Abstract
Osmotic regulation of proU expression in the enterobacteria is achieved, at least in part, by a repression mechanism involving the histone-like nucleoid protein H-NS. By the creation of binding sites for the TyrR regulator protein in the vicinity of the ς70-controlled promoter of proU in Escherichia coli, we were able to demonstrate a superposed TyrR-mediated activation by l-phenylalanine (Phe), as well as repression by l-tyrosine, of proU expression in vivo. Based on the facts that pronounced activation in the presence of Phe was observed even at a low osmolarity and that the affinity of binding of TyrR to its cognate sites on DNA is not affected by Phe, we argue that H-NS-mediated repression of proU at a low osmolarity may not involve a classical silencing mechanism. Our data also suggest the involvement of recruited RNA polymerase in the mechanism of antirepression in E. coli.
The proU operon in Escherichia coli and Salmonella typhimurium encodes a binding-protein-dependent transport system that mediates the active uptake of the compatible solutes glycine betaine and l-proline during growth in media of elevated osmolarities. Under such growth conditions, the expression of proU is induced 400-fold at the level of initiation of transcription, but the underlying regulatory mechanisms are not clearly understood (reviewed in references 6 and 14).
Analyses of the cis regulatory regions necessary for osmotic induction of proU have identified an extended sequence (more than 500 bp long) designated the negative regulatory element (NRE), whose proximal end is situated approximately 70 bp downstream of the ς70-controlled promoter (P2) and which is required for the full repression of proU at a low osmolarity (7, 10, 22, 28, 29). Genetic and biochemical data suggest that the NRE mediates the repressor function of the histone-like nucleoid protein H-NS on proU (approximately 20- to 25-fold) (7, 10, 22, 29). Nevertheless, the NRE does not serve as a portable cassette for osmotic regulation when placed downstream of heterologous promoters (7, 28), indicating that sequences around and upstream of P2 are also required for its function. Furthermore, in mutants lacking H-NS or the NRE, or both, a residual 8- to 10-fold osmotic inducibility of proU is observed (7, 10, 22, 28, 29); this inducibility has been interpreted to represent a second distinct mechanism acting directly on the cis element(s) in the close vicinity of P2 (7, 14). Finally a ςs-controlled promoter, P1 (situated 190 bp upstream of P2), has also been identified which, at least in S. typhimurium, is cryptic and whose relevance in proU regulation is as yet unclear (7, 33).
It has been suggested that H-NS-mediated repression of proU at a low osmolarity is achieved by promoter “silencing” and that relief of repression at a high osmolarity is the consequence of cytoplasmic potassium glutamate accumulation (6, 28). In the silencing model, the NRE serves as a position-independent silencer locus (10, 28, 49) akin to that described for the regulation of several eukaryotic genes (5, 27). The following features have been cited in support of this model. (i) H-NS is not a typical sequence-specific regulator protein (for reviews, see references 3 and 47), nor is the NRE a typical operator sequence. Indeed, there exist two regions of curved DNA in the vicinity of proU P2 (see Fig. 1B), one falling within the proU NRE and the other located about 150 bp upstream of the promoter (13, 29, 40, 41), to both of which H-NS exhibits preferential binding (22, 29, 40). (ii) The separation and phase angle of the NRE from proU P2 can be varied over a distance of 200 bp without affecting its ability to mediate repression (10, 18, 28). (iii) NRE-mediated repression is also observed for several different variants of the P2 promoter (19, 49). (iv) A role for H-NS binding has been implicated in the only locus (bgl) in E. coli where silencing has been unequivocally established (26, 38, 39); the protein has also been postulated to silence several other genes in the organism (12, 23). One question as yet unanswered is whether the repressive action, consequent to the binding of H-NS to proU, is direct (43) or indirect (18, 19, 29).
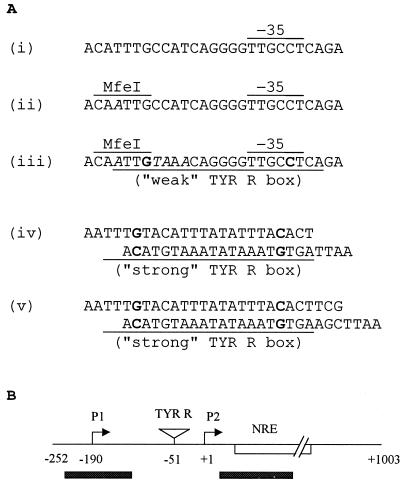
FIG. 1.
Introduction of the TYR R box(es) near proU P2. (A) The nucleotide sequence upstream of the −35 region of the wild-type proU P2 promoter (i) and those following sequential site-directed mutagenesis to create first an MfeI site (at position −51 relative to the start site of P2 transcription) (ii) and then a weak TYR R box (iii) are shown. The MfeI and the −35 hexamer sequences are indicated, and the mutated base residues are in italics. Also shown are the pairs of annealed oligonucleotide sequences (iv and v) that were used to generate the double-stranded TYR R strong box sequences (identical to that in tyrP) flanked with 5′-AATT overhangs for construction of the repression tester and activation tester variants, following insertion into the MfeI sites shown in sequences iii and ii, respectively. The 22-bp TYR R box sequences (whose consensus is the palindrome 5′-N2TGTAAAN6TTTACAN2-3′, in which the residues shown in bold are invariant) are underlined, and the invariant residues are in bold. (B) Schematic depiction of the position of insertion of the TYR R strong box sequences in the two proU variants, relative to P1, P2, and the NRE. For nucleotide numbering, the start site of P2 transcription has been taken to be +1. Shaded bars indicate the regions of DNA curvature to which H-NS exhibits preferential binding.
In vitro tests of the silencing model are rendered difficult by the fact that no accepted method for the reconstitution of proU osmotic regulation in a cell-free system exists. One prediction of the silencing model, which would help distinguish it from other mechanisms of H-NS-mediated repression of proU, is that the silencing effect would extend sufficiently upstream of the proU promoter to interfere also with the recognition of closely linked binding sites for other DNA-binding proteins. This is the hypothesis which we have sought to test in this study, by first creating specific sites for binding of the regulator protein TyrR adjacent to the proU P2 promoter and then addressing the question of whether TyrR binds these sites in vivo at low and high osmolarities. Our results indicate that TyrR-mediated regulation can be superimposed on osmotic regulation of proU transcription and suggest in particular that the chromatin architecture in the proU P2 promoter region, even at a low osmolarity, is permissive to the binding of and activation by TyrR. To that extent, therefore, it appears to be unlikely that a silencing mechanism operates to achieve proU repression at a low osmolarity.
Overview of and rationale for choosing the TyrR regulation system.
The TyrR protein in E. coli mediates the transcriptional regulation of several operons involved in the biosynthesis and transport of the aromatic amino acids (reviewed in references 30 and 31). The protein can act as either a repressor or an activator depending upon the promoter and the particular coeffector to which it is bound. We chose to work with the TyrR system primarily because of the fact that neither the affinity of the protein for its cognate binding sites on DNA nor the footprint obtained on such binding is altered in the presence of its coactivator l-phenylalanine (Phe) (2, 4, 31, 35). The need for imposing this constraint on our choice of system is explained below. In the case of other well-characterized activator proteins such as CRP (20), AraC (37), MalT (37), and the LysR family of proteins (36), the association of the proteins with their respective coactivators leads to an alteration of the DNA-binding characteristics of the proteins.
TyrR-mediated repression is achieved in the presence of the coeffector l-tyrosine (Tyr). Genes whose expression is repressed by TyrR often have two adjacent TyrR binding sites (TYR R boxes), one of which overlaps the promoter. (The TYR R box is 22 bp long, and its consensus sequence is described in the legend to Fig. 1.) The box overlapping the promoter has a relatively weak affinity for TyrR (weak TYR R box) and is bound only when it is close to and on the same face of the helix as the other box, which has a stronger binding affinity for the protein (strong TYR R box). For instance, in the case of the tyrP gene (encoding a Tyr-specific permease), which is repressed by TyrR-Tyr, the weak box overlaps the −35 region of the promoter, whereas the strong box is upstream of and separated from the weak box by 1 bp. It has been shown that, in the presence of Tyr, the protein self-associates to form a hexamer and that it binds cooperatively to both boxes to cause repression.
Transcriptional activation by TyrR in the presence of Phe requires only the presence of a strong box suitably positioned upstream of the promoter. A spacing of 18 bp between the TYR R box and the −35 hexamer is optimal for the purpose. The TyrR dimer remains constitutively bound to the strong box, and upon binding Phe it acquires the ability to activate transcription by the process of RNA polymerase recruitment (15, 32); under these specific conditions, TyrR has been shown to increase the affinity of binding of RNA polymerase to the adjacent promoter and to stimulate open-complex formation (16) by functioning as a class I transcription activator (21, 48).
In the case of native tyrP, the 1-bp separation between the two TYR R boxes (which is necessary for repression control) places the strong box 15 bp away from the −35 hexamer, which distance is suboptimal for Phe-mediated activation (1). The greatest activation effect at tyrP is observed for that template in which the strong box has been moved upstream by another 3 bp; in the latter situation, addition of Tyr also leads to an activation rather than a repression of tyrP expression (1).
Creation of the TYR R box(es) near proU P2.
In this study, we chose to simulate at proU P2 the regulatory features described above for the tyrP gene (1, 2). In order to test whether TyrR could repress proU, it was necessary (i) to use site-directed mutagenesis to create a weak box overlapping the −35 region of the P2 promoter and (ii) to introduce a strong box sequence upstream of and 1 bp away from the weak box, as is the case in native tyrP. In order to test the ability of TyrR to activate proU, it was necessary to introduce a strong box sequence 3 bp farther upstream than in the previous construct and to leave the remainder of the proU regulatory region unaltered. For convenience, these two sets of alterations are referred to below as the repression tester and activation tester variant sequences, respectively.
The template used for the mutagenesis reactions was an M13 phage derivative bearing a proU fragment comprising P1, P2, and the NRE. This 1.26-kb proU fragment (see Fig. 1B) is identical to that earlier described for plasmid pHYD272 (7) and is known to carry all the cis elements involved in proU osmoresponsivity. Recombinant DNA manipulations were performed essentially as described previously (34). Site-directed substitution mutations were introduced by the method of Vandeyar et al. (44), using a commercially available kit from United States Biochemical Corp.
The sequence of wild-type proU P2 in the region of interest is shown in Fig. 1A, sequence i. To facilitate the introduction of the strong box sequence, a unique MfeI site, CAATTG, was created by introducing a T-to-A substitution (underlined) 14 nucleotides upstream of the −35 hexamer (Fig. 1A, sequence ii). A weak TYR R box overlapping the −35 region was then created by site-directed mutagenesis of a CCAT sequence to TAAA (Fig. 1A, sequence iii). (The −35 hexameric sequence itself was left unaltered, although there is a natural match with the right arm of the palindromic TYR R box consensus at three of six positions and with the −35 region of tyrP at four of six positions.) The introduction of a strong TYR R box sequence flanked with 5′-AATT overhangs (shown in Fig. 1A, sequence iv) into the MfeI site of sequence iii in Fig. 1A led to the creation of the repression tester variant, that is, with two TYR R boxes separated by 1 bp. On the other hand, the introduction of the strong TYR R box sequence flanked with 5′-AATT overhangs and containing an additional 3 bp (Fig. 1A, sequence v) into the MfeI site of sequence ii in Fig. 1A resulted in the construction of the activation tester variant, that is, with the strong TYR R box positioned 18 bp upstream of the −35 hexamer.
After each step of mutagenesis, M13 phage clones carrying the correct mutation were identified by appropriate single-nucleotide sequence tracking. The complete sequence of the P2 promoter region for each of the two variants finally obtained was verified by automated DNA sequence analysis (data not shown). The structure and disposition of the regulatory elements in the proU variants constructed in this study are schematically depicted in Fig. 1B.
In order to undertake in vivo expression studies, the variant proU sequences were then subcloned upstream of the lacZ reporter gene in the very-low-copy-number trimethoprim resistance plasmid pMU2385 (46). The resultant plasmids were designated pMU6442 (with the activation tester variant sequence) and pMU6443 (with the repression tester variant sequence). As a control, plasmid pMU6441 was also constructed as a derivative of pMU2385 carrying the 1.26-kb wild-type proU regulatory region.
Effects of the TYR R box(es) on proU regulation in hns+ and hns derivatives.
The plasmids pMU6441, pMU6442, and pMU6443 were each transformed into a pair of isogenic tyrR+ and tyrR366 strains, JP7740 and JP8042, respectively, for lacZ expression studies. Both strains are prototrophic, Δlac, and recA (46). In light of the role suggested for the H-NS protein in proU silencing, we also transformed the three plasmids into strains JP10938 (tyrR+ hns-205::Tn10) and JP10939 (tyrR366 hns-205::Tn10), which are the recA+ hns derivatives of JP7740 and JP8042, respectively. (The hns mutations were introduced by phage P1 transduction, with strain PD145 [8] serving as the donor.) The transformant derivatives were cultured in defined low- and high-osmolarity media supplemented when necessary with Tyr or Phe, and the specific activity of β-galactosidase in each culture was determined by the method of Miller (24). Each value reported is the mean of at least three independent measurements.
In order to test for the cis effects of the introduced sequence variations on proU osmotic regulation, we first determined the values for lacZ expression in the tyrR mutant derivatives (hns+ and hns), that is, in which the possibility of a confounding effect caused by binding of TyrR was excluded. The results are presented in Table 1. In these tyrR host strains, supplementation of the culture medium with Tyr or Phe had no effect on β-galactosidase expression from any of the three plasmids (data not shown).
TABLE 1.
β-Galactosidase expression in tyrR366 strains from lac fusions to proU and its variantsa
| Plasmid | β-Galactosidase activity for:
|
|||
|---|---|---|---|---|
|
hns+
|
hns-205
|
|||
| −NaCl | +NaCl | −NaCl | +NaCl | |
| pMU6441 | 0.5 | 1,450 | 34 | 1,582 |
| pMU6442 | 1 | 351 | 68 | 720 |
| pMU6443 | <0.5 | 135 | 33 | 624 |
Cultures of strains JP8042 (tyrR366 hns+) and JP10939 (tyrR366 hns-205::Tn10) carrying the indicated plasmid derivatives were grown, for β-galactosidase assays, to mid-exponential phase in one-quarter-strength medium 56 (containing glucose [0.2%], thiamine [1 μg/ml], and trimethoprim [10 μg/ml]) without and with 0.3 M NaCl supplementation (−NaCl and +NaCl, respectively). Medium 56 was modified from the medium described by Monod et al. (25) and contains the following, per liter: K2HPO4 (10.6 g), NaH2PO4 · 12H2O (6.1 g), (NH4)2SO4 (2 g), MgSO4 · 7H2O (0.2 g), Ca(NO3)2 (10 mg), and FeSO4 · 7H2O (0.5 mg) (pH adjusted to 7.0). Enzyme specific activity values are reported in Miller units (24).
Under the conditions of growth and assay used in this study, we observed a >1,000-fold osmotic induction of wild-type proU expression in the tyrR hns+ strain (Table 1). Neither proU variant (in pMU6442 or pMU6443) was affected in low-osmolarity-medium repression in the hns+ strain; on the other hand, the expression levels in the NaCl-supplemented medium were lower than that for wild-type proU itself (Table 1). Nevertheless, at least a 250-fold osmotic inducibility was still observed for both mutant derivatives in the hns+ strain. A possible explanation for this partial loss of osmoresponsivity is that the strong TYR R box insertions in pMU6442 and pMU6443 (27 and 24 bp, respectively) fortuitously introduce half-integral turns of the DNA helix in the region, which in an earlier study was shown also to be correlated with reduced expression of proU at a high osmolarity (41).
In the tyrR hns background, the expression profiles for the three plasmids were more or less similar to one another (Table 1). Consistent with the data from earlier studies (7, 10, 18, 22, 29), absence of the H-NS protein led to a moderate increase in proU-lacZ expression in low-osmolarity medium. All three plasmids exhibited residual osmotic inducibility in the hns strain, although once again the absolute values for lacZ expression from pMU6442 and pMU6443 in the NaCl-supplemented medium were less than that for the wild-type proU control.
We then measured the levels of lacZ expression from the three plasmids in the tyrR+ strains (hns+ and hns) to determine the regulatory role of TyrR on the mutant proU promoters. The results are presented in Table 2. In concord with earlier practice (2), the magnitude of TyrR-mediated regulation by the two coeffectors was calculated as the ratio of β-galactosidase activity in the tyrR mutant to that in the tyrR+ strain in the presence of the particular coeffector (repression) or its reciprocal (activation).
TABLE 2.
β-Galactosidase expression in tyrR+ strains from lac fusions to proU and its variantsa
| Plasmid(s) | Coeffector | β-Galactosidase activity for:
|
|||
|---|---|---|---|---|---|
|
hns+
|
hns-205
|
||||
| −NaCl | +NaCl | −NaCl | +NaCl | ||
| pMU6441 (wild type) | None | 1.3 | 1,471 | 29 | 1,683 |
| pMU6442 (activation tester) | None | 1 | 309 | 51 | 708 |
| Tyr | 22 | 1,320 | 82 | 1,274 | |
| Phe | 198 | 2,352 | 626 | 2,411 | |
| pMU6443 (repression tester) | None | <0.5 | 133 | 17 | 448 |
| Tyr | <0.5 | 61 | 11 | 215 | |
| Phe | <0.5 | 166 | NDb | ND | |
| pMU6443 and pMU1065 (repression tester and multicopy tyrR+) | None | <0.5 | 100 | ND | ND |
| Tyr | <0.5 | 20 | ND | ND | |
Methods for growth and enzyme assays were as described in the footnote to Table 1. The plasmids were present in strain JP7740 (tyrR+ hns+) or JP10938 (tyrR+ hns-205::Tn10). The coeffectors were present at a final concentration of 1 mM. Growth media for the strain derivatives carrying plasmid pMU1065 (46) were supplemented with kanamycin at 20 μg per ml. Enzyme specific activity values are reported in Miller units (24).
ND, not determined.
As expected, lacZ expression from the wild-type proU regulatory region was not affected by TyrR at low or high osmolarities in either the hns+ or hns background (compare the values for pMU6441 in Tables 1 and 2). Furthermore, even for the plasmids pMU6442 and pMU6443 in both the hns+ and hns derivatives, TyrR did not exert any significant regulatory effect during growth in the low- or high-osmolarity minimal media that were not supplemented with Phe or Tyr.
In the case of plasmid pMU6442 (bearing the activation tester variant of proU), we found that β-galactosidase expression was activated by TyrR in the presence of Phe, and less so in the presence of Tyr, in both the low- and the high-osmolarity media (Table 2). In the low-osmolarity medium, the magnitudes of activation mediated by TyrR-Phe for the hns+ and hns strains were approximately 200- and 10-fold, respectively. The corresponding values for activation mediated by TyrR-Tyr were around 22- and 1.5-fold, respectively. The marked TyrR-mediated activation for pMU6442 could not be demonstrated for another related plasmid variant (designated pMU6445) in which the strong TYR R box was positioned 3 bp closer to the P2 promoter (data not shown).
A moderate level of TyrR-mediated repression in the presence of Tyr (around twofold) was demonstrated for plasmid pMU6443 (bearing the repression tester variant of proU) in the hns+ strain at a high osmolarity and the hns mutant at both low and high osmolarities (Table 2). Repression in the hns+ strain at a low osmolarity could not be demonstrated because of the very low levels of basal expression in these cultures. Repression was rendered more pronounced (6.8-fold) in the hns+ strain additionally carrying a multicopy tyrR+ plasmid pMU1065 (46) (Table 2). As expected, growth in the presence of Phe did not repress lacZ expression from pMU6443 in the tyrR+ strain (Table 2).
Absence of correlation between intrinsic promoter strength and degree of Phe-mediated activation.
The level of activation by TyrR-Phe of proU in plasmid pMU6442 is at least an order of magnitude higher than that reported earlier for tyrP or other genes for aromatic amino acid metabolism (even after optimization of spacing between the strong TYR R box and the −35 region). We considered the possibility that this difference (in degree of activation) merely reflects the fact that the promoter for proU is inherently weaker than the TyrR-activable promoters of the native TyrR regulon. This hypothesis is rendered more plausible by the data in Table 2, which reveal that even in proU the degree of activation is most pronounced when the level of basal expression is the lowest (that is, in the hns+ strain grown in low-osmolarity medium).
We sought to test this hypothesis by creating a down-promoter mutation in tyrP and then examining the degree of activation by TyrR at the mutated promoter. For this purpose, the A residue (underlined) in the −35 hexamer (TTGACG) of tyrP was converted to the noncanonical C, which is found in proU P2 (Fig. 1A, sequence i), by site-directed mutagenesis. The tyrP template into which this mutation was introduced is identical to one described in an earlier study (48) that has the strong TYR R box situated 18 bp upstream of the −35 region (that is, at a location optimal for studying activation).
The expression of the lacZ reporter gene on each of two isogenic plasmids, pMU6449 and pMU2055, carrying the mutant and wild-type tyrP promoter sequences, respectively, was then determined in transformants of JP7740 (tyrR+) and JP8042 (tyrR). Consistent with the results of earlier work (1), the wild-type tyrP promoter was activated 12- and 6.5-fold by Phe and Tyr, respectively, in the tyrR+ host (Table 3). The mutant tyrP promoter exhibited a 16-fold reduction in basal expression in the tyrR strain, but the levels of activation supported by TyrR (8- and 4-fold with Phe and Tyr, respectively) were more or less similar to those for the wild-type promoter (Table 3). We therefore conclude that there is no correlation, at least in tyrP, between promoter strength and the magnitude of TyrR-mediated activation.
TABLE 3.
β-Galactosidase expression from tyrP-lac fusions on plasmids pMU2055 and pMU6449a
| Plasmid | tyrP −35 region (sequence) | β-Galactosidase activity for:
|
|||
|---|---|---|---|---|---|
| tyrR366 |
tyrR+
|
||||
| MM | MM + Tyr | MM + Phe | |||
| pMU2055 | Wild type (TTGACG) | 130 | 138 | 850 | 1,560 |
| pMU6449 | Mutant (TTGCCG) | 8 | 4 | 31 | 67 |
The plasmids were present in strain JP8042 (tyrR366) or JP7740 (tyrR+). Methods for growth and enzyme assays were as described in the footnote to Table 1, with the modification that the minimal salts medium (MM) used was prepared from half-strength medium 56. Tyr or Phe supplementation was at 1 mM. Enzyme specific activity values are reported in Miller units (24).
Conclusions.
In this study, we have successfully designed and created modified proU regulatory regions that have now acquired an additional facet of activation or repression control by the TyrR protein and that still retain substantial osmoresponsivity in the tyrR mutant background. These results establish, for the first time, that appropriately positioned TYR R boxes are sufficient to confer TyrR-mediated regulation on a heterologous promoter in vivo.
Although the proU regulatory region used in this study carries two promoters, several lines of evidence suggest that osmoresponsivity and TyrR control are both exerted at promoter P2. (i) As mentioned above and reviewed earlier (6, 14), no role for P1 in normal proU osmotic regulation has yet been established. Mutations that abolish P2 promoter activity abolish proU expression. Conversely, rpoS mutations that abolish P1 promoter activity do not affect normal proU regulation. Furthermore, there is no evidence that transcription from P1 traverses past P2 into the NRE region (33). (ii) The placement of the TYR R box(es) in the activation tester and repression tester variant plasmids pMU6442 and pMU6443, respectively, was designed specifically to exert regulation at the P2 promoter. (iii) Finally, the osmoresponsivity of lacZ expression from plasmids pMU6442 and pMU6443 was not affected in an rpoS::Tn10 mutant (data not shown), thereby excluding a role for the P1 promoter in such regulation.
The striking finding in this study was the 200-fold stimulation of proU expression at a low osmolarity achieved with TyrR-Phe in the activation tester variant. The fact that the binding of the TyrR protein dimer to the strong TYR R box is constitutive, that is, independent of Phe (2, 4, 31, 35), with the latter merely serving to convert the bound protein into an active conformation for the recruitment of RNA polymerase, allows us to make two inferences: (i) the strong TYR R box upstream of P2 is accessible for TyrR protein binding even at a low osmolarity, and (ii) TyrR binding by itself (in the absence of Phe) has no effect on proU repression under these conditions. Our results therefore indicate that if silencing does occur at the proU P2 promoter, it does not extend to this upstream TYR R box region.
Our findings may also be important for an understanding of antirepression as a mechanism of activation of gene expression in E. coli. An antirepressor may be operationally defined as a factor which promotes transcription by interfering with a system of repression. Antirepression may be said to exist when the magnitude of transcriptional activation mediated by the factor is higher in the presence of a particular repressing condition than in its absence. Examples of transcriptional activation by RNA polymerase recruitment and antirepression may not be mutually exclusive.
Several instances in which DNA-binding regulator proteins act as antirepressors of H-NS in mediating transcriptional activation are known. These include cyclic AMP-cyclic AMP receptor protein for the divergently transcribed promoters in the pap locus (11) and perhaps too for bgl (26, 39), CfaD for the promoter of the cfaABCE operon (17), IHF for the early promoter of phage Mu (45), and FIS for the P1 promoter of each of the rRNA operons (42) and perhaps for the hns promoter itself (9). In each case, it has been assumed that binding of the specific regulator protein to DNA directly alters the nucleoprotein topology in a manner that renders H-NS incapable of repression.
Earlier results obtained with TyrR also suggest that the protein acts as an antirepressor of HU and IHF in mediating activation at the mtr and tyrP promoters (48). In the present study as well, we found that the magnitude of TyrR-mediated activation of proU in pMU6442 at a low osmolarity, in the presence of either Tyr or Phe, is much higher in the hns+ strain (where H-NS serves to repress proU expression) than in the hns mutant (Table 2). Therefore, TyrR fulfils the operational definition of an antirepressor of H-NS in this situation. Yet, as argued above, TyrR binding by itself (in the absence of Phe) does not alter the repressive nucleoprotein topology at proU during growth in low-osmolarity medium. Therefore, our findings implicate, for the first time, recruited RNA polymerase as a component in the mechanism of antirepression.
Finally, the results in Table 3 also indicate that the substantially enhanced magnitude of stimulation at proU by TyrR-Phe may not simply be a consequence of proU bearing a weaker promoter than that of tyrP. One could speculate, therefore, that this difference is a reflection of the relative degrees of basal repression to which different promoters, including those of the native TyrR regulon (48), are subjected by the binding of the nucleoid proteins.
Acknowledgments
We thank all the members of the Pittard laboratory for their advice and stimulating discussions.
Financial support for the study was provided by the Australian Research Council and the Bilateral Science and Technology Collaboration Program (to A.J.P.) and by the award of a CSIR Raman Research Fellowship (to J.G.). J.G. is an Honorary Senior Fellow of the Jawaharlal Nehru Centre for Advanced Scientific Research.
REFERENCES
- 1.Andrews A E, Dickson B, Lawley B, Cobbett C, Pittard A J. Importance of the position of TYR R boxes for repression and activation of the tyrP and aroF genes in Escherichia coli. J Bacteriol. 1991;173:5079–5085. doi: 10.1128/jb.173.16.5079-5085.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews A E, Lawley B, Pittard A J. Mutational analysis of repression and activation of the tyrP gene in Escherichia coli. J Bacteriol. 1991;173:5068–5078. doi: 10.1128/jb.173.16.5068-5078.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 4.Baseggio N, Davies W D, Davidson B E. Identification of the promoter, operator, and 5′ and 3′ ends of the mRNA of the Escherichia coli K-12 gene aroG. J Bacteriol. 1990;172:2547–2557. doi: 10.1128/jb.172.5.2547-2557.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brand A H, Breeden L, Abraham J, Sternglanz R, Nasmyth K. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985;41:41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- 6.Csonka L N, Epstein W. Osmoregulation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1210–1223. [Google Scholar]
- 7.Dattananda C S, Rajkumari K, Gowrishankar J. Multiple mechanisms contribute to osmotic inducibility of proU operon expression in Escherichia coli: demonstration of two osmoresponsive promoters and of a negative regulatory element within the first structural gene. J Bacteriol. 1991;173:7481–7490. doi: 10.1128/jb.173.23.7481-7490.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dersch P, Kneip S, Bremer E. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K-12 to a cold environment. Mol Gen Genet. 1994;245:255–259. doi: 10.1007/BF00283274. [DOI] [PubMed] [Google Scholar]
- 9.Falconi M, Brandi A, La Teana A, Gualerzi C O, Pon C L. Antagonistic involvement of FIS and H-NS proteins in the transcriptional control of hns expression. Mol Microbiol. 1996;19:965–975. doi: 10.1046/j.1365-2958.1996.436961.x. [DOI] [PubMed] [Google Scholar]
- 10.Fletcher S A, Csonka L N. Fine-structure deletion analysis of the transcriptional silencer of the proU operon of Salmonella typhimurium. J Bacteriol. 1995;177:4508–4513. doi: 10.1128/jb.177.15.4508-4513.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forsman K, Sondén B, Göransson M, Uhlin B E. Antirepression function in Escherichia coli for the cAMP-cAMP receptor protein transcriptional activator. Proc Natl Acad Sci USA. 1992;89:9880–9884. doi: 10.1073/pnas.89.20.9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Göransson M, Sondén B, Nilsson P, Dagberg B, Forsman K, Emanuelsson K, Uhlin B E. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature. 1990;344:682–685. doi: 10.1038/344682a0. [DOI] [PubMed] [Google Scholar]
- 13.Gowrishankar J. Nucleotide sequence of the osmoregulatory proU operon of Escherichia coli. J Bacteriol. 1989;171:1923–1931. doi: 10.1128/jb.171.4.1923-1931.1989. . (Erratum, 172:1165, 1990.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gowrishankar J, Manna D. How is osmotic regulation of transcription of the Escherichia coli proU operon achieved? A review and a model. Genetica. 1996;97:363–378. doi: 10.1007/BF00055322. [DOI] [PubMed] [Google Scholar]
- 15.Hochschild A, Dove S L. Protein-protein contacts that activate and repress prokaryotic transcription. Cell. 1998;92:597–600. doi: 10.1016/s0092-8674(00)81126-5. [DOI] [PubMed] [Google Scholar]
- 16.Hwang, J. S., and A. J. Pittard. Unpublished data.
- 17.Jordi B J A M, Dagberg B, de Haan L A M, Hamers A M, van der Zeijst B A M, Gaastra W, Uhlin B E. The positive regulator CfaD overcomes the repression mediated by histone-line protein H-NS (H1) in the CFA/I fimbrial operon of Escherichia coli. EMBO J. 1992;11:2627–2632. doi: 10.1002/j.1460-2075.1992.tb05328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordi B J A M, Fielder A E, Burns C M, Hinton J C D, Dover N, Ussery D W, Higgins C F. DNA binding is not sufficient for H-NS-mediated repression of proU expression. J Biol Chem. 1997;272:12083–12090. doi: 10.1074/jbc.272.18.12083. [DOI] [PubMed] [Google Scholar]
- 19.Jordi B J A M, Owen-Hughes T, Hulton C S, Higgins C F. DNA twist, flexibility and transcription of the osmoregulated proU promoter of Salmonella typhimurium. EMBO J. 1995;14:5690–5700. doi: 10.1002/j.1460-2075.1995.tb00256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 21.Lawley B, Fujita N, Ishihama A, Pittard A J. The TyrR protein of Escherichia coli is a class I transcription activator. J Bacteriol. 1995;177:238–241. doi: 10.1128/jb.177.1.238-241.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lucht J M, Dersch P, Kempf B, Bremer E. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J Biol Chem. 1994;269:6578–6586. [PubMed] [Google Scholar]
- 23.McGovern V, Higgins N P, Chiz R S, Jaworski A. H-NS over-expression induces an artificial stationary phase by silencing global transcription. Biochimie. 1994;76:1019–1029. doi: 10.1016/0300-9084(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 25.Monod J, Cohen-Bazire G, Cohn M. Sur la biosynthèse de la β-galactosidase (lactase) chez Escherichia coli. La spécificité de l’induction. Biochim Biophys Acta. 1951;7:585–599. doi: 10.1016/0006-3002(51)90072-8. [DOI] [PubMed] [Google Scholar]
- 26.Mukerji M, Mahadevan S. Characterization of the negative elements involved in silencing the bgl operon of Escherichia coli: possible roles for DNA gyrase, H-NS, and CRP-cAMP in regulation. Mol Microbiol. 1997;24:617–627. doi: 10.1046/j.1365-2958.1997.3621725.x. [DOI] [PubMed] [Google Scholar]
- 27.Ogbourne S, Antalis T M. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem J. 1998;331:1–14. doi: 10.1042/bj3310001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Overdier D G, Csonka L N. A transcriptional silencer downstream of the promoter in the osmotically controlled proU operon of Salmonella typhimurium. Proc Natl Acad Sci USA. 1992;89:3140–3144. doi: 10.1073/pnas.89.7.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owen-Hughes T A, Pavitt G D, Santos D S, Sidebotham J M, Hulton C S J, Hinton J C D, Higgins C F. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell. 1992;71:255–265. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- 30.Pittard A J. Biosynthesis of the aromatic amino acids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 458–484. [Google Scholar]
- 31.Pittard A J, Davidson B E. TyrR protein of Escherichia coli and its role as repressor and activator. Mol Microbiol. 1991;5:1585–1592. doi: 10.1111/j.1365-2958.1991.tb01904.x. [DOI] [PubMed] [Google Scholar]
- 32.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 33.Rajkumari K, Ishihama A, Gowrishankar J. Evidence for transcription attenuation rendering cryptic a ςS-dependent promoter of the osmotically regulated proU operon of Salmonella typhimurium. J Bacteriol. 1997;179:7169–7173. doi: 10.1128/jb.179.22.7169-7173.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Sarsero J P, Pittard A J. Molecular analysis of the TyrR protein-mediated activation of mtr gene expression in Escherichia coli K-12. J Bacteriol. 1991;173:7701–7704. doi: 10.1128/jb.173.23.7701-7704.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 37.Schleif R. Two positively regulated systems, ara and mal. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D. C: American Society for Microbiology; 1996. pp. 1300–1309. [Google Scholar]
- 38.Schnetz K. Silencing of Escherichia coli bgl promoter by flanking sequence elements. EMBO J. 1995;14:2545–2550. doi: 10.1002/j.1460-2075.1995.tb07252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnetz K, Wang J C. Silencing of the Escherichia coli bgl promoter: effects of template supercoiling and cell extracts on promoter activity in vitro. Nucleic Acids Res. 1996;24:2422–2428. doi: 10.1093/nar/24.12.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka K, Muramatsu S, Yamada H, Mizuno T. Systematic characterization of curved DNA segments randomly cloned from Escherichia coli and their functional significance. Mol Gen Genet. 1991;226:367–376. doi: 10.1007/BF00260648. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka K, Ueguchi C, Mizuno T. Importance of stereospecific positioning of the upstream cis-acting DNA element containing a curved DNA structure for the functioning of the Escherichia coli proV promoter. Biosci Biotechnol Biochem. 1994;58:1097–1101. doi: 10.1271/bbb.58.1097. [DOI] [PubMed] [Google Scholar]
- 42.Tippner D, Afflerbach H, Bradaczek C, Wagner R. Evidence for a regulatory function of the histone-like Escherichia coli protein H-NS in ribosomal RNA synthesis. Mol Microbiol. 1994;11:589–604. doi: 10.1111/j.1365-2958.1994.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 43.Ueguchi C, Mizuno T. The Escherichia coli nucleoid protein H-NS functions directly as a transcriptional repressor. EMBO J. 1993;12:1039–1046. doi: 10.1002/j.1460-2075.1993.tb05745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandeyar M A, Weiner M P, Hutton C J, Batt C A. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene. 1988;65:129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- 45.van Ulsen P, Hillebrand M, Zulianello L, van de Putte P, Goosen N. Integration host factor alleviates the H-NS-mediated repression of the early promoter of bacteriophage Mu. Mol Microbiol. 1996;21:567–578. doi: 10.1111/j.1365-2958.1996.tb02565.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang P, Yang J, Pittard A J. Promoters and transcripts associated with the aroP gene of Escherichia coli. J Bacteriol. 1997;179:4206–4212. doi: 10.1128/jb.179.13.4206-4212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams R M, Rimsky S. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks. FEMS Microbiol Lett. 1997;156:175–185. doi: 10.1111/j.1574-6968.1997.tb12724.x. [DOI] [PubMed] [Google Scholar]
- 48.Yang J, Camakaris H, Pittard A J. In vitro transcriptional analysis of TyrR-mediated activation of the mtr and tyrP+3 promoters of Escherichia coli. J Bacteriol. 1996;178:6389–6393. doi: 10.1128/jb.178.21.6389-6393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Fletcher S A, Csonka L N. Site-directed mutational analysis of the osmotically regulated proU promoter of Salmonella typhimurium. J Bacteriol. 1996;178:3377–3379. doi: 10.1128/jb.178.11.3377-3379.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]