Abstract
The level of the vitamin B12 transport protein BtuB in the outer membrane of Escherichia coli is strongly reduced by growth in the presence of cobalamins. Previous analyses of regulatory mutants and of btuB-lacZ fusions indicated that the primary site of btuB gene regulation was at the translational level, and this required sequences throughout the 240-nucleotide (nt) leader region. Cobalamin-dependent regulation of transcriptional fusions was of a lesser magnitude but required, in addition to the leader, sequences within the first 100 nt of the coding sequence, termed the translated regulatory region (TRR). To analyze the process of transcription-level regulation of btuB in E. coli, the levels and metabolism of btuB RNA were analyzed by S1 nuclease protection assays, and mutations that alter the coupling of translational and transcriptional control were analyzed. Expression of transcriptional fusions was found to correlate with changes in the level of intact btuB RNA and was related to changes in the metabolic stability of the normally long-lived RNA. Mutational analysis showed that the btuB start codon and a hairpin structure that can sequester the Shine-Dalgarno sequence are necessary for cobalamin-dependent regulation and that translation of the TRR is necessary for extended RNA stability and for expression of the transcriptional fusion. The absence of regulation at the stage of transcription initiation was confirmed by the findings that several truncated btuB RNA fragments were expressed in a constitutive manner and that the normal regulatory response occurred even when the btuB promoter and upstream sequences were replaced by the heterologous bla and lac promoters. Transcription driven by phage T7 RNA polymerase was not regulated by cobalamins, although some regulation at the translational level was retained. Cobalamin-dependent changes in RNA structure were suggested from the RNase III-dependent production of a transcript fragment that is made only in the presence of cobalamin and is independent of the regulatory outcome. These results indicate that the primary control of btuB expression by cobalamin occurs at the level of translation initiation, which directly affects the level and stability of btuB RNA in a process that requires the presence of the intact translated regulatory region.
Expression of the btuB genes of Escherichia coli and Salmonella enterica serovar Typhimurium, encoding the outer membrane cobalamin (Cbl) transporter BtuB, and of the cob operon of S. typhimurium, encoding the biosynthetic pathway for Cbls, is markedly depressed during growth in the presence of Cbls, such as vitamin B12 (CN-Cbl) (6, 9, 25). The properties of this Cbl-dependent regulatory system deduced from the expression of btuB-lacZ fusions suggest the operation of a novel mode of gene regulation. The regulatory system responds to the intracellular level of 5′-deoxyadenosyl-Cbl (Ado-Cbl), which is formed from CN-Cbl by the action of the btuR or cobA gene product (13, 20). Unlike control systems that depend on repressor action, Cbl-mediated regulation is independent of gene copy number and affects expression of btuB-lacZ fusions at both the transcriptional and translational levels. The modulation of translational fusions by Cbl is at least 25-fold, while that of transcriptional fusions is modest: about 5-fold in E. coli (14) and even less in Salmonella typhimurium (17). The transcripts of the btuB gene or the cbiA gene, which is the first gene of the large cob operon, contain a 241- or 468-nucleotide (nt) leader before the start of the respective coding region. Regulatory mutations selected for increased expression under repressive conditions occur at numerous sites throughout these leader regions (14, 18, 19). No candidate regulatory proteins have been identified, since all unlinked mutations yet obtained that confer altered regulation affect either entry of CN-Cbl into the cell or its conversion to Ado-Cbl (6a, 20).
Analysis of transcriptional and translational fusions carrying various portions of the respective regulatory regions has suggested the involvement of separate elements in different stages of Cbl-dependent control. The primary site of regulation appears to be at translation initiation and requires the integrity of most or all of the leader region. Cbl-dependent control of transcriptional lac fusions requires the integrity of both the leader region and the initial part of the btuB coding region out to around residue +350 (7, 14, 17). This translated regulatory region (TRR), between nt 241 and 350, includes a putative Rho-independent terminator, a G+C-rich stem and loop followed by a series of U residues, whose integrity is essential for transcriptional control (7). Unlike most attenuators, candidate terminators for btuB and cbiA lie past the site of translation initiation, in the early portion of the protein-coding sequences (11). Control of translational fusions occurs in the absence of the translated btuB sequences and appears to be moderated by formation of an RNA hairpin that includes or is near the Shine-Dalgarno sequence (17, 19). Expression of btuB requires the presence of the B12 box, a 17-nt element that is conserved in the leaders of the btuB and cbiA genes (18). Precise deletion of the B12 box sequences resulted in a very low and unregulated level of btuB expression (7).
Although the extent of regulation by Cbl of btuB-lacZ transcriptional fusions in S. typhimurium is slight (17), an appreciable level of control of btuB in E. coli and of cob in S. typhimurium occurs (14, 18). This control depends on the presence of each respective TRR, which has been suggested to function as an attenuator, although no evidence for this proposal was obtained. To investigate the complex regulation seen with gene fusions, we analyzed the changes in btuB RNA levels detected by S1 nuclease protection assays under a variety of conditions that affect the frequency of btuB translation or transcription initiation or RNA stability. Prompted by the finding that alteration of translation dramatically affected expression of transcriptional fusions and RNA levels and stability, we concluded that the TRR is specifically responsible for changes in btuB RNA levels, rather than that this apparent regulation is a nonspecific reflection of the lability of untranslated RNA. We propose that the negative action of the TRR is prevented by the passage of ribosomes through it, which is modulated by the primary site of regulation at the stage of ribosome binding.
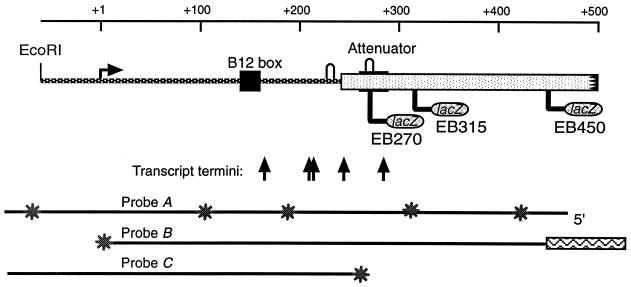
The btuB regulatory region, with the location of the DNA hybridization probes and the sites of reporter fusions used in this study, is schematically shown in Fig. 1.
FIG. 1.
Schematic representation of btuB regulatory region. The DNA regions carried in the plasmids used in this study are shown under the coordinate numbers relative to the start of transcription. The locations of the BamHI sites used as junctions of fusion to lacZ in the three fusions are indicated. The locations of the B12 box, conserved in Cbl-dependent promoters, the potential stem-loop structures in the transcript that comprise the Shine-Dalgarno sequence and its complement, and the attenuator are marked. The vertical arrows indicate the locations of the 3′ termini of the partial-length transcript fragments. On the bottom are indicated the extent of the three probes used in this study, aligned with the 5′ end on the right; the position of the label is indicated by the symbols.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains, plasmids, and oligonucleotide primers used in this study are listed in Tables 1 and 2. In most experiments, the host strain was RK5173, a metE derivative of MC4100. Strain JM109 was used as the host of plasmids pXN26 and pXN27, which were very unstable in RK5173. For plasmids carrying btuB-lacZ fusions transcribed from the T7 late promoter, the host strain was RK3515, a btuB+ transductant of BL21(DE3), in which production of T7 RNA polymerase is induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were routinely grown in Luria-Bertani medium. Ampicillin (100 μg/ml) was added to maintain selection for plasmids. When indicated, Ado-Cbl was added to a concentration of 1 μM to establish repressive conditions. Unless indicated, cells were grown at 37°C with vigorous aeration.
TABLE 1.
List of bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| CA244 | lacZ trp relA spoT | 10 |
| JM109 | Promega | |
| MC1061 | F−araD139 galU galK ΔlacX74 Δ(ara-leu)7697 rpsL hsdR | J. Beckwith |
| MG1655 | Wild type | 24 |
| MG1693 | F−thyA715λ− | 2 |
| RK3513 | BL21(DE3) btuB+ zij::Tn10 | This study |
| RK5173 | Δ(argF-lac)U169 araD139 fib5301 ptsF25 relA1 rpsL150 rbsR22 deoC1 gyrA219 non-9 metE70 | 13 |
| SK5665 | MG1693 rne-1 | 2 |
| SK6867 | MC1061 rnc-105 Tcr | 2 |
| SK7621 | MC1061 Δrnc38::Km | 2 |
| SK7669 | MG1693 rne-3071 | 2 |
| TX2817 | MG1655 hfq2::ΩKm | 24 |
| Plasmids | ||
| pAG1 | pBR322 btuB+; Ap | |
| pRS414 | Translational lac fusion vector; Ap | 22 |
| pRS415 | Transcriptional lac fusion vector; Ap | 22 |
| pEB450X | pRS415 with −60 to +450 of btuB-lacZ; Ap | 7 |
| pEB450L | pRS414 with −60 to +450 of btuB-lacZ; Ap | 7 |
| pEB315X | pRS415 with −60 to +315 of btuB-lacZ; Ap | 7 |
| pEB270X | pRS415 with −60 to +270 of btuB-lacZ; Ap | 7 |
| pXN5 | pBluescript SK+ with +1 to +450 of btuB; Ap | This study |
| pXN17 | pRS415 with T7P, +1 to +450 of btuB-lacZ; Ap | This study |
| pXN18 | pRS414 with T7P, +1 to +450 of btuB-lacZ; Ap | This study |
| pXN24 | pRS415 with lacP, +1 to +450 of btuB-lacZ; Ap | This study |
| pXN25 | pRS414 with lacP, +1 to +450 of btuB-lacZ; Ap | This study |
| pXN26 | pRS415 with blaP, +1 to +450 of btuB-lacZ; Ap | This study |
| pXN27 | pRS414 with blaP, +1 to +450 of btuB-lacZ; Ap | This study |
TABLE 2.
Sequence of primers used in this study
| Primer | Sequence |
|---|---|
| T7btu | CTGTAGCATCCACTTGAATTCTAATACGACTCACTATAGCCGGTCCTGTGAGTTAATAGG |
| XNO-2 | CCGTCAGCAGGGAAGCTTTT |
| XNO-4 | CCGCTTTCATCGGTTGTCC |
| XNO-9 | AAACTCTCACGGATCTTACC |
| XNO-10 | AAACGGCCGGTCCTGTGAGTTAAT |
| XNO-14 | AAGAATTCGGCGCCTTCCTGTTTTTGCTCA |
| XNO-15 | TTGTGAGCGGATAACAATTTC |
| XNO-16 | AAAGGATCCCCGTTTTGGGTGATATCG |
| XNO-26 | GTAATATTGATGAAACCTTCGCGATCCTTCTTCTATTG |
| XNO-27 | CTTCTTCTATTGTGGATCGATTACAATGATTAAAAAAGC |
| XNO-43 | CAGCGAAGCTTTTTTAATCGATGTAAAGCA |
| XNO-44 | CAATGATTAAAAAAGCTTAGCTGCTGACGGCGT |
| XNO-45 | CAGCGAAGCTTTTATTAATCATTGTAATGC |
| XNO-47 | GATACTCTCGTCGTACTGCTAACCGTTTTG |
| XNO-49 | AAGAATTCAGCAGGCACGACAG |
| XNO-50 | AAGAATTCAGTACAATCTGCT |
| XNO-51 | CGTATGTTGTGTGGGCCGGTCCTGTGAG |
| XNO-52 | GAGACAATAACCCTGCCGGTCCTGTGAG |
| XNO-57 | AAAAAAGTCTCGCGCTGATGGCGT |
All plasmids used in this study are derived from pRS414 or pRS415, which allow formation of translation or transcription fusions to lacZ, respectively (22). Inserts were introduced as EcoRI-BamHI fragments. The insert’s EcoRI site occurs at −60 in the wild-type btuB sequence, and the BamHI sites were introduced as linker insertions or substitutions, as previously described (7). The BamHI site at each fusion junction occurs in the same translational reading frame. In the translational fusions, the btuB coding sequences are coupled to the 9th codon of lacZ (22). In the transcriptional fusions, the lacZ gene is separated from the btuB junction by about 100 bp of trp-lac DNA and possesses its own ribosome-binding site. Construction of the plasmids carrying btuB-lacZ fusions was performed as described previously (7).
Construction of btuB mutants and fusions.
All nucleotide coordinates are numbered relative to the btuB transcription start site. Plasmid DNAs were purified and introduced into host cells by standard protocols (21). All restriction endonucleases, DNA polymerases, and DNA ligase were used according to the recommendations of the manufacturer.
Mutations were introduced into the btuB leader region by a one-step or two-step PCR process. The first step used mutagenic oligonucleotides (XNO-26, XNO-27, or XNO-47) and oligonucleotide XNO-4, which anneals downstream of the BamHI site in plasmid pRS415, as primers and plasmid pEB450X DNA as a template. This product was purified by gel electrophoresis and used as an adapter in the second PCR step. In this step, oligonucleotide XNO-15, which anneals to plasmid pAG1 DNA at vector sequences upstream of the EcoRI site, and oligonucleotide XNO-4 were used as primers, and plasmid pAG1 was used as a template, in the presence of the product of the first PCR. Thus, the product of the first PCR step, which contains the desired mutation, was further extended and amplified, resulting in a DNA fragment containing EcoRI and BamHI sites and the desired mutation. The EcoRI-BamHI fragment was generated, purified, and ligated into the corresponding sites of pRS414 and pRS415, and the presence of the expected mutation was verified by sequencing. Mutants M43, M44, M45, and M57 were constructed by one-step PCR mutagenesis and cloned into pEB450 plasmids as EcoRI-HindIII or HindIII-BamHI fragments.
The two-step PCR scheme was also used to construct btuB-lacZ fusions under control of heterologous promoters. Oligonucleotides XNO-51 for lac and XNO-52 for bla are composed of the 14 nt upstream of the site of transcription initiation of the appropriate promoter followed by the first 14 nt of btuB transcribed sequences. These primers were used with oligonucleotide XNO-4 as the primer and plasmid pEB450X DNA as the template in the first-step PCR. In the second step, oligonucleotides XNO-49 for lac and XNO-50 for bla, which anneal upstream of the appropriate promoter region and contain an EcoRI site, were used along with oligonucleotide XNO-4 as primers and pGEM-3Z DNA (Promega) as a template, in the presence of the first PCR product as an adapter. The resulting EcoRI-BamHI fragment was subcloned as described above. To construct the btuB-lacZ fusions under control of the T7 promoter, oligonucleotide T7-btu, which contains an EcoRI site and 16 nt of the T7 consensus sequence fused to the first 22 nt of btuB-transcribed sequences, was used in PCR with XNO-4 as the primer and pEB450X DNA as the template, and the resulting EcoRI-BamHI fragment was subcloned as described above.
RNA extraction and S1 nuclease protection analysis.
Total cell RNA was prepared by the hot phenol extraction method described by Emory and Belasco (5), as modified. About 109 logarithmic-phase cells were collected by centrifugation for 20 s at 4°C, and the cell pellet was immediately frozen until all samples for the experiment were collected. The cells were suspended in 450 μl of lysis buffer (150 mM sucrose, 10 mM sodium acetate [pH 4.5]) on ice and mixed with 50 μl of 10% sodium dodecyl sulfate (SDS). Samples were heated at 70°C for 3 min and then extracted three times with phenol at 70°C. Nucleic acids were precipitated by addition of 1/10 volume of sodium acetate and 2 volumes of ethanol. The sample was air dried and suspended in 100 μl of DNase buffer (20 mM sodium acetate [pH 4.5], 20 mM NaCl, 10 mM MgCl2) and treated with 1 to 2 U of DNase I for 30 min at 25°C. This digestion was followed by phenol extraction and ethanol precipitation, and the product was suspended in 15 μl of water and stored at −20°C.
Probe A was synthesized by PCR with primers XNO-15 and XNO-16 and with plasmid pAG1 DNA as the template. This product extends from plasmid vector sequences upstream of the site of insertion of the btuB-lacZ fusion to position +470 of the wild-type btuB sequence. Probe A was internally labeled by asymmetric PCR with primer XNO-16, in a labeling reaction mixture containing 3 μM [α-32P]dCTP, 25 μM unlabeled dCTP, and 125 μM other deoxynucleoside triphosphates (dNTPs). Probe B is a restriction fragment generated by digestion of plasmid pXN5 with EagI and PvuII. In plasmid pXN5, an EagI restriction site (C′GGCCG) replaced the sequence at the start of btuB transcription (TTGCCG) by PCR mutagenesis. A fill-in reaction catalyzed by the Klenow fragment of DNA polymerase I with [α-32P]dCTP and dGTP resulted in labeling of the 3′ end of the DNA strand complementary to btuB RNA, and the position of the labeled nucleotide hybridized to the transcript position at +1. Probe C was synthesized by PCR with primers XNO-15 and 5′-labeled XNO-2 with plasmid pAG1 DNA as template. This product extends from upstream vector sequences to position +269. The PCR primers were labeled at their 5′ ends with [γ-32P]ATP and polynucleotide kinase. The bla-specific fragment was synthesized and labeled by PCR with XNO-14 and 5′-labeled XNO-9 as primers and plasmid pAG1 DNA as a template.
For the S1 nuclease protection assay, modified from that described by Emory and Belasco (5), 1 μl of extracted RNA was mixed with 16 μl of formamide and 2 μl of hybridization buffer (400 mM PIPES [pH 6.4], 400 mM NaCl, 10 mM EDTA) along with the indicated 32P-labeled DNA probes in a volume of 1 μl. This mixture was heated to 80°C for 4 min and then held at 50°C for 4 to 16 h. To the sample was added 300 μl of S1 buffer (280 mM NaCl, 50 mM sodium acetate [pH 4.5], 4.5 mM ZnSO4) containing 5 μg of denatured herring sperm DNA and 40 U of S1 nuclease. Following incubation for 90 min at 37°C, the reaction was stopped by addition of 80 μl of stop buffer (4 M ammonium acetate, 20 mM EDTA), and the DNA was precipitated with 5 μg of yeast tRNA and 1 ml of ethanol. The precipitate was suspended in sequencing gel loading buffer, resolved on a 6% polyacrylamide sequencing gel, and analyzed with a Molecular Dynamics PhosphorImager.
β-Galactosidase assays.
For assays of β-galactosidase expression, cells were grown in minimal salts medium A with 0.5% glycerol and 0.5% casein hydrolysate. The level of β-galactosidase was determined as previously described (7), by measuring the rate of hydrolysis of 2 mM o-nitrophenyl-β-d-galactopyranoside in cells permeabilized with SDS-CHCl3. All assays were performed in triplicate, and most were repeated.
RESULTS
Hairpin structure at the ribosome-binding site affects regulation.
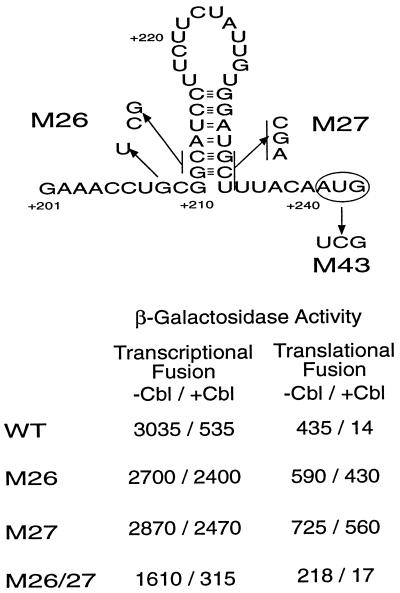
Previous work concluded that translational control requires the presence of sequences up to the start of translation, whereas transcriptional control requires, in addition, 60 to 100 nt of the TRR. We examined the involvement in btuB expression of a potential hairpin structure formed by the Shine-Dalgarno sequence and a complementary sequence 12 bp upstream (Fig. 2). Mutations were introduced that alter each stem of this hairpin. In mutant M26, the complementary sequence was changed from GCATCC to CGATCC (an additional G-to-T substitution at +208 created an NruI site to facilitate mutant identification). Mutant M27 changed the Shine-Dalgarno sequence GGATGCT to GGATCGA, which has little effect on the match to the consensus. The double mutant M26/27 combined both sets of changes and restored the capacity for pairing of the Shine-Dalgarno sequence and its upstream complement. The effect of these changes was measured in EB450 transcriptional and translational btuB-lacZ fusions carrying the btuB promoter followed by 450 nt of transcribed sequences before fusion to the lacZ reporters (Fig. 1).
FIG. 2.
Hairpin structure comprising the Shine-Dalgarno sequence and its upstream complement and the mutations that affect either the stem or the initiation codon. The translation initiation codon is indicated in the oval. The base changes introduced by the mutations are indicated. The units of β-galactosidase activities of transcriptional and translational EB450 fusions are presented. WT, wild type.
Translational fusions expressing the M26 or M27 substitutions showed a slight increase in expression but almost complete loss of Cbl-dependent regulation (Fig. 2). The 1.4- to 1.7-fold increase in expression may reflect increased access of the ribosome-binding site because of its decreased sequestration by the upstream complement. The combination of both substitutions in the compensatory M26/27 mutant resulted in restoration of the wild-type degree of regulation, indicating that the pairing of these two sequences must occur as part of the process of translational control, as had been previously proposed from similar studies with the S. typhimurium btuB and cbiA genes (17, 19).
Strikingly, the M26 and M27 substitutions resulted in almost complete loss of Cbl-dependent regulation of the transcriptional fusion, although they had no effect on the level of expression (Fig. 2). The compensatory M26/27 combination restored wild-type regulation. Using the S1 nuclease protection assay described below, it was seen that the relative levels of btuB RNA paralleled the changes in reporter activity. Thus, the ability to form the hairpin that includes the ribosome-binding site is crucial for both translational and transcriptional regulation. This apparent coordination of translational and transcriptional activity could reflect the participation of this hairpin in formation of an RNA conformation that leads to RNA attenuation or cleavage, to the increased lability of untranslated RNA, or to a specific effect of translation on the activity of the TRR.
The extent of translated sequences affects transcription.
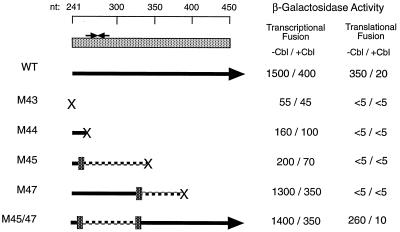
To test whether btuB translation affects transcription, the AUG start codon was converted to UCG in the M43 substitution, and assayed for effect on expression in EB450 transcriptional and translational fusions. As expected, this substitution resulted in complete loss of expression of the translational fusion. Surprisingly, there was a drastic reduction in expression of the transcriptional fusion to about 2% of the wild-type level (Fig. 3). The amount of full-length btuB RNA was also greatly reduced but was decreased further by Ado-Cbl (data not shown).
FIG. 3.
Effect of mutations that affect btuB translation on regulation of btuB-lacZ fusions. The initial portion of the btuB coding region is shown at the top, along with nucleotide coordinates. The paired arrows show the location of the putative transcription terminator. Symbols indicate the consequences of the mutations, as follows: X, translation termination or blockage; vertical bar, frameshift mutation; solid line, wild-type BtuB amino acid sequence; dashed line, frameshifted amino acid sequence. The expression of units of β-galactosidase activity from EB450 transcriptional and translational fusions in cells grown in the absence or presence of Ado-Cbl is presented for each mutant.
To test whether the decrease in transcription required the presence of the TRR, the M43 mutation was placed in the context of the EB270 transcriptional fusion. This fusion contains the btuB promoter and transcribed region to +270. It is deleted for most of the TRR and exhibits high-level constitutive expression, but almost fully repressible behavior as a translational fusion (repression ratio of 13) (7). Expression from this EB270-M43 fusion was comparable to that of the wild-type EB270 fusion and was increased by the presence of Ado-Cbl (1,700 U without Ado-Cbl and 2,900 U with Ado-Cbl). These results showed that the decreased transcription activity that occurred in the EB450 fusion upon blockage of translation depends on the integrity of the translated regulatory region, suggesting that translation of the TRR might overcome its negative effect on transcription.
The ability of translating or stalled ribosomes to affect attenuator function (reviewed in reference 11) or endonucleolytic digestion of RNA (3, 16) is well known. To test whether the extent of translation or the presence of specific polypeptide sequences in the translated product affected transcriptional activity, several translation termination mutations were introduced early in the btuB coding sequence and analyzed in EB450 transcriptional and translational fusions (Fig. 3). The potential attenuator element in the btuB TRR extends from codon 7 to codon 17 (i.e., nt +260 to +290). Mutation M44 introduced a chain-terminating UAG sequence at codon 6. It eliminated expression of the translational fusion and strongly reduced both expression and regulation of the transcriptional fusion. Thus, ribosome attachment and initiation of translation are not sufficient to alleviate the inhibition of transcription.
Mutation M45 introduced a +1 frameshift at codon 3, resulting in a change in the distal reading frame until termination occurs at codon 33. This mutant showed reduced transcriptional activity, but a substantial degree of Cbl-mediated regulation (repression ratio of around 3) (Fig. 3). Mutation M47 introduced a −1 frameshift at codon 30, resulting in a change in the amino acid sequence until termination occurs at codon 49. This mutant showed almost wild-type levels of transcriptional expression and regulation, even though the translational fusion was negative. Thus, loss of ribosome movement past codon 49 of btuB does not affect transcriptional activity. The M45/47 combination allows translation of the entire btuB gene, although the amino acid sequence from codon 3 to codon 30 is changed by the frameshift. This combination gave wild-type levels of expression and regulation of the transcriptional fusion and nearly wild-type expression of the translational fusion, showing that the sequence of the translated product is not a factor in determining the level and regulation of transcription. Similar results were obtained with a −1 frameshift at codon 7, which terminates translation at codon 8. Its low and unregulated transcriptional expression was fully restored by combination with M45 (+1 shift at codon 3). These results show that transcriptional activity is controlled by translation through the TRR between positions 240 and 330, rather than through formation of transcript secondary structures or the general lability of untranslated RNA.
Differential regulation of btuB RNA fragments.
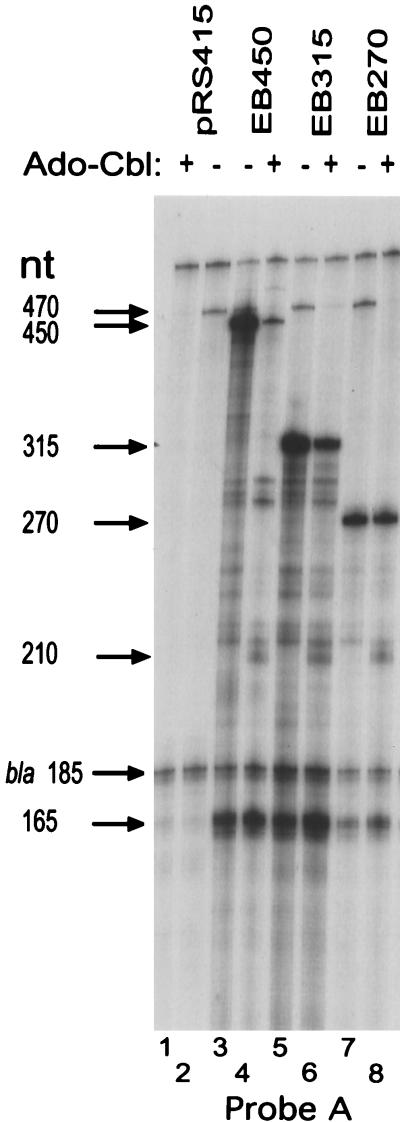
The transcripts produced from Cbl-regulated genes have not been previously described. S1 nuclease protection was used to detect changes in btuB RNA levels, length, or stability. Three hybridization probes were used to identify the origin of RNA transcribed from three btuB-lacZ fusions, designated EB450, EB315, and EB270 (Fig. 1). These fusions contain the btuB promoter region and 450, 315, or 270 nt, respectively, of btuB-transcribed sequence joined at an introduced BamHI site as a transcriptional fusion to trp-lacZYA in the plasmid vector pRS415 (7). These inserts exhibit comparable degrees of Cbl-dependent regulation when present as translational fusions, with repression ratios of 19, 21, and 13, respectively. In transcriptional fusions, they exhibit repressible, partially repressible, or constitutive behavior, respectively, attributed to the progressive deletion of portions of the TRR in the two shorter inserts. Examination of these nested fusions with different regulatory behaviors allowed the simplest test of whether changes in the patterns of transcripts are associated with regulation. Expression from plasmid-borne btuB-lacZ fusions was used to increase the amount of btuB RNA. Elevated gene copy number does not appear to affect btuB regulation by Cbls (1, 25). The transcript of the chromosomal btuB allele was detectable and showed changes similar to those of the plasmid-borne EB450 fusion. RNA was extracted from cells grown in the absence and presence of 1 μM Ado-Cbl, hybridized to one of the three btuB probes and a bla DNA probe, subjected to S1 nuclease digestion, resolved by electrophoresis, and detected by autoradiography or phosphorimaging. The bla transcript served as a control for RNA loading.
Hybridization to probe A, extending from upstream vector sequences to +470 of btuB sequence and internally labeled by asymmetric PCR synthesis, allowed detection of all btuB-derived RNA species. Cells carrying the vector plasmid, pRS415, yielded the 470-nt product derived from the chromosomal btuB allele; its levels were strongly depressed by growth with Ado-Cbl (Fig. 4, lanes 1 and 2). Each of the btuB-lacZ fusion templates gave rise to multiple RNA species with different regulatory responses (lanes 3 to 8). The major transcripts were the expected full-length species of 450, 315, and 270 nt, respectively. The amount of the 450-nt species expressed from EB450 was strongly reduced during growth with Ado-Cbl, whereas the 315-nt species from EB315 was less strongly decreased, and the 270-nt species from EB270 showed only a slight change. Quantification of band intensities showed that the repression ratios for the full-length species (amount in absence of Ado-Cbl/amount in presence of Ado-Cbl; normalized to the amount of the 185-nt bla transcript fragment) from EB450, EB315, and EB270 were 4.6, 1.7, and 1.3, respectively. These values agreed with the repression ratios for β-galactosidase expressed from these transcriptional fusions: 4.9, 2.0, and 1.1, respectively (7). This correspondence indicates that the expression from btuB-lacZ transcriptional fusions is a valid measure of btuB RNA levels.
FIG. 4.
S1 nuclease protection analysis of transcripts expressed from three btuB-lacZ fusions. RNA was extracted from cells of strain RK5173 carrying the plasmids with the indicated btuB-lacZ fusions. Cells were grown as indicated in the absence or presence of 1 μM Ado-Cbl and extracted RNA was hybridized with 32P-labeled probe A. In all assays, a constant amount of labeled probe complementary to the bla gene transcript was included. The sizes of the protected DNA fragments are indicated.
The other major btuB RNA species were a group of fragments ca. 165 nt long. High-resolution electrophoresis of these fragments next to a sequencing ladder showed that these fragments were 162 to 167 nt in length (data not shown). The amount of these fragments did not change significantly under repressive conditions, indicating that the btuB promoter is expressed in a constitutive manner. Several RNA species were present in smaller amounts. Products migrating between 280 and 300 nt and at 215 nt were constitutively produced. Fragments of 240 to 250 nt were depressed by Ado-Cbl, and a 210-nt product was present only in repressed cells. The formation of the 210-nt fragment in the presence of Ado-Cbl occurred with all three btuB-lacZ fusions, indicating that its production is not directly correlated with transcriptional activity. Products smaller than 150 nt were not detected.
The origin of the sequences represented by these fragments was determined by hybridization to two other probes (Fig. 1). Probe B was labeled on its 3′ end at the residue complementary to +1 of the btuB transcript, and thus its label is protected only by btuB transcripts that contain the authentic 5′ end. All RNA species detected by probe B were identical to those detected with the internally labeled probe A (data not shown). Probe C was labeled on its 5′ end at the position complementary to nt +269 and can detect btuB RNA species that extend beyond +269, including any that might be generated by endonucleolytic cleavage. The only significant protected btuB species was the 269-nt fragment representing the full-length product of the three templates (data not shown). These results showed that all of the RNA fragments detected with probe A are sense-strand btuB products and start at nucleotide +1. The truncated fragments could result from transcription termination or endonucleolytic cleavage, but if formed by nuclease action, the distal fragment must be much more labile than the promoter-proximal fragment.
Kinetics of repression of btuB RNA levels.
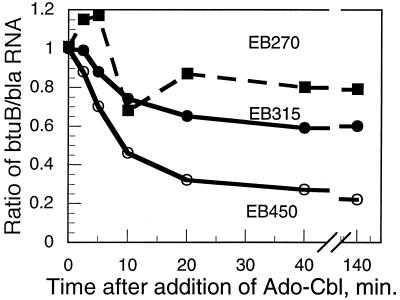
The S1 nuclease protection assay was used to determine the rate of change in the levels of the btuB transcripts following addition of 1 μM Ado-Cbl. The ratio of the full-length btuB transcript to the bla transcript was calculated by ImageQuant analysis (Fig. 5). The level of intact btuB RNA from the repressible EB450 fusion began to decline within minutes to a steady-state level of 22% of the derepressed level. The lag probably reflects the time for accumulation of Ado-Cbl to effective levels in the cell. The 470-nt RNA from the chromosomal allele also showed a similar decline. It took about 20 min before the new steady-state level of btuB RNA was achieved, which was longer than expected if this RNA had a lifetime typical of mRNA in E. coli.
FIG. 5.
Time course of decrease of btuB RNA levels following addition of Ado-Cbl. RNA was extracted from cells of strain RK5173 carrying the indicated fusion plasmids. Cells were grown in minimal medium A to mid-log phase. Ado-Cbl was added to 1 μM, and samples were removed at the indicated times for the S1 protection assay by using internally labeled probe A. The amount of full-length btuB RNA, normalized to the intensity of the bla transcript, is plotted as a function of the time after addition of Ado-Cbl.
The full-length btuB RNA level in the partially repressible EB315 strain fell to 60% of the initial level with kinetics similar to those in the EB450 strain. The RNA level in the poorly repressed EB270 strain fell slightly, to 80% of the initial level. In all three strains, the levels of the unregulated 290- and 165-nt species were not substantially affected, and the 210-nt species increased under repressive conditions, but only after a lag of 5 to 10 min, i.e., with slower kinetics than the repression of the full-length species.
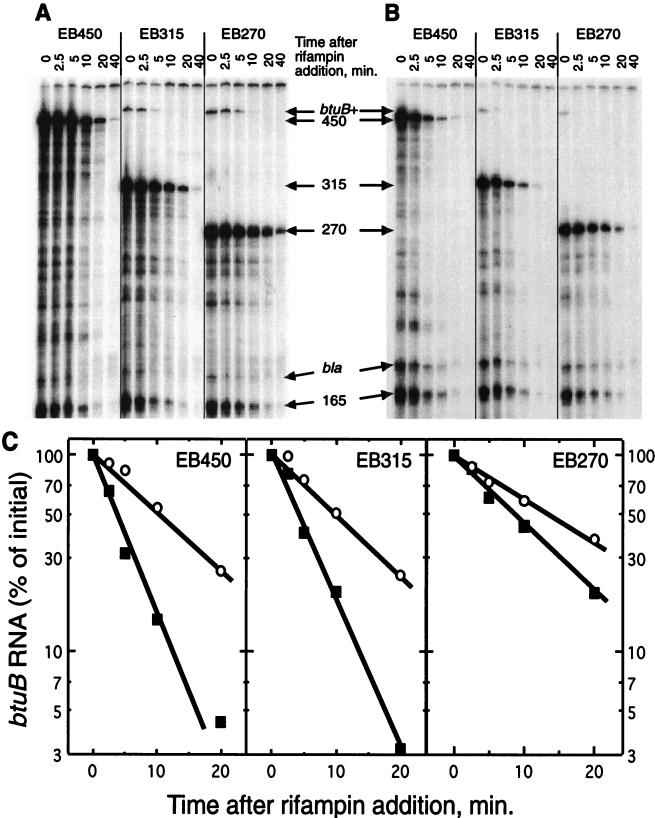
Decreased stability of btuB RNA during repression.
The metabolic stability of btuB RNA was estimated from the rate of its decay following the addition of rifampin to cells growing in the absence (Fig. 6A) or upon simultaneous addition of Ado-Cbl (Fig. 6B). The amounts of the btuB RNA species were determined by S1 nuclease protection assays and quantified with the ImageQuant program (Fig. 6C). The full-length btuB RNA expressed from all three btuB-lacZ fusions in the absence of Ado-Cbl was long-lived, with half-lives (t1/2s) in the range of 9.4 to 13.4 min. All of the partial-length btuB transcripts were also long-lived, suggesting that their stability is determined within the first 160 nt. The bla RNA exhibited the short t1/2 of 2 to 4 min, typical of most bacterial mRNAs (8).
FIG. 6.
Metabolic stability of btuB RNA in the presence or absence of Cbl following the addition of rifampin. RNA was extracted from cells of RK5173 carrying plasmid pRS415 with the indicated btuB::lacZ fusion-encoding inserts at the indicated times after the addition of rifampin and was subjected to S1 nuclease protection of internally labeled probe A. (A) Cells in minimal medium. (B) Cells that received Ado-Cbl at the same time as rifampin. (C) Amount of full-length btuB transcript, as a fraction of the initial value, derived from the three templates, for cells in the absence (open symbols) and presence (solid symbols) of Ado-Cbl.
In the presence of Ado-Cbl, the stability of the full-length products of EB450 and EB315 and of the chromosome-encoded btuB transcript was markedly reduced, with t1/2s in the range of 2 to 4 min (Fig. 6B and C). The t1/2 for the EB270 transcript decreased slightly to around 8.3 min. The stability of the constitutively expressed 165-nt fragments was not appreciably affected by Ado-Cbl. Thus, the decreased amount of intact btuB RNA under repressive conditions is associated with its decreased stability. This is a seemingly paradoxical situation, since addition of Ado-Cbl results in increased RNA lability in the presence of rifampin but a slow decrease in RNA levels when transcription occurs. This behavior can be explained by the constitutive synthesis of a long-lived RNA whose level and stability is affected by Cbl only after it has reached a length beyond nt 300.
Effect of RNases on regulation and stability of btuB RNA.
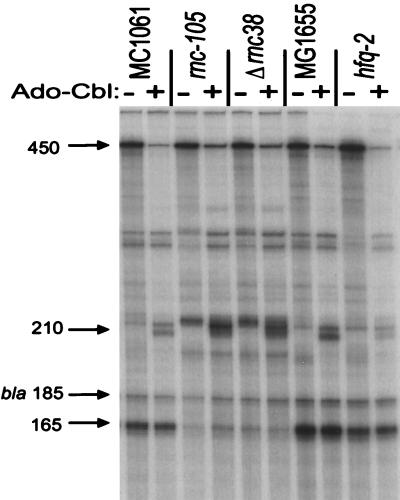
The formation of the several truncated species of btuB RNA and the increase in its lability in the presence of Ado-Cbl could result from transcription attenuation or endonucleolytic cleavage. The effect of mutations that block expression of various RNases involved in RNA processing on the distribution and regulation of the transcripts from the EB450 fusion was analyzed by S1 nuclease protection assay. Two rnc mutants defective in RNase III activity displayed a marked decrease in the 165-nt fragments and an increase in the Cbl-induced 210-nt fragment (Fig. 7), but they had no significant effect on the regulation of the full-length transcript or on lacZ expression. These results suggest that an RNA secondary structure is formed that is cleaved by RNase III at position 165 immediately downstream of the B12 box. Since RNase E is essential for viability, its involvement was tested following thermal inactivation of alleles that confer a temperature-sensitive growth phenotype. We found there was no substantial effect on the distribution of the RNA species or on the regulation of full-length RNA levels or lacZ expression as a result of individual inactivation of RNase E, II, D, PH, BN, or D or of polynucleotide phosphorylase (data not shown). Many of these RNases have redundant activities, and defects in processing are observed only when most of them have been inactivated (10, 12).
FIG. 7.
Effect of mutations affecting RNase III and Hfq-1 activities on the pattern of btuB-specific transcripts. RNA was extracted from the indicated parental and mutant cells carrying the EB450 fusion and grown in the absence or presence of Ado-Cbl. The RNA was used for S1 nuclease protection of internally labeled probe A.
The hfq gene encodes an RNA-binding protein, host factor-1, that can disrupt RNA secondary structure and allow increased translation or replication of rpoS mRNA or phage Qβ RNA, respectively (15, 24). An hfq null mutant carrying the EB450 fusion displayed normal regulation of β-galactosidase levels and of full-length btuB RNA (Fig. 7), indicating that Hfq function is not needed for transcription elongation or BtuB regulation. However, the amount of the Cbl-induced 210-nt fragment was strongly reduced, indicating a possible role for changes in RNA secondary structure in the generation of this fragment.
Regulation of transcription driven by heterologous promoters.
To test whether promoter sequences and the nature of the transcription complex play any role in regulation, the btuB promoter in the EB450 transcriptional and translational fusions was replaced with the heterologous lac, bla, and phage T7 RNA polymerase-dependent φ10 late promoters. The wild-type transcription start site was retained in all cases (data not shown). The levels of β-galactosidase differed in response to promoter strengths and the degree of induction with IPTG (Table 3). Two different regulatory outcomes were seen. Expression of transcriptional and translational fusions driven by the lac and bla promoters was reduced by Ado-Cbl to a degree similar to that with the btuB promoter. S1 nuclease protection assays revealed no qualitative difference in the distributions or relative amounts of the btuB transcript fragments (data not shown). Thus, the promoter and upstream sequences play no detectable role in Cbl-dependent regulation.
TABLE 3.
LacZ expression from plasmids carrying btuB-lacZ fusions expressed from heterologous promoters
| Host strain | Promoter | Presence of 0.5 mM IPTG | β-Galactosidase activity (U) with transcriptional fusion
|
RRa | β-Galactosidase activity (U) with translational fusion
|
RRa | ||
|---|---|---|---|---|---|---|---|---|
| −Ado-Cbl | +Ado-Cbl | −Ado-Cbl | +Ado-Cbl | |||||
| RK5173 | btuB | − | 3,035 | 535 | 5.7 | 435 | 14 | 31 |
| RK5173 | blaP | − | 2,925 | 625 | 4.7 | 640 | 36 | 17.8 |
| JM109 | lacP | − | 5,380 | 1,695 | 3.2 | 1,085 | 25 | 43 |
| JM109 | lacP | + | 7,250 | 2,200 | 3.3 | 2,775 | 60 | 46 |
| RK5173 | T7 | − | 70 | 78 | 1.0 | 10 | 2 | 5 |
| RK3513 | T7 | − | 450 | 365 | 1.2 | 45 | 12 | 3.8 |
| RK3513 | T7 | + | 5,015 | 4,790 | 1.1 | 975 | 470 | 2.6 |
RR, repression ratio (activity −Ado-Cbl/activity +Ado-Cbl).
Different behavior was seen when transcription was driven by T7 RNA polymerase, which is widely used for gene expression, because of its high rate of elongation, single-subunit composition, and diminished response to transcriptional pause and termination signals (4, 23). Plasmids pXN17 and pXN18 contain the upstream region of the T7 φ10 promoter joined at the +1 position to the EB450 transcriptional and translational fusions, respectively. Although the T7 promoter extends into the transcribed region, T7 RNA polymerase started transcription at the same position as that used by the wild-type promoter. Expression of β-galactosidase in strain RK5173 was very low, indicating the dependence on T7 RNA polymerase (Table 3). Expression in strain RK3513, a btuB+ transductant of the T7 RNA polymerase-producing strain, BL21(DE3), was increased more than 10-fold by induction of T7 RNA polymerase synthesis with 0.5 mM IPTG for 1.5 h. The transcriptional fusion was not appreciably affected by Ado-Cbl, with repression ratios of <1.3. Although the levels of the full-length transcript were not decreased following growth with Ado-Cbl, the 210-nt fragment was still induced (data not shown). Many of the other btuB fragments were still present following T7 polymerase-driven transcription, but those around 280 and 295 nt were absent. Cbl-dependent regulation of the translational fusion still occurred, but to a diminished degree, with repression ratios of >2.5. These results suggest that transcriptional regulation depends on the nature and properties of the transcription complex, such as its response to termination signals, rather than a feature of the RNA transcript, such as a secondary structure that is cleaved by an RNase.
DISCUSSION
Previous studies using reporter fusions to the E. coli btuB gene (7, 14) and to the S. typhimurium btuB and cbiA genes (17–19) revealed a similar and unexpected pattern of dependence on the length of the transcribed sequences for Cbl-dependent regulation at both the transcriptional and translational levels. In all three Cbl-regulated genes, translational control appears to be the major site of control over gene expression. The results presented here agreed with studies of the other Cbl-regulated genes that the hairpin formed over or near the ribosome-binding site is a major component in translational control. This paper provides for the first time information relevant to the process of transcriptional control and changes in the transcript during Cbl-dependent regulation. Ravnum and Andersson (17) concluded that expression of btuBSt was not subject to a substantial degree of transcriptional control, based on the behavior of lac fusions. However, the other Cbl-regulated genes exhibit an appreciable degree of apparent transcriptional control, and the results presented here indicate that this is a specific feature of btuBEc expression. This conclusion is based on our demonstration of changes in btuB RNA levels. The fact that btuB-lacZ fusions of different lengths exhibit different regulatory behaviors allowed analysis of the role of the TRR and correlations of changes in the transcripts with changes in the regulatory behavior of the fusions.
There was concern that the apparent regulation of transcriptional fusions might be an artifactual consequence of readthrough into the lacZ gene from ribosomes that initiated translation in the btuB sequence. Several lines of evidence eliminated this concern. First, introduction of a translational stop at codon 49 completely blocked btuB translation, but had no effect on the level or regulation of the btuB-lacZ transcriptional fusion or of btuB RNA. Second, the changes in the level of full-length btuB RNA matched well the behavior of transcriptional fusions. Third, Cbl-dependent regulation is associated with changes in the stability of full-length btuB RNA. Finally, two different btuB-lacZ transcriptional fusions were constructed with an RNase III cleavage site upstream of the lacZ gene to dissect the translation of lacZ from that of btuB. The repression ratios in the two constructs were 4.9 and 6.0, which are very similar to those in the pRS415 fusion vector. Thus, the possibility that transcriptional regulation is a consequence of translational readthrough from btuB sequences is untenable.
Cbl-dependent regulation of transcriptional fusions and of btuB RNA levels could reflect the general or nonspecific lability of untranslated RNA, such that when btuB translation is prevented, the distal sequences are subject to endonucleolytic degradation. We found that transcriptional control of btuB requires the integrity of the TRR. The EB270 fusion, which showed greatly reduced transcriptional regulation, still possesses 30 nt of btuB sequence before the fusion and 100 nt of trp-lac sequences before the start of the lacZ gene, which should provide a suitable target for RNA turnover. In addition, transcriptional fusions to sequences upstream of the start of translation show the same activity of β-galactosidase as fusions after the start of translation, suggesting that translation does not provide an inherent increase in stability (7). However, it is difficult to eliminate the impact of the lability of untranslated RNA.
If the TRR confers a specific mechanism involved in btuB RNA turnover under repressive conditions, this region might function either as a transcriptional attenuator or as a site for endonuclease cleavage. The existence of an attenuator was suggested by Lundrigan et al. (14) and Ravnum and Andersson (17), based on the presence of a potential G+C-rich stem and loop followed by 4 U residues in the transcript. Deletions to position +270 or further upstream, which affect this structure, completely eliminated transcriptional control (7). Further deletions between +285 to +303 resulted in reduced regulation, whereas deletions ending at +345 of distal showed normal regulation. Transcript fragments ending at +280 to +300 should result from termination at the attenuator, but their amounts increased only modestly under repressive conditions, indicating either that attenuation is not operative or that the attenuated products are labile. Base changes in the putative attenuator element partially reduced transcriptional regulation (unpublished data). Thus, we can conclude that the putative attenuator sequence is important for transcriptional control, but downstream sequences out to between +315 and +345 are also involved to some degree. Note that deletions that result in complete loss of transcriptional control retain translation-level regulation (7). We found no evidence for the action of RNase E or III in regulation, although RNase III cleavage of the btuB transcript was observed. The involvement of a Cbl-regulated turnover mechanism in addition to attenuator action is indicated from the kinetics of RNA turnover. The full-length RNA is long-lived, but becomes labile in the presence of Ado-Cbl. This result cannot be explained by the operation of an attenuation event as the sole regulatory factor affecting btuB RNA, since the lability is imposed upon completed RNA molecules or even nascent transcripts past the site of attenuation.
There is no control process operative at the level of transcription initiation. Normal regulation was seen when transcription was driven by heterologous sigma 70-dependent promoters, as long as the proper start site was retained. In addition, some btuB transcripts are expressed in a constitutive manner, notably the 165-nt fragments. Dissociation of transcriptional and translational regulation occurred with T7 RNA polymerase, which is less responsive to transcriptional pausing and termination signals than is E. coli RNA polymerase (4). This result suggests that transcriptional regulation depends on an activity of the transcription complex, such as polymerase pausing or termination, rather than on the formation of an RNA structure whose cleavage by an RNase triggers RNA degradation. The retention of translational regulation indicates that this level of control is independent of the properties of the transcription complex and may be determined by RNA secondary structure, perhaps involving access to the ribosome-binding site.
Specific transcript fragments were produced, whose 3′ ends occur at potentially relevant sites. The 165-nt fragments end at the downstream side of the B12 box and were greatly decreased in RNase III-deficient mutants. The 3′ ends of the constitutively expressed 235- to 250-nt fragments occur just past the stem-loop structure formed by the Shine-Dalgarno hairpin, and the 3′ end of the Cbl-induced 210-nt fragment occurs just before this hairpin. However, in no case was there an invariant correlation between the level of any fragment and the level of btuB expression. The intact transcript and most of its fragments were long-lived under nonrepressing conditions. The inability to observe fragments shorter than 165 nt suggests that the first 165 nt of the transcript, which includes the B12 box, are needed for the enhanced stability. As discussed above, the presence of the TRR sequences between +270 and +315 is needed for the decrease in RNA stability. This decrease in stability appears to be unaffected by the absence of RNase E or RNase III. Stabilizing elements, such as double-stranded structures at the 5′ ends of RNA, have been documented (8).
Thus, all of the results obtained here indicate that translating ribosomes prevent the decreases in btuB RNA levels and stability that are brought about by the presence of the TRR. This behavior appears to represent a novel and specific mechanism of gene regulation. Attenuators and RNase-cleavage sites that are affected by translation are well known (3, 11), but there are few cases known in which regulated translation of a coding sequence is directly coupled to these determinants of RNA stability. Most attenuators act at a site before the regulated structural gene. The placement of the transcriptional regulatory region distal to the start of translation allows the translational regulatory process to simultaneously affect the transcriptional process.
The mechanism of Cbl-dependent translation regulation remains a subject worthy of further study. It is known from previous studies to require the integrity of most of the leader region, including the proper transcription start site (unpublished data and reference 7) and the potential for formation of the hairpin around the ribosome-binding site. However, no candidate regulatory proteins that might influence the structure of the RNA in a Cbl-dependent manner have surfaced yet. Changes in the conformation of the transcript can be deduced from the appearance of the Cbl-inducible 210-nt fragment. We are attempting to define changes in btuB RNA secondary structure in the presence of Ado-Cbl and to test for specific binding of Ado-Cbl to the RNA.
We conclude that the translated region is essential for transcriptional control and the Cbl-dependent decrease in RNA stability. The activity of this region is directly regulated by the Cbl-inhibited translation of the btuB gene, such that translation through this region must occur to prevent termination and RNA turnover. The presence of Ado-Cbl may cause changes in RNA secondary structures that affect accessibility of the ribosome-binding site. Questions under study address the precise role of the attenuator and distal sequences, the relationship of translation to attenuator action, and, most importantly, how Ado-Cbl is sensed and affects RNA structure or translation initiation.
ACKNOWLEDGMENTS
We thank Sidney Kushner, Malcolm Winkler, and Murray Deutscher for provision of strains and Joanna Goldberg and Igor Olekhnovich for helpful comments.
This work was supported by grant GM19078 from the National Institutes of Health.
REFERENCES
- 1.Aufrère R, Tempête M, Bohin J-P. Regulation of expression of the gene for vitamin B12 receptor cloned on a multicopy plasmid in Escherichia coli. Mol Gen Genet. 1986;205:358–365. doi: 10.1007/BF00430451. [DOI] [PubMed] [Google Scholar]
- 2.Babitzke P, Granger L, Olszewski J, Kushner S R. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J Bacteriol. 1993;175:229–239. doi: 10.1128/jb.175.1.229-239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun F, Le Derout J, Régnier P. Ribosomes inhibit an RNase E cleavage which induces the decay of rpsO mRNA of Escherichia coli. EMBO J. 1998;17:4790–4797. doi: 10.1093/emboj/17.16.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chevrier-Miller M, Jacques N, Raibaud O, Dreyfus M. Transcription of single-copy hybrid lacZ genes by T7 RNA polymerase in Escherichia coli: mRNA synthesis and degradation can be uncoupled from translation. Nucleic Acids Res. 1990;18:5787–5792. doi: 10.1093/nar/18.19.5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emory S A, Belasco J G. The ompA 5′ untranslated RNA segment functions in Escherichia coli as a growth-rate-regulated mRNA stabilizer whose activity is unrelated to translational efficiency. J Bacteriol. 1990;172:4472–4481. doi: 10.1128/jb.172.8.4472-4481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escalante-Semerena J C, Roth J R. Regulation of cobalamin biosynthetic operons in Salmonella typhimurium. J Bacteriol. 1987;169:2251–2258. doi: 10.1128/jb.169.5.2251-2258.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Franklund, C. V. Unpublished observations.
- 7.Franklund C V, Kadner R J. Multiple transcribed elements control expression of the Escherichia coli btuB gene. J Bacteriol. 1997;179:4039–4042. doi: 10.1128/jb.179.12.4039-4042.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen M J, Chen L-H, Fejzo M L S, Belasco J G. The ompA 5′ untranslated region impedes a major pathway for mRNA degradation in Escherichia coli. Mol Microbiol. 1994;12:707–716. doi: 10.1111/j.1365-2958.1994.tb01058.x. [DOI] [PubMed] [Google Scholar]
- 9.Kadner R J. Repression of synthesis of the vitamin B12 receptor in Escherichia coli. J Bacteriol. 1978;136:1050–1057. doi: 10.1128/jb.136.3.1050-1057.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly K O, Deutscher M P. The presence of only one of five exoribonucleases is sufficient to support the growth of Escherichia coli. J Bacteriol. 1992;174:6682–6684. doi: 10.1128/jb.174.20.6682-6684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landick R, Turnbough C L, Jr, Yanofsky C. Transcription attenuation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. Cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1263–1286. [Google Scholar]
- 12.Li Z, Deutscher M P. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell. 1996;86:503–512. doi: 10.1016/s0092-8674(00)80123-3. [DOI] [PubMed] [Google Scholar]
- 13.Lundrigan M D, Kadner R J. Altered cobalamin metabolism in Escherichia coli btuR mutants affects btuB regulation. J Bacteriol. 1989;171:154–161. doi: 10.1128/jb.171.1.154-161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundrigan M D, Köster W, Kadner R J. Transcribed sequences of the Escherichia coli btuB gene control its expression and regulation by vitamin B12. Proc Natl Acad Sci USA. 1991;88:1479–1483. doi: 10.1073/pnas.88.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein, HF-1, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 16.Petersen C. Translation and mRNA stability in bacteria: a complex relationship. In: Belasco J G, Brawerman G, editors. Control of messenger RNA stability. San Diego, Calif: Academic Press; 1993. pp. 117–145. [Google Scholar]
- 17.Ravnum S, Andersson D I. Vitamin B12 repression of the btuB gene in Salmonella typhimurium is mediated via a translational control which requires leader and coding sequences. Mol Microbiol. 1997;23:35–42. doi: 10.1046/j.1365-2958.1997.1761543.x. [DOI] [PubMed] [Google Scholar]
- 18.Richter-Dahlfors A A, Andersson D I. Cobalamin (vitamin B12) repression of the Cob operon in Salmonella typhimurium requires sequences within the leader and the first translated open reading frame. Mol Microbiol. 1992;6:743–749. doi: 10.1111/j.1365-2958.1992.tb01524.x. [DOI] [PubMed] [Google Scholar]
- 19.Richter-Dahlfors A A, Ravnum S, Andersson D I. Vitamin B12 repression of the cob operon in Salmonella typhimurium: translational control of the cbiA gene. Mol Microbiol. 1994;13:541–553. doi: 10.1111/j.1365-2958.1994.tb00449.x. [DOI] [PubMed] [Google Scholar]
- 20.Roth J R, Lawrence J G, Bobik T A. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 23.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 24.Tsui H-C, Leung H-C E, Winkler M E. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 25.Wei B-Y, Bradbeer C, Kadner R J. Conserved structural and regulatory regions in the Salmonella typhimurium btuB gene for the outer membrane vitamin B12 transport protein. Res Microbiol. 1992;143:459–466. doi: 10.1016/0923-2508(92)90091-2. [DOI] [PubMed] [Google Scholar]