/M cell cycle checkpoints to promote the drug sensitivity of ovarian cancer to Olaparib.
Figure 4.
(A) Sensitivity of cell cycle checkpoint NBS1−/HR− type ovarian cancer to Olaparib treatment, (B) sensitivity of NBS1−/HR− type ovarian cancer to Olaparib treatment detected by TUNEL assay, and (C) NBS1 activates pathway proteins of G2/M phase drug sensitivity.
Abbreviations: NBS1, Nijmegen Breakage Syndrome 1; HR, homologous recombination; TUNEL, terminal deoxynucleotidyl transferase dUTP nick-end labeling.
Inhibiting NBS1 and Activating CyclinB Promote the Olaparib Sensitivities of Ovarian Cancer to In Vivo
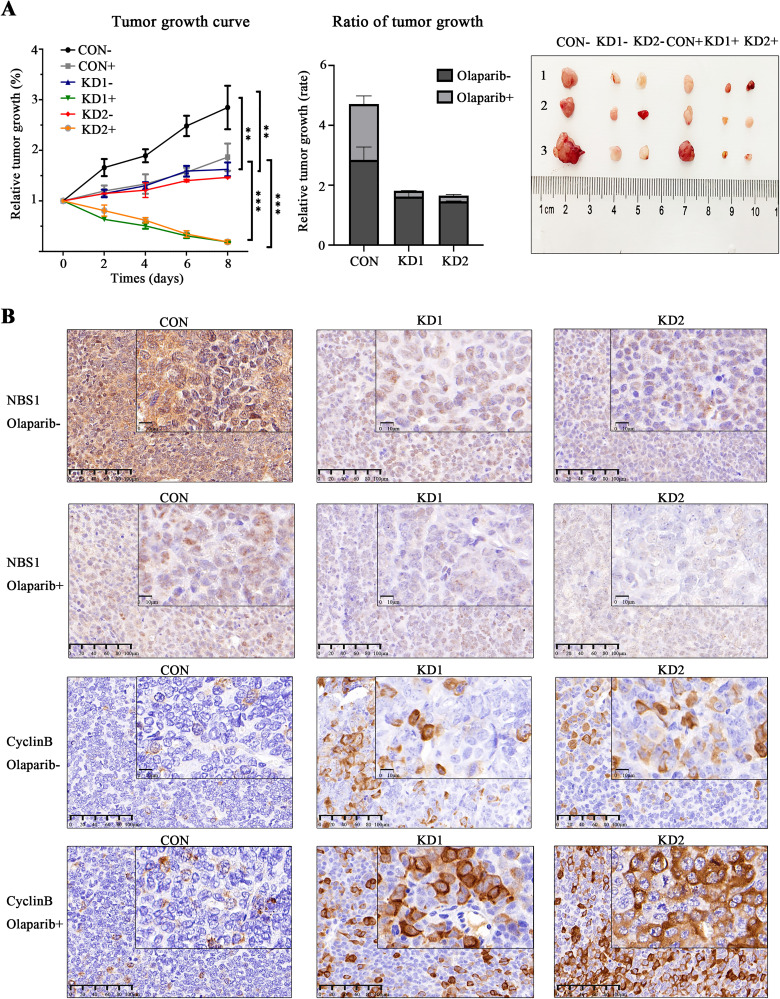
Prior in vitro results have demonstrated that inhibition of NBS1 can decrease HR repair activities, and NBS1−/HR− ovarian cancer cells are more responsive to PARP inhibitors than wild-type ovarian cancer cells. We further inoculated CON and KD1/2 cells into BALB/c nude mice via subcutaneous injection to establish control and NBS1-deficient ovarian cancer mouse models and assessed the impact of Olaparib on NBS1-deficient mice and its drug mechanism of action. Our findings showed that the tumor growth rate in NBS1-null mice was significantly slower than that in the control group, and after 1 week of Olaparib treatment, the tumor tissue in the NBS1-null group significantly shrunk (Figure 5A). Subsequently, we aimed to investigate the function of NBS1 in regulating Olaparib drug sensitivity and its connection to the activation of the CyclinB signaling pathway. We examined the expressions of NBS1 and CyclinB proteins using immunohistochemistry and compared them to the control group. We observed an increase in CyclinB expression in mice with NBS1 down-regulation, indicating higher sensitivity to Olaparib than the control group, and the expression of CyclinB protein was substantially higher than that in the control group (Figure 5B). These results were verified by in vivo experiments, indicating that inhibition of NBS1 increased the drug sensitivity of ovarian cancer to Olaparib. In HR-deficient ovarian cancer, NBS1-targeted inhibition can act synergistically with Olaparib to inhibit cancer growth, advancing the clinical application of PARP inhibitors in ovarian cancer.
Figure 5.
(A) Olaparib inhibits tumor tissue growth in Nijmegen Breakage Syndrome 1 (NBS1) down-regulated nude mice and (B) tumor tissue immunohistochemistry.
Discussion
Adjuvant chemotherapy is currently used to improve the clinical outcome of ovarian cancer patients following surgery. However, chemotherapy resistance, particularly to platinum-based drugs, leads to relapse and death in many patients. 20 Proteomics analysis of ovarian cancer and its adaptive responses to therapy can indeed provide valuable insights into the disease and lead to the discovery of new therapeutic options. Proteomic analysis can reveal novel drug targets by identifying proteins that are crucial for cancer cell survival and growth. 21 Personalized treatment with PARP inhibitors is a promising approach for ovarian cancer. Initially discovered in the synthetic lethal screening of BRCA2 mutant cells, PARP inhibitors effectively treat ovarian cancer with BRCA mutations. 22 However, since most ovarian cancer patients carry wild-type BRCA1/2, PARP inhibitors have limited clinical benefit. Therefore, further research and the development of new therapeutic strategies are necessary for PARP inhibitors to effectively treat ovarian cancer. Identifying the gene action target of PARP inhibitors is a crucial step toward achieving this goal.
The MRE11-RAD50-NBS1 (MRN) complex is a crucial sensor in DSBs, 23 and NBS1 is a critical component of this complex. 24 Clinical studies have demonstrated that NBS1 is an independent predictor of EOC recurrence and is linked to positive clinicopathological characteristics. 25 NBS1 plays a vital role in repairing DSBs through NHEJ and HR repair, 26 and preclinical evidence also suggests that NBS1 is a critical factor in the chemosensitivity of ovarian cancer.27,28 Targeting HR repair deficiency (HRD) is a vital approach for anticancer treatments, and several modalities have shown varying degrees of effectiveness, including chemotherapy and PARP inhibitors. NBS1 has been identified as a biomarker for HRD 29 and proposes expanding the potential utility of PARP inhibitors in therapeutic applications beyond BRCA-mutated tumors. 30 Our findings validate that targeted inhibition of NBS1 enhances the drug sensitivity of Olaparib in treating ovarian cancer. Although this result offers a new theoretical rationale for the clinical treatment of Olaparib, further clinical trials are still required in the future.
The promotion of DSB repair in human cells is facilitated by ataxia-telangiectasia mutated (ATM) activity and various other factors, including the MRN complex and CtBP-interacting protein (CtIP), which is regulated by the cell cycle.31,32 Targeting the PI3K pathway is a strategy used in cancer treatment to inhibit the uncontrolled growth and survival of cancer cells. The inhibition of the PI3K may lead to genomic instability and mitotic catastrophe through a decrease of the activity of the spindle assembly checkpoint protein Aurora kinase B and consequently increase the occurrence of lagging chromosomes during prometaphase. 33 However, the connection between the DNA damage repair process and cell cycle regulation has not been well-established. The impairment of NBS1 phosphorylation not only negatively impacts HR repair but also hinders the recovery of DNA replication after replication fork inactivation. Therefore, CDK1-mediated NBS1 phosphorylation functions as a molecular switch that controls the choice of DSB repair mode. 34 Our findings suggest that NBS1 is phosphorylated by CDK1/CyclinB, which leads to the coordinated inhibitory effect of the activation of the G2/M phase cell cycle checkpoints and HRD. This provides an important foundation for the treatment of BRCA-WT (wild-type) tumors with PARP inhibitors. However, further research is required to explore the interaction site between NBS1 and CyclinB protein. Sensitizing BRCA-WT cancers with PARP inhibitor combination therapy is an important area of research in oncology. Combining PARP inhibitors with targeted therapies, such as angiogenesis inhibitors or kinase inhibitors, can disrupt multiple signaling pathways in cancer cells. This synergy may lead to increased sensitivity to treatment and better outcomes in BRCA-WT cancer. 35
This study found that targeted inhibition of NBS1 can serve as a co-sensitizer for PARP inhibitors, synergistically activating the G2/M phase checkpoint response, and enhancing the sensitivity of ovarian cancer to radiotherapy and chemotherapy. However, these research findings still need to be validated in clinical trials in the future, and the development of small-molecule inhibitors specifically targeting NBS1 or its interactions with other DNA repair proteins is an active area of research. The potential for toxicity is a major concern with NBS1 inhibitors. Because they target DNA repair mechanisms, they can interfere with the repair of DNA damage in normal cells, leading to unwanted side effects. Balancing the therapeutic effect on cancer cells with the avoidance of normal tissue toxicity is a significant challenge. In conclusion, while NBS1 targeted inhibition combined with PARPi holds promise as a potential cancer treatment, their clinical applications are currently limited by issues related to specificity, toxicity, resistance, and the need for more clinical data. Addressing these limitations through continued research, improved drug development, and patient selection strategies will be critical to realizing the full potential of combination therapies in the future.
Conclusions
The presence or absence of NBS1 can impact chromosomal stability and cell cycle dynamics. It is also involved in activating DNA repair and cell cycle checkpoint signal pathways. In comparison to NBS1-WT cells, NBS1-KD cells lead to increased sensitivity to PARP agents and alterations in the balance between DNA damage repair and programmed cell death. This promotes sensitivity to radiotherapy through a synergistic inhibitory effect with PARP inhibitors by activating the Cyclin B regulation. The present study revealed that NBS1 can be potentially used as a new target for ovarian cancer therapy, providing new insights into targeted therapy for ovarian cancer.
Acknowledgments
All authors contributed to the study’s conception and design. Material preparation, data collection, and analysis were performed by Ailing Zhong, Chien-shan Cheng, and Renquan Lu. The first draft of the manuscript was written by Ailing Zhong and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Abbreviations
- DSBs
DNA double-strand breaks
- EOC
epithelial ovarian cancer
- HR
homologous recombination
- HRD
HR repair deficiency
- IR
ionizing radiation
- PARP
poly(ADP-ribose) polymerase
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: The study was approved by the Laboratory Animal Management and Use Committee of Fudan University, Shanghai Cancer Center. The approval number is FUSCC-IACUC-S2022-0151.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Science and Technology Commission of Shanghai Municipality (grant number 21YF1408500).
ORCID iD: Ailing Zhong https://orcid.org/0000-0002-4646-0438
References
- 1.Chatterjee N, Walker GC. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen. 2017;58(5):235-263. doi: 10.1002/em.22087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071-1078. doi: 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah S, Cheung A, Kutka M, Sheriff M, Boussios S. Epithelial ovarian cancer: providing evidence of predisposition genes. Int J Environ Res Public Health. 2022;19(13):8113. doi: 10.3390/ijerph19138113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Syed A, Tainer JA. The MRE11-RAD50-NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu Rev Biochem. 2018;87:263-294. doi: 10.1146/annurev-biochem-062917-012415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Zhou J, Lim CU. The role of NBS1 in DNA double strand break repair, telomere stability, and cell cycle checkpoint control. Cell Res. 2006;16(1):45-54. doi: 10.1038/sj.cr.7310007 [DOI] [PubMed] [Google Scholar]
- 6.Tauchi H, Kobayashi J, Morishima K, et al. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature. 2002;420(6911):93-98. doi: 10.1038/nature01125 [DOI] [PubMed] [Google Scholar]
- 7.Matsuura S, Kobayashi J, Tauchi H, Komatsu K. Nijmegen breakage syndrome and DNA double strand break repair by NBS1 complex. Adv Biophys. 2004;38:65-80. [PubMed] [Google Scholar]
- 8.Tauchi H, Matsuura S, Kobayashi J, Sakamoto S, Komatsu K. Nijmegen breakage syndrome gene, NBS1, and molecular links to factors for genome stability. Oncogene. 2002;21(58):8967-8980. doi: 10.1038/sj.onc.1206136 [DOI] [PubMed] [Google Scholar]
- 9.Girard PM, Riballo E, Begg AC, Waugh A, Jeggo PA. Nbs1 promotes ATM dependent phosphorylation events including those required for G1/S arrest. Oncogene. 2002;21(27):4191-4199. doi: 10.1038/sj.onc.1205596 [DOI] [PubMed] [Google Scholar]
- 10.Buscemi G, Savio C, Zannini L, et al. Chk2 activation dependence on Nbs1 after DNA damage. Mol Cell Biol. 2001;21(15):5214-5222. doi: 10.1128/MCB.21.15.5214-5222.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paull TT. Mechanisms of ATM activation. Annu Rev Biochem. 2015;84:711-738. doi: 10.1146/annurev-biochem-060614-034335 [DOI] [PubMed] [Google Scholar]
- 12.Gogineni VR, Nalla AK, Gupta R, Dinh DH, Klopfenstein JD, Rao JS. Chk2-mediated G2/M cell cycle arrest maintains radiation resistance in malignant meningioma cells. Cancer Lett. 2011;313(1):64-75. doi: 10.1016/j.canlet.2011.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poveda A, Floquet A, Ledermann JA, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a final analysis of a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2021;22(5):620-631. doi: 10.1016/S1470-2045(21)00073-5 [DOI] [PubMed] [Google Scholar]
- 14.Banerjee S, Moore KN, Colombo N, et al. Maintenance olaparib for patients with newly diagnosed advanced ovarian cancer and a BRCA mutation (SOLO1/GOG 3004): 5-year follow-up of a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2021;22(12):1721-1731. doi: 10.1016/S1470-2045(21)00531-3 [DOI] [PubMed] [Google Scholar]
- 15.Moore K, Colombo N, Scambia G, et al. Maintenance Olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 27 2018;379(26):2495-2505. doi: 10.1056/NEJMoa1810858 [DOI] [PubMed] [Google Scholar]
- 16.Ledermann JA, Drew Y, Kristeleit RS. Homologous recombination deficiency and ovarian cancer. Eur J Cancer. 2016;60:49-58. doi: 10.1016/j.ejca.2016.03.005 [DOI] [PubMed] [Google Scholar]
- 17.Revythis A, Limbu A, Mikropoulos C, et al. Recent insights into PARP and immuno-checkpoint inhibitors in epithelial ovarian cancer. Int J Environ Res Public Health. 2022;19(14):8577. doi: 10.3390/ijerph19148577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah PD, Wethington SL, Pagan C, et al. Combination ATR and PARP inhibitor (CAPRI): a phase 2 study of ceralasertib plus olaparib in patients with recurrent, platinum-resistant epithelial ovarian cancer. Gynecol Oncol. 2021;163(2):246-253. doi: 10.1016/j.ygyno.2021.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Secord A A, O'Malley DM, Sood AK, Westin SN, Liu JF. Rationale for combination PARP inhibitor and antiangiogenic treatment in advanced epithelial ovarian cancer: a review. Gynecol Oncol. 2021;162(2):482-495. doi: 10.1016/j.ygyno.2021.05.018 [DOI] [PubMed] [Google Scholar]
- 20.Stordal B, Davey R. A systematic review of genes involved in the inverse resistance relationship between cisplatin and paclitaxel chemotherapy: role of BRCA1. Curr Cancer Drug Targets. 2009;9(3):354-365. doi: 10.2174/156800909788166592 [DOI] [PubMed] [Google Scholar]
- 21.Ghose A, Gullapalli SVN, Chohan N, et al. Applications of proteomics in ovarian cancer: dawn of a new era. Proteomes. 2022;10(2):16. doi: 10.3390/proteomes10020016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fong PC, Yap TA, Boss DS, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28(15):2512-2519. doi: 10.1200/JCO.2009.26.9589 [DOI] [PubMed] [Google Scholar]
- 23.Oh J, Symington LS. Role of the Mre11 complex in preserving genome integrity. Genes (Basel). 2018;9(12):589. doi: 10.3390/genes9120589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antoccia A, Kobayashi J, Tauchi H, Matsuura S, Komatsu K. Nijmegen breakage syndrome and functions of the responsible protein, NBS1. Genome Dyn. 2006;1:191-205. doi: 10.1159/000092508 [DOI] [PubMed] [Google Scholar]
- 25.Lee YK, Park NH, Lee H. Clinicopathological values of NBS1 and DNA damage response genes in epithelial ovarian cancers. Exp Mol Med. 2015;47(11):e195. doi: 10.1038/emm.2015.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi J, Antoccia A, Tauchi H, Matsuura S, Komatsu K. NBS1 and its functional role in the DNA damage response. DNA Repair (Amst). 2004;3(8-9):855-861. doi: 10.1016/j.dnarep.2004.03.023 [DOI] [PubMed] [Google Scholar]
- 27.Alblihy A, Alabdullah ML, Ali R, et al. Clinicopathological and functional evaluation reveal NBS1 as a predictor of platinum resistance in epithelial ovarian cancers. Biomedicines. 2021;9(1):56. doi: 10.3390/biomedicines9010056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy-Leo C, Darwiche F, Tainsky MA. DNA repair mechanisms, protein interactions and therapeutic targeting of the MRN complex. Cancers (Basel). 2022;14(21):5278. doi: 10.3390/cancers14215278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoppe MM, Sundar R, Tan DSP, Jeyasekharan AD. Biomarkers for homologous recombination deficiency in cancer. J Natl Cancer Inst. 2018;110(7):704-713. doi: 10.1093/jnci/djy085 [DOI] [PubMed] [Google Scholar]
- 30.Swisher EM, Lin KK, Oza AM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18(1):75-87. doi: 10.1016/S1470-2045(16)30559-9 [DOI] [PubMed] [Google Scholar]
- 31.Jazayeri A, Falck J, Lukas C, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8(1):37-45. doi: 10.1038/ncb1337 [DOI] [PubMed] [Google Scholar]
- 32.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284(14):9558-9565. doi: 10.1074/jbc.M808906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aliyuda F, Moschetta M, Ghose A, et al. Advances in ovarian cancer treatment beyond PARP inhibitors. Curr Cancer Drug Targets. 2023;23(6):433-446. doi: 10.2174/1568009623666230209121732 [DOI] [PubMed] [Google Scholar]
- 34.Falck J, Forment JV, Coates J, et al. CDK targeting of NBS1 promotes DNA-end resection, replication restart and homologous recombination. EMBO Rep. 2012;13(6):561-568. doi: 10.1038/embor.2012.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellare GP, Patro BS. Resveratrol sensitizes breast cancer to PARP inhibitor, talazoparib through dual inhibition of AKT and autophagy flux. Biochem Pharmacol. 2022;199:115024. doi: 10.1016/j.bcp.2022.115024 [DOI] [PubMed] [Google Scholar]