Abstract
The first identification and characterization of a prokaryotic gene (spsA) encoding sucrose-phosphate synthase (SPS) is reported for Synechocystis sp. strain PCC 6803, a unicellular non-nitrogen-fixing cyanobacterium. Comparisons of the deduced amino acid sequence and some relevant biochemical properties of the enzyme with those of plant SPSs revealed important differences in the N-terminal and UDP-glucose binding site regions, substrate specificities, molecular masses, subunit compositions, and regulatory properties.
Sucrose is the major product of photosynthesis in most plants and is exported from leaves to all heterotrophic tissues. Sucrose-phosphate synthase (SPS) (UDP-glucose: d-fructose-6-phosphate 2-α-d-glucosyltransferase) (EC 2.4.1.14), one of the key enzymes in the control of sucrose synthesis, has been studied extensively in various plant species. Importantly, SPS activity is controlled by allosteric effectors (glucose-6-phosphate and orthophosphate) and by reversible covalent modification (6).
On the other hand, the knowledge of sucrose metabolism in unicellular organisms is very limited. Biochemical properties of SPSs isolated from several species of green algae (21) were similar to those of plant enzymes. The first evidence of the biosynthesis of sucrose through the action of SPS and sucrose-phosphate phosphatase in prokaryotes was shown by Porchia and Salerno (15), who described the isolation and characterization of two forms of SPS from Anabaena sp. strain PCC 7119, a filamentous heterocystous cyanobacterium. Biochemical properties of these enzymes were strikingly different from those of plant SPSs. We report here that SPS is also present in a unicellular non-nitrogen-fixing cyanobacterium and prove that the Synechocystis sp. strain PCC 6803 open reading frame (ORF) sll0045 (19) encodes a protein with SPS activity. This enzyme has distinct biochemical regulatory properties and molecular structure in comparison with those of plant SPSs.
(Part of this research was conducted by L. Curatti and P. Desplats in partial fulfillment of the requirements for Ph.D. degrees from Universidad Nacional de Mar del Plata, Mar del Plata, Argentina.)
Detection of SPS activity in Synechocystis sp. strain PCC 6803 extracts.
The presence of SPS in unicellular cyanobacteria has not been demonstrated until now. Therefore, we decided to examine the occurrence of SPS activity in Synechocystis, a unicellular non-nitrogen-fixing prokaryote. When Synechocystis cell extracts were subjected to ion-exchange chromatography on DEAE-Sephacel, a single peak of SPS activity eluted at a NaCl concentration similar to that of SPS-I from Anabaena sp. strain PCC 7119 (15). The reaction products were identified as UDP (27) and sucrose-6-phosphate (15). The apparent molecular mass (MM) of the native enzyme was about 85 kDa, as estimated by gel filtration through a Sephadex G-100 column (15; also data not shown).
Expression of the Synechocystis sp. strain PCC 6803 ORF sll0045 in Escherichia coli.
Recently, Kaneko and coworkers determined the complete sequence of the Synechocystis sp. strain PCC 6803 genome (9). Sequence comparison analysis revealed the presence of an ORF (sll0045) which shares about 30% identity with plant SPSs. To ascertain if this ORF encodes a protein with SPS activity, we constructed the plasmid pSySPS containing the putative SPS ORF flanked by 500 bp of upstream sequence and 1,200 bp of downstream sequence by standard protocols (25). Extracts from E. coli harboring pSySPS harvested in late exponential phase showed an SPS activity of about 0.25 μmol · min−1 · mg of protein−1 (14, 15, 19), indicating that the ORF sll0045 is contained in an SPS gene (spsA) and codes for an active SPS protein. Under similar conditions, E. coli harboring pBluescript II SK(+) had no SPS activity.
spsA gene disruption.
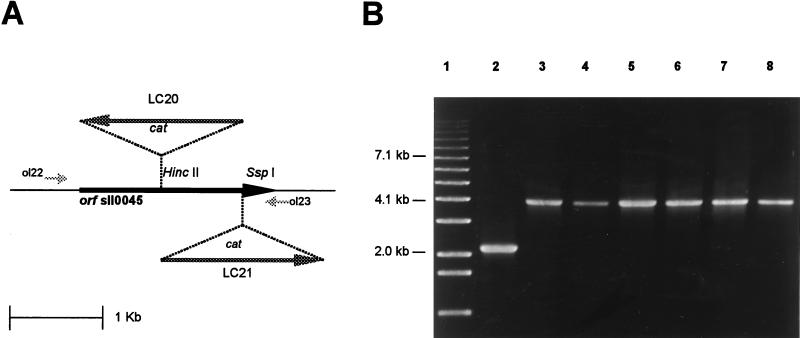
Two insertion mutants were constructed to further confirm the identity of the ORF sll0045. Several complete segregant clones were obtained after transformation of Synechocystis sp. strain PCC 6803 with pLC20 or pLC21 to generate strain LC20 or LC21 (16), respectively (Fig. 1A). The segregation was confirmed by PCR analysis (Fig. 1B). No SPS activity could be detected in the mutant cells, providing additional evidence that the ORF sll0045 encodes an SPS protein. The growth curves of Synechocystis sp. strain PCC 6803, LC20, and LC21 under standard conditions (BG-11 liquid medium, 28°C, continuous illumination) did not show significant differences, suggesting that SPS activity is not essential for their growth.
FIG. 1.
Insertion inactivation of Synechocystis sp. strain PCC 6803 ORF sll0045. (A) Integration sites of the cat cartridge used to obtain the insertion mutants LC20 and LC21. The arrows indicate the direction of transcription. (B) PCR analysis done with primers ol22 and ol23 (shown in panel A) and genomic DNA from Synechocystis sp. strain PCC 6803 (lane 2), LC20 (lanes 3 to 5), and LC21 (lanes 6 to 8). Lane 1, 1-kb ladder of molecular size markers (Gibco BRL).
Biochemical characterization of Synechocystis sp. strain PCC 6803 SPS expressed in E. coli.
To determine the biochemical properties of Synechocystis SPS, the enzyme was partially purified from extracts of E. coli harboring pSySPS. When extracts were chromatographed onto a DEAE-Sephacel column (15), a single peak of SPS activity was detected at an elution position similar to that of the enzyme isolated from Synechocystis cells. The concentrated SPS fraction at this stage (purified ca. 25-fold; free of phosphoglucose isomerase [13] and inorganic phosphatases [15]) was used to analyze some biochemical properties of the enzyme.
SPS activity showed a broad pH dependence, with a maximum around pH 7.5 to 8.5. The addition of divalent cations to the incubation mixture (10 mM MgCl2 or MnCl2) increased enzyme activity about 80 to 90%.
Specificity for the glucosyl donor was then investigated, and the kinetic constant values from Wolf’s plots were determined. As shown in Table 1, the enzyme was not specific for UDP-glucose: it could also use ADP-glucose and, to a minor extent, GDP-glucose as substrates. The apparent Km for fructose-6-phosphate was 1.5 mM. The enzymic preparation was not able to yield sucrose from fructose and UDP-glucose (or ADP-glucose).
TABLE 1.
Kinetic constants calculated for Synechocystis sp. strain PCC 6803 SPSa
| Substrate | Km (mM) | Vmax (U · μl−1) | Vmax/Km (U · mM−1 · μl−1) |
|---|---|---|---|
| ADP-glucose | 1.6 | 1.78 | 1.11 |
| UDP-glucose | 3.5 | 3.04 | 0.87 |
| GDP-glucose | 2.4 | 0.41 | 0.17 |
The data are representative of at least two independent experiments performed in duplicate. The standard deviations in all determinations did not exceed 10% of the mean. One unit corresponds to the production of one μmol of sucrose per min. Km values are apparent Kms.
The actions of the two allosteric effectors of plant SPS (glucose-6-phosphate [activator] and orthophosphate [inhibitor] [6]) on enzyme activity were assayed. Glucose-6-phosphate did not activate the enzyme, and orthophosphate caused only a 17% reduction of enzyme activity at concentrations as high as 20 mM. A similar result was obtained for Anabaena SPS (14). By comparison, 2 to 5 mM glucose-6-phosphate increases activity about fivefold, whereas 10 mM orthophosphate inhibits SPS activity from rice leaves by 80 to 90%, (20), indicating that the regulation of plant and cyanobacterium SPSs may be different.
Sequence analysis of the Synechocystis SPS protein.
The Synechocystis ORF sll0045 encodes a protein with a predicted MM of 81,421 Da. This value is similar to the apparent MM calculated for the native SPS isolated from Synechocystis extracts (see above). These results indicate that Synechocystis SPS protein is composed of a single polypeptide. Our recent studies showed that Anabaena SPS is also a monomer, with an apparent MM of 45 to 47 kDa (15). The different MMs of both cyanobacterial enzymes could be due either to species differences or to proteolysis of Anabaena SPS during the isolation procedure. By contrast, plant SPSs are either dimers or tetramers of 116- to 138-kDa subunits, depending on the experimental procedure used to determine the relative MM of the holoenzyme (1, 20, 22).
A comparison of the deduced amino acid sequences of Synechocystis and plant SPSs (Fig. 2) revealed two important differences: (i) Synechocystis SPS lacks the N-terminal 178-amino-acid-residue sequence of plant SPSs, which contains the phosphorylation site reported to be critical for the regulation of the enzyme activity (Ser-158 in spinach [12]; Ser-162 in maize [7]), and (ii) the UDP-glucose binding site determined in spinach SPS by photoaffinity labeling (23) and highly conserved in plant species is remarkably divergent in Synechocystis SPS (amino acid residues 59 to 72). We speculate that this divergence may be responsible for the observed differences in substrate specificity: plant SPSs are specific for UDP-glucose, whereas Synechocystis (Table 1) and Anabaena (15) SPSs are not. The putative fructose-6-phosphate (the other SPS substrate) binding site (residues 199 to 206 in maize SPS [24]) is highly conserved in the Synechocystis SPS sequence (Fig. 2).
FIG. 2.
Alignment of the N-terminal regions of the predicted SPS amino acid sequences from Synechocystis sp. strain PCC 6803 (residues 1 to 83) (9), Zea mays (residues 1 to 256) (30), and Spinacia oleracea (residues 1 to 256) (10). This alignment was generated with the program Megalign of the DNAStar package, using the Clustal method with PAM 250 residue weight table. Positions that are identical in different proteins are indicated in bold type. Stippled boxes indicate the UDP-glucose (box 2) and putative fructose-6-phosphate (box 1) binding domains. The sites of phosphorylation in plant SPSs (Ser-162 for maize and Ser-158 for spinach) (6) are underlined and indicated by arrowheads. Gaps introduced to maximize alignment are indicated by dashes.
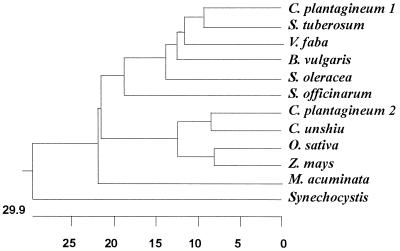
A dendogram generated using the deduced amino acid sequences reported for various SPSs shows that the Synechocystis enzyme clearly diverges from plant SPSs (Fig. 3).
FIG. 3.
Phylogenetic tree of SPSs. SPSs from Craterostigma plantagineum (8), Solanum tuberosum (2), Vicia faba (4), Beta vulgaris (5), Spinacia oleracea (10), Zea mays (30), Saccharum officinarum (GenBank accession no. AB001338), Oryza sativa (28), Citrus unshiu (11), Musa acuminata (GenBank accession no. U59489), and Synechocystis sp. strain PCC 6803 (9) were used to construct this tree. The tree was generated with the program Megalign of the DNAStar package, using the Clustal method with PAM 250 residue weight table.
By using Southern blot analysis, we failed to detect spsA homologous sequences in other cyanobacteria corresponding to different taxonomical groups (18): Synechoccocus sp. strain PCC 7942 (group I); Anabaena sp. strain PCC 7119, Anabaena sp. strain PCC 7120, and Anabaena variabilis (group IV); and Calothrix sp. strain PCC 7601 (group V).
Conclusions.
We demonstrate by expression in E. coli and insertion inactivation that the Synechocystis sp. strain PCC 6803 ORF sll0045 encodes a protein with SPS activity. This is the first identification of a prokaryotic SPS gene (spsA). However, the role of sucrose in cyanobacteria is still a point of discussion. Its presence in these organisms has usually been associated with responses to different environmental stresses (3, 17), and it was hypothesized that it could be the carbon carrier substance from vegetative cells to heterocysts in filamentous cyanobacteria (26, 29). However, the fact that SPS is present in a unicellular non-nitrogen-fixing cyanobacterium such as Synechocystis even under standard growth conditions suggests that sucrose may not be exclusively associated with carbon transport or stress tolerance.
Acknowledgments
We thank S. Tabata (Kazusa DNA Research Institute, Chiba, Japan) for kindly providing the cosmid CSO415, H. G. Pontis for helpful discussions, and C. Fernández and C. Rodríguez for technical assistance.
G.S. is a Career Investigator of the Consejo Nacional de Investigaciones Cientificas y Técnicas (CONICET). This work was supported partly by grants from the CONICET, the Agencia Nacional de Promoción Cientifica y Tecnológica from Argentina, and the Rockefeller Foundation.
REFERENCES
- 1.Bruneau J-M, Worrell A C, Cambou B, Lando D, Voelker T A. Sucrose phosphate synthase, a key enzyme for sucrose biosynthesis in plants. Plant Physiol. 1991;96:473–478. doi: 10.1104/pp.96.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frommer W B, Sonnewald U. Molecular analysis of carbon partitioning in solanaceous species. J Exp Bot. 1995;46:587–607. [Google Scholar]
- 3.Hagemann M, Erdmann N. Environmental stresses. In: Ashwani K R, editor. Cyanobacterial nitrogen metabolism and environmental biotechnology. New Delhi, India: Narosa Publishing House; 1997. pp. 156–220. [Google Scholar]
- 4.Heim U, Weber H, Wobus U. Cloning and characterization of full length cDNA encoding sucrose-phosphate synthase from faba bean. Gene. 1996;178:201–203. doi: 10.1016/0378-1119(96)00373-3. [DOI] [PubMed] [Google Scholar]
- 5.Hesse H, Sonnewald U, Willmitzer L. Cloning and expression analysis of sucrose-phosphate synthase from sugar beet (Beta vulgaris L.) Mol Gen Genet. 1995;247:515–520. doi: 10.1007/BF00293155. [DOI] [PubMed] [Google Scholar]
- 6.Huber S C, Huber J L. Role and regulation of sucrose-phosphate synthase in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:431–444. doi: 10.1146/annurev.arplant.47.1.431. [DOI] [PubMed] [Google Scholar]
- 7.Huber S C, McMichael R W, Jr, Huber J L, Bachmann M, Yamamoto Y T, Conkling M A. Light regulation of sucrose synthesis: role of protein phosphorylation and possible involvement of cytosolic [Ca2+] Curr Top Plant Physiol Am Soc Plant Physiol Ser. 1995;13:35–44. [Google Scholar]
- 8.Ingram J, Chandler J W, Gallagher L, Salamini F, Bartels D. Analysis of cDNA clones encoding sucrose-phosphate synthase in relation to sugar interconversions associated with dehydration in the resurrection plant Craterostigma plantagineum Hochst. Plant Physiol. 1997;115:113–121. doi: 10.1104/pp.115.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Osumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 10.Klein R R, Crafts-Brandner S J, Salvucci M E. Cloning and developmental expression of the sucrose-phosphate synthase gene from spinach. Planta. 1993;190:498–510. doi: 10.1007/BF00224789. [DOI] [PubMed] [Google Scholar]
- 11.Komatsu A, Takanokura Y, Omura M, Akihama T. Cloning and molecular analysis of cDNAs encoding three sucrose-phosphate synthase isoforms from citrus fruit (Citrus unshiu Marc) Mol Gen Genet. 1996;252:346–351. doi: 10.1007/BF02173781. [DOI] [PubMed] [Google Scholar]
- 12.McMichael R W, Jr, Kochansky J, Klein R R, Salvucci M E, Huber S C. Identification of the major regulatory phosphorylation site in sucrose-phosphate synthase. Arch Biochem Biophys. 1993;307:248–252. doi: 10.1006/abbi.1993.1586. [DOI] [PubMed] [Google Scholar]
- 13.Pontis H G, Babio J R, Salerno G L. Reversible unidirectional inhibition of sucrose synthase activity by disulfides. Proc Natl Acad Sci USA. 1981;78:6667–6669. doi: 10.1073/pnas.78.11.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porchia A C. Ph.D. thesis. Mar del Plata, Argentina: Universidad Nacional de Mar del Plata; 1998. [Google Scholar]
- 15.Porchia A C, Salerno G L. Sucrose biosynthesis in a prokaryotic organism: presence of two sucrose-phosphate synthases in Anabaena with remarkable differences compared with the plant enzymes. Proc Natl Acad Sci USA. 1996;93:13600–13604. doi: 10.1073/pnas.93.24.13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porter R D. DNA transformation. Methods Enzymol. 1988;167:703–712. doi: 10.1016/0076-6879(88)67081-9. [DOI] [PubMed] [Google Scholar]
- 17.Reed R H, Richardson D L, Warr S L, Stewart W D P. Carbohydrate accumulation and osmotic stress in cyanobacteria. J Gen Microbiol. 1984;130:5–25. [Google Scholar]
- 18.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 19.Salerno G L. Measurement of enzymes related to sucrose metabolism in permeabilized Chlorella vulgaris cells. Physiol Plant. 1985;64:259–264. [Google Scholar]
- 20.Salerno G L, Pagnussat G C, Pontis H G. Studies on sucrose-phosphate synthase from rice leaves. Cell Mol Biol. 1998;44:407–416. [PubMed] [Google Scholar]
- 21.Salerno G L, Porchia A C, Sánchez N. Biosynthesis of sucrose in lower organisms. Curr Top Plant Physiol Am Soc Plant Physiol Ser. 1995;14:34–39. [Google Scholar]
- 22.Salvucci M E, Drake R R, Haley B E. Purification and photoaffinity labeling of sucrose phosphate synthase from spinach leaves. Arch Biochem Biophys. 1990;281:212–218. doi: 10.1016/0003-9861(90)90434-z. [DOI] [PubMed] [Google Scholar]
- 23.Salvucci M E, Klein R R. Identification of the uridine-binding domain of sucrose-phosphate synthase. Plant Physiol. 1993;102:529–536. doi: 10.1104/pp.102.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salvucci M E, van de Loo F J, Klein R R. The structure of sucrose-phosphate synthase. Curr Top Plant Physiol Am Soc Plant Physiol Ser. 1995;14:25–33. [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Schilling N, Ehrnsperger K. Cellular differentiation of sucrose metabolism in Anabaena variabilis. Z Naturforsch. 1985;40:776–779. [Google Scholar]
- 27.Stitt M, Wilke J, Feil R, Heldt H W. Coarse control of sucrose-phosphate synthase in leaves: alterations of the kinetic properties in response to the rate of photosynthesis and the accumulation of sucrose. Planta. 1988;174:217–230. doi: 10.1007/BF00394774. [DOI] [PubMed] [Google Scholar]
- 28.Valdez-Alarcón J J, Ferrando M, Salerno G, Jimenez-Moraila B, Herrera-Estrella L. Characterization of a rice sucrose-phosphate synthase encoding gene. Gene. 1996;170:217–222. doi: 10.1016/0378-1119(95)00854-3. [DOI] [PubMed] [Google Scholar]
- 29.Wolk P C, Ernst A, Elhai J. Heterocyst metabolism and development. Adv Photosynth. 1994;1:769–823. [Google Scholar]
- 30.Worrell A C, Bruneau J M, Summerfelt K, Boersig M, Voelker T A. Expression of a maize leaf sucrose-phosphate synthase in tomato alters leaf carbohydrate partitioning. Plant Cell. 1991;3:1121–1130. doi: 10.1105/tpc.3.10.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]