Abstract
In Escherichia coli, high-abundance chemoreceptors are present in cellular amounts approximately 10-fold greater than low-abundance chemoreceptors. Cells containing only low-abundance receptors exhibit abnormally low tumble frequencies and do not migrate effectively in spatial gradients. These defects reflect an inherent activity difference between the two receptor classes. We used in vitro assays to investigate this difference. The low-abundance receptor Trg mediated an ∼100-fold activation of the kinase CheA, only twofold less than activation by the high-abundance receptor Tar. In contrast, Trg was less than 1/20 as active as Tar for in vitro methylation. As observed for high-abundance receptors, kinase activation by Trg varied with the extend of modification at methyl-accepting sites; low methylation corresponded to low kinase activation. Thus, in Trg-only cells, low receptor methylation would result in low kinase activation, correspondingly low content of phospho-CheY, and a decreased dynamic range over which attractant binding could modulate kinase activity. These features could account for the low tumble frequency and inefficient taxis exhibited by Trg-only cells. Thus, the crucial functional difference between the receptor classes is likely to be methyl-accepting activity. We investigated the structural basis for this functional difference by introducing onto the carboxy terminus of Trg a CheR-binding pentapeptide, usually found only at the carboxy termini of high-abundance receptors. This addition enhanced the in vitro methyl-accepting activity of Trg 10-fold.
Transmembrane, methyl-accepting receptor proteins mediate chemotaxis by Escherichia coli (13, 18, 37). Many related proteins have been detected in other eubacterial and archaeal species by antigenic cross-reaction (30) or sequence comparisons (23, 49). These proteins define an extensive family of sensory receptors that mediate bacterial and archaeal taxis. The hallmarks of the family are a highly conserved region (∼50 residues) crucial for intracellular signaling and adjacent regions containing methyl-accepting glutamyl residues that are covalently modified in the process of sensory adaptation. In E. coli and Salmonella typhimurium, the highly conserved regions have been shown to be involved in the control of a noncovalently associated histidine kinase, CheA (reviewed in reference 37), which is a well-characterized member of a large family of homologous histidine kinases that serve not only taxis systems but also a vast array of two-component, environment-sensing systems in eubacteria, archaea, and eukaryotes (1, 3).
The influence of a chemoreceptor on an associated kinase has two distinct aspects: (i) basal activation and (ii) modulation of activity in response to changes in receptor occupancy (6–8, 32). Both require formation of a ternary complex consisting of a receptor, CheA, and an accessory protein, CheW (17, 41). This complex is stable over times relevant for sensory response and adaptation (17). Basal activation of CheA by an interacting receptor establishes a steady-state activity of the kinase that determines the cellular content of the phosphorylated response regulator CheY (phospho-CheY). Phospho-CheY interacts with the flagellar switch to induce clockwise (CW) rotation of an otherwise counterclockwise (CCW)-rotating motor. Proper basal activation establishes a phospho-CheY content in a normal cell that creates a balance between CCW and CW flagellar rotation, which produces a corresponding alternation between smooth swimming and tumbling. The pattern causes a swimming cell to move in a random walk. Modulation of kinase activity from its level of basal activation is effected by receptors that have experienced a change in ligand occupancy but have not yet adapted. The modulation results in an altered content of phospho-CheY, which changes the CCW-to-CW balance and thus the tumble frequency, biasing the random walk to direct the cell in a favorable direction.
The four well-characterized receptors of E. coli have a common organization (see references 13 and 18 for details and specific references). In each monomer of a receptor homodimer, an amino-terminal periplasmic domain of ∼150 residues and a carboxy-terminal cytoplasmic domain of ∼300 residues are connected by two transmembrane segments. Residue identity among the aligned sequences of the four periplasmic domains is minimal but is nearly 60% for the cytoplasmic domain. The cytoplasmic domain includes the highly conserved region and the methyl-accepting sites. The receptors Tsr, Tar, Trg, and Tap mediate taxis toward serine, aspartate and maltose, ribose and galactose, and dipeptides, respectively. A recently discovered fifth receptor, Aer, lacks a substantial periplasmic domain but mediates responses to oxygen and to perturbations of membrane energetics by utilizing a bound flavin (4, 40). Among the four extensively characterized receptors, two high-abundance chemoreceptors, Tsr and Tar, are present in cellular amounts 5- to 10-fold greater than two low-abundance receptors, Trg and Tap (19). In the absence of high-abundance receptors, cells exhibit abnormally low tumble frequencies and greatly compromised abilities to migrate in spatial gradients of attractants recognized by the remaining low-abundance receptors (14, 20, 45, 46). These defects are not corrected by increasing cellular amounts of the low-abundance receptor (14, 46). Thus high-abundance and low-abundance receptors are distinguished not simply by different amounts in a wild-type cell but also by an inherent difference in activity. Characterization of hybrids between a high-abundance receptor and a low-abundance receptor, either Tsr and Trg (14) or Tar and Tap (46), revealed that this inherent difference in activity resides in the cytoplasmic domain, even though it is in this domain that residue identity among receptors is most conserved.
What is the nature of the difference between high-abundance and low-abundance receptors that allows the former but not the latter to establish a physiologically useful tumble frequency and to mediate effective taxis as the sole receptor in a cell? A clear possibility is that low-abundance receptors are ineffective in basal activation of the kinase CheA. Ineffective activation would mean a low level of steady-state phosphorylation that would result in a low cellular concentration of phospho-CheY and thus a low tumble frequency. Also, low basal activation of CheA could affect signaling by reducing the dynamic range over which kinase activity could be modulated in response to increases in receptor occupancy. A difference in kinase activation would be consistent with the observation that the inherent difference between high- and low-abundance receptors resides in the cytoplasmic domain. Thus, we undertook the study of kinase activation by the low-abundance receptor Trg, using in vitro assays for phosphorylation. In these assays Trg was almost as effective as the high-abundance receptor Tar in activating CheA, implying that kinase activation was unlikely to be the crucial functional difference between the two receptor types. However, we observed a striking difference in vitro in the methyl-accepting activities of the two receptors. The low-abundance receptor Trg was significantly less effective than the high-abundance receptor Tar as an acceptor for in vitro methylation. This appears to be the central functional difference between the two receptor types.
MATERIALS AND METHODS
Bacterial strains and plasmids.
CP553 (10) and RP3098 (38) are strains of E. coli K-12. The former carries chromosomal deletions of trg, tsr, tar, tap, cheR, and cheB. The latter, provided by J. S. Parkinson (University of Utah), carries a deletion from flhA through flhD and thus lacks the genes for all Che proteins. pGB1 (10) and pNT201 (7) carry trg and tar, respectively, under the control of a tac promoter. pAL11 is a derivative of pHSe5 (31), in which tandem tac promoters control the expression of trgt, an altered form of trg extended by the final 19 codons of tsr. pCW and pCW/cheA carry cheW and cheA, respectively, under the control of tandem tac promoters (16, 17) and were obtained from F. W. Dahlquist (University of Oregon). pLR22 and pLR22ΔcheY (29), which carry both cheY and cheZ or only cheZ, respectively, were gifts from P. Matsumura (University of Illinois at Chicago). pME43 (43), which contains cheR, was obtained from J. Stock (Princeton University).
Protein purification and quantification.
CheA, CheW, CheY, and CheZ were produced in RP3098 harboring the appropriate plasmid and purified as described by Hess et al. (22) or Matsumura et al. (29). The first three were obtained at >95% purity, and CheZ was >80% pure. Over 70% of the purified CheA was the long form (44). Concentrations of pure proteins and of protein in cell extracts were determined by the Bio-Rad assay using bovine serum albumin as the standard. Total protein in membrane samples was determined by the Peterson modification of the Lowry assay (39), the amount of receptor was determined by quantitative immunoblots (14) in which test samples and pure standards of Trg or Tar were present on the same immunoblot, and intensities were quantified with a densitometer (Molecular Dynamics, Inc.).
Preparation of membranes containing chemoreceptors.
Membranes containing chemoreceptors were prepared essentially as described by Bogonez and Koshland (5). CP553 or RP3098 cells harboring an appropriate plasmid were grown in 10 to 20 ml of Luria broth at 35°C with agitation. At an optical density at 600 nm of 0.4, isopropylthio-β-d-galactoside (IPTG) was added to 1 mM, and 3.5 h later cells were harvested by centrifugation, washed with 50 mM Tris-HCl (pH 7.5)–0.5 mM EDTA–2 mM dithiothreitol–10% glycerol (TEDG), suspended in 1 ml of 50 mM Tris-HCl (pH 7.5)–10% (wt/vol) glycerol–10 mM EDTA–1 mM 1,10-phenanthroline–1 mM phenylmethylsulfonyl fluoride (PMSF), and put in a 5-ml plastic scintillation vial. The vial was placed in an ice-salt bath, and the suspension was sonicated for six 5-s pulses (25-s intervals between pulses) with a Tekmar TM-250 sonic disrupter (9-mm-diameter horn; 60% maximum power). The suspension was centrifuged 10 min at 14,000 × g, 4°C in an Eppendorf Microcentrifuge. The supernatant was removed and centrifuged 24 min at 100,000 rpm in Beckman TLA100.2 rotor (350,000 × g) to pellet membrane vesicles. Vesicles were suspended in 1 ml of TEDG containing 2 M KCl and centrifuged as described above. Pelleted vesicles were suspended in 50 μl of TEDG, distributed in 5-μl portions into small plastic centrifuge tubes flushed with nitrogen, quick frozen in −20°C ethanol, and stored at −70°C.
Protein phosphorylation.
A coupled phosphorylation assay was used to monitor the formation of phospho-CheY by the receptor-CheW-CheA ternary complex under conditions under which phosphorylation of CheA was the rate-limiting step (9). CheA (5 pmol), CheW (80 pmol), CheY (200 pmol), and receptor-containing membranes were incubated in 15 μl of 50 mM Tris-HCl (pH 7.5)–50 mM KCl–10% (wt/vol) glycerol–1 mM PMSF–0.5 mM EDTA–5 mM MgCl2 at room temperature for 1 h. In some experiments, 12 pmol of CheZ or 9 μg of protein from a CheR-containing cell lysate (providing a final CheR concentration of 1 to 2 μM) was also in the mixture. Reactions were initiated at room temperature by addition of 5 μl of [γ-32P]ATP (∼2,000 cpm/pmol; Amersham) to 0.15 mM and stopped after 10 s by addition of electrophoresis sample buffer containing EDTA. Samples were processed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (14% polyacrylamide) (33). Gels were briefly stained, destained, and dried immediately. Phospho-CheY was quantified with a PhosphorImager (Molecular Dynamics, Inc.).
Receptor methylation.
A cell extract enriched in the CheR methyltransferase was prepared as described by Shapiro and Koshland (42), dialyzed extensively against TEDG, and stored at −70°C. Isolated membranes containing receptor (or no receptor for a control) were incubated at room temperature for 30 min in 150 mM potassium phosphate (pH 7.0)–10% (wt/vol) glycerol–1 mM EDTA–0.5 mM PMSF. In some experiments CheA and CheW were included at concentrations equimolar to the receptor, CheY was present at 10 μM, and incubation times were extended to 1 h. Reactions were initiated by addition of the CheR-containing cell extract to which had been added S-adenosyl-[3H-methyl]-l-methionine (AdoMet) (∼380 cpm/pmol; Amersham) to produce final concentrations of 1 to 2 μM CheR in total protein (0.3 mg/ml) (12) and 50 μM AdoMet. At various times, 10-μl samples were removed, mixed with 17 μl of double-strength electrophoresis sample buffer, and boiled for 30 s. A 24-μl aliquot of boiled solution was analyzed by SDS-PAGE (33). Regions including receptor bands were excised from stained, dried gels, and the extent of receptor methylation was quantified by using alkali hydrolysis to release methylesters as methanol and vapor-phase equilibrium to capture the volatile, radiolabeled methanol (11).
RESULTS
Kinase activation.
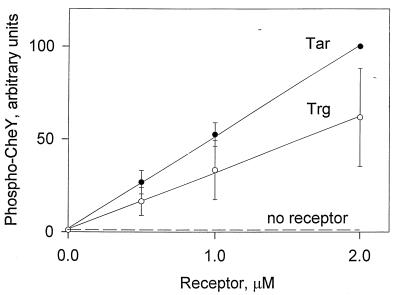
We compared the abilities of the low-abundance receptor Trg and the high-abundance receptor Tar to stimulate the kinase activity of CheA in vitro by a coupled assay in which phosphorylation of CheA was the rate-limiting step for appearance of phospho-CheY (9). Isolated membranes containing no receptor, Trg, or Tar were incubated with purified CheA, CheW, and CheY to allow complex formation, [γ-32P]ATP was added, and the initial rate of production of 32P-labeled phospho-CheY assessed by SDS-PAGE and phosphorimaging. Quantitative comparisons of kinase activation by the two different types of receptors required attention to specific details of technical manipulations and experimental design. We found that the activity of a membrane-embedded receptor in the coupled assay could be affected by the age and storage history of the membrane preparation. Thus, comparison of activities of Trg and Tar required isolating membranes at the same time and doing manipulations and assays in parallel. Since phospho-CheY is susceptible to hydrolysis as gels are run and processed and losses can vary from gel to gel (9), we placed samples to be compared on the same gel and determined by internal controls that losses did not vary with the location of the gel lane. In order to assess possible differences in kinase activation, it was important to adjust protein stoichiometries in the assay mixture so that production of phospho-CheY was a function of the amount of receptor; otherwise, differences between the receptor types could be masked. As shown in Fig. 1, we were able to define the desired assay conditions. Under these conditions, addition of membranes containing the low-abundance receptor Trg resulted in a substantial stimulation of kinase activity to a level above that detected for membrane-lacking chemoreceptor, a stimulation similar in magnitude to that mediated by the high-abundance receptor Tar (Fig. 1). As documented previously for Tar (7), kinase activation by membrane-embedded Trg was strongly dependent on the presence of both CheA and CheW (data not shown). We increased the amount of added receptor up to 8 μM under the same assay conditions (data not shown) and observed increased production of phospho-CheY up to approximately 4 μM Tar or Trg. Above that concentration the amount of phospho-CheY detected decreased, perhaps reflecting a reduction in the number of complete ternary complexes as excess receptor bound CheW (present at 4 μM) but not CheA (present at 0.25 μM), or perhaps reflecting inhibition due to increasing amounts of crude membrane. In any case, at all receptor concentrations tested from 0.5 to 8 μM, kinase activation by Trg was comparable to but slightly lower than activation by Tar.
FIG. 1.
Concentration dependence of kinase activation by Trg and Tar. Isolated membranes containing no receptor, Trg, or Tar were incubated with CheA, CheW, CheY, and [γ-32P]ATP under conditions under which phosphorylation of CheA was the rate-limiting step for the appearance of phospho-CheY (9). 32P-labeled phospho-CheY produced over 10 s was quantified by SDS-PAGE and phosphorimaging. The dotted line represents the small amount of 32P-labeled phospho-CheY formed through the low activity of CheA autophosphorylation in the absence of activating receptor. In experiments not shown here, addition of 1 mM aspartate to mixtures containing Tar reduced phospho-CheY ∼50-fold. The data are averages from four independent experiments, but several other experiments showed the same patterns. The error bars represent standard errors. Prior to averaging, the data sets were normalized by using the values for 2 μM Tar, and thus that point does not have error bars.
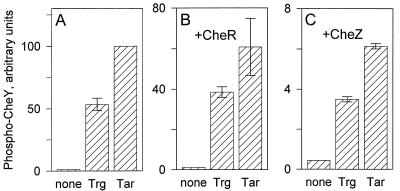
We chose a receptor concentration of 1 μM for repeated experimental comparisons of kinase activation by Trg and Tar and found a consistent pattern in which Trg-mediated activation of CheA increased phospho-CheY production at least 100-fold over the level in the absence of receptor, to a value within a factor of 2 of the Tar-mediated activation. Figure 2A presents representative data from several such independent experiments. We considered the possibility that other components of the chemosensory system might alter the relative abilities of the two receptors to activate kinase. Tar has a high-affinity binding site for the methyl-transferase, CheR (47), that is missing in Trg, and thus complexes of receptor, CheW, and CheA in vivo would also include bound CheR for high-abundance receptors but not for low-abundance ones. We tested the effect of CheR on Trg- and Tar-mediated stimulation of kinase by adding to our usual in vitro assay a cell extract enriched in CheR. The presence of the extract somewhat reduced the amount of phospho-CheY detected, probably as the result of ATPase activity in the extract, but the relative levels of stimulation by the two receptors were not significantly altered (Fig. 2B). CheZ accelerates auto-dephosphorylation of phospho-CheY, and its possible role in signaling is an area of active investigation. As expected (7) the presence of CheZ in the assay resulted in a reduction of ∼20-fold in phospho-CheY. However, there was no significant change in the relative activities of the two receptor types (Fig. 2C).
FIG. 2.
Relative kinase activation by Trg and Tar. Assays were performed as described in the legend to Fig. 1, using 1 μM receptor. Isolated membranes containing no receptor, Trg, or Tar were incubated with CheA, CheW, CheY, and [γ-32P]ATP with no other additions (A), with the addition of a cell lysate, providing 1 to 2 μM CheR (B), or with the addition of 0.6 μM CheZ (C). The data are averages from two independent experiments, but many other experiments showed similar patterns. The error bars represent standard errors. Prior to averaging, the data sets were normalized by using the values for Tar-mediated production in the absence of CheR or CheZ. For this reason there are no error bars for the Tar value in panel A.
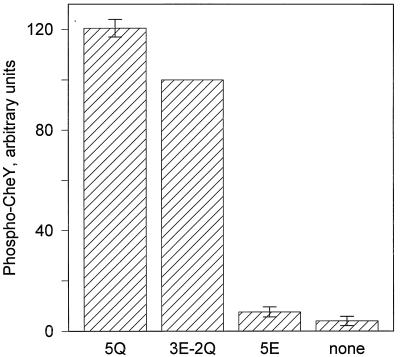
In vitro studies of the high-abundance receptor Tar have demonstrated that receptor-mediated activation of the kinase CheA is modulated by the state of its methyl-accepting sites (6). The sites can be negatively charged (glutamates) or neutral (glutamyl methyl esters or glutamines). The former state reduces kinase activation; the latter enhances it. The trg gene codes for three glutamates and two glutamines at the five methyl-accepting sites (the glutamines are subsequently deamidated to produce methyl-accepting glutamates) (33), and it was this form of Trg (three glutamates, two glutamines [3E-2Q]) that was used in the experiments described above and compared with the gene-encoded form of Tar (2E-2Q). In considering possible differences in kinase activation by low- and high-abundance receptors it was important to determine whether the effects of receptor modification on kinase activation by the low-abundance receptor were similar to the pattern documented for the high-abundance receptor. Figure 3 shows that Trg with uncharged side chains at all five modification sites, Trg (5Q), mediates greater kinase stimulation than the gene-encoded form Trg (3E-2Q) and that a form of Trg with all charged side chains, Trg (5E), mediates almost no stimulation. This is the same pattern observed for Tar (6) and is consistent with the activities of the three forms of Trg in vivo (36).
FIG. 3.
Kinase activation by Trg (5Q), Trg (3E-2Q), and Trg (5E). Assays were performed as described in the legend to Fig. 1, using 1 μM receptor. The data are averages from three independent experiments, including one with several replicates. Prior to averaging, the average values from independent experiments were normalized by using the value for Trg (3E-2Q). For this reason there are no error bars for that value. The error bars represent standard errors.
Methyl-accepting activity.
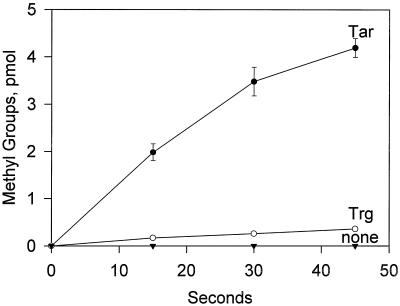
We compared the activities of Trg and Tar as substrates for methylation in vitro. Isolated membranes containing no receptor, Trg, or Tar were incubated with a cell extract enriched in CheR in the presence of AdoMet, and samples of the mixture were analyzed for carboxyl methylation of the receptors by SDS-PAGE and quantification of [3H]methanol released from alkali-treated slices of gels containing the receptor protein. There was a striking difference in the methyl-accepting activities of Trg and Tar. As shown in Fig. 4, the initial rate of Trg methylation was approximately 5% of the value for Tar. Preincubation with CheA, CheW, and CheY to form signaling complexes did not alter the difference in methyl-accepting activity between the two receptors (data not shown).
FIG. 4.
Time courses of methylation in vitro. Isolated membranes containing no receptor, Trg, or Tar were incubated with a cellular extract containing ∼1 μM CheR and 50 μM AdoMet at receptor concentrations of ∼5 μM. At the indicated times samples containing ∼50 pmol of receptor were removed, processed, and analyzed as described in Materials and Methods. The data are averages from three independent experiments, and the error bars show standard errors. Points without bars had standard errors within the size of the symbol.
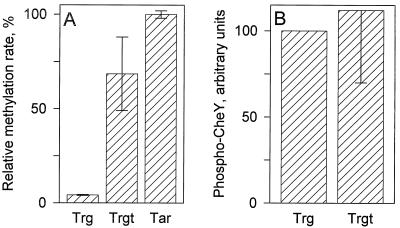
What is the origin of the difference in methyl-accepting activity in vitro by Trg and Tar? Wu et al. (47) identified a CheR-binding site in the form of a 5-amino-acid sequence (NWETF) that is present at the extreme carboxy terminus of high-abundance receptors but absent in low-abundance receptors. We added a 19-codon sequence corresponding to the final 19 residues of the high-abundance receptor Tsr to the 3′ end of the trg gene, creating a form of Trg with the CheR-binding site, NWETF, attached by a linker segment to the carboxy terminus of the low-abundance receptor. The details of this construction and characterizations of the activities of the hybrid receptor in vivo are described elsewhere (15). In the present study, we used this hybrid, which we call Trgt (for Trg plus the 19-residue Tsr tail), to test to what degree the low methyl-accepting activity of Trg in vitro reflected the absence of the carboxy-terminal CheR binding site. As shown in Fig. 5A, Trgt exhibited 10-fold more methyl-accepting activity in vitro than native Trg and thus was within a factor of 2 of Tar. This indicates that most of the difference between the methyl-accepting activities of the native forms of a low-abundance and a high-abundance receptor can be attributed to the respective absence and presence of the NWETF binding site for the methyltransferase. It was possible that addition onto Trg of a carboxy-terminal sequence from a high-abundance receptor would also alter the activation of the CheA kinase by this low-abundance receptor. The data shown in Fig. 5B indicate that this was not the case.
FIG. 5.
Effects of adding to Trg the 19-residue, carboxy-terminal tail of Tsr. Tar, Trg, and the hybrid receptor Trgt were tested for methyl-accepting activity (A) and kinase activation (B) as described in the legends to Fig. 4 and 2, respectively. Methylation rates were determined after a 1-min incubation and were expressed as a percentage relative to the rate for Tar. Kinase activation was expressed as a percentage relative to the Trg-mediated value. The data are averages from two independent experiments, and the error bars show standard errors.
DISCUSSION
It was plausible that the inability of a low-abundance receptor to mediate effective taxis in the absence of other receptors reflected an inherent inefficiency in basal activation of the kinase CheA. With inefficient basal activation by low-abundance receptors, cells with only such receptors would have a decreased steady-state level of phospho-CheY and a decreased dynamic range over which to modulate kinase activity. These features would result in the experimentally observed low frequency of tumbles and ineffective tactic migration. We tested this hypothesis by investigating in vitro the activation of CheA by the low-abundance chemoreceptor Trg and found that Trg stimulated the kinase almost as well as the high-abundance receptor Tar. Thus, kinase activation per se is unlikely to be the inherent and substantial functional difference between high- and low-abundance receptors. Instead, we feel the explanation involves the consequences of the significant difference we observed in methyl-accepting activity in vitro between the two receptor classes. In brief, since adaptational methylation is inefficient for low-abundance receptors in the absence of high-abundance receptors, modest increases in ligand occupancy would result in abnormally extended periods of low kinase activity, correspondingly low levels of phospho-CheY, low tumble frequencies, and a decreased dynamic range over which to modulate kinase activity. Issues related to these ideas are considered in more detail in the following paragraphs.
Basal activation of kinase CheA.
Like the high-abundance receptor Tar, the low-abundance receptor Trg effectively stimulated the kinase activity of CheA and the linked phosphorylation of CheY (Fig. 1 and 2). For high-abundance receptors, kinase activation is known to be mediated by formation of ternary complexes with CheA and CheW (17, 41). Kinase stimulation by Trg implies that this low-abundance receptor also forms ternary complexes with CheA and CheW. The requirement for CheW in Trg-mediated stimulation of CheA in our in vitro assays supports this implication. The existence of comparable interactions with CheA and CheW for both high- and low-abundance receptors is reasonable, since the greatest sequence identity among these receptors is in the region implicated in interaction with CheA and CheW (2, 26). It appears that Trg, and probably other low-abundance receptors, interact physically with CheA and CheW to create basal activation of the kinase.
If Trg and Tar each activate CheA through formation of ternary complexes, why is the activation mediated by the two receptors in vitro not identical? At least three factors might contribute, two related to the assay and one inherent in the proteins: (i) protein stability, (ii) modification ratios, and (iii) nonconserved residues. (i) With regard to protein stability, in the course of isolation and purification Trg is more susceptible to proteolysis than Tar (unpublished observations), implying that it is more prone to partial unfolding, a tendency that could cause a greater proportion of Trg in a membrane preparation to be incapable of kinase activation. (ii) With regard to modification ratios, kinase activation by a receptor varies from almost no activation if all methyl-accepting sites carry negative side chains (glutamates) to maximal activation if all carry neutral side chains (glutamines or glutamyl methyl esters) (reference 6; Fig. 3), but it is not known whether the crucial parameter is the number of neutral sites or the ratio of neutral to charged sites. If the ratio were the controlling factor, Trg (3E-2Q), the form of Trg tested in Fig. 1 and 2, would exhibit lower kinase activation than Tar (2E-2Q), the form of Tar to which it was compared. (iii) With regard to nonconserved residues, the sequences of Tar and Trg differ at several positions near the highly conserved core. These could subtly reduce kinase activation exhibited by Trg. If this third factor were important, could it be the underlying basis of the low tumble frequency and inefficient taxis in Trg-only cells? The data argue against this notion. In vitro, the rate of production of phospho-CheY from Tar-activated CheA can be matched with Trg-activated kinase by adding less than twofold more Trg (Fig. 1). If this relationship holds in vivo, which seems likely since the receptor concentrations tested in Fig. 1 are in the range of those estimated to exist in a wild-type cell (19), then increasing the Trg dosage should correct the problem. However, increasing Trg dosage across a 100-fold range does not improve taxis (14). Thus, it seems unlikely that the modest difference in kinase activation detected in vitro accounts for the differences in receptor action in vivo.
What are alternative explanations? Specifically, how can our observation in vitro of significant phospho-CheY production from Trg-activated CheA be reconciled with the implication from cellular behavior that Trg-only cells are deficient in phospho-CheY? It was possible that some important feature of the in vivo state was absent from our in vitro phosphorylation assays, for instance a component of the chemosensory system besides CheA, CheW, and CheY. We tested whether the presence of CheR or CheZ would drastically reduce Trg-mediated production of phospho-CheY relative to Tar-mediated production (Fig. 2B and C) but found no indication of such an effect. Alternatively, higher-order interactions that might not be present in vitro could create differential effects on high- and low-abundance receptors in vivo that would alter kinase activation. Such higher order interactions could include receptor clustering (27, 28), action on one receptor by enzymes bound to a neighboring receptor (24, 25), or interactions of some combination of Che proteins with the ternary complex plus CheY. We have not yet investigated those possibilities, but our studies of in vitro methylation suggest an alternative that can account for the data.
Methyl-accepting activity.
We found in vitro that the low-abundance receptor Trg was dramatically less efficient as a methyl-accepting substrate than was the high-abundance receptor Tar. This observation in vitro parallels numerous observations in vivo. In cells lacking high-abundance receptors, adaptation to Trg-mediated stimuli is so slow that for large temporal changes in receptor occupancy, adaptation does not occur even over extended periods of observation (20). In Trg-only cells, steady-state methylation is significantly lowered and increases in methylation following stimulation are only just detectable (14, 20, 21, 35, 48). These defects in vivo can be understood as direct consequences of a low rate of Trg methylation and thus imply that what we observed in vitro is also the case in vivo. A low rate of methylation would mean slow adaptation and reduced steady-state levels of methylation because of a shifted balance between methylation and demethylation. Thus, unlike the issue of kinase activation by Trg for which in vivo observations implied a different result than that found by in vitro assays, there is a consistency between in vivo and in vitro assessments of methyl-accepting activity by this low-abundance receptor. Moreover, our characterization of the effects of adaptational modifications on kinase activation by Trg (Fig. 3) suggests a resolution of the apparent inconsistency between observations about kinase activation in vivo and in vitro. The data in Fig. 3 show that kinase activation in vitro is not an invariant property of Trg but instead is a function of the extent of modification at the methyl-accepting sites. Low levels of methylation mean low kinase activation. Thus, for Trg-only cells, in which Trg methylation is at a low level, the in vitro results in Fig. 3 would predict little activation of the kinase. This is consistent with the low tumble frequency and inefficient taxis exhibited by such cells.
Since a low methyl-accepting activity appears to be the fundamental functional difference between the low-abundance receptor Trg and its high-abundance cousins, it was important to investigate what structural features contributed to this low activity. These could include one or more of the following: the methyl-accepting sites themselves, the absence of a CheR-binding site at the carboxy terminus, or other Trg-specific features of the cytoplasmic domain. The results shown in Fig. 5A argue strongly that the absence of the CheR-binding site is a crucial factor. Trg carrying the NWETF pentapeptide attached to its carboxy terminus by a linker segment is over 10-fold more active for methylation in vitro than wild-type Trg. This enhanced activity, within a factor of 2 of the high-abundance receptor Tar, provides a quantitative measure of the contribution the CheR-binding pentapeptide (47) makes to methylation rate. Several studies have shown that mutant forms of high-abundance receptors with altered or missing pentapeptides are defective in methylation in vivo (34) or in vitro (24, 25). Our characterization of Trgt demonstrates that the Tsr tail containing the CheR-binding pentapeptide can provide enhanced functional interaction in vitro with the methyltransferase for a receptor otherwise only marginally effective as a methyl acceptor. This defines a quantitative foundation for in vivo investigations of the consequences of pentapeptide-mediated association of CheR with chemoreceptors. Such investigations, described elsewhere (15), demonstrate enhanced function of Trgt in vivo.
ACKNOWLEDGMENTS
This work was supported by grant GM29963 from the National Institutes of Health to G.L.H.
We thank Angela Lilly for construction of pAL11 and our colleagues cited in Materials and Methods for bacterial strains and plasmids.
REFERENCES
- 1.Alex L A, Simon M I. Protein histidine kinases and signal transduction in prokaryotes and eukaryotes. Trends Genet. 1994;10:133–138. doi: 10.1016/0168-9525(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 2.Ames P, Yu Y A, Parkinson J S. Methylation segments are not required for chemotactic signaling by cytoplasmic fragments of Tsr, the methyl-accepting serine chemoreceptor of Escherichia coli. Mol Microbiol. 1996;19:737–746. doi: 10.1046/j.1365-2958.1996.408930.x. [DOI] [PubMed] [Google Scholar]
- 3.Appleby J L, Parkinson J S, Bourret R B. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- 4.Bibikov S I, Biran R, Rudd K E, Parkinson J S. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogonez E, Koshland D E., Jr Solubilization of a vectorial transmembrane receptor in a functional form: aspartate receptor of chemotaxis. Proc Natl Acad Sci USA. 1985;82:4891–4895. doi: 10.1073/pnas.82.15.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borkovich K A, Alex L A, Simon M I. Attenuation of sensory receptor signaling by covalent modification. Proc Natl Acad Sci USA. 1992;89:6756–6760. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borkovich K A, Kaplan N, Hess J F, Simon M I. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci USA. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borkovich K A, Simon M I. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990;63:1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- 9.Borkovich K A, Simon M I. Coupling of receptor function to phosphate-transfer reactions in bacterial chemotaxis. Methods Enzymol. 1991;200:205–214. doi: 10.1016/0076-6879(91)00140-r. [DOI] [PubMed] [Google Scholar]
- 10.Burrows G G, Newcomer M E, Hazelbauer G L. Purification of receptor protein Trg by exploiting a property common to chemotactic transducers of Escherichia coli. J Biol Chem. 1989;264:17309–17315. [PubMed] [Google Scholar]
- 11.Chelsky D, Gutterson N I, Koshland D E., Jr A diffusion assay for detection and quantitation of methyl-esterified proteins on polyacrylamide gels. Anal Biochem. 1984;141:143–148. doi: 10.1016/0003-2697(84)90437-8. [DOI] [PubMed] [Google Scholar]
- 12.Chervitz S A, Lin C M, Falke J J. Transmembrane signalling by the aspartate receptor: engineered disulfides reveal static regions of the subunit interface. Biochemistry. 1995;34:9722–9733. doi: 10.1021/bi00030a010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falke J J, Bass R B, Butler S L, Chervitz S A, Danielson M A. The two-component signalling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng X, Baumgartner J W, Hazelbauer G L. High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J Bacteriol. 1997;179:6714–6720. doi: 10.1128/jb.179.21.6714-6720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng, X., A. A. Lilly, and G. L. Hazelbauer. Enhanced function conferred on low-abundance receptor Trg by a methyltransferase docking site. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 16.Gegner J A, Dahlquist F W. Signal transduction in bacteria: CheW forms a reversible complex with the protein kinase CheA. Proc Natl Acad Sci USA. 1991;88:750–754. doi: 10.1073/pnas.88.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gegner J A, Graham D R, Roth A F, Dahlquist F W. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- 18.Hazelbauer G L. Bacterial chemoreceptors. Curr Opin Struct Biol. 1991;2:505–510. [Google Scholar]
- 19.Hazelbauer G L, Engström P, Harayama S. Methyl-accepting chemotaxis protein III and transducer gene trg. J Bacteriol. 1981;145:43–49. doi: 10.1128/jb.145.1.43-49.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazelbauer G L, Engström P. Parallel pathways for transduction of chemotactic signals in Escherichia coli. Nature. 1980;238:98–100. doi: 10.1038/283098a0. [DOI] [PubMed] [Google Scholar]
- 21.Hazelbauer G L, Park C, Nowlin D M. Adaptational “cross-talk” and the crucial role of methylation in chemotactic migration by Escherichia coli. Proc Natl Acad Sci USA. 1989;86:1448–1452. doi: 10.1073/pnas.86.5.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hess J F, Bourret R B, Simon M I. Phosphorylation assays for proteins of the two-component regulatory system controlling chemotaxis in E. coli. Methods Enzymol. 1991;200:188–204. doi: 10.1016/0076-6879(91)00139-n. [DOI] [PubMed] [Google Scholar]
- 23.Le Moual H, Koshland D E., Jr Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J Mol Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- 24.Le Moual H, Quang T, Koshland D E., Jr Methylation of the Escherichia coli chemotaxis receptors: intra- and interdimer mechanisms. Biochemistry. 1997;36:13441–13448. doi: 10.1021/bi9713207. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Li G, Weis R M. The serine chemoreceptor from Escherichia coli is methylated through an inter-dimer process. Biochemistry. 1997;36:11851–11857. doi: 10.1021/bi971510h. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Parkinson J S. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J Bacteriol. 1991;173:4941–4951. doi: 10.1128/jb.173.16.4941-4951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Levit M, Lurz R, Surette M G, Stock J B. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. EMBO J. 1997;16:7231–7240. doi: 10.1093/emboj/16.24.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddock J R, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 29.Matsumura P, Rydel J J, Linzmeier R, Vacante D. Overexpression and sequence of the Escherichia coli cheY gene and biochemical activities of the CheY protein. J Bacteriol. 1984;160:36–41. doi: 10.1128/jb.160.1.36-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan D G, Baumgartner J W, Hazelbauer G L. Proteins antigenically related to methyl-accepting chemotaxis proteins of Escherichia coli detected in a wide range of bacterial species. J Bacteriol. 1993;175:133–140. doi: 10.1128/jb.175.1.133-140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muchmore D C, McIntosh L P, Russell C B, Andersen D E, Dahlquist F W. Expression and nitrogen-15 labeling of proteins for proton and nitrogen-15 nuclear magnetic resonance. Methods Enzymol. 1989;177:44–73. doi: 10.1016/0076-6879(89)77005-1. [DOI] [PubMed] [Google Scholar]
- 32.Ninfa E G, Stock A, Mowbray S, Stock J. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J Biol Chem. 1991;266:9764–9770. [PubMed] [Google Scholar]
- 33.Nowlin D M, Bollinger J, Hazelbauer G L. Sites of covalent modification in Trg, a sensory transducer of Escherichia coli. J Biol Chem. 1987;262:6039–6045. [PubMed] [Google Scholar]
- 34.Okumura H, Nishiyama S-I, Sasaki A, Homma M, Kawagishi I. Chemotactic adaptation is altered by changes in the carboxy-terminal sequence conserved among the major methyl-accepting chemoreceptors. J Bacteriol. 1998;180:1862–1868. doi: 10.1128/jb.180.7.1862-1868.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oosawa K, Imae Y. Glycerol and ethylene glycol: members of a new class of repellents of Escherichia coli chemotaxis. J Bacteriol. 1993;154:104–112. doi: 10.1128/jb.154.1.104-112.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park C, Dutton D P, Hazelbauer G L. Effects of glutamines and glutamates at sites of covalent modification of a methyl-accepting transducer. J Bacteriol. 1990;172:7179–7187. doi: 10.1128/jb.172.12.7179-7187.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parkinson J S. Signal transduction schemes in bacteria. Cell. 1993;72:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 38.Parkinson J S, Houts S E. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson G L. A simplification of the protein assay method of Lowry et al., which is more generally applicable. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 40.Rebbapragada A M, Johnson M S, Harding G P, Zuccarelli A J, Fletcher H M, Zhulin I B, Taylor B L. The Aer protein and serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuster S C, Swanson R V, Alex L A, Bourret R B, Simon M I. Assembly and function of a quaternary signal transduction complex monitored by surface plasmon resonance. Nature. 1993;365:343–347. doi: 10.1038/365343a0. [DOI] [PubMed] [Google Scholar]
- 42.Shapiro M J, Koshland D E., Jr Mutagenic studies of the interaction between the aspartate receptor and methyltransferase from Escherichia coli. J Biol Chem. 1994;269:11054–11059. [PubMed] [Google Scholar]
- 43.Simms S A, Stock A M, Stock J B. Purification and characterization of the S-adenosylmethionine:glutamyl methyltransferase that modifies membrane chemoreceptor proteins in bacteria. J Biol Chem. 1987;262:8537–8543. [PubMed] [Google Scholar]
- 44.Smith R A, Parkinson J S. Overlapping genes at the cheA locus of E. coli. Proc Natl Acad Sci USA. 1980;77:5370–5374. doi: 10.1073/pnas.77.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Springer M S, Goy M F, Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci. 1977;74:3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weerasuriya S, Schneider B M, Manson M D. Chimeric chemoreceptors in Escherichia coli: signalling properties of Tar-Tap and Tap-Tar hybrids. J Bacteriol. 1998;180:914–920. doi: 10.1128/jb.180.4.914-920.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu J, Li J, Li G, Long D G, Weis R M. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto K, Macnab R M, Imae Y. Repellent response functions of the Trg and Tap chemoreceptors of Escherichia coli. J Bacteriol. 1990;172:383–388. doi: 10.1128/jb.172.1.383-388.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W, Brooun A, McCandless J, Banda P, Alam M. Signal transduction in the archaeon Halobacterium salinarium is processed through three subfamilies of 13 soluble and membrane-bound transducer proteins. Proc Natl Acad Sci USA. 1996;93:4649–4654. doi: 10.1073/pnas.93.10.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]