Abstract
First studies on the structure-function relationship of the S-layer protein from B. stearothermophilus PV72/p2 revealed the coexistence of two binding domains on its N-terminal part, one for peptidoglycan and another for a secondary cell wall polymer (SCWP). The peptidoglycan binding domain is located between amino acids 1 to 138 of the mature S-layer protein comprising a typical S-layer homologous domain. The SCWP binding domain lies between amino acids 240 to 331 and possesses a high serine plus glycine content.
Bacillus stearothermophilus is a strictly aerobic, thermophilic, endospore-forming species of gram-positive bacteria which is frequently endowed with a crystalline bacterial cell surface layer (S-layer; for reviews see references 3, 8, 21, 23, and 24) as the outermost cell envelope component. The S-layer lattice from B. stearothermophilus PV72/p6 shows hexagonal symmetry and is composed of subunits with a molecular weight of 130,000 (25). The gene encoding this S-layer protein (SbsA) has been cloned and sequenced (10). A different S-layer protein, SbsB, is produced by an oxygen-induced variant strain (B. stearothermophilus PV72/p2); this protein has a molecular weight of 97,000 and assembles into an oblique lattice type (20). These S-layer proteins are encoded by different genes (9, 10). Chemical analysis of native peptidoglycan-containing sacculi revealed that both organisms have an identical peptidoglycan chemotype but possess a secondary cell wall polymer (SCWP) of different chemical composition (6, 17). The SCWP from B. stearothermophilus PV72/p2 is mainly composed of N-acetylglucosamine and N-acetylmannosamine in a molar ratio of 2 to 1, shows a molecular weight of 24,000, and is covalently linked to the peptidoglycan backbone (17). Recently, the SCWP was found to inhibit the in vitro self-assembly of the guanidine hydrochloride (GHCl)-extracted SbsB protein by keeping it in the water-soluble state (19). Moreover, the isolated SCWP significantly enhanced the stability of the S-layer protein under proteolytic attack (19). Previous studies revealed that the SCWP plays an important role in anchoring the S-layer protein via the N-terminal region to the rigid cell wall layer (17).
By sequence comparison, S-layer homologous (SLH) domains (13) were identified on the N-terminal part of several S-layer proteins (4, 5, 7, 15, 16, 26) or at the very C-terminal end of other cell-associated exoproteins (11, 12, 14). In general, SLH domains were suggested to anchor these proteins permanently or transiently to the cell surface. An SLH domain was identified on the N-terminal part of SbsB but not on SbsA (9, 10). Experiments performed in the present study were carried out to clarify whether binding domains for peptidoglycan and for SCWP coexist on the N-terminal part of SbsB, the S-layer protein from B. stearothermophilus PV72/p2.
Characterization of proteolytic cleavage fragments formed with endoproteinase Glu-C in the absence and in the presence of the SCWP.
Proteolytic degradation of the S-layer protein was performed with the endoproteinase Glu-C, which under the applied experimental conditions attacks S-layer proteins after glutamic acid (17). When limited proteolysis was carried out in 0.1% sodium dodecyl sulfate (SDS) in 50 mM Tris-HCl buffer (pH 7.2), identical cleavage patterns were obtained in the absence and in the presence of the SCWP (Fig. 1A, B, and C). As determined by Edman degradation, the cleavage fragments showing molecular weights of 90,000 and 85,000 on SDS gels carried the N terminus of the mature S-layer protein, whereas the 66,000-molecular-weight cleavage fragment had the N-terminal sequence L-T-S-S-N-T-N-T-V. Comparison with the amino acid sequence of the whole S-layer protein (9) revealed that cleavage had occurred beyond glutamic acid at position 299. In contrast to the results obtained in 0.1% SDS, proteolysis of the S-layer protein in 2 M GHCl was clearly influenced by the SCWP. In the absence of the SCWP most of the S-layer protein was attacked by endoproteinase Glu-C, leading to formation of two major cleavage fragments with molecular weights of 85,000 and 55,000 and one minor cleavage fragment with a molecular weight of 66,000 (Fig. 1B). The two major cleavage fragments had an identical N-terminal sequence, V-T-K-G-T-P-T-S-F, showing that the S-layer protein was attacked beyond glutamic acid at position 138. The 66,000-molecular-weight fragment carried the N terminus of the mature S-layer protein. In the presence of the SCWP only about half of the S-layer protein was attacked by endoproteinase Glu-C, leading to the formation of an N-terminal 66,000-molecular-weight cleavage fragment (Fig. 1C). Sequencing of a small protein band with an apparent molecular weight of 30,000 led to the N-terminal sequence I-D-V-N-A. Judged by the N-terminal sequence which started with isoleucine at position 611 and by the molecular weight, this cleavage fragment was most probably formed together with the N-terminal 66,000-molecular-weight fragment and represented the C-terminal part of the S-layer protein.
FIG. 1.
SDS-polyacrylamide gel electrophoresis pattern of cleavage fragments formed by limited proteolysis of the S-layer protein from B. stearothermophilus PV72/p2 with endoproteinase Glu-C in 0.1% SDS (lane A) and 2 M GHCl (lanes B and C) in 50 mM Tris-HCl buffer (pH 7.2) in the absence (lane B) and the presence (lane C) of the SCWP. Conditions for proteolytic cleavage were as follows. One milligram of S-layer protein was dissolved in 1 ml of 0.1% SDS or 2 M GHCl, 40 μg of endoproteinase Glu-C was added, and samples were incubated for 1 h at 37°C. The concentration of the SCWP was 250 μg per 1 mg of S-layer protein. Proteolytic cleavage fragments subjected to N-terminal sequencing are indicated by arrows.
Affinity studies with proteolytic cleavage fragments with known N-terminal sequences and native and HF-extracted peptidoglycan-containing sacculi.
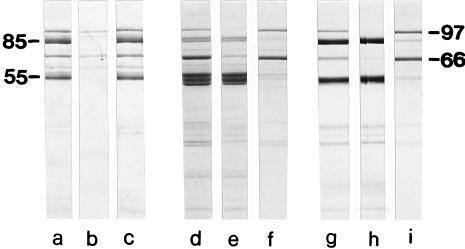
In order to obtain information on the location of peptidoglycan and SCWP binding domains, affinity binding studies were carried out with native sacculi as well as HF-extracted, peptidoglycan-containing sacculi that were completely devoid of the SCWP. Moreover, a potential SCWP binding domain on the S-layer protein was blocked by the addition of SCWP (17, 19). In 0.1% SDS in the absence or in the presence of SCWP, the whole S-layer protein and the two high-molecular-weight N-terminal cleavage fragments (90,000 and 85,000) were bound to the native or HF-extracted peptidoglycan-containing sacculi (Fig. 2A and B). On the other hand, the 66,000-molecular-weight cleavage fragment missing the N-terminal 299 amino acids remained unbound (Fig. 2C). These results confirmed data from previous studies indicating that the N-terminal 299 amino acids play an important role in anchoring the S-layer subunits to the rigid cell wall layer (17). After proteolytic degradation of the S-layer protein in 2 M GHCl and removal of GHCl by dialysis, the whole S-layer protein and all high-molecular-weight cleavage fragments could bind to native peptidoglycan-containing sacculi (Fig. 3a, b, and c). Using HF-extracted peptidoglycan-containing sacculi as an affinity matrix, only the whole S-layer protein and the N-terminal 66,000-molecular-weight cleavage fragment were enriched in the pellet, whereas cleavage fragments missing the N-terminal 138 amino acids remained in the supernatant (Fig. 3d, e, and f). Since HF-extracted peptidoglycan-containing sacculi represented pure peptidoglycan of the A1γ-chemotype (2, 22), attachment of the whole S-layer protein and the N-terminal cleavage fragment could have occurred only via a peptidoglycan binding domain which must be located within the segment of the N-terminal 138 amino acids. For further proof regarding the coexistence of a peptidoglycan and an SCWP binding domain on the N-terminal part of the S-layer protein, proteolytic cleavage fragments were prepared in 2 M GHCl and then SCWP (250 μg per mg of S-layer protein) was added to block a potential binding domain. After removal of GHCl by dialysis and incubation with native peptidoglycan-containing sacculi, the whole S-layer protein and the N-terminal cleavage fragment were detected in the pellet (Fig. 3g, h, and i), whereas cleavage fragments missing the N-terminal 138 amino acids remained in the supernatant. Thus, blocking of the SCWP binding domain confirmed that a peptidoglycan binding domain exists within the N-terminal 138 amino acids (Fig. 4).
FIG. 2.
Affinity studies performed with proteolytic cleavage fragments prepared with endoproteinase Glu-C in 0.1% SDS in 50 mM Tris-HCl buffer (pH 7.2) and native peptidoglycan-containing sacculi. Shown are proteolytic cleavage fragments before incubation with native peptidoglycan-containing sacculi (lane A), remaining in the clear supernatant recognizing as a binding site native peptidoglycan-containing sacculi (lane B), and (lane C). Lane B, two minor cleavage fragments with apparent molecular weights of 57,000 and 52,000 (V-P-V-Q-V and T-K-P-V-D-F) starting with either amino acid 355 or amino acid 409 of the mature S-layer protein. For the affinity studies, 1 mg of peptidoglycan-containing sacculi per mg of S-layer protein was added. Lane D, molecular weight standard (molecular weights given are multipliers of 1,000). Proteolytic cleavage fragments subjected to N-terminal sequencing are indicated by arrows.
FIG. 3.
Affinity studies with proteolytic cleavage fragments prepared in 2 M GHCl in 50 mM Tris-HCl buffer (pH 7.2) and native (lanes a through c and lanes g through i) or HF-extracted (lanes d through f) peptidoglycan-containing sacculi. In lanes g through i, the polymer binding domain on the S-layer protein was blocked by addition of SCWP (250 μg/mg of S-layer protein). Shown are proteolytic cleavage fragments before incubation with peptidoglycan-containing sacculi (lanes a, d, and g), remaining in the clear supernatant (lanes b, e, and h), and recognizing native (lanes c and i) and HF-extracted (lane f) peptidoglycan-containing sacculi as a binding site. Molecular weights given are multipliers of 1,000.
FIG. 4.
Schematic drawing of the S-layer protein from B. stearothermophilus PV72/p2 showing the location of the peptidoglycan and SCWP binding domain and the different endoproteinase Glu-C cleavage sites (positions of glutamic acid residues are given). The mature S-layer protein consists of 889 amino acids. ±, with or without SCWP.
Location of the SCWP binding domain.
For determining the location of the SCWP binding domain, S-layer self-assembly products (10 mg) were dissolved in 2 M GHCl, an excess amount of SCWP (10 mg) was added to keep the S-layer protein in the water-soluble state after removal of GHCl by dialysis (19), and the S-layer protein was digested with 200 μg of pronase E for 5 h at 37°C in the presence of 10 mM CaCl2. Separation of the pronase-E-digested material by gel permeation chromatography led to a product with two distinct peaks (not shown). The first peak (fraction I) gave a strong reaction when examined with the UV detector, eluted at a molecular weight of >200,000, and consisted of the whole S-layer protein and a series of proteolytic cleavage fragments. The second peak (fraction II) eluted at a molecular weight of 30,000 and gave only a distinct peak when examined with the refraction index detector, indicating that the major part of this fraction was SCWP. Amino acid analysis of the material collected in fraction II revealed that in comparison to those in the whole S-layer protein, the serine and glycine contents were significantly increased (11.3% versus 6.0% for serine and 10.2% versus 6.0% for glycine) and that serine and glycine occurred in a molar ratio of 1 to 0.9 (Table 1). As derived from the amino acid sequence of the whole S-layer protein, glycine is more regularly distributed whereas serine is concentrated at two distinct regions. The first serine-rich segment is located between amino acids 262 to 318 of the mature S-layer protein. The region containing serine and glycine in a molar ratio of 1 to 0.9 lies between amino acids 242 to 328. The second serine-rich segment was found on the C-terminal part of the S-layer protein, between amino acids 681 to 726. In this segment, the molar ratio of serine to glycine was 9 to 1 and even if the range was extended to amino acids 664 to 738, a molar ratio of only 2.25 to 1 was obtained for serine to glycine.
TABLE 1.
Amino acid composition of the whole S-layer protein and of the peptide remaining attached to the SCWP after digestion of the S-layer protein with pronase E for 5 h at 37°C
| Amino acid | Amino acid composition (mol%) of:
|
Calculated no. of amino acids on the peptide remaining attached to the SCWPa | No. of amino acids in S-layer proteinb | |
|---|---|---|---|---|
| Mature S-layer protein according to the sequence (9)/ amino acid analysis | Peptide remaining attached to the SCWP | |||
| Asp/Asn | 14.3/13.1 | 11.7 | 10.3 | 10 |
| Thr | 10.2/9.4 | 10.7 | 9.5 | 9 |
| Ser | 6.0/5.7 | 11.3 | 10.0 | 10 |
| Glu/Gln | 6.2/6.7 | 9.4 | 8.3 | 8 |
| Gly | 6.0/6.5 | 10.2 | 9.0 | 9 |
| Ala | 8.5/9.3 | 10.5 | 9.3 | 9 |
| Val | 11.5/11.7 | 13.9 | 12.3 | 15 |
| Ile | 3.2/4.3 | 2.9 | 2.6 | 2 |
| Leu | 5.0/6.0 | 6.8 | 6.0 | 6 |
| Phe | 4.1/4.2 | 1.7 | 1.5 | 1 |
| Lys | 8.0/8.8 | 10.9 | 9.6 | 10 |
In relation to 10 amino acids for serine.
Sequence between amino acids 240 and 331.
On the basis of N-terminal sequencing (A-T-G-I-K), the first amino acid of the peptide remaining attached to the SCWP was alanine at position 240. The amino acid composition was identical to that of the serine- and glycine-rich N-terminal segment extending from amino acids 240 to 331 of the mature S-layer protein (Table 1). Thus, the SCWP binding domain must be located in the sequence between amino acids 240 to 331, on which three double serine sequences have been identified (9). Interestingly, the existence of segments showing a high serine plus threonine plus glycine content was also reported for cell-associated exoenzymes (14). According to secondary structure predictions the high serine plus glycine plus threonine content indicated the presence of loops (18).
Amino acid analysis of fraction II further showed that the molar ratio of glucosamine to serine had increased to 10 to 1. Based on the estimated molecular weight (24,000) and the composition of the SCWP, one polymer chain consists of 120 monosaccharides, which corresponds to 80 glucosamine residues. Since the peptide remaining attached to the SCWP contained 10 serine residues (Table 1), the molar ratio between the SCWP and the peptide representing the SCWP binding domain was 1.25 to 1.
Sequence comparison.
For comparison of the S-layer protein from B. stearothermophilus PV72/p2 with other S-layer proteins, amino acid sequence similarity searches were performed with the BLAST program (1). For the very N-terminal part of the S-layer protein (amino acids 1 to 138) representing the peptidoglycan binding domain and comprising the SLH domain (amino acids 29 to 78), the highest scores of identity were found with the S-layer protein EA1 from Bacillus anthracis (32%) (7), with the S-layer proteins from Bacillus licheniformis (26) (34%) and Bacillus sphaericus 2362 (5) (30%), and with the S-layer protein Sap from B. anthracis (15) (38%). Interestingly, no identity to other S-layer proteins was detected for the segment between amino acids 240 and 331, representing the SCWP binding domain.
Conclusion.
The results from the affinity studies revealed that the N-terminal part of SbsB, the S-layer protein from B. stearothermophilus PV72/p2, carries two binding domains, one for peptidoglycan and another for SCWP. The peptidoglycan binding domain comprising a typical SLH domain showed the highest scores of identity with S-layer proteins from other Bacillus spp., whereas the SCWP binding domain was not related to other S-layer proteins. Since the peptidoglycan A1γ-chemotype is widely distributed among Bacillaceae (2), the SCWP can be considered as the cell envelope component endowing the peptidoglycan-containing layer with specific (physico)chemical properties and may even mask the peptidoglycan. According to this consideration, SbsB did not recognize as a binding site native peptidoglycan-containing sacculi from B. stearothermophilus wild-type strains, which possess an identical peptidoglycan chemotype but have an SCWP of different chemical composition (20). By contrast, the S-layer proteins from two B. stearothermophilus wild-type strains have an identical N-terminal region which is responsible for anchoring the S-layer subunits via an identical SCWP to the rigid cell wall layer, thereby enabling heterologous recrystallization (6). Interestingly, the N-terminal part of the S-layer proteins from B. stearothermophilus wild-type strains does not comprise an SLH domain and does not show identity to other S-layer proteins, strongly indicating the absence of a peptidoglycan-binding domain.
Acknowledgments
This work was supported by the Austrian Science Foundation, project P-12938, and by the Ministry of Research and Transport.
We thank Sonja Zayni for sugar analyses.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archibald A R, Hancock I C, Harwood C R. Cell wall structure, synthesis, and turnover. In: Sonenshein A, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 381–410. [Google Scholar]
- 3.Beveridge T J. Bacterial S-layers. Curr Opin Struct Biol. 1994;4:204–212. [Google Scholar]
- 4.Beveridge T J, Pouwels P H, Sára M, Kotiranta A, Lounatmaa K, Kari K, Kerosuo E, Happasalo M, Egelseer E M, Schocher I, Sleytr U B, Morelli L, Callegari M-L, Nomellini J F, Bingle W H, Smit J, Leibovitz E, Lemaire M, Miras I, Salamitou S, Beguin P, Ohayon H, Gounon P, Matuschek M, Sahm K, Bahl H, Grogono-Thomas R, Dworkin J, Blaser M J, Woodland R M, Newell D G, Kessel M, Koval S F. Functions of S-layers. FEMS Microbiol Rev. 1997;20:99–149. doi: 10.1111/j.1574-6976.1997.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 5.Bowditch R D, Baumann P, Yousten A A. Cloning and sequencing of the gene encoding a 125-kilodalton surface-layer protein from Bacillus sphaericus 2362 and of a related cryptic gene. J Bacteriol. 1989;171:4178–4188. doi: 10.1128/jb.171.8.4178-4188.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egelseer E M, Leitner K, Jarosch M, Hotzy C, Zayni S, Sleytr U B, Sára M. The S-layer proteins of two Bacillus stearothermophilus wild-type strains are bound via their N-terminal region to a secondary cell wall polymer of identical chemical composition. J Bacteriol. 1998;180:1488–1495. doi: 10.1128/jb.180.6.1488-1495.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Etienne-Toumelin I, Sirard J-C, Duflot E, Mock M, Fouet A. Characterization of the Bacillus anthracis S-layer: cloning and sequencing of the structural gene. J Bacteriol. 1995;177:614–620. doi: 10.1128/jb.177.3.614-620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.König H, Messner P (Guest Ed.) Structure, biochemistry, molecular biology and applications of microbial S-layers. FEMS Microbiol Rev. 1997;20:1–178. [Google Scholar]
- 9.Kuen B, Koch A, Asenbauer E, Sára M, Lubitz W. Molecular characterization of the Bacillus stearothermophilus PV72 S-layer gene sbsB induced by oxidative stress. J Bacteriol. 1997;179:1664–1670. doi: 10.1128/jb.179.5.1664-1670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuen B, Sleytr U B, Lubitz W. Sequence analysis of the sbsA gene encoding the 130 kDa surface layer protein of Bacillus stearothermophilus PV72. Gene. 1994;145:115–120. doi: 10.1016/0378-1119(94)90332-8. [DOI] [PubMed] [Google Scholar]
- 11.Leibovitz E, Ohayon H, Gounon P, Béguin P. Characterization and subcellular localization of the Clostridium thermocellum scaffolding dockerin binding protein SdbA. J Bacteriol. 1997;179:2519–2523. doi: 10.1128/jb.179.8.2519-2523.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemaire M, Ohayon H, Gounon P, Fujino T, Beguin P. OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J Bacteriol. 1995;177:2451–2459. doi: 10.1128/jb.177.9.2451-2459.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lupas A, Engelhardt H, Peters J, Santarius U, Volker S, Baumeister W. Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J Bacteriol. 1994;176:1224–1233. doi: 10.1128/jb.176.5.1224-1233.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matuschek M, Burchhardt G, Sahm K, Bahl H. Pullulanase of Thermoanaerobacterium thermosulfurigenes EM1 (Clostridium thermosulfurogenes): molecular analysis of the gene, composite structure of the enzyme, and a common model for its attachment to the cell surface. J Bacteriol. 1994;176:3295–3302. doi: 10.1128/jb.176.11.3295-3302.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mesnage S, Tosi-Couture E, Mock M, Gounon P, Fouet A. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol Microbiol. 1997;23:1147–1155. doi: 10.1046/j.1365-2958.1997.2941659.x. [DOI] [PubMed] [Google Scholar]
- 16.Olabarría G, Carrascosa J L, de Pedro M A, Berenguer J. A conserved motif in S-layer proteins is involved in peptidoglycan binding in Thermus thermophilus. J Bacteriol. 1996;178:4765–4772. doi: 10.1128/jb.178.16.4765-4772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ries W, Hotzy C, Schocher I, Sleytr U B, Sára M. Evidence that the N-terminal part of the S-layer protein of Bacillus stearothermophilus PV72/p2 recognizes a secondary cell wall polymer. J Bacteriol. 1997;179:3892–3898. doi: 10.1128/jb.179.12.3892-3898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rost B, Schneider R, Sander C. Progress in protein structure prediction? Trends Biochem Sci. 1993;18:120–123. doi: 10.1016/0968-0004(93)90017-h. [DOI] [PubMed] [Google Scholar]
- 19.Sára M, Dekitsch C, Mayer H F, Egelseer E M, Sleytr U B. Influence of the secondary cell wall polymer on the reassembly, recrystallization, and stability properties of the S-layer protein from Bacillus stearothermophilus PV72/p2. J Bacteriol. 1998;180:4146–4153. doi: 10.1128/jb.180.16.4146-4153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sára M, Kuen B, Mayer H F, Mandl F, Schuster K C, Sleytr U B. Dynamics in oxygen-induced changes in S-layer protein synthesis from Bacillus stearothermophilus PV72 and the S-layer deficient variant T5 in continuous culture and studies of the cell wall composition. J Bacteriol. 1996;178:2108–2117. doi: 10.1128/jb.178.7.2108-2117.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sára M, Sleytr U B. Crystalline bacterial cell surface layers (S-layers): from cell structure to biomimetics. Prog Biophys Mol Biol. 1996;65:83–111. doi: 10.1016/s0079-6107(96)00007-7. [DOI] [PubMed] [Google Scholar]
- 22.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sleytr U B, Messner P, Pum D, Sára M, editors. Crystalline bacterial cell surface proteins. Austin, Tex: Landes Company Academic Press; 1996. [Google Scholar]
- 24.Sleytr U B, Sára M. Bacterial S-layer proteins: structure-function relationship and their biotechnological applications. Trends Biotechnol. 1997;15:20–26. doi: 10.1016/S0167-7799(96)10063-9. [DOI] [PubMed] [Google Scholar]
- 25.Sleytr U B, Sára M, Küpcü Z, Messner P. Structural and chemical characterization of S-layers of selected strains of Bacillus stearothermophilus and Desulfotomaculum nigrificans. Arch Microbiol. 1986;146:19–24. doi: 10.1007/BF00690152. [DOI] [PubMed] [Google Scholar]
- 26.Zhu X, McVeigh R R, Malathi P, Gosh B K. The complete nucleotide sequence of the Bacillus licheniformis NM105 S-layer encoding gene. Gene. 1996;173:189–194. doi: 10.1016/0378-1119(96)00233-8. [DOI] [PubMed] [Google Scholar]