Abstract
Plasmids with the aadA gene from plasmid R100, which confers resistance to the aminoglycosides spectinomycin and streptomycin in Escherchia coli, can be introduced into wild-type Myxococcus xanthus, strain DK1622, by electroporation. Recombinant M. xanthus strains with integrated plasmids carrying the aadA gene acquire resistance to high levels of these antibiotics. Selection for aadA in M. xanthus can be carried out independently of, or simultaneously with, selection for resistance to kanamycin. The kinds and frequencies of recombination events observed between integrative plasmids with aadA and the M. xanthus chromosome are similar to those observed after the transformation of yeast. Cleavage of integrative plasmid DNA at a site adjacent to a region of homology between the plasmid and the M. xanthus genome favors the targeted disruption of M. xanthus genes by allele replacement.
The soil bacterium Myxococcus xanthus is a model prokaryotic system for the study of intercellular communication. Cell-cell interactions play central roles in both gliding motility and an elaborate multicellular developmental cycle initiated by starvation. Development leads to the morphogenesis of fruiting bodies containing differentiated, heat-resistant spores. Although the change from rod-shaped vegetative cells to spherical spores is rapid, genetic methods have been used successfully to arrange the early steps of this process into an ordered series of molecular events. Many mutations that block M. xanthus development do not affect vegetative growth. Consequently, methods for the analysis of mutations that block development have relied on genetic manipulations with vegetative cells.
Following the discovery that transposon Tn5 is active in M. xanthus (14), Tn5-lac, among the first hybrid transposons used to generate reporter gene fusions, was constructed and used in M. xanthus (11). The isolation of transposon-generated fusions of lacZ with developmentally induced promoters has resulted in a set of temporal signposts that distinguish different developmental stages (12, 13). In addition, M. xanthus has generalized transducing phages (5, 18). which permit fine-structure mapping. One of these phages, Mx8, is temperate and integrates as a prophage at a preferred chromosomal locus (attB). Because plasmid origins of replication that function in other gram-negative bacteria do not function as origins in M. xanthus, these plasmids can be used as integrative vectors if they carry either a region of homology with the M. xanthus genome (19, 24, 26) or the Mx8 int-attP region (15, 20).
Because M. xanthus has a high frequency of homologous recombination, the integration of plasmids by homologous recombination can be used to generate merodiploids and exchange engineered alleles with chromosomal alleles of targeted genes (19). Thus, plasmids with cloned M. xanthus inserts can be used in the same way that yeast integrative plasmids (yIPs) are used to manipulate the Saccharomyces cerevisiae genome (8, 21). Alternatively, plasmids can be integrated at the attB locus to construct more-stable merodiploids for complementation tests (24, 26). Plasmids can be introduced into M. xanthus efficiently by using the improved method for electroporation we developed (10).
The rules for homologous recombination between a plasmid and the M. xanthus genome appear to be similar to those for recombination between yIPs and the S. cerevisiae genome (21). Both circular and linear plasmid DNAs give rise to recombinants, and electroporation with linear plasmid DNA often results in the integration of a circular form of the plasmid, presumably mediated by gap repair of double-stranded ends. Gap repair of the ends of linear plasmids poses a significant problem for the construction of gene disruptions, because single crossover events between a repaired plasmid and the genome, leading to cointegrate formation, can be more frequent than double crossover events leading to allele replacement.
To date, only the kanamycin resistance (Kmr) determinants derived from Tn5 (14) or Tn903 (25) have worked well in both Escherichia coli and M. xanthus hosts. Unfortunately, M. xanthus is naturally resistant to gentamicin. Although tetracycline resistance determinants have been shown to function in M. xanthus (2), these determinants work poorly when it is grown in its preferred, rich medium supplemented with Mg2+. Growth in the presence of Mg2+ likely facilitates a natural mechanism for tetracycline export and thereby leads to a high background of tetracycline-resistance phenocopies. Although resistance to tetracycline mediated by the Tn10 tetA determinant works well in M. xanthus, selection for this gene on high-copy-number plasmids in E. coli is lethal (4), unless the cells are plated on media with oxytetracycline. The tetA determinant of plasmid pBR322, which works well in high copy numbers in E. coli, works relatively poorly in M. xanthus (our unpublished observations). Li and Shimkets have used the trimethoprim resistance determinant for plasmid R338 successfully in M. xanthus (16). We have found that its success in M. xanthus depends on the relative concentration of competing folates in rich, Casitone-containing medium and that selection for this marker in many E. coli hosts can be problematic.
As part of our search for additional antibiotic resistance determinants that function well in both M. xanthus and E. coli, we tested whether the aadA gene of plasmid R100 might confer resistance to both spectinomycin (Spr) and streptomycin (Smr) in M. xanthus. Campos and coworkers have shown that M. xanthus is sensitive to both antibiotics (5). Consistent with this, we found that wild-type M. xanthus (strain DK1622) (9) plated with low efficiencies (8 × 10−7) on rich CTPM (28) medium supplemented with either 0.8 mg of spectinomycin per ml or 1.0 mg of streptomycin per ml. On medium with both antibiotics, M. xanthus plated with an even lower efficiency (4 × 10−7), and usually >90% of the colonies formed after 5 days at 32°C did not grow when repurified on the same medium. Presumably, the potency of these antibiotics decreased during the long (5- to 7-day) incubation times required for the growth of M. xanthus colonies. Consistent with this idea, the frequency of resistant phenocopies is greater on plates that have been stored at room temperature for several days before use.
To test whether the aadA gene confers antibiotic resistance on M. xanthus, we constructed plasmid pAY952 (17). This plasmid was constructed by adding the 2.2-kb fragment of Mx8 DNA with the int and attP genes to plasmid pGB2 (6), which has both the aadA gene and an origin of replication derived from R100. Upon electroporation of pAY952 into DK1622, Spr Smr recombinants arose at a frequency of about 200-fold above the backgroud frequency of spontaneous resistant mutants (Table 1), indicating that the aadA gene functions in M. xanthus.
TABLE 1.
Efficiencies of electroporation of int-attP plasmids into wild-type M. xanthus (strain DK1622).
| Plasmid | Refer-ence | Size (kb) | Marker(s)a | EOE (μg−1)b |
|---|---|---|---|---|
| pBGS18 | 25 | 3.6 | Kmr | 0 |
| pAY721 | 23 | 5.8 | Kmrint-attP region | 2.6 ± 104 |
| pGB2 | 6 | 4.0 | Spr Smr | 1.6 ± 102 |
| pAY952 | 17 | 6.2 | Spr Smrint-attP region | 3.6 ± 104 |
Plasmids carry either the npt! gene (Kmr determinant) from transposon Tn903 or the aadA gene (Spr Smr determinant) from conjugative plasmid R100; a subset of plasmids also encode the int-attP site-specific recombination functions from temperate M. xanthus phage Mx8.
Efficiencies of electroporation (EOE) were determined from the average results of at least four independent experiments, in which 100 to 200 ng of DNA was used to electroporate 109 wild-type cells as described previously (10). The value of 1.6 ± 102electroporants/μg for pGB2 DNA represents the background of spontaneous Spr Smr mutants; all of these (23 of 23 tested) did not repurify as resistant colonies. Spontaneous resistant mutants can be also distinguished from recombinants carrying integrated plasmids by amplifying purified M. xanthus DNA from resistant strains with the primers TM5 (CCCCAAGCTTGGTACCACTAGTTATTTGCCGACTACCTTGGTGA) and TM6 (AAAAAAGCTTCCATGGTTTCATGGCTTGTTATGACTG), specific for the aadA gene and its promoter, respectively. In addition, these primers permit amplification of the aadA gene and its promoter from plasmid templates for direct cloning into sites within M. xanthus genes.
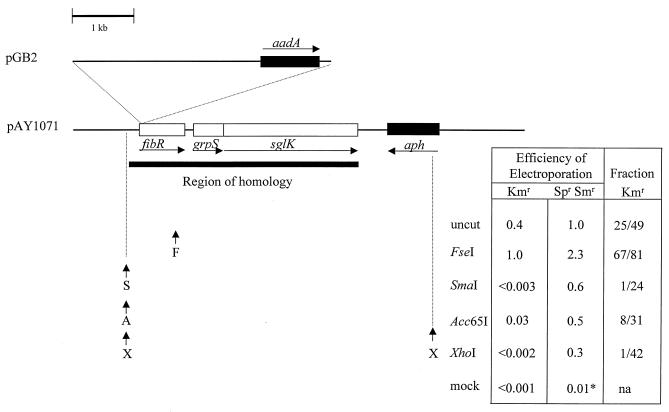
To demonstrate that this Spr Smr marker can be used in combination with selection for kanamycin resistance, we constructed a cointegrate plasmid, pAY1074, with an insertion of pGB2 in the fibR gene (28). The fibR gene is located within a 3.5-kb region of the M. xanthus genome which we subcloned from plasmid pAY694 (28) into plasmid vector pBGS18, which has the Kmr determinant of transposon Tn903 (25), to make pAY1071. To build pAY1071, the 3.5-kb BglII-HindIII fragment of pAY694 was ligated to the BamHI and HindIII sites of pBGS18. To build pAY1074, pAY1071 was cleaved at a unique AgeI site within fibR, pGB2 was cleaved at its unique XmaI site in its polylinker, and the plasmids were ligated together.
We electroporated both circular plasmid DNA and plasmid DNA treated with several different restriction endonucleases into DK1622, selected for Kmr Spr Smr or Spr Smr recombinants, and screened the Spr Smr recombinants for the Kmr phenotype. Four conclusions can be drawn from the results shown in Fig. 1. First, when a closed circular plasmid carrying a Spr Smr insertion in a 3.5-kb region of homology with the M. xanthus genome is electroporated into M. xanthus, about half of the recombinants are Kmr cointegrates, and the other half are Kms strains in which an allele replacement event has occurred. Second, cleavage of the plasmid at a unique site within the region of homology (FseI) stimulates recombination between the plasmid and chromosome. This result suggests that, as in yeast (21), DNA ends are recombinogenic in M. xanthus. However, cleavage at a unique site within the region of homology (such as the FseI site of pAY1074) does not discourage the formation of cointegrates. Third, cleavage of plasmid pAY1074 at a unique site past the end of the region of homology (SmaI or Acc65I) does not stimulate the yield of recombinants, as it does for some yIPs (21). However, it favors the recovery of Spr Smr strains resulting from allele replacement events, and this bias is more pronounced with SmaI, which generates a blunt-ended linear module. Fourth, cleavages of the plasmid at two sites (with XhoI), one at the junction of the homologous region and a second with the Kmr determinant of the plasmid vector backbone, have similar effects on the frequency and types of recombinants. In both M. xanthus and S. cerevisiae, illegitimate recombination events between homologous substrates and the genome are almost never observed, and the similarities between the recombinational fates of circular and linear plasmid DNAs in these hosts are striking. Thus, the simplest interpretation of our results is that, as in S. cerevisiae, double-stranded DNA ends are the preferred substrates for the M. xanthus recombinational machinery.
FIG. 1.
Cleavage of plasmid DNA prior to electroporation into M. xanthus affects the frequency and nature of recombination events between the plasmid and chromosome. Shown to approximate scale in the upper left is the structure of plasmid pAY1074, which is Kmr (aph) plasmid pAY1071 with an insertion of Spr Smr (aadA) plasmid pGB2 in the M. xanthus fibR gene. The region of homology carried by the plasmid is 3.5 kb in length (28). The efficiencies of electroporation of closed circular plasmid pAY1074 DNA (uncut) and pAY1074 DNA cleaved at the indicated sites are compared on the right. The first two columns are numbers representing the independent measurements of Kmr and Spr Smr recombinants arising from the same electroporations and are the averages of six independent determinations. Fractions shown in the third column were obtained by testing individual Spr Smr electroporants obtained from two independent determinations for a Kmr phenotype. All efficiencies of electroporation are given realtive to the efficiency of Spr Smr recombinants arising with uncut pAY1074 DNA (4.5 × 103 ± 2.7 × 103 μg−1). Cleavage of pAY1074 DNA at its unique FseI site within the region of homology stimulated the formation of recombinants, whereas cleavage at sites at the junction of, or past, the region of homology reduced the frequency of Spr Smr recombinants. Prior cleavage at sites outside the region of homology reduced the frequency of recovery of Kmr Spr Smr conintegrates between the plasmid and chromosome. Kms Spr Smr recombinants were favored, presumably after double-recombination events flanking the pGB2 insertion resulted in the desired allele replacement event. The asterisk indicates that more than 90% of the colonies obtained on medium with both antibiotics, spectinomycin and streptomycin, after mock electroporation (with no plasmid DNA) did not grow when repurified on the same medium, na, not applicable.
These results show that a simple procedure can be used to cross an insertion marked by an antibiotic resistance determinant onto the M. xanthus chromosome. A plasmid carrying the insertion should be cleaved at one or both ends of its region of homology with the chromosome prior to electroporation to discourage the formation of cointegrates. We have obtained similar results after cleavage and electroporation of plasmids with several different subcloned regions of the M. xanthus genome. In each case, cleavage of a plasmid at one or both ends of the region of homology with the chromosome favors the recovery of recombinants in which allele replacement events have occurred. This method for generating allele replacements on the M. xanthus genome is simpler than the previous methods, which have relied on two sequential selections for the integration and excision of plasmids carrying the selectable Kmr determinant and the counterselectable E. coli galK (27) or Bacillus subtilis sacBR (29) genes.
We note that the electroporation of linear DNA molecules derived from circular plasmids into several other bacteria has been used successfully to generate allele replacements. Linearization of a plasmid carrying a subcloned Haemophilus ducreyi gene interrupted by an antibiotic resistance determinant also appears to stimulate allele exchange (7). The more recent demonstrations that linear DNA molecules permit allelic exchange in Mycobacterium bovis BCG (1), Mycobacterium tuberculosis (3), and Borrelia burgdorferi (22) will facilitate the genetic analysis of these pathogens in the near future.
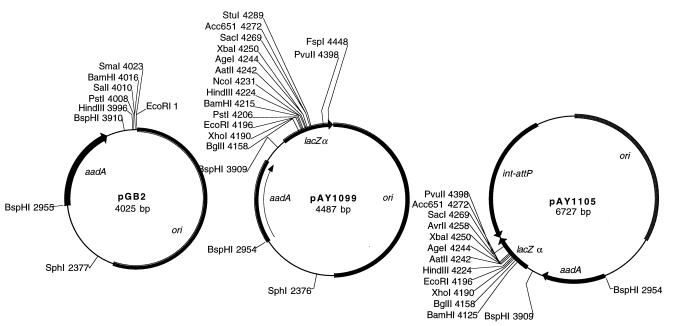
To increase the versatility of Spr Smr plasmids as shuttle vectors for use in both E. coli and M. xanthus, we constructed a derivative of pGB2, pAY1099, with the α-complementing fragment of the lacZ gene from plasmid pLTMUS28 (New England Biolabs). Primers 5′ GGAGGGTGGCCAAATGTGAGTTAGCTCACTCA and GCCGGCCAATTGTTATTACCAAGCGAAGCGCC were used to amplify bp 2315 to 2798 of pLITMUS28, and the amplified product was cleaved with MscI and MfeI and ligated to the EcoRI and filled-in HindIII sites of pGB2 (6). This amplified fragment has many unique cloning sites within a polylinker located at the 5′ end of the lacZ gene. Many recombinant plasmids with inserts in this polylinker can be screened by α-complementation in an appropriate E. coli host, such as JM107 (30), in the presence of chromogenic substrates for β-galactosidase.
We also made a derivative of pGB2 with both the α-complementing fragment and the Mx8 int-attP region present in pAY952. pAY952 was cleaved with EcoRI and Acc65I, ends were filled in, and the plasmid backbone was ligated to make plasmid pAY1103. The amplified fragment of pLITMUS28 was cleaved with MscI and MfeI and ligated to the filled-in HindIII site of pAY1103 to make pAY1105. When derivatives of pAY1105 are electroporated into M. xanthus, they prefer to integrate at the attB bacterial attachment site for prophage Mx8, even if they carry a region of homology with the M. xanthus genome (data not shown). The structures of these integrative shuttle vectors, which may have more general uses in other myxobacteria or gram-negative hosts sensitive to the aminoglycosides spectinomycin and streptomycin, are shown in Fig. 2.
FIG. 2.
Structures of plasmids pGB2, pAY1099, and pAY1105. The aadA gene is flanked by a pair of BspHI sites in each vector, and its translation initiates at an ATG codon within one of these sites. Also shown are unique sites within the polylinker of pGB2 derived from pUC8 and within the fragment of the lacZ gene carried by plasmids pAY1099 and pAY1105 derived from pLITMUS28 (New England Biolabs). The positions of the aadA gene from plasmid R100, the region from R100 including its origin of replication (ori) (also present in Salmonella plasmid pSC101), the region of the E. coli lac operon encoding the β-complementing fragment of LacZ, and the int and attP genes from myxophage Mx8 are indicated. Text files of the sequences of these plasmids and detailed notes on their construction are available by e-mail upon request.
Acknowledgments
We thank George Churchward for the gift of plasmid pGB2 DNA and Ellie Graham for technical assistance.
This work was supported by grants GM50962 and GM53392 to P.L.H. and P.Y., respectively, from the National Institutes of Health and by EPSCoR grant OSR-9350539 to P.L.H.
REFERENCES
- 1.Aldovini A, Husson R N, Young R A. The uraA locus and homologous recombination in Mycobacterium bovis BCG. J Bacteriol. 1993;175:7282–7289. doi: 10.1128/jb.175.22.7282-7289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avery L, Kaiser D. In situ transposon replacement and isolation of a spontaneous tandem genetic duplication. Mol Gen Genet. 1983;191:99–109. doi: 10.1007/BF00330896. [DOI] [PubMed] [Google Scholar]
- 3.Balasubramanian V, Pavelka M S, Jr, Bardarov S S, Martin J, Weisbrod T R, McAdam R A, Bloom B R, Jacobs W R., Jr Allelic exchange in Mycobacterium tuberculosis with long linear recombination substrates. J Bacteriol. 1996;178:273–279. doi: 10.1128/jb.178.1.273-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg C M, Liu L, Wang B, Wang M-D. Rapid identification of bacterial genes that are lethal when cloned on multicopy plasmids. J Bacterial. 1988;170:468–470. doi: 10.1128/jb.170.1.468-470.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos J M, Geisselsoder J, Zusman D R. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978;119:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- 6.Churchward G, Belin D, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 7.Hansen E J, Latimer J L, Thomas S E, Helminen M, Albritton W L, Radolf J D. Use of electroporation to construct isogenic mutants of Haemophilus ducreyi. J Bacteriol. 1992;174:5442–5449. doi: 10.1128/jb.174.16.5442-5449.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinnen A, Hicks J B, Fink G R. Transformation of yeast. Proc Natl Acad Sci USA. 1978;75:1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kashefi K, Hartzell P L. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol Microbiol. 1995;15:483–494. doi: 10.1111/j.1365-2958.1995.tb02262.x. [DOI] [PubMed] [Google Scholar]
- 11.Kroos L, Kaiser D. Construction of Tn5-lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc Natl Acad Sci USA. 1984;81:5816–5820. doi: 10.1073/pnas.81.18.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroos L, Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987;1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- 13.Kroos L, Kuspa A, Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 14.Kuner J, Kaiser A D. Introduction of transposon Tn5 into Myxococcus for analysis of developmental and other non-selectable mutants. Proc Natl Acad Sci USA. 1981;78:425–429. doi: 10.1073/pnas.78.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Shimkets L J. Site-specific integration and expression of a developmental promoter in Myxococcus xanthus. J Bacteriol. 1988;170:5552–5556. doi: 10.1128/jb.170.12.5552-5556.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S-F, Shimkets L J. Effect of dsp mutations on the cell-to-cell transmission of CsgA in Myxococcus xanthus. J Bacteriol. 1993;175:3648–3652. doi: 10.1128/jb.175.11.3648-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magrini, V., C. Creighton, M. L. Storms, and P. Youderian. Site-specific recombination of temperate Myxococcus xanthus phage Mx8: genetic elements required for integration. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 18.Martin S, Sodergren E, Masuda T, Kaiser A D. Systematic isolation of transducing phages for Myxococcus xanthus. Virology. 1978;88:44–53. doi: 10.1016/0042-6822(78)90108-3. [DOI] [PubMed] [Google Scholar]
- 19.O’Connor K A, Zusman D R. Coliphage P1-mediated transduction of cloned DNA from Escherichia coli to Myxococcus xanthus: use for complementation and recombinational analyses. J Bacteriol. 1983;155:317–329. doi: 10.1128/jb.155.1.317-329.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orndorff P, Stellwag E, Starich T, Dworkin M, Zissler J. Genetic and physical characterization lysogeny by bacteriophage MX8 in Myxococcus xanthus. J Bacteriol. 1983;154:772–779. doi: 10.1128/jb.154.2.772-779.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orr-Weaver T L, Szostak J W, Rothstein R J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci USA. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosa P, Samuels D S, Hogan D, Stevenson B, Casjens C, Tilly K. Directed insertion of a selectable marker into a circular plasmid of Borrelia burgdorferi. J Bacteriol. 1996;178:5946–5953. doi: 10.1128/jb.178.20.5946-5953.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salmi D, Magrini V, Hartzell P L, Youderian P. Genetic determinants of immunity and integration of temperate Myxococcus xanthus phage Mx8. J Bacteriol. 1998;180:614–621. doi: 10.1128/jb.180.3.614-621.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimkets L J, Asher S J. Use of recombination techniques to examine the structure of the csg locus of Myxococcus xanthus. Mol Gen Genet. 1988;211:63–71. doi: 10.1007/BF00338394. [DOI] [PubMed] [Google Scholar]
- 25.Spratt B G, Hedge P J, te Heesen S, Edelman, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 26.Stephens K, Kaiser A D. Genetics of gliding motility in Myxococcus xanthus: molecular cloning of the mgl locus. Mol Gen Genet. 1987;207:256–266. [Google Scholar]
- 27.Ueki T, Inouye S, Inouye M. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene. 1996;183:153–157. doi: 10.1016/s0378-1119(96)00546-x. [DOI] [PubMed] [Google Scholar]
- 28.Weimer R M, Creighton C, Stassinopoulos A, Youderian P, Hartzell P L. A chaperone in the HSP70 family controls production of extracellular fibrils in Myxococcus xanthus. J Bacteriol. 1998;180:5357–5368. doi: 10.1128/jb.180.20.5357-5368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu S S, Kaiser D. Markerless deletions of pil genes in Myxococcus xanthus generated by counterselection with the Bacillus subtilis sacB gene. J Bacteriol. 1996;178:5817–5821. doi: 10.1128/jb.178.19.5817-5821.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]