Abstract
Spray-induced gene silencing (SIGS) is a powerful and eco-friendly method for crop protection. Based off the discovery of RNA uptake ability in many fungal pathogens, the application of exogenous RNAs targeting pathogen/pest genes results in gene silencing and infection inhibition. However, SIGS remains hindered by the rapid degradation of RNA in the environment. As extracellular vesicles are used by plants, animals, and microbes in nature to transport RNAs for cross-kingdom/species RNA interference between hosts and microbes/pests, nanovesicles and other nanoparticles have been used to prevent RNA degradation. Efforts examining the effect of nanoparticles on RNA stability and internalization have identified key attributes that can inform better nanocarrier designs for SIGS. Understanding sRNA biogenesis, cross-kingdom/species RNAi, and how plants and pathogens/pests naturally interact are paramount for the design of SIGS strategies. Here, we focus on nanotechnology advancements for the engineering of innovative RNA-based disease control strategies against eukaryotic pathogens and pests.
Introduction
Plant pathogens and pests are a significant and devastating problem for the agricultural industry, resulting in average crop losses between 10 and 40% across major food security crops, even with conventional disease mitigation methods [1]. Due to global warming, the range of ecological niches and spread of plant-pathogenic fungi and pests are increasing [2]. Therefore, the development of novel, robust, and eco-friendly antifungals and insecticides is urgently needed.
Effective pathogen control strategies have been developed by exploiting naturally occurring phenomena in host–pathogen interactions. RNA interference (RNAi) has been identified as a critical regulatory mechanism in eukaryotes in which small RNAs (sRNAs), including microRNAs (miRNAs) and small interfering RNAs (siRNAs), silence the expression of target genes with certain complementary sequences. sRNAs are processed by the Ribonuclease III-like enzyme Dicer or Dicer-like (DCL) proteins and loaded into Argonaute (AGO) proteins to enable silencing of target genes by mRNA cleavage and degradation, translational inhibition, or transcriptional gene silencing [3-5].
In addition to endogenous regulation, sRNAs are transported between interacting organisms in a mechanism deemed “cross-kingdom RNAi”. Cross-kingdom RNAi is part of plant defense responses in which plants produce and transfer sRNAs to silence pathogen genes and inhibit infection. Host-induced gene silencing (HIGS) is to engineer plants to express double stranded RNAs (dsRNAs) or sRNAs that target essential or virulence-related genes in pathogens/pests [6]. Although effective, generation of HIGS-modified crops is time-consuming, challenging, and unfeasible for some plant species. HIGS plants are also classified as genetically modified organisms, which have an arduous regulatory approval process and are unfavorable in some consumer markets [6,7].
Recently, it was discovered that many organisms, including fungi and oomycetes, can efficiently take up RNA from the environment [8]. Exogenous dsRNAs can be processed by native host or pathogen/pest RNAi machinery into siRNAs that silence genes in the pathogen or pest [9-11], a process known as ‘environmental RNAi’. This prompted the development of spray-induced gene silencing (SIGS), in which sRNA or dsRNA targeting pathogen/pest genes are topically applied to plant surfaces for specific and effective crop protection. Since then, dsRNA has been applied for plant protection through various methods including spraying, infiltrating, root soaking, spreading on leaves, and injection with success [11,12]. In addition to the targeting of pathogen, pest, and viral sequences, SIGS approaches have been used to modify endogenous plant genes and phenotypes [13-15]. This ability to alter gene expression can provide a powerful tool for investigating plant genetics.
Although promising, sRNA and dsRNA molecules have a short period of environmental stability and variable uptake rates in pathogens and pests [8]. To improve the efficacy of SIGS, recent advances have been made in nanoparticle-based protection of RNAs that drastically increase the feasibility of RNA-based crop protection strategies. Here, we discuss important considerations for generating successful SIGS strategies against plant pests and pathogens that emphasize both nanoparticle and dsRNA attributes. These include analysis of effective gene targets and their pathways, and new insights into nanoparticle characteristics and dsRNA properties that highly influence cellular uptake and silencing efficiency.
The conserved nature of cross-kingdom RNAi
Cross-kingdom RNA trafficking is an important facet of host–microbe interactions as shown by numerous discoveries of functional RNA transfer between interacting species. In the plant kingdom, cross-kingdom RNAi was observed between the model plant Arabidopsis thaliana and the fungal pathogen Botrytis cinerea, in which fungal sRNAs enter host cells and hijack Arabidopsis AGO1 to silence plant immunity genes, weakening host immunity to promote fungal infection [16]. Like B. cinerea, other plant pathogens such as Fusarium oxysporum [17], Verticillium dahliae [18,19], the oomycete Hyaloperonospora arabidopsidis [20], and the rust fungus Puccinia striiformis [21,22] all send sRNAs into plant host cells to silence host immunity target genes and facilitate infection. Conversely, plant hosts such as A. thaliana [23], cotton [24], tomato [25,26], and wheat [27] transport sRNAs to silence pathogen virulence-related genes and inhibit infection, emphasizing the bidirectional nature of cross-kingdom RNAi.
In addition, the mechanisms of cross-kingdom RNAi seem heavily conserved. Many of the sRNA effectors from plant pathogens utilize host AGO proteins, such as AGO1 and AGO4, to silence plant immunity genes as evidenced by studies of B. cinerea [16], V.dahliae [18,19], F. oxysporum [17,28] and H. arabidopsidis [20]. Moreover, the parasitic plant Cuscuta campestris delivers sRNAs into host plants during parasitism [29]. Cross-kingdom RNAi occurs in symbiotic plant-microbe relationships as well. The ectomycorrhizal fungus Pisolithus microcarpus delivers a microRNA into the host Eucalyptus grandis and induces cross-kingdom gene silencing during symbiosis [30]. Strikingly, although bacteria lack traditional RNAi machinery, tRNA-derived RNA fragments (tRFs) from nodule-forming Rhizobia bacterium are transferred into host soybean root cells, bind to soybean AGO1, and induce silencing of plant host nodulation-related genes [31].
Cross-kingdom RNAi has also been observed in several animal pathogen/parasite interactions such as between the nematode parasite Heligmosomoides polygyrus and mouse cells [32] and between the yeast pathogen Candida albicans and human monocyte cells [33]. As well, the insect pathogenic fungus Beauveria bassiana transfers sRNAs that bind to host mosquito AGO1 to silence host genes [34]. Cross-kingdom RNAi is conserved among a wide range of interacting organisms, and many microbial sRNAs function similarly by loading into host AGO proteins to silence host genes, either to promote infection or maintain symbiosis. Despite the prevalence of RNA translocation between species, how these RNAs move through extracellular spaces in different organisms is not yet fully understood.
Plant extracellular vesicles are a critical mechanism for sRNA transport
There is significant evidence for the role of extracellular vesicles (EVs) in RNA translocation between interacting organisms. In mammalian systems, diverse cell types including monocytes [33], adipocytes [35], cancer cells [36,37] and immune cells [38,39] all use EVs to protect and transport sRNAs, mRNAs, and other RNAs to specific target cells. Plant fungal pathogen B. cinerea also transports sRNAs in fungal EVs, which are taken up by A. thaliana cells through clathrin-mediated endocytosis [40]. Furthermore, bacteria, algae, and nematodes utilize EVs for RNA transport to interacting microbes or host tissues [32,41,42]. In plants, the presence of many ribonucleases in the apoplast [43] suggests that extracellular RNAs must have a means of protection. Indeed, Arabidopsis EVs, especially TET8-positive EVs have been shown to protect their RNA cargo from degradation from both proteases and RNases, illustrating how plant EVs are an efficacious route of RNA protection and transport within plants [23,47]. It has been speculated that RNA-binding proteins may protect RNAs from degradation. However, most RNA binding proteins only bind to specific motifs of RNA, which is evidenced by methods that use RNase treatment to cleave RNA in RNA-protein complexes to identify RNA-binding sites [44]. The apoplast also contains many proteases that can disrupt the stability of RNA-protein complexes [45,46]. Therefore, it is questionable whether protein binding is a prevalent or sufficient strategy for extracellular RNA protection in plants. Similar observations in plant, microbial and mammalian cell systems demonstrate how conserved the use of EVs for RNA protection and transport is across kingdoms [37,40,48-50].
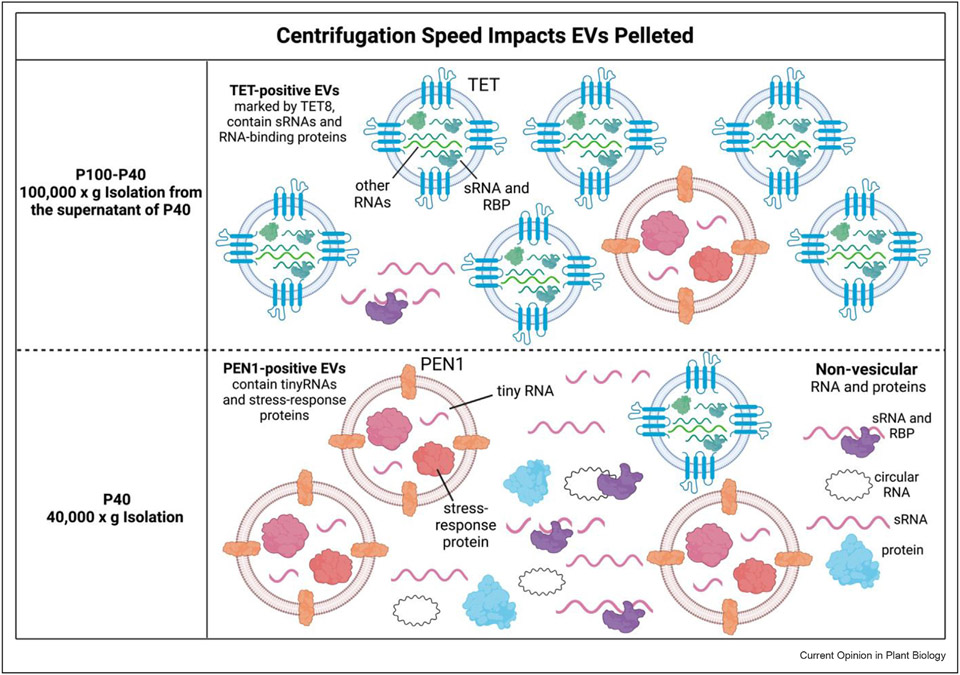
The isolation of high quality EVs is critical for studying EVs and their RNA cargo. Three methods have been used for plant EV isolation: differential centrifugation followed by ultracentrifugation, density gradient ultracentrifugation, or immunoaffinity capture. Of the three methods, differential centrifugation followed by ultracentrifugation results in the highest yield of EVs while the other two methods enable isolation of specific EV densities or subtypes. Importantly, the use of swinging-bucket rotors during ultracentrifugation is recommended for collecting high-quality intact EVs, as this type of rotor minimizes vesicle disruption [35,36]. Two ultracentrifugation speeds have been used to pellet plant EVs: 40,000 ×g (P40 fraction) and 100,000×g (P100 fraction) (Figure 1). The P40 fraction enriches for PEN1-positive EVs, which contain tiny RNAs and proteins [51,52], and non-vesicular RNAs with unknown mechanisms of degradation protection [43,45,46]. Centrifugation of the supernatant of P40 at 100,000×g (P100–P40 fraction) collects TET-positive EVs, which are enriched in functional sRNAs and proteins [23,47,53]. In density gradient ultracentrifugation, different EV subtypes are separated by density [47,51]. Most reliably, immunoaffinity purification can isolate specific EV subtypes by using antibodies against EV subtype-specific protein markers [46,47]. Unfortunately, few plant EV markers have been characterized to date, so there is a strong need to identify additional markers.
Figure 1. Centrifugation speed impacts the EV subtype that is isolated from plants.
Different ultracentrifugation speeds differentially pellet distinct plant EV subtypes. In Arabidopsis, 40,000×g collects the P40 fraction which is enriched in PEN1-positive EVs, extravesicular RNAs, and RNA-protein complexes, and misses a large portion of TET-positive EVs [51,53,54]. PEN1-positive EVs are enriched in tiny RNAs and stress-response proteins [53]. Centrifugation at 100,000×g (P100) is necessary to pellet plant exosomes such as TET-positive EVs [51]. The supernatant of the P40 fraction can be centrifuged at 100,000×g to collect TET-positive EVs in the P100–P40 fraction [51]. TET-positive EVs are enriched in sRNAs involved in cross-kingdom RNAi and RNA-binding proteins [38].
Differences in plant conditions and extraction methods of apoplastic washing fluid can greatly affect the EVs that are captured. Collection of apoplastic washing fluid from detached leaves rather than whole plants alone can reduce contamination from cell debris and cytoplasmic content [47]. Therefore, it is important to avoid generalizations about the function of all plant EV subtypes based on individual methods of capture. Studies of native extracellular RNA protection are paramount in the design of effective RNA application for SIGS. Indeed, mimicking plant EVs using artificial vesicles to protect dsRNA was shown to prolong SIGS-based plant protection against B. cinerea [54].
Leveraging RNAi to control plant pathogens and pests
Fungi and oomycetes are highly amenable to RNAi-based control strategies. More than 50 proof-of-concept studies have demonstrated the viability of HIGS-mediated control of ascomycetes, basidiomycetes, and oomycetes, with an average disease resistance of 60% [6]. The success of HIGS is positively correlated with the quality and quantity of sRNA transport from plant cells [55] and uptake by pathogens/pests [6]. Efforts to elucidate these mechanisms are ongoing. Sustainably produced dsRNA/siRNA in HIGS leads to long-lasting RNAi, which is important for organisms that do not have RNAi amplification or transitivity mechanisms [56]. HIGS also eliminates the need for repeated dsRNA application as is the case with SIGS [57]. However, for some phloem-sucking insects and necrotrophic pathogens, dsRNA processing by plant RNAi machinery can limit HIGS efficacy [58,59]. Several studies have highlighted how dsRNA is more effective for gene silencing and inducing phenotypic changes in pests/pathogens than siRNA [60,61]. As such, SIGS has been demonstrated to have similar or higher efficacy than HIGS against some fungi and insects [61-63]. It may be partially due to the lower stability of sRNAs than long dsRNAs.
The generation and secretion of sRNA effectors is important for virulence in many fungal and oomycete pathogens, so RNAi pathway components, such as DCL genes, represent effective SIGS targets. Although a few examples of Dicer-independent sRNA biogenesis in fungi and mammals have also been reported [64-66], successful disease control through SIGS of pathogen DCL genes has been demonstrated with B. cinerea [19,67], Fusarium graminearum [68,69], and Plasmopara viticola [70]. As well, mutation of fungal DCL genes attenuated the virulence of F. graminearum [69], Penicillium italicum [71], P. viticola [70] and Valsa mali [72], and both the virulence and growth of B. cinerea [16,19,67] and Colletotrichum gloeosporioides [73]. Fungal-derived phased siRNAs were recently discovered at the Blumeria hordei-barley interface, suggesting a role in endogenous sRNA-directed fungal gene regulation through the fungal RNAi pathway during infection [74]. Phased siRNAs are mostly generated by DCLs and RNA-dependent RNA polymerases (RdRPs), suggesting that RdRPs could be good potential targets for SIGS. However, in SIGS against Fusarium asiaticum [75] and V. dahliae [56], RdRPs were found to be less critical for secondary amplification than in plants, indicating the importance of targeting genes from other pathways in tandem. Genes involved in fungal vesicle trafficking such as vacuolar protein sorting 51 (VPS51), dynactin complex large subunit (DCTN1), and suppressor of actin (SAC1) [54,76], as well as other essential genes for fungal growth and development have emerged as effective dsRNA targets [9,77].
Improving RNAi-based plant protection through smarter dsRNA designs
Selecting the appropriate target genes for HIGS or SIGS is the most essential factor for successful pest/pathogen control; however, additional features such as dsRNA stability, transport, length, and processing are key for improving efficacy. Clathrin-mediated endocytosis (CME) is a primary mechanism for dsRNA uptake in fungal pathogens and insect pests [78-80]. Consequently, it has become apparent that chimeric dsRNA designs, rather than dsRNA cocktails, are more effective for SIGS and should be considered for use in future studies [81-83]. Chimeric dsRNAs avoid the potential oversaturation of endocytic components and competitive inhibition of target genes observed during combinatorial delivery of multiple dsRNAs [82] and can enable multiplexed targeting of different genes, pathways and organisms in a single construct [81,83].
Co-opting clathrin-independent endocytosis pathways in addition to CME can further drive SIGS improvement. Design of short (23 nt) “paperclip” RNAs with partially closed ends bypassed CME inhibition in the yellow fever mosquito Aedes aegypti to enter cells via a clathrin-independent pathway and facilitate transcript knockdown [79]. Suggested resistance mechanisms that pests/pathogens may evolve to evade SIGS includes mutations of RNA uptake pathways. Therefore, accessing multiple uptake routes is important. Chemically modifying dsRNA can affect how it is enzymatically processed, leading to increased resistance to nuclease degradation and improved RNAi [84]. dsRNA lengths of ~500 nucleotides are also the most optimal for SIGS in balancing efficient cellular uptake/processing and minimizing potential degradation and off-target effects [85,86]. Off-target RNAi has been observed with dsRNA that contains matching sequences of 29–32 bp with non-target genes [87], so rational design of dsRNA sequences to minimize these effects and maximize silencing efficiency is of utmost importance.
Nanoparticles as RNA carriers for protection
The vulnerability of dsRNA to environmental factors (e.g., rainfall, UV light), enzymatic degradation, and undesirable processing by the plant RNAi machinery substantially limits SIGS as a technology [88-90]. However, nanoparticles, which are materials with dimensions between 1 and 100 nm in diameter, can assist in overcoming these challenges and are becoming increasingly common in plant protection schemes. Nanoparticles can also be designed with unique physicochemical and biological properties that further augment the efficacy of SIGS (Figure 2).
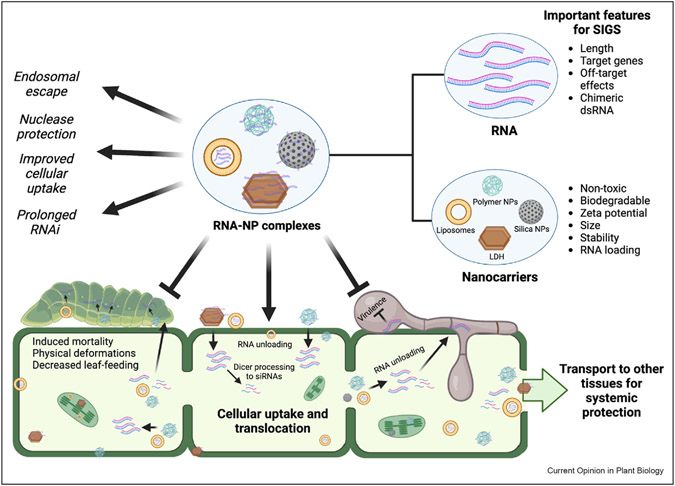
Figure 2. RNA-nanoparticle complexes provide enhanced RNA stability and uptake, leading to greater efficiency in SIGS-based crop protection strategies.
This schematic illustrates how RNA can be encapsulated in various nanocarriers to form RNA-nanoparticle complexes. Important features for dsRNA and nanocarrier design that could improve SIGS efficacy are highlighted along the side. Use of RNA-nanoparticle complexes in SIGS results in improved and prolonged gene silencing relative to naked dsRNA due to better endosomal escape, nuclease protection, and cellular uptake. RNA-nanoparticle complexes can be topically applied to plant surfaces where they are directly taken up by pests and pathogens or internalized by plant cells. After internalization, RNA is released from RNA-nanoparticle complexes and taken up by plants or pests/pathogens, translocated, and/or processed to siRNAs. RNA-nanoparticle complexes can also be directly transported to other plant tissues to provide systemic protection.
For some pathogens and pests, undesirable processing of dsRNA into siRNA by plant machinery can result in lower HIGS and SIGS efficacy. Several examples have demonstrated how direct application of exogenous RNA is sufficient for targeted suppression of transgenes in Arabidopsis [91,92]. Sprayed dsRNA can be internalized by plant cells and processed into siRNAs by plant RNAi machinery or left intact and translocated to other tissues [93]. It is currently unknown what factors dictate the amount of internalized dsRNA that is translocated rather than processed into siRNAs. In RNAi efforts against agricultural pests, noted variations in RNAi efficacy have been attributed to differences in the levels of unprocessed dsRNA available for pest/pathogen uptake [59]. Plant chloroplasts lack RNAi machinery and accordingly, the use of chloroplast-mediated RNAi was more effective against chewing [94] and nonchewing insects [95] versus traditional HIGS approaches using nuclear-expressed dsRNA. Artificial miRNAs containing insect pre-miRNA backbones also eluded processing by plant Dicer, culminating in enhanced gene silencing and mortality of Helicoverpa armigera relative to pre-miRNAs with plant backbones [96].
Furthermore, it has been shown that necrotrophic fungi, at least F. graminearum, can take up long, unprocessed dsRNA precursors from plant tissues in addition to the dsRNAs sprayed on the surface [93,97]. This may explain why SIGS is more effective than HIGS in the barley-F. graminearum pathosystem [61,86]. Like in some insects, dsRNA must be processed by pathogens to result in gene silencing [97,98]. Supportively, mutation of fungal DCL genes in F. graminearum desensitized the fungus to dsRNA-mediated gene silencing, but susceptibility to siRNAs was unaffected [97]. For pathogenic fungi, nitrogen and phosphate fertilization has been shown to promote their growth relative to mutualists and saprotrophs [99]. Several bacterial species can also utilize extracellular DNA as a carbon and phosphate source [100,101]. Therefore, fungal pathogens could be motivated to take up exogenous RNA as nutrients through mechanisms such as endocytosis [78], although this remains to be experimentally demonstrated.
Use of nanoparticles to stabilize dsRNA can prevent detrimental dsRNA internalization, enzymatic degradation, and processing by plant cells [45,46]. In several cases, RNA release from nanoparticles is facilitated by acidic conditions that are induced by pathogens upon invasion, enabling direct and targeted delivery of dsRNA to the pathogen [102,103]. Moreover, nanoparticle compositions can be engineered for RNA delivery to specific tissues and organs [104-106]. Efficient RNA delivery can also be achieved with non-cell-internalizing nanoparticles as siRNA can be released from the nanoparticle surface by surrounding biofluids [107]. Therefore, nanoparticles can improve RNAi efficacy by increasing the local concentration and residence time of dsRNA available for cellular uptake by preventing its cytoplasmic processing or degradation.
Use of nanoparticles in insects overcomes endosomal accumulation and extreme pHs
Efforts to control insect pests using naked dsRNA have had variable results due to the challenges of RNA instability, limited cellular uptake, overcoming endosomal accumulation after ingestion, and extreme pH modalities in the insect that lead to dsRNA degradation. As such, encapsulation of dsRNA in nanoparticles can improve SIGS efficacy by addressing these challenges. This is critical since dsRNA is often taken up by cells through endocytosis, and if it cannot exit endosomes, the dsRNA will eventually be degraded in lysosomes, resulting in inefficient RNAi [108,109]. Consequently, RNA loaded into layered double hydroxide (LDH) clay nanosheets [81,110], chitosan [111-113], carbon-based materials [114,115], liposomes [116-118] or star polycations [119,120] resulted in more severe physical deformations, retarded larval growth, and increased mortality up to 90% relative to naked dsRNA. Nanoparticle use further critically prevented salivary degradation of dsRNA [117,118,121] and degradation in pHs up to 11 [122]. Although for some insects, additional surface modifications are required to facilitate effective SIGS. In A. aegypti, conjugation of chitosan nanoparticles with sodium tripolyphosphate (TPP) was required to induce a 20–65% mortality rate [123]. Addition of a phenolic coating on dsRNA-biodegradable polymers similarly enhanced endosomal escape and increased cellular uptake in Spodoptera frugiperda (Sf9) cells to facilitate RNAi [118]. These modifications imparted greater size control, a more positive charge, and stronger dsRNA binding that provided greater stability and internalization [118,123]. Nanoparticle-dsRNA treatment can also upregulate endocytosis-related genes in plant [124] and insect [125] cells relative to naked dsRNA, which further contributes to the enhanced RNA delivery and gene silencing effects observed.
Nanoparticles in SIGS against microbial pathogens
Although not as extensively studied as insects, progress is being made in utilizing nanoparticles in SIGS-based control of fungal and microbial pathogens [54,76,126-128]. Topical application of LDH containing dsRNA targeting sRNA effector biogenesis genes DCL 1 & 2 or vesicle trafficking genes VPS51, DCTN1, and SAC1 (VDS) genes extended pathogen protection on pre-harvest materials for up to 4 weeks and on postharvest materials for up to 10 days [54,76,129]. Nanoparticle use can also overcome the constraint of limited RNA uptake rates shown by a subset of fungal/oomycete pathogens [8], which extends SIGS-based control strategies to previously inaccessible organisms. Using carbon dots (CD), Wang et al. enabled SIGS-based control of wild-type and fungicide-resistant Phytophthora pathogens by improving dsRNA stability, release, and internalization through CME [127]. In Rhizoctonia solani, star polymer-complexed chitosan nanoparticles outperformed other formulations, including unmodified chitosan nanoparticles, in terms of dsRNA stability and cellular uptake with plant protection observed up to 20 days later [128]. Importantly, these efforts highlight the importance of nanoparticles and their surface chemistry for extending SIGS as a control strategy to a broader range of pests/pathogens.
Metal nanoparticles have previously been used alone or in combination with conventional fungicides to control fungicide-sensitive and -resistant pathogens such as B. cinerea and Alternaria alternata [130-132]. Therefore, combining these nanoparticles with RNA delivery could enable multipronged control of fungal pathogens by integrating RNAi-based control with traditional fungicides. In voriconazole fungicide-resistant Aspergillus flavus, treatment with Lipofectamine™3000 +siRNA complexes lowered the minimum inhibitory concentration of voriconazole 2- to 4-fold [133]. Similarly, complimenting fungicide application with dsRNA-CD treatment reduced the amount of fungicide needed to achieve protection from Phytophthora by 90% compared to the fungicide alone [127].
Systemic and sustained protection can be achieved by tuning nanoparticle properties
In 2017, a seminal study demonstrated how dsRNA-LDH application enabled dsRNA persistence on plant leaves for up to 30 days and protection against plant viruses for up to 20 days on sprayed and newly emerged leaves [102]. Concomitantly, a single foliar spray of lipid-modified polyethylenimine (lmPEI) loaded with siRNA targeting grapevine leafroll associated virus-3 (GLRaV-3) was sufficient to reduce GLRaV-3 titers in grapevine. Furthermore, multiple doses of the treatment could facilitate recovery of infected grapevines and berries [134]. This ability of SIGS to provide protection to newly emerged and distant plant tissues is critical for long-term protection and is supported by studies demonstrating long-distance trafficking of RNA through the vasculature [135]. Therefore, understanding how nanoparticle-RNA complexes are transported within plants and taken up by plant pathogens and pests will greatly advance the development of systemic RNAi-based protection methods.
Investigations on LDH, CDs, CeO2, and silica nanoparticles have all illustrated how small (<50 nm), hydrophilic, and more positively charged nanoparticles exhibited the highest foliar delivery efficiencies into organelles, diverse cell types, and the extracellular space [105,136,137]. As such, size-dependent silencing effects can be observed after spray application with carbon dots [138]. Furthermore, foliar application of smaller (~6 nm) star polymer nanocarriers favored symplastic unloading in young leaves, while larger particles (~35 nm) improved apoplastic unloading in roots [139]. Modulating nanoparticle compactness and bending stiffness can enable nanomaterials to easily move through the plant cell wall to further improve plant uptake [107,140]. Finally, nanoparticle zeta potential (ζ) strongly influences gene silencing efficiency and impacts internalization and mobility as well. Conditions that induced a net negative zeta potential of chitosan-dsRNA or star polycation complexes, including high pH [>7] media conditions or high natural organic matter, abolished dsRNA-mediated gene knockdown in Caenorhabditis elegans [141] and limited nanocarrier mobility to young leaves and roots [139].
Many of these nanoparticles are likely translocated via the vascular system through energy-dependent and independent processes, as their small size allows them to pass through the plant cell wall [105,137]. This mechanism also extends to trees, where trunk injection of fluorine- and ruthenium-labelled polymer nanoparticles resulted in their translocation throughout the trunk tissue via the xylem sap [142]. Therefore, targeted protection of specific plant tissues will be achieved by designing nanomaterials that promote translocation through the plant vasculature or co-opt specific uptake routes. For example, spray application of siRNA complexed with cell-penetrating peptides enabled efficient and targeted gene silencing through stomata-dependent-uptake and delivery to plant nuclei and chloroplasts [106].
Bacterial-based systems facilitate sustained SIGS in pests and pathogens
In contrast to the nanomaterial systems described above, the use of bacterial-based systems for RNAi can allow for simultaneous synthesis and delivery of dsRNA. This can lower the cost of SIGS by providing sustained dsRNA delivery, eliminating the need for repeated dsRNA application. Furthermore, since insects lack RdRPs, engineering bacterial symbionts for dsRNA delivery can provide sustained RNAi throughout the host’s life and overcome challenges associated with feeding or injecting dsRNA. Bacterial-based RNAi already shows great promise against viruses and fungal pathogens with topical treatment of Escherichia coli-encapsulated dsRNA, or the dsRNA-producing bacteria itself, protecting plant material against infection as effectively as chemically synthesized naked dsRNA [143,144]. In addition, in vitro application of E. coli-derived anucleated minicells loaded with dsRNA targeting chitin synthase class III (Chs3a, Chs3b) and the DCL1 and DCL2 genes of Botryotinia fuckeliana halted disease progression on strawberries for up to 12 days in greenhouse conditions [145]. Efforts to utilize bacterial symbiont-mediated RNAi (smRNAi) for insect control, however, have yielded mixed results.
In thrips and kissing bugs, smRNAi was effective in reducing gene expression and providing greater protection of cucumber seedlings [146], and in honeybees, smRNAi improved bee survival against parasitic mites [147]. The engineered symbionts could be detected more than 250 days after initial uptake and were horizontally transmitted [146,147]. In contrast, application of smRNAi to aphids using E. coli and the native symbiont, Serratia symbiotica CWBI-2.3T, was unable to induce reproducible aphid phenotypes or even gene knockdown [148]. This discrepancy could be partially attributed to selection of an appropriate symbiont. Genetically tractable laboratory strains like E. coli are foreign to the host, which can induce immune responses and reduce colonization of these transgenic bacteria. Other important caveats are the need for bacterial RNase III mutants, which promote stable dsRNA production, and localization of the symbiont within the host as how dsRNA is delivered by the symbiont during smRNAi remains unknown [148].
A similar but parallel approach for insect control termed pathogen-mediated RNAi has also emerged. Building upon their discovery of cross-kingdom RNAi between B. bassiana and mosquito [34], Cui et al. engineered B. bassiana to produce immunosuppressive miRNAs against A. aegypti and Galleria mellonella [149]. Topical application of transgenic B. bassiana expressing A. aegypti miR-8 and miR-375, negative regulators of the Toll immune signalling pathway, resulted in a 20–30% reduction in survival time for insecticide-resistant A. aegypti and G. mellonella larvae [149]. Importantly, miR-8 and miR-375 are conserved in several agricultural insect pests so this technology can be easily translated for control of other organisms. While the use of bacterial-based systems is relatively new compared to topical application of exogenous dsRNA, there is immense promise in this technology.
Conclusions and future directions
Nanotechnology is becoming increasingly popular in modern agriculture as a tool to address major food security challenges but has only recently been leveraged with powerful RNAi-based strategies such as SIGS to control and combat plant pests and pathogens. Specifically, the use of nanocarriers in SIGS has alleviated previous limitations with RNA uptake and stability in the environment and after ingestion (Figure 2). Many hosts and interacting microbes actively exchange sRNAs for reciprocal gene silencing, but these sRNAs must be protected from degradation to remain intact and functional. As characterized across different kingdoms of life, the use of EVs is a prevalent strategy for RNA transport and protection, and considerations of these mechanisms of native host RNA trafficking are crucial in guiding the design of effective and robust SIGS applications.
The use of nanocarriers for RNAi-based control will greatly improve the efficacy of SIGS; however, before this technology can be wholly adopted, studies must be performed examining the lifespan and fate of nanocarriers in the environment, the impact on non-target organisms, and the potential transfer to other plant hosts. A study on the rice-hopper-spider food chain found that ingestion of treated plant tissues or guttation droplets was sufficient to trigger both targeted and off-target RNAi in consumer hoppers and predator spiders [87]. Some reports have made headway in including non-target organisms when testing new dsRNA constructs or dsRNA-nanoparticle complexes, and this practice should be incorporated into future SIGS studies [81,112,113]. Modelling the fate of nanocarrier compounds after degradation and their interactions with soil and water will also complement these efforts [150,151].
Encapsulation of RNA in nanocarriers provides extensive protection from environmental conditions and nuclease degradation. Many of these nanocarriers further promote cellular uptake in target organisms by facilitating endosomal escape which significantly improves RNAi efficiency. Importantly, recent studies have generated not only preventative plant protection using RNA-nanocarrier complexes but also curative treatment of ongoing, systemic viral infections. It will be interesting to see if curative protection against microbial pathogens can be achieved. Material features including a positive zeta potential, small size, and hydrophilicity have been identified as key parameters that can be tuned to optimize RNA delivery. Thus, the development of novel and effective SIGS approaches can be accomplished by complementing these nanocarrier advances with improvements in dsRNA design and target gene selection. The costs of these technologies, both financial and environmental should continue to be considered. As well, additional work is needed to understand the environmental fate of dsRNA and nanocarriers during and after plant protection. Overall, the current interest and emphasis on nanocarrier design strongly supports the potential of nanotechnology to propel SIGS to be a comprehensive plant protection system against multiple pests and pathogens.
Acknowledgements
We apologize for not being able to cite many related and interesting studies due to space limitations. The work in Dr. Jin’s lab was supported by grants from National Institutes of Health (R35GM136379), National Science Foundation (IOS 2020731), United States Department of Agriculture (2021-67013-34258), the Australian Research Council Research Hub for Sustainable Crop Protection (IH190100022) and the CIFAR ‘Fungal Kingdom’ fellowship. A.C is supported by the USDA National Institute of Food and Agriculture, AFRI project 2022-09774.The figures were created using BioRender.com.
Footnotes
Declaration of competing interest
The authors declare they have no competing financial interests or personal relationships that would inappropriately bias the work reported in this paper.
Data availability
No data was used for the research described in the article.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
* * of outstanding interest
- 1.Savary S, Willocquet L, Pethybridge SJ, Esker P, McRoberts N, Nelson A: The global burden of pathogens and pests on major food crops. Nat Ecol Evol 2019, 3:430–439. [DOI] [PubMed] [Google Scholar]
- 2.Fausto A, Rodrigues ML, Coelho C: The still underestimated problem of fungal diseases worldwide. Front Microbiol 2019, 10:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosa C, Kuo YW, Wuriyanghan H, Falk BW: RNA interference mechanisms and applications in plant pathology. Annu Rev Phytopathol 2018, 56:581–610. [DOI] [PubMed] [Google Scholar]
- 4.Huang C, Wang H, Hu P, Hamby R, Jin H: Small RNAs – big players in plant-microbe interactions. Cell Host Microbe 2019, 26:173–182. [DOI] [PubMed] [Google Scholar]
- 5.Lopez-Gomollon S, Baulcombe DC: Roles of RNA silencing in viral and non-viral plant immunity and in the crosstalk between disease resistance systems. Nat Rev Mol Cell Biol 2022, 23:645–662. [DOI] [PubMed] [Google Scholar]
- 6.Koch A, Wassenegger M: Host-induced gene silencing – mechanisms and applications. New Phytol 2021, 231:54–59. [DOI] [PubMed] [Google Scholar]
- 7.Collinge DB, Sarrocco S: Transgenic approaches for plant disease control: status and prospects 2021. Plant Pathol 2022, 71:207–225. [Google Scholar]
- 8.*. Qiao L, Lan C, Capriotti L, Ah-fong A, Sanchez JN, Hamby R, Heller J, Zhao H, Glass NL, Judelson HS, et al. : Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol J 2021, 19: 1756–1768. By comparing the uptake rates and the susceptibility of different fungal plant pathogen species to SIGS applications, the researchers showed that RNA uptake and therefore silencing efficiency differed greatly between different fungal pathogens. This highlights the need for improved nanoparticle design to enhance RNA uptake, and gives overarching information on which fungal pathogen species are or are not viable targets for control by SIGS methods.
- 9.Hernández-Soto A, Chacón-Cerdas R: RNAi crop protection advances. Int J Mol Sci 2021, 22, 12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoang BTL, Fletcher SJ, Brosnan CA, Ghodke AB, Manzie N, Mitter N: RNAi as a foliar spray: efficiency and challenges to field applications. Int J Mol Sci 2022, 23:6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilir Ö, Göl D, Hong Y, McDowell JM, Tör M: Small RNA-based plant protection against diseases. Front Plant Sci 2022, 13, 951097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das PR, Sherif SM: Application of exogenous dsRNAs-induced RNAi in agriculture: challenges and triumphs. Front Plant Sci 2020, 11:946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiselev KV, Suprun AR, Aleynova OA, Ogneva ZV, Kalachev AV, Dubrovina AS: External dsrna downregulates anthocyanin biosynthesis-related genes and affects anthocyanin accumulation in arabidopsis thaliana. Int J Mol Sci 2021, 22:6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nerva L, Guaschino M, Pagliarani C, De Rosso M, Lovisolo C, Chitarra W: Spray-induced gene silencing targeting a glutathione S-transferase gene improves resilience to drought in grapevine. Plant Cell Environ 2022, 45:347–361. [DOI] [PubMed] [Google Scholar]
- 15.Simon I, Persky Z, Avital A, Harat H, Schroeder A, Shoseyov O: Foliar application of dsRNA targeting endogenous potato (Solanum tuberosum) isoamylase genes ISA1, ISA2, and ISA3 confers transgenic phenotype. Int J Mol Sci 2023, 24:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, Huang H Da, Jin H: Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342:118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji HM, Mao HY, Li SJ, Feng T, Zhang ZY, Cheng L, Luo SJ, Borkovich KA, Ouyang SQ: Fol-milR1, a pathogenicity factor of Fusarium oxysporum, confers tomato wilt disease resistance by impairing host immune responses. New Phytol 2021, 232: 705–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B Sen, Li YC, Guo HS, Zhao JH: Verticillium dahliae secretes small RNA to target host MIR157d and retard plant floral transition during infection. Front Plant Sci 2022, 13, 847086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M, Weiberg A, Lin FM, Thomma BPHJ, Huang H Da, Jin H: Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat Plants 2016, 2, 1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.**. Dunker F, Trutzenberg A, Rothenpieler JS, Kuhn S, Pröls R, Schreiber T, Tissier A, Kemen A, Kemen E, Hückelhoven R, et al. : Oomycete small RNAs bind to the plant RNA-induced silencing complex for virulence. Elife 2020, 9, e56096. By designing a novel Csy4/GUS repressor reporter system that can visualize local gene silencing alongside Hyaloperonospora arabidopsidis hyphae within plant tissues, this paper demonstrated that H. arabiopdisidis trafficks RNAs into plant cells and suppress host target genes. This is a novel example of cross-kingdom RNAi and an innovative method to study the translocation and functionality of RNA between interacting organisms.
- 21.Mueth NA, Ramachandran SR, Hulbert SH: Small RNAs from the wheat stripe rust fungus (Puccinia striiformis f.sp. tritici). BMC Genom 2015, 16:718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B, Sun Y, Song N, Zhao M, Liu R, Feng H, Wang X, Kang Z: Puccinia striiformis f. sp. tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol 2017, 215:338–350. [DOI] [PubMed] [Google Scholar]
- 23.Cai Q, Qiao L, Wang M, He B, Lin F, Palmquist J, Jin H: Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360:1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang T, Zhao YL, Zhao JH, Wang S, Jin Y, Chen ZQ, Fang YY, Hua Cl, Ding SW, Guo HS: Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat Plants 2016, 2, 16153. [DOI] [PubMed] [Google Scholar]
- 25.Wu F, Huang Y, Jiang W, Jin W: Genome-wide identification and validation of tomato-encoded sRNA as the cross-species antifungal factors targeting the virulence genes of Botrytis cinerea. Front Plant Sci 2023, 14, 1072181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng X, Jin W, Wu F: Novel tomato miRNA miR1001 initiates cross-species regulation to suppress the conidiospore germination and infection virulence of Botrytis cinerea in vitro. Gene 2020, 759, 145002. [DOI] [PubMed] [Google Scholar]
- 27.Jiao J, Peng D: Wheat microrna1023 suppresses invasion of fusarium graminearum via targeting and silencing FGSG_ 03101. J Plant Interact 2018, 13:514–521. [Google Scholar]
- 28.He B, Cai Q, Weiberg A, Li W, Cheng A-P, Ouyang S, Borkovich K, Stajich J, Abreu-Goodger C, Jin H: Botrytis cinerea small RNAs are associated with tomato AGO1 and silence tomato defense-related target genes supporting cross-kingdom RNAi. bioRxiv 2023, 10.1101/2022.12.30.522274. [DOI] [Google Scholar]
- 29.Shahid S, Kim G, Johnson NR, Wafula E, Wang F, Coruh C, Bernal-Galeano V, Phifer T, Depamphilis CW, Westwood JH, et al. : MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 2018, 553:82–85. [DOI] [PubMed] [Google Scholar]
- 30.Wong-Bajracharya J, Singan VR, Monti R, Plett KL, Ng V, Grigoriev IV, Martin FM, Anderson IC, Plett JM: The ectomycorrhizal fungus Pisolithus microcarpus encodes a microRNA involved in cross-kingdom gene silencing during symbiosis. Proc Natl Acad Sci USA 2022, 119, e2103527119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren B, Wang X, Duan J, Ma J: Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 2019, 365:919–922. [DOI] [PubMed] [Google Scholar]
- 32.Buck AH, Coakley G, Simbari F, McSorley HJ, Quintana JF, Le Bihan T, Kumar S, Abreu-Goodger C, Lear M, Harcus Y, et al. : Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun 2014, 5:5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halder LD, Babych S, Palme DI, Mansouri-Ghahnavieh E, Ivanov L, Ashonibare V, Langenhorst D, Prusty B, Rambach G, Wich M, et al. : Candida albicans induces cross-kingdom miRNA trafficking in human monocytes to promote fungal growth. mBio 2022, 13, e03563–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui C, Wang Y, Liu J, Zhao J, Sun P, Wang S: A fungal pathogen deploys a small silencing RNA that attenuates mosquito immunity and facilitates infection. Nat Commun 2019, 10:4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Martin R, Wang G, Brandão BB, Zanotto TM, Shah S, Kumar Patel S, Schilling B, Kahn CR: MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2022, 601:446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, Schiffelers RM, De Wit E, Berenguer J, Ellenbroek SIJ, et al. : In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 2015, 161:1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yokoi A, Yoshioka Y, Yamamoto Y, Ishikawa M, Ikeda SI, Kato T, Kiyono T, Takeshita F, Kajiyama H, Kikkawa F, et al. : Malignant extracellular vesicles carrying MMP1 mRNA facilitate peritoneal dissemination in ovarian cancer. Nat Commun 2017, 8, 14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MA, Bernad A, Sánchez-Madrid F: Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2011, 2:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dosil SG, Lopez-Cobo S, Rodriguez-Galan A, Fernandez-Delgado I, Ramirez-Huesca M, Milan-Rois P, Castellanos M, Somoza A, Gómez MJ, Reyburn HT, et al. : Natural killer (NK) cell-derived extracellular-vesicle shuttled microRNAs control T cell responses. Elife 2022, 11, e76319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He B, Wang H, Liu G, Chen A, Calvo A, Cai Q, Jin H: Fungal small RNAs ride in extracellular vesicles to enter plant cells through clathrin-mediated endocytosis. Nat Commun 2023, 14:4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahmadi Badi S, Bruno SP, Moshiri A, Tarashi S, Siadat SD, Masotti A: Small RNAs in outer membrane vesicles and their function in host-microbe interactions. Front Microbiol 2020, 11:1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schatz D, Schleyer G, Saltvedt MR, Sandaa RA, Feldmesser E, Vardi A: Ecological significance of extracellular vesicles in modulating host-virus interactions during algal blooms. ISME J 2021, 15:3714–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borniego ML, Molina MC, Guiamét JJ, Martinez DE: Physiological and proteomic changes in the apoplast accompany leaf senescence in arabidopsis. Front Plant Sci 2020, 10:1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. : Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 2016, 13:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delaunois B, Colby T, Belloy N, Conreux A, Harzen A, Baillieul F, Clément C, Schmidt J, Jeandet P, Cordelier S: Large-scale proteomic analysis of the grapevine leaf apoplastic fluid reveals mainly stress-related proteins and cell wall modifying enzymes. BMC Plant Biol 2013, 13:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Wang Y, Wang Y: Apoplastic proteases: powerful weapons against pathogen infection in plants. Plant Commun 2020, 1, 100085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y, Wang S, Cai Q, Jin H: Effective methods for isolation and purification of extracellular vesicles from plants. J Integr Plant Biol 2021, 63:2020–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma M, Morgado P, Zhang H, Ehrenkaufer G, Manna D, Singha U: Characterization of extracellular vesicles from entamoeba histolytica identifies roles in intercellular communication that regulates parasite growth and development. Infect Immun 2020, 88, e00349–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon S, Rupp O, Brachmann A, Blum CF, Kraege A, Goesmann A, Feldbrügge M: mRNA inventory of extracellular vesicles from ustilago maydis. J Fungi 2021, 7:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ciullo A, Li C, Li L, Ungerleider KC, Peck K, Marbán E, Ibrahim AGE: Biodistribution of unmodified cardiosphere-derived cell extracellular vesicles using single RNA tracing. J Extracell Vesicles 2022, 11, e12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rutter BD, Innes RW: Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol 2017, 173:728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zand Karimi H, Baldrich P, Rutter BD, Borniego L, Zajt KK, Meyers BC, Innes RW: Arabidopsis apoplastic fluid contains sRNA- and circular RNA–protein complexes that are located outside extracellular vesicles. Plant Cell 2022, 34:1863–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He B, Cai Q, Qiao L, Huang C, Wang S, Miao W, Ha T, Wang Y, Jin H: RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat Plants 2021, 7:342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiao L, Niño-Sánchez J, Hamby R, Capriotti L, Chen A, Mezzetti B, Jin H: Artificial nanovesicles for dsRNA delivery in spray-induced gene silencing for crop protection. Plant Biotechnol J 2023, 21:854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z, Gao X, Zhong S, Li Y, Shi M, Zhang B, Zhang S, Shen H, Liu X: Host-induced gene silencing of PcCesA3 and PcOSBP1 confers resistance to Phytophthora capsici in Nicotiana benthamiana through NbDCL3 and NbDCL4 processed small interfering RNAs. Int J Biol Macromol 2022, 222:1665–1675. [DOI] [PubMed] [Google Scholar]
- 56.Zhang T, Zhao JH, Fang YY, Guo HS, Jin Y: Exploring the effectiveness and durability of trans-kingdom silencing of fungal genes in the vascular pathogen Verticillium dahliae. Int J Mol Sci 2022, 23:2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willow J, Soonvald L, Sulg S, Kaasik R, Silva AI, Taning CNT, Christiaens O, Smagghe G, Veromann E: RNAi efficacy is enhanced by chronic dsRNA feeding in pollen beetle. Commun Biol 2021, 4:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qi T, Guo J, Peng H, Liu P, Kang Z, Guo J: Host-induced gene silencing: a powerful strategy to control diseases of wheat and barley. Int J Mol Sci 2019, 20:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaffer L: RNA-based pesticides aim to get around resistance problems. Proc Natl Acad Sci U S A 2020, 117:32823–32826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, Wu M, Wang B, Han Z: Comparison of the RNA interference effects triggered by dsRNA and siRNA in Tribolium castaneum. Pest Manag Sci 2013, 69:781–786. [DOI] [PubMed] [Google Scholar]
- 61.Koch A, Höfle L, Werner BT, Imani J, Schmidt A, Jelonek L, Kogel KH: SIGS vs HIGS: a study on the efficacy of two dsRNA delivery strategies to silence Fusarium FgCYP51 genes in infected host and non-host plants. Mol Plant Pathol 2019, 20:1636–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.*. Hu D, Chen ZY, Zhang C, Ganiger M: Reduction of Phakopsora pachyrhizi infection on soybean through host- and spray-induced gene silencing. Mol Plant Pathol 2020, 21:794–807. Through identifying novel gene silencing targets and using a virus-based HIGS approach, the authors compared SIGS and HIGS efficacy on an economically important wheat rust fungus, demonstrating the greater success of SIGS for control of this fungal infection. The paper outlines the workflow for developing SIGS strategies in a novel pathosystem.
- 63.Li X, Mu K, Yang S, Wei J, Wang C, Yan W, Yuan F, Wang H, Han D, Kang Z, et al. : Reduction of rhizoctonia cerealis infection on wheat through host- and spray-induced gene silencing of an orphan secreted gene. Mol Plant Microbe Interact 2022, 35:803–813. [DOI] [PubMed] [Google Scholar]
- 64.Lee HC, Li L, Gu W, Xue Z, Crosthwaite SK, Pertsemlidis A, Lewis ZA, Freitag M, Selker EU, Mello CC, et al. : Diverse pathways generate MicroRNA-like RNAs and dicer-independent small interfering RNAs in fungi. Mol Cell 2010, 38:803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Herrera-Carrillo E, Berkhout B: Dicer-independent processing of small RNA duplexes: mechanistic insights and applications. Nucleic Acids Res 2017, 45:10369–10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hill AC, van Leeuwen D, Schlösser V, Behera A, Mateescu B, Hall J: Chemically synthesized, self-assembling small interfering RNA-prohead RNA molecules trigger dicer-independent gene silencing. Chem Eur J 2022, 28, e202103995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Duanis-Assaf D, Galsurker O, Davydov O, Maurer D, Feygenberg O, Sagi M, Poverenov E, Fluhr R, Alkan N: Double-stranded RNA targeting fungal ergosterol biosynthesis pathway controls Botrytis cinerea and postharvest grey mould. Plant Biotechnol J 2022, 20:226–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Werner BT, Gaffar FY, Schuemann J, Biedenkopf D, Koch AM: RNA-Spray-Mediated silencing of Fusarium graminearum AGO and DCL genes improve barley disease resistance. Front Plant Sci 2020, 11:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Werner BT, Koch A, Šečić E, Engelhardt J, Jelonek L, Steinbrenner J, Kogel K-H: Fusarium graminearum DICER-like-dependent sRNAs are required for the suppression of host immune genes and full virulence. PLoS One 2021, 16, e0252365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.*. Haile ZM, Gebremichael DE, Capriotti L, Molesini B, Negrini F, Collina M, Sabbadini S, Mezzetti B, Baraldi E: Double-stranded RNA targeting dicer-like genes compromises the pathogenicity of Plasmopara viticola on grapevine. Front Plant Sci 2021, 12, 667539. Through the development of a new SIGS based control for downy mildew on grape, this paper showed the versatility of SIGS approaches for targeting new pathogen species through the use of homologous gene targets. This paper shows that once target genes are identified in one pathogen, the homologs in other pathogens could be ideal gene targets for novel strategies.
- 71.Yin C, Zhu H, Jiang Y, Shan Y, Gong L: Silencing dicer-like genes reduces virulence and sRNA generation in Penicillium italicum, the cause of citrus blue mold. Cells 2020, 9:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feng H, Xu M, Liu Y, Dong R, Gao X, Huang L: Dicer-like genes are required for H2O2 and KCl stress responses, pathogenicity and small RNA generation in Valsa Mali. Front Microbiol 2017, 8:1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Q, An B, Hou X, Guo Y, Luo H, He C: Dicer-like proteins regulate the growth, conidiation, and pathogenicity of Colletotrichum gloeosporioides from Hevea brasiliensis. Front Microbiol 2018, 8:2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.**. Kusch S, Singh M, Thieron H, Spanu PD, Panstruga R: Site-specific analysis reveals candidate cross-kingdom small RNAs, tRNA and rRNA fragments, and signs of fungal RNA phasing in the barley-powdery mildew interaction. Mol Plant Pathol 2023, 24:570–587. This study suggests that fungal genomes, similar to plant and mammalian genomes, could be subject to sRNA-directed post-transcriptional gene regulation due to the discovery of Blumeria hordei-derived phased siRNAs at the barley infection site that map to transposons within the B. hordei genome. The paper also characterizes B. hordei sRNAs in haustoria that are predicted to target barley genes. The researchers also showed that EVs from the B. hordei-barley infection site are enriched in B. hordei-derived micro-like RNAs, which have a potential role in host gene silencing. This is a novel example of potential cross-kingdom RNAi, and suggests a new endogenous fungal gene regulatory mechanism using native RNAi machinery.
- 75.Song XS, Gu KX, Duan XX, Xiao XM, Hou YP, Duan YB, Wang JX, Yu N, Zhou MG: Secondary amplification of siRNA machinery limits the application of spray-induced gene silencing. Mol Plant Pathol 2018, 19:2543–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niño-Sánchez J, Sambasivam PT, Sawyer A, Hamby R, Chen A, Czislowski E, Li P, Manzie N, Gardiner DM, Ford R, et al. : Bio-Clay™ prolongs RNA interference-mediated crop protection against Botrytis cinerea. J Integr Plant Biol 2022, 64: 2187–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Degnan RM, McTaggart AR, Shuey LS, Pame LJS, Smith GR, Gardiner DM, Nock V, Soffe R, Sale S, Garrill A, et al. : Exogenous double-stranded RNA inhibits the infection physiology of rust fungi to reduce symptoms in planta. Mol Plant Pathol 2023, 24:191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wytinck N, Sullivan DS, Biggar KT, Crisostomo L, Pelka P, Belmonte MF, Whyard S: Clathrin mediated endocytosis is involved in the uptake of exogenous double-stranded RNA in the white mold phytopathogen Sclerotinia sclerotiorum. Sci Rep 2020, 10, 12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abbasi R, Heschuk D, Kim B, Whyard S: A novel paperclip double-stranded RNA structure demonstrates clathrin-independent uptake in the mosquito Aedes aegypti. Insect Biochem Mol Biol 2020, 127, 103492. [DOI] [PubMed] [Google Scholar]
- 80.McGraw E, Roberts JD, Kunte N, Westerfield M, Streety X, Held D, Avila LA: Insight into cellular uptake and transcytosis of peptide nanoparticles in spodoptera frugiperda cells and isolated midgut. ACS Omega 2022, 7:10933–10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jain RG, Fletcher SJ, Manzie N, Robinson KE, Li P, Lu E, Brosnan CA, Xu ZP, Mitter N: Foliar application of clay-delivered RNA interference for whitefly control. Nat Plants 2022, 8:535–548. [DOI] [PubMed] [Google Scholar]
- 82.Shen X, Peng Y, Song H, Wang J, Zhao J, Tang P, Han Z, Wang K: Key factors determining competitions between double-stranded RNAs in Tribolium castaneum. Pestic Biochem Physiol 2022, 181, 105009. [DOI] [PubMed] [Google Scholar]
- 83.Wang K, Cheng H, Chen J, Zhu G, Tang P, Han Z: Chimeric double-stranded RNAs could act as tailor-made pesticides for controlling storage insects. J Agric Food Chem 2021, 69: 6166–6171. [DOI] [PubMed] [Google Scholar]
- 84.Howard JD, Beghyn M, Dewulf N, De Vos Y, Philips A, Portwood D, Kilby PM, Oliver D, Maddelein W, Brown S, et al. : Chemically modified dsRNA induces RNAi effects in insects in vitro and in vivo: a potential new tool for improving RNA-based plant protection. J Biol Chem 2022, 298, 102311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang K, Peng Y, Fu W, Shen Z, Han Z: Key factors determining variations in RNA interference efficacy mediated by different double-stranded RNA lengths in Tribolium castaneum. Insect Mol Biol 2019, 28:235–245. [DOI] [PubMed] [Google Scholar]
- 86.Höfle L, Biedenkopf D, Werner BT, Shrestha A, Jelonek L, Koch A: Study on the efficiency of dsRNAs with increasing length in RNA-based silencing of the Fusarium CYP51 genes. RNA Biol 2020, 17:463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.*. Zhang H, Chen J, Gao J, Zhang Q, Liu X, Han Z: New insights into transmission pathways and possible off-target effects of insecticidal dsRNA released by treated plants. Pestic Biochem Physiol 2022, 188, 105281. The authors examined dsRNA transmission through the rice-hopper-spider food chain and observed targeted and off-target RNAi in plant tissues, consumers and predators. dsRNA sequences with 29–32 bp matching non-target genes could trigger off-target RNAi. This paper provides a good framework for how researchers can evaulate the potential off-target effects of dsRNA in the field.
- 88.Šečić E, Kogel KH: Requirements for fungal uptake of dsRNA and gene silencing in RNAi-based crop protection strategies. Curr Opin Biotechnol 2021, 70:136–142. [DOI] [PubMed] [Google Scholar]
- 89.Niu D, Hamby R, Sanchez JN, Cai Q, Yan Q, Jin H: RNAs — a new frontier in crop protection. Curr Opin Biotechnol 2021, 70: 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rank AP, Koch A: Lab-to-Field transition of RNA spray applications – how far are we? Front Plant Sci 2021, 12, 755203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dubrovina AS, Aleynova OA, Kalachev AV, Suprun AR, Ogneva ZV, Kiselev KV: Induction of transgene suppression in plants via external application of synthetic dsRNA. Int J Mol Sci 2019, 20:1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kiselev KV, Suprun AR, Aleynova OA, Ogneva ZV, Kostetsky EY, Dubrovina AS: The specificity of transgene suppression in plants by exogenous dsRNA. Plants 2022, 11:715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brosnan CA, Sawyer A, de Felippes FF, Carroll BJ, Waterhouse PM, Mitter N: Intact double stranded RNA is mobile and triggers RNAi against viral and fungal plant pathogens. bioRxiv 2021, 10.1101/2021.11.24.469772. [DOI] [Google Scholar]
- 94.Bally J, Fishilevich E, Bowling AJ, Pence HE, Narva KE, Waterhouse PM: Improved insect-proofing: expressing double-stranded RNA in chloroplasts. Pest Manag Sci 2018, 74:1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.*. Wu M, Dong Y, Zhang Q, Li S, Chang L, Vanessa Loiacono F, Ruf S, Zhang J, Bock R: Efficient control of western flower thrips by plastid-mediated RNA interference. Proc Natl Acad Sci U S A 2022, 119, e2120081119. In this study, the authors demonstrate how even non-chewing insects like western flower thrips can be controlled using plastid-mediated RNAi with efficient gene knockdown and high insect mortality. This work also reveals how dsRNA cassettes are more suitable for HIGS given the observation that hairpin RNA cassettes can cause genomic instability.
- 96.Bally J, Fishilevich E, Doran RL, Lee K, de Campos SB, German MA, Narva KE, Waterhouse PM: Plin-amiR, a pre-microRNA-based technology for controlling herbivorous insect pests. Plant Biotechnol J 2020, 18:1925–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Koch A, Biedenkopf D, Furch A, Weber L, Rossbach O, Abdellatef E, Linicus L, Johannsmeier J, Jelonek L, Goesmann A, et al. : An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog 2016, 12, e1005901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gaffar FY, Imani J, Karlovsky P, Koch A, Kogel K-H: Different components of the RNA interference machinery are required for conidiation, ascosporogenesis, virulence, deoxynivalenol production, and fungal inhibition by exogenous double-stranded RNA in the head blight pathogen Fusarium graminearum. Front Microbiol 2019, 10:1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lekberg Y, Arnillas CA, Borer ET, Bullington LS, Fierer N, Kennedy PG, Leff JW, Luis AD, Seabloom EW, Henning JA: Nitrogen and phosphorus fertilization consistently favor pathogenic over mutualistic fungi in grassland soils. Nat Commun 2021, 12:3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pinchuk GE, Ammons C, Culley DE, Li SMW, McLean JS, Romine MF, Nealson KH, Fredrickson JK, Beliaev AS: Utilization of DNA as a sole source of phosphorus, carbon, and energy by Shewanella spp.: ecological and physiological implications for dissimilatory metal reduction. Appl Environ Microbiol 2008, 74:1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mcdonough E, Kamp H, Camilli A: Vibrio cholerae phosphatases required for the utilization of nucleotides and extracellular DNA as phosphate sources. Mol Microbiol 2016, 99: 453–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mitter N, Worrall EA, Robinson KE, Li P, Jain RG, Taochy C, Fletcher SJ, Carroll BJ, Lu GQ, Xu ZP: Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat Plants 2017, 3, 16207. [DOI] [PubMed] [Google Scholar]
- 103.Huang W, Wang M, Hu Z, Yang T, Pei H, Zhang F: Multifunctional metal-organic framework with pH-response for co-delivery of prochloraz and siRNA to synergistic control pathogenic fungi. Colloids Surfaces A Physicochem Eng Asp 2023, 670, 131563. [Google Scholar]
- 104.Molesini B, Pennisi F, Cressoni C, Vitulo N, Dusi V, Speghini A, Pandolfini T: Nanovector-mediated exogenous delivery of dsRNA induces silencing of target genes in very young tomato flower buds. Nanoscale Adv 2022, 4:4542–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yong J, Zhang R, Bi S, Li P, Sun L, Mitter N, Carroll BJ, Xu ZP: Sheet-like clay nanoparticles deliver RNA into developing pollen to efficiently silence a target gene. Plant Physiol 2021, 187:886–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.*. Thagun C, Horii Y, Mori M, Fujita S, Ohtani M, Tsuchiya K, Kodama Y, Odahara M, Numata K: Non-transgenic gene modulation via spray delivery of nucleic acid/peptide complexes into plant nuclei and chloroplasts. ACS Nano 2022, 16: 3506–3521. Using cell-penetrating peptides (CPP) complexed with siRNA, the authors demonstrate efficient gene silencing in plant nuclei and chloroplasts through foliar spray application. This study introduces a new high-throughput method for targeted genetic engineering without the use of transgenes.
- 107.**. Zhang H, Goh NS, Wang JW, Pinals RL, González-Grandío E, Demirer GS, Butrus S, Fakra SC, Del Rio Flores A, Zhai R, et al. : Nanoparticle cellular internalization is not required for RNA delivery to mature plant leaves. Nat Nanotechnol 2022, 17: 197–205. Using AuNPs of various sizes and shapes, the authors demonstrate how nanoparticle size is important for biomolecule delivery in plants, but that efficient cargo delivery is possible even without nanoparticle internalization. This paper lays the foundation for nanomaterial designs that place more importance on facilitating nanomaterial-plant cell wall interactions rather than just internalization.
- 108.Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O’Farrell PH, Andino R: The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol 2006, 8: 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yoon JS, Gurusamy D, Palli SR: Accumulation of dsRNA in endosomes contributes to inefficient RNA interference in the fall armyworm, Spodoptera frugiperda. Insect Biochem Mol Biol 2017, 90:53–60. [DOI] [PubMed] [Google Scholar]
- 110.Worrall EA, Bravo-Cazar A, Nilon AT, Fletcher SJ, Robinson KE, Carr JP, Mitter N: Exogenous application of RNAi-inducing double-stranded RNA inhibits aphid-mediated transmission of a plant virus. Front Plant Sci 2019, 10:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gurusamy D, Mogilicherla K, Palli SR: Chitosan nanoparticles help double-stranded RNA escape from endosomes and improve RNA interference in the fall armyworm, Spodoptera frugiperda. Arch Insect Biochem Physiol 2020, 104, e21678. [DOI] [PubMed] [Google Scholar]
- 112.Kolge H, Kadam K, Galande S, Lanjekar V, Ghormade V: New frontiers in pest control: chitosan nanoparticles-shielded dsRNA as an effective topical RNAi spray for gram podborer biocontrol. ACS Appl Bio Mater 2021, 4:5145–5157. [DOI] [PubMed] [Google Scholar]
- 113.Kolge H, Kadam K, Ghormade V: Chitosan nanocarriers mediated dsRNA delivery in gene silencing for Helicoverpa armigera biocontrol. Pestic Biochem Physiol 2023, 189, 105292. [DOI] [PubMed] [Google Scholar]
- 114.Das S, Debnath N, Cui Y, Unrine J, Palli SR: Chitosan, carbon quantum dot, and silica nanoparticle mediated dsRNA delivery for gene silencing in Aedes aegypti: a comparative analysis. ACS Appl Mater Interfaces 2015, 7:19530–19535. [DOI] [PubMed] [Google Scholar]
- 115.Wang K, Peng Y, Chen J, Peng Y, Wang X, Shen Z, Han Z: Comparison of efficacy of RNAi mediated by various nanoparticles in the rice striped stem borer (Chilo suppressalis). Pestic Biochem Physiol 2020, 165, 104467. [DOI] [PubMed] [Google Scholar]
- 116.Castellanos NL, Smagghe G, Sharma R, Oliveira EE, Christiaens O: Liposome encapsulation and EDTA formulation of dsRNA targeting essential genes increase oral RNAi-caused mortality in the Neotropical stink bug Euschistus heros. Pest Manag Sci 2019, 75:537–548. [DOI] [PubMed] [Google Scholar]
- 117.Gurusamy D, Mogilicherla K, Shukla JN, Palli SR: Lipids help double-stranded RNA in endosomal escape and improve RNA interference in the fall armyworm, Spodoptera frugiperda. Arch Insect Biochem Physiol 2020, 104, e21678. [DOI] [PubMed] [Google Scholar]
- 118.Dhandapani RK, Gurusamy D, Palli SR: Development of catechin, poly- l -lysine, and double-stranded RNA nanoparticles. ACS Appl Bio Mater 2021, 4:4310–4318. [DOI] [PubMed] [Google Scholar]
- 119.Niu L, Yan H, Sun Y, Zhang D, Ma W, Lin Y: Nanoparticle facilitated stacked-dsRNA improves suppression of the Lepidoperan pest Chilo suppresallis. Pestic Biochem Physiol 2022, 187, 105183. [DOI] [PubMed] [Google Scholar]
- 120.Wang Z, Li M, Kong Z, Wang E, Zhang B, Lv J, Xu X: Star polycation mediated dsRNA improves the efficiency of RNA interference in phytoseiulus persimilis. Nanomaterials 2022, 12:3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yao Y, Lin DJ, Cai XY, Wang R, Hou YM, Hu CH, Gao SJ, Wang J Da: Multiple dsRNases involved in exogenous dsRNA degradation of fall armyworm spodoptera frugiperda. Front Physiol 2022, 13, 850022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Christiaens O, Tardajos MG, Reyna ZLM, Dash M, Dubruel P, Smagghe G: Increased RNAi efficacy in Spodoptera exigua via the formulation of dsRNA with guanylated polymers. Front Physiol 2018, 9:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dhandapani RK, Gurusamy D, Howell JL, Palli SR: Development of CS-TPP-dsRNA nanoparticles to enhance RNAi efficiency in the yellow fever mosquito, Aedes aegypti. Sci Rep 2019, 9: 8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang J, Yan S, Xie S, Yin M, Shen J, Li Z, Zhou Y, Duan L: Construction and application of star polycation nanocarrier-based microRNA delivery system in Arabidopsis and maize. J Nanobiotechnol 2022, 20:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.*. Ma Z, Zheng Y, Chao Z, Chen H, Zhang Y, Yin M, Shen J, Yan S: Visualization of the process of a nanocarrier-mediated gene delivery: stabilization, endocytosis and endosomal escape of genes for intracellular spreading. J Nanobiotechnol 2022, 20: 124. This paper reveals how clathrin-mediated endocytosis is activated by star-polycations to enable dsRNA uptake and transmission from the SPc nanocarrier in insects. It is one of the first studies to provide direct visualization of the cellular delivery process and insight into the mechanisms of nanocarrier-mediated RNAi, specifically how nanocarriers upregulate expression of genes involved in CME.
- 126.Mosa MA, Youssef K: Topical delivery of host induced RNAi silencing by layered double hydroxide nanosheets: an efficient tool to decipher pathogenicity gene function of Fusarium crown and root rot in tomato. Physiol Mol Plant Pathol 2021, 115, 101684. [Google Scholar]
- 127.**. Wang Z, Li Y, Zhang B, Gao X, Shi M, Zhang S, Zhong S, Zheng Y, Liu X: Functionalized carbon dot-delivered RNA nano fungicides as superior tools to control Phytophthora pathogens through plant RdRP1 mediated spray-induced gene silencing. Adv Funct Mater 2023, 33, 2213143. In this study, the authors demonstrate how using carbon dots (CDs) can extend SIGS to organisms such as Phytophthora, which exhibit limited dsRNA uptake. They find that complexing dsRNA with CDs improves dsRNA internalization and stability to drastically lower the amount of fungicide needed for pathogen control. This paper is one of the first to demonstrate how nanoparticles can be used to facilitate SIGS in pathogens that were previously recalcitrant to SIGS efforts using naked dsRNA.
- 128.Wang Y, Yan Q, Lan C, Tang T, Wang K, Shen J, Niu D: Nanoparticle carriers enhance RNA stability and uptake efficiency and prolong the protection against Rhizoctonia solani. Phytopathol Res 2023, 5:2. [Google Scholar]
- 129.Duanis-Assaf D, Shlar I, Galsurker O, Davydov O, Maurer D, Feygenberg O, Poverenov E, Fluhr R, Alkan N: Nano-clay, layered-double hydroxide (LDH), improves the efficacy of double-stranded RNA in controlling postharvest decay. Postharvest Biol Technol 2022, 193, 112051. [Google Scholar]
- 130.Malandrakis AA, Kavroulakis N, Chrysikopoulos CV: Synergy between Cu-NPs and fungicides against Botrytis cinerea. Sci Total Environ 2020, 703, 135557. [DOI] [PubMed] [Google Scholar]
- 131.Luksiene Z, Rasiukeviciute N, Zudyte B, Uselis N: Innovative approach to sunlight activated biofungicides for strawberry crop protection: ZnO nanoparticles. J Photochem Photobiol B Biol 2020, 203, 111656. [DOI] [PubMed] [Google Scholar]
- 132.El-Gazzar N, Ismail AM: The potential use of Titanium, Silver and Selenium nanoparticles in controlling leaf blight of tomato caused by Alternaria alternata. Biocatal Agric Biotechnol 2020, 27, 101708. [Google Scholar]
- 133.Nami S, Baradaran B, Mansoori B, Kordbacheh P, Rezaie S, Falahati M, Khosroshahi LM, Safara M, Zaini F: The utilization of RNA silencing technology to mitigate the voriconazole resistance of aspergillus flavus; lipofectamine-based delivery. Adv Pharmaceut Bull 2017, 7:53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.**. Avital A, Muzika NS, Persky Z, Bar G, Michaeli Y, Fridman Y, Karny A, Shklover J, Shainsky J, Savaldi-Goldstein S, et al. : Foliar delivery of siRNA particles for treating viral infections in agricultural grapevines. Adv Funct Mater 2021, 31, 2101003. In this paper, the authors demonstrate how foliar application of lipid-modified polyethylenimine (lmPEI)-dsRNA complexes can lower viral titers and trigger recovery of infected vines and berries. This study is one of the first to demonstrate systemic treatment of an ongoing viral infection in plants.
- 135.Liu L, Chen X: Intercellular and systemic trafficking of RNAs in plants. Nat Plants 2018, 4:869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.**. Hu P, An J, Faulkner MM, Wu H, Li Z, Tian X, Giraldo JP: Nanoparticle charge and size control foliar delivery efficiency to plant cells and organelles. ACS Nano 2020, 14:7970–7986. The authors systemically investigate the contribution of different nanoparticle on cellular uptake and delivery to different plant tissues. They find that small, hydrophilic, and positively-charged nanoparticles exhibit the highest delivery efficiencies. This paper provides an excellent example of an effort to understand plant nano-kinetics.
- 137.Yong J, Wu M, Zhang R, Bi S, Mann CWG, Mitter N, Carroll BJ, Xu ZP: Clay nanoparticles efficiently deliver small interfering RNA to intact plant leaf cells. Plant Physiol 2022, 190: 2187–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schwartz SH, Hendrix B, Hoffer P, Sanders RA, Zheng W: Carbon dots for efficient small interfering RNA delivery and gene silencing in plants. Plant Physiol 2020, 184:647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang Y, Fu L, Li S, Yan J, Sun M, Giraldo JP, Matyjaszewski K, Tilton RD, Lowry GV: Star polymer size, charge content, and hydrophobicity affect their leaf uptake and translocation in plants. Environ Sci Technol 2021, 55:10758–10768. [DOI] [PubMed] [Google Scholar]
- 140.Zhang H, Demirer GS, Zhang H, Ye T, Goh NS, Aditham AJ, Cunningham FJ, Fan C, Landry MP: DNA nanostructures co-ordinate gene silencing in mature plants. Proc Natl Acad Sci U S A 2019, 116:7543–7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lichtenberg SS, Nuti K, DeRouchey J, Tsyusko OV, Unrine JM: Efficacy of chitosan/double-stranded RNA polyplex nanoparticles for gene silencing under variable environmental conditions. Environ Sci Nano 2020, 7:1582–1592. [Google Scholar]
- 142.*. Beckers SJ, Staal AHJ, Rosenauer C, Srinivas M, Landfester K, Wurm FR: Targeted drug delivery for sustainable crop protection: transport and stability of polymeric nanocarriers in plants. Adv Sci 2021, 8, 2100067. Using 19F-magnetic resonance imaging or induced coupled plasma – optical emission spectroscopy, the authors track the distribution and fate of polymeric nanoparticles after trunk injection. This paper illustrates how these techniques can be used to clearly address questions regarding nanoparticle translocation and distribution in plants.
- 143.Necira K, Makki M, Sanz-García E, Canto T, Djilani-Khouadja F, Tenllado F: Topical application of escherichia coli-encapsulated dsrna induces resistance in nicotiana benthamiana to potato viruses and involves rdr6 and combined activities of dcl2 and dcl4. Plants 2021, 10:644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Niño-Sánchez J, Chen LH, De Souza JT, Mosquera S, Stergiopoulos I: Targeted delivery of gene silencing in fungi using genetically engineered bacteria. J Fungi 2021, 7:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Islam MT, Davis Z, Chen L, Englaender J, Zomorodi S, Frank J, Bartlett K, Somers E, Carballo Sm, Kester M, et al. : Minicell-based fungal RNAi delivery for sustainable crop protection. Microb Biotechnol 2021, 14:1847–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Whitten MMA, Facey PD, del Sol R, Fernández-Martínez LT, Evans MC, Mitchell JJ, Bodger OG, Dyson PJ: Symbiont-mediated RNA interference in insects. Proc R Soc B Biol Sci 2016, 283, 20160042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Leonard SP, Powell JE, Perutka J, Geng P, Heckmann LC, Horak RD, Davies BW, Ellington AD, Barrick JE, Moran NA: Engineered symbionts activate honey bee immunity and limit pathogens. Science 2020, 367:573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.*. Elston KM, Maeda GP, Perreau J, Barrick JE: Addressing the challenges of symbiont-mediated RNAi in aphids. PeerJ 2023, 11, e14961. The authors seek to apply symbiont-mediated RNAi for use in aphids using two different bacterial strains. Despite logical and thorough experimental designs, they failed to observe any reproducible gene silencing phenotypes. This study does an excellent job of highlighting critical considerations for smRNAi efficacy that demonstrate how RNAi is not a one size fits all technique.
- 149.Cui C, Wang Y, Li Y, Sun P, Jiang J, Zhou H, Liu J, Wang S: Expression of mosquito miRNAs in entomopathogenic fungus induces pathogen-mediated host RNA interference and increases fungal efficacy. Cell Rep 2022, 41, 111527. [DOI] [PubMed] [Google Scholar]
- 150.Kah M, Johnston LJ, Kookana RS, Bruce W, Haase A, Ritz V, Dinglasan J, Doak S, Garelick H, Gubala V: Comprehensive framework for human health risk assessment of nanopesticides. Nat Nanotechnol 2021, 16:955–964. [DOI] [PubMed] [Google Scholar]
- 151.Williams RJ, Harrison S, Keller V, Kuenen J, Lofts S, Praetorius A, Svendsen C, Vermeulen LC, van Wijnen J: Models for assessing engineered nanomaterial fate and behaviour in the aquatic environment. Curr Opin Environ Sustain 2019, 36:105–115. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.