Abstract
We previously identified a novel regulator of the exotoxin A gene (toxA) in Pseudomonas aeruginosa, PtxR, that belongs to the LysR family of prokaryotic regulatory proteins. Preliminary data also suggest that PtxR affects the expression of siderophores in P. aeruginosa. Because toxA expression and siderophore production in this organism are coordinately regulated by the ferric uptake regulator (Fur) and the Fur-regulated alternative sigma factor PvdS, regulation of ptxR itself in the context of these regulators was examined. RNase protection analyses of ptxR transcription revealed that there are two independent transcription initiation sites (T1 and T2). While transcription from the promoter of T1 is constitutive throughout the growth cycle of PAO1, transcription from the second promoter (P2) is negatively affected by iron. Transcription from the P2 promoter is constitutive in a fur mutant under microaerobic conditions but still iron regulated during aerobic growth. High concentrations (>100 nM) of the ferric uptake regulatory protein (Fur) failed to bind to either of the promoter regions of ptxR in either gel mobility shift assays or DNase I footprint experiments. These results indicate that Fur indirectly regulates the iron-dependent expression of ptxR. Iron-regulated transcription of ptxR from the P2 promoter, but not constitutive expression from the P1 promoter, was dependent on the Fur-regulated alternative sigma factor gene pvdS, even under aerobic conditions. Consequently, there are two levels of iron-regulated expression of ptxR. The iron-regulated expression of ptxR under microaerobic conditions from the P2 promoter of ptxR is mediated indirectly by Fur through the iron-regulated expression of pvdS. In contrast, pvdS-mediated iron regulation of ptxR under aerobic conditions is Fur independent.
Exotoxin A is one of a growing number of virulence factors produced by Pseudomonas aeruginosa that are regulated by multiple environmental factors (7, 14, 18, 20). Not only is the toxA gene influenced by the environmental levels of iron, but it is also affected by the presence of certain nucleotides in the growth medium, as well as oxygen tension and temperature (7, 18, 20). Several genes that have an impact on the regulation of exotoxin A production by P. aeruginosa have now been described. These genes include regA, vfr, fur, and pvdS (1, 3, 6, 9, 16, 19). However, some of these genes have differential effects on the iron-dependent expression of toxA and siderophores. Barton et al. reported that Fur affects the iron-regulated expression of toxA only under microaerobic conditions (1). That is, a Fur mutant of P. aeruginosa was unaffected in iron-regulated expression of toxA under aerobic growth conditions, but expression of toxA under microaerobic conditions was constitutive in this mutant. In contrast, expression of siderophores was constitutive under aerobic conditions in Fur mutants.
Recently we described a gene, ptxR, that also appears to regulate exotoxin A production in P. aeruginosa at the transcriptional level (4). The presence of a multicopy plasmid carrying ptxR in P. aeruginosa results in a four- to fivefold increase in exotoxin A production. Some preliminary evidence suggests that ptxR enhances toxA transcription through regA, a gene encoding a positive regulator of toxA. Nonetheless, it is not clear whether these effects are direct or indirect. PtxR has substantial homology to several members of the LysR family of transcriptional activators (13). There is a helix-turn-helix motif typical of LysR regulators (and DNA binding proteins in general) at the amino terminus of PtxR. Since PtxR, however, has not yet been purified, DNA binding studies have not yet been done on the regA or toxA promoters. It is also not clear whether ptxR is involved in the iron-regulated phase of toxA expression (2). No studies on the regulation of the ptxR gene itself have been reported. Preliminary observations from our laboratory indicate that multiple copies of ptxR also have an impact on siderophore expression in P. aeruginosa (5). Previously, we cogently demonstrated that the iron-directed, coordinate expression of toxA and siderophores occurs, at least in part, through the ferric uptake regulator (Fur) and the Fur-regulated pvdS gene, which encodes an alternative sigma factor (1, 9). Consequently, it is worthwhile to delineate the impact that the levels of environmental iron might have on the expression of ptxR itself and to assess whether Fur and PvdS influence the expression of this regulatory gene.
RNase protection assays were used to determine whether transcription of ptxR is affected by the same relative levels of iron that negatively regulate the expression of toxA, the alternative sigma factor gene pvdS, and siderophores in P. aeruginosa. The riboprobes required for these assays were synthesized by using suitable oligonucleotide primers. The primers were utilized to amplify certain parts within either the ptxR open reading frame or its upstream region. The probe for the 5′ region of ptxR comprises bp 52 to 607 of the ptxR sequence entered in GenBank (accession no. U35068), while the internal probe encompasses bp 740 through 1066. The amplified DNA fragments were cloned into the pCRII vector (Invitrogen Corporation, Carlsbad, Calif.). RNA probes were then generated from these fragments by runoff transcription from either the SP6 or T7 promoter using the Riboprobe kit (Promega). The riboprobe for mapping the transcriptional start sites of ptxR extended from 504 bp 5′ to the ptxR initiation codon to the first 61 bp of ptxR and carries an additional ∼100 bases of pCRII vector sequences. An additional riboprobe used in some of these studies (see Fig. 1C) included the region that encodes an internal portion of PtxR as well as additional pCRII vector sequences. RNase protection experiments were done as previously described (1, 9). Autoradiographs of the dried gels were scanned and imported into Adobe Photoshop for adjustment of brightness and contrast (Adobe Systems Incorporated, San Jose, Calif.). Quantitative analysis of the autoradiographs was performed with NIH Image software, version 1.55.
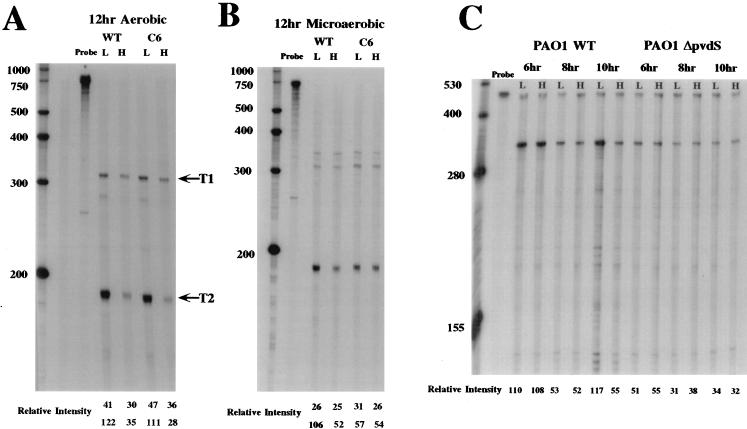
FIG. 1.
RNase protection assays of ptxR transcription. (A) RNase protection analysis of ptxR transcription in the PAO1 wild type and the Fur mutant PAO1C6 grown under aerobic conditions as previously described (1). RNA was isolated at 12 h from cells grown under low (L)- or high (H)-iron conditions (1). The riboprobe used in these experiments encompasses the 5′ region of ptxR plus its translational initiation codon. This probe detects both the T1 and T2 transcripts. (B) RNase protection analysis of ptxR transcription in the PAO1 wild type and the Fur mutant PAO1C6 grown under microaerobic conditions as previously described (1). (C) RNase protection analysis of ptxR mRNA in the PAO1 wild type and its pvdS mutant, PAO1ΔpvdS. The ptxR internal RNA probe was used in these experiments. The higher band in each lane represents an excess of undigested probe. The lower band in each panel represents ptxR mRNA, including both T1 and T2 transcripts. The cultures were grown aerobically with (H) and without (L) iron. Samples were obtained at the times indicated. The RI for each band was determined by using NIH Image software, version 1.55. An RNA size standard ladder is on the left of each panel. The lanes labeled probe contain the labeled probe used in these studies that was not digested with the RNase cocktail. WT, wild type; C6, PAO1C6.
RNase protection experiments were first performed with the 565-bp ptxR-specific RNA probe that extends 5′ to the translational initiation codon of ptxR. Ostensibly, this probe extends beyond the transcriptional initiation site and promoter region of ptxR. In Fig. 1A, detection of two protected fragments in the 12-h RNA sample from cells grown aerobically under low-iron conditions suggests that ptxR has two transcriptional initiation sites under these conditions, T1 and T2, but at this point it was also possible that T2 was a product of the processing of the T1 transcript. Another RNase protection analysis of RNA isolated from 6- and 10-h samples corroborated these data (data not shown). Transcription from the T1 site, the putative P1 promoter, does not appear to be strongly influenced by iron and appears constitutive during all growth phase periods (i.e., 6, 8, 10, and 12 h) examined. Based on the size of the protected fragment in these experiments, the T2, iron-dependent, transcription start site is ∼166 ± 3 bp 5′ to the ptxR translation initiation codon.
Next, the issue of whether iron-regulated expression of ptxR is dependent on Fur was addressed by examination of ptxR expression in the well-characterized P. aeruginosa Fur mutant, PAO1C6 (1). Figure 1A shows that the iron-regulated pattern of ptxR expression under aerobic conditions from the P2 promoter in the Fur mutant is essentially identical to that observed in the wild-type strain (PAO1WT) (relative intensities [RIs] of PAO1WT of 122 [low iron] versus 35 [high iron]; compared to RIs of PAO1C6 of 111 [low iron] versus 28 [high iron]). In contrast, in the PAO1C6 Fur mutant grown under microaerobic conditions, iron-dependent transcription of ptxR from this promoter was constitutive (RIs of 57 [low iron] versus 54 [high iron] [Fig. 1B]) but still significantly influenced by iron in PAO1WT (RIs of 106 [low iron] versus 52 [high iron]). These data suggest that the T1 and T2 transcripts are independent and that the T2 transcript is not merely processed from the T1 transcript since the T2 transcript is differentially expressed during both the growth phase and in response to the levels of environmental iron, as well as to Fur.
The initial observations regarding the iron-regulated expression of ptxR were supported by examining the expression of β-galactosidase from a ptxR::lacZ translational fusion carried on a plasmid in PAO1WT and in the ΔpvdS mutant (PAO1ΔpvdS). The level of β-galactosidase activity expressed from this fusion was determined as previously described (8, 9). However, for reasons that are not clear at the present time, the level of expression of β-galactosidase in these studies was too low to be completely certain that there was a significant difference between expression of the ptxR::lacZ fusion in the wild-type strain and the ΔpvdS mutant. Nevertheless, there was a reproducible greater-than-twofold higher level of β-galactosidase expression in PAO1WT carrying the ptxR::lacZ fusion when it was grown under iron-limiting conditions than there was in PAO1ΔpvdS carrying this fusion when it was grown under the same conditions. There was no difference in the levels of β-galactosidase in these strains when they were grown in iron-replete medium (data not shown). These data correlate well with the RNase studies described in this report and together with the RNase protection experiments support our contention that there is a PvdS-dependent iron-regulated expression of ptxR and a PvdS-independent expression of this gene which is not iron regulated.
We also performed additional RNase protection experiments using RNA preparations from different times in the growth phase of PAO1 and an independent riboprobe composed only of ptxR coding sequences. This ptxR-specific RNA probe contains the region encoding ∼110 amino acids of PtxR. The experimental results shown in Fig. 1C were obtained with RNA samples taken at 6, 8, and 10 h from cells grown aerobically in the presence or absence of added iron. Quantitative analysis of the autoradiograph revealed no significant differences in the amount of ptxR mRNA detected in cells grown for 6 or 8 h under high- or low-iron conditions (RI at 6 h, 110 [low iron] versus 108 [high iron]; RI at 8 h, 53 [low iron] versus 52 [high iron]). However, at the 10-h time point, cells grown under iron-limiting conditions produced levels of ptxR-specific mRNA more than twofold higher than those from cells grown under iron-replete conditions (RI, 117 [low iron] versus 55 [high iron]). It should be noted that the internal probe used in these studies does measure both the T1 and T2 transcripts detected in the previous experiments. Accordingly, at least some of the transcriptional activity detected at 10 h in the cells grown in iron-replete medium is likely the result of expression from the P1 promoter. To confirm and more precisely identify the transition to iron-regulated transcription, additional RNA samples were obtained from cells grown for 6 and 12 h (RNase protection autoradiograph not shown). Again, at 6 h, no difference in the amount of ptxR mRNA was detected in cells grown in the presence or absence of iron (RI at 6 h, 120 [low iron] versus 114 [high iron]). By contrast, at 12 h, significantly higher levels of ptxR-specific message were detected in cells grown in low-iron medium than in cells grown in high-iron medium (RI, 121 [low iron] versus 52 [high iron]). These results confirm the RNase protection studies using the 5′ probe, which indicated that there are most likely two independent transcriptional initiation sites for this gene. One is constitutively expressed throughout the growth cycle, while the other is negatively influenced by iron in the late logarithmic and early stationary phases of growth.
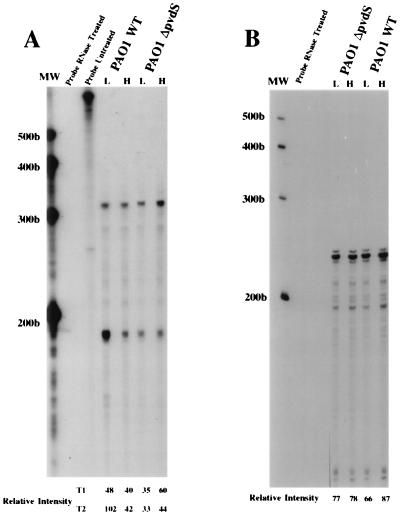
Additional data strongly supporting these assertions are shown in Fig. 2. In these experiments, we used the probe that encompasses both promoters of the ptxR gene (see above). While there were no significant differences in the expression of either the T1 or T2 transcript at the 6-h time point with respect to whether the cells were grown under iron-deficient or iron-replete conditions (data not shown), there was a significant difference in the amount of the T2 transcript detected in the PAO1 wild-type strain grown for 12 h under iron-limiting conditions or iron-replete conditions (RI, 102 [low iron] versus 42 [high iron]). In contrast, there was no significant difference in the amount of the T2 transcript detected in the ΔpvdS mutant grown under iron-replete or iron-deficient conditions (RI, 33 [low iron] versus 44 [high iron]). Also in support of our hypothesis that the ptxR gene has an iron-dependent and an iron-independent promoter, the levels of the T1 transcript remained relatively constant regardless of the level of iron. There does appear to be a slightly increased expression of the T1 transcript in the ΔpvdS mutant, but this occurs under iron-replete conditions only. In the experiment illustrated in Fig. 2B, we also provided a control for the amount of mRNA in the samples analyzed in this study.
FIG. 2.
RNase protection analysis in the PAO1 wild type (PAO1WT) and its pvdS mutant, PAO1ΔpvdS. (A) The probe from the promoter region of the ptxR gene described in the text was used in these experiments. Included are the probe without any added mRNA, digested with the RNase cocktail, and the undigested probe. The cultures were grown aerobically with (H) and without (L) iron. All samples were obtained at 12 h. (B) The RNA samples were examined with the probe generated from the constitutively expressed lipoprotein gene (oprX) described in the text and elsewhere (1) and entered in GenBank under accession no. AF050676. Although included in the analysis, the undigested probe is not shown in this figure. The RI for each band was determined by using NIH Image software, version 1.55. MW, molecular size standards (measured in bases).
We previously reported that a gene encoding an outer membrane-associated lipoprotein (oprX; GenBank accession no. AF050676), which is divergently transcribed from the fur gene, is constitutively expressed in P. aeruginosa with regard to growth phase and to levels of iron in the growth media (1). In this assay, the same RNA samples used to generate the results shown in Fig. 2A were examined by RNase protection assays using the probe derived from the constitutively expressed lipoprotein gene. As shown in Fig. 2B, the relative levels of this transcript are constant for each of the strains or conditions examined. (It is important to note that the order of the PAO1WT and PAO1ΔpvdS samples are reversed in Fig. 2A and B.) The data from these RNase protection assays using the probe from the constitutively expressed lipoprotein gene strongly indicate that the differences we have observed for the ptxR T2 transcript are not caused by errors in the determination of the amount of RNA used in the different samples. Moreover, in support of the view that the band we designate as the T1 transcript is not the result of incomplete digestion of unhybridized RNA, we have provided data in Fig. 2 showing that the probes in the absence of any mRNA sample are completely digested by the RNase cocktail used in these experiments.
Based on these RNase protection data and our preliminary ptxR::lacZ fusion studies (see above), ptxR appears to be expressed at a low level in PAO1. This low level of expression is a characteristic feature of several genes encoding LysR-type regulators (6).
The ferric uptake regulator (Fur) in P. aeruginosa and in numerous other prokaryotic organisms is the central regulatory protein influencing the expression of iron-regulated genes. This is accomplished by iron-bound Fur interacting with an operator site (Fur box) in iron-regulated genes and directly repressing their synthesis by inhibiting transcription initiation. However, it is also recognized that Fur may exert its effect indirectly by altering the expression of activators or sigma factors required for the expression of iron-regulated genes. To test whether the former scenario applies to ptxR expression, we examined whether Fur purified from P. aeruginosa could directly bind to the promoter region of ptxR at biologically relevant concentrations (e.g., <1 μM). Both gel mobility shift assays and DNase I footprint assays performed as previously described were used for this purpose (1, 10, 11). A region that extends more than 500 bp 5′ to the initiation codon of ptxR was examined in these assays. This noncoding region does contain two sequences that have a match in 11 of 19 nucleotides with a consensus Fur box and we have previously shown that a match in 10 of 19 nucleotides can constitute a functional Fur binding operator in P. aeruginosa (11). Nevertheless, despite this similarity to a consensus Fur binding site, in both assays concentrations of Fur purified from P. aeruginosa as high as 1 μM failed to bind to any sequence in the entire intergenic region between ptxR and another gene 5′ to ptxR that is divergently transcribed from ptxR (12); in contrast, control DNA fragments that are known to bind Fur, including those containing synthetic consensus Fur binding sites, were always positive in these assays. These data suggest that Fur does not regulate the expression of ptxR under microaerobic conditions by directly binding to the region 5′ to ptxR and by blocking the access of RNA polymerase to either the P1 or P2 promoter.
The pvdS gene encodes a putative alternative sigma factor that, in addition to regulating the production of the siderophore pyoverdine, regulates the expression of toxA and regA in P. aeruginosa (9). Expression of the pvdS gene appears to be directly influenced by the ferric uptake regulatory protein Fur, which binds to the pvdS promoter and blocks its transcription in cells grown under iron-replete conditions (9). Because Fur does not have a direct impact on the iron-regulated phase of ptxR expression by binding to either of its promoters, it seems likely that it could impart its effect indirectly through another gene, perhaps pvdS. This alternative scenario was examined by analyzing the transcription of ptxR in a wild-type PAO1 background and in a previously described mutant of PAO1 carrying a specific deletion in the pvdS gene. This PAO1 ΔpvdS mutant strain has growth curve characteristics identical to the PAO1 wild-type parental strain (9). The expression of ptxR in the PAO1 wild type and in the PAO1 ΔpvdS mutant was examined by RNase protection experiments and by analysis of β-galactosidase expression in these strains carrying the ptxR::lacZ fusion described above. Our data also offer compelling evidence that the iron-regulated P2 promoter of ptxR is dependent on the expression of pvdS and that Fur regulates the latter phase of ptxR expression indirectly in cells grown in a microaerobic environment but not in P. aeruginosa PAO1 grown under aerobic conditions.
An increasingly complex regulatory circuit directing the coordinate expression of toxA expression and siderophore production in P. aeruginosa is emerging. At least five separate regulatory genes (regA, fur, vfr, pvdS, and ptxR) have been demonstrated to have a substantial (i.e., greater-than-fivefold) impact on the production of exotoxin A in this opportunistic organism, although it is not yet clear how all of these regulatory factors coordinately exert their effect on production of this virulence determinant or whether they do so all at the same time (1, 3, 6, 9, 16, 19). It has been known for a number of years that the iron-regulated phase of regA, which encodes a positive activator of toxA transcription, is intertwined with the iron-regulated phase of toxA (1–3, 6, 16, 17). More recently, we demonstrated that Fur is required for this iron-dependent control of regA and, ultimately, toxA (1–3, 6, 16, 17). However, we found that Fur regulates the expression of toxA only under microaerobic conditions; under aerobic conditions, the iron-regulated expression of toxA is unaffected in the Fur mutant employed in the present studies (see below). We conjectured that there is an additional iron-dependent regulator controlling the expression of these genes under aerobic conditions (1, 9). In support of this hypothesis, we discerned that the gene encoding the putative alternative sigma factor PvdS is situated between Fur and the iron-dependent regulation of regA and toxA in the regulatory pathway leading to optimal production of exotoxin A (1, 9). Concerning other possible regulators of toxA, West et al. demonstrated that the product of the vfr gene, which is highly homologous to the catabolite repressor protein (Crp) of Escherichia coli, is required for the optimal expression of both regA and ultimately toxA (19). Vfr seems to exert its effect by directly binding to the toxA and regA promoters, enhancing their transcription. However, the issue of whether environmental or physiological conditions (e.g., levels of cyclic nucleotides) exert an effect on binding and regulation by Vfr is not clear at the present time. There does not appear to be any significant direct association between Vfr and the iron-regulated expression of regA or toxA.
In our previous and present reports, we provide insight into another level of complexity in the processes regulating exotoxin A expression. Previous experimental data suggest that ptxR also imparts a significant effect on the transcription of toxA (4). Optimal enhancement of toxA transcription by ptxR is detectable in P. aeruginosa as soon as the early to mid-logarithmic phase of growth, a pattern that is similar to the enhancement of toxA transcription by regA (2, 3). We therefore examined how ptxR expression might be affected by the environmental levels of iron and by proteins known to play key roles in the iron-regulated expression of toxA, PvdS and Fur. Such experiments could help us understand the possible role that ptxR might play in the cascade of events leading to the production of exotoxin A. The pattern of regulation of ptxR expression by iron and these regulators parallels the pattern of the response of regA to these same factors; that is, there is a biphasic pattern to the expression of ptxR similar to that observed with regA (2, 3). The early phase of transcription from the ptxR P1 promoter is constitutive. However, during the late logarithmic stages of growth, transcription of ptxR becomes negatively influenced by iron to a significant degree (Fig. 1 and 2). Interestingly, the timing of this biphasic pattern is virtually coincidental with the biphasic pattern of regA expression. That is, at some point during the late logarithmic phase to the early stationary phase of growth (at 6 to 10 h), the expression of transcription of both ptxR and regA becomes significantly influenced by the environmental levels of iron in the growth medium. However, in contrast to the late expression of regA, which is not detectable in iron-replete medium, a low constitutive level of ptxR transcription is detected during the later stages of the growth cycle.
Because we had previously implicated Fur and the alternative sigma factor PvdS in the iron-regulated pathways for the expression of regA and toxA, we postulated that either or both of these factors could also influence the expression of ptxR. The data presented in this report clearly link PvdS, and indirectly Fur, to the iron-regulated expression of ptxR, although the mechanism by which ptxR exerts its effect on the expression of regA and toxA is still obscure. PtxR also appears to have an effect on siderophore production. Multiple copies of ptxR abrogate iron-regulated expression of siderophores on chrome azure S plates. Moreover, ptxR is located immediately 3′ to an operon involved in the production of the chromophore of pyoverdine, although it is transcribed in the opposite direction from this operon (15) (Genbank accession no. AF002222).
Determination of the factors that influence the DNA binding activity of this putative LysR-type regulator is a critical issue that needs to be addressed. While the members of this class of regulators are thought to bind DNA, via the DNA binding motif located in their amino-terminal regions, they apparently respond to different factors that have a crucial impact on their ability to bind to the promoter region of a target gene (13). Accordingly, until such a factor is identified for PtxR, it will be difficult to obtain a completely lucid image of the mechanism by which PtxR exerts its effects on the expression of toxA or other genes, including those involved in siderophore production. Examination of the ability of PtxR to bind to the promoters toxA and other iron-regulated genes will ultimately be required to understand how PtxR influences production of exotoxin A and siderophores. Moreover, although we demonstrated that pvdS is required for the iron-dependent phase of ptxR expression, it is not clear from the available data whether PvdS directly regulates the P2 promoter of ptxR by binding and activating its transcription under iron-deficient conditions or whether there is yet another regulatory gene positioned between pvdS and ptxR. At the present time, there is not enough information about what constitutes a binding site for PvdS. In the case of the regulation of toxA by iron, it appears that an additional regulator (besides PvdS and Fur), perhaps PtxR, is required for the iron-regulated production of exotoxin A under aerobic conditions. In this regard, it is interesting that we found that Fur affects the iron-regulated expression of both toxA and ptxR only under microaerobic conditions but that Fur has an effect on siderophore expression under aerobic conditions (1, 9).
The data presented in this paper add to the already complicated mechanism for the regulation of exotoxin A production by iron and by oxygen levels. Why such a complex circuitry might be needed for the expression of a single extracellular virulence factor is the most perplexing aspect of this regulation. The answer to this question may be related to the complexity of P. aeruginosa and its remarkable ability to survive in diverse environments. It is of interest that ptxR, regA, and toxA, in contrast to fur and pvdS, apparently do not exist in the other pseudomonads most closely related to P. aeruginosa, i.e., P. fluorescens and P. putida. We performed Southern blot hybridization studies using probes from all of these genes against genomic DNA from diverse P. aeruginosa isolates as well as isolates of P. fluorescens and P. putida. While the probes derived from fur and pvdS strongly hybridize with genomic DNA from all the species mentioned above even under high-stringency conditions, probes derived from ptxR, regA, and toxA hybridized to genomic DNA from the P. aeruginosa strains examined but failed to hybridize to the genomic DNA from either P. fluorescens or P. putida, even under low-stringency conditions (12). It is reasonable to suppose that those genes which contribute exotoxin A production are of some importance to P. aeruginosa in environments where other fluorescent pseudomonads fail to effectively compete, such as in a compromised mammalian host. It may be crucial for P. aeruginosa to maintain multiple activators and repressors for the expression of exotoxin A so that the variety of different environments (e.g., aerobic and microaerobic) that this organism may encounter in such a host do not completely abrogate expression of this virulence determinant. Further experimental analysis of the molecular processes by which the increasing number of regulatory genes such as ptxR and pvdS interact may provide additional insight into these interesting questions.
Acknowledgments
This work was supported by NIH Public Health Service grants AI 15490 (to M. L. Vasil) and AI 33386 (to A. Hamood) from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Barton H A, Johnson Z, Vasil A I, Vasil M L. Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol Microbiol. 1996;21:1001–1017. doi: 10.1046/j.1365-2958.1996.381426.x. [DOI] [PubMed] [Google Scholar]
- 2.Frank D W, Iglewski B H. Kinetics of toxA and regA mRNA accumulation in Pseudomonas aeruginosa. J Bacteriol. 1988;170:4477–4483. doi: 10.1128/jb.170.10.4477-4483.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frank D W, Storey D G, Hindahl M S, Iglewski B H. Differential regulation by iron of regA and toxA transcript accumulation in Pseudomonas aeruginosa. J Bacteriol. 1989;171:5304–5313. doi: 10.1128/jb.171.10.5304-5313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamood A N, Colmer J A, Ochsner U A, Vasil M L. Isolation and characterization of a Pseudomonas aeruginosa gene, ptxR, which positively regulates exotoxin A production. Mol Microbiol. 1996;21:97–110. doi: 10.1046/j.1365-2958.1996.6251337.x. [DOI] [PubMed] [Google Scholar]
- 5.Hamood, A. N., J. A. Colmer, U. A. Ochsner, and M. L. Vasil. 1998. Unpublished observations.
- 6.Hedstrom R C, Funk C R, Pavlovskis O R, Galloway D R. Cloning of a gene involved in regulation of exotoxin A expression in Pseudomonas aeruginosa. Infect Immun. 1986;51:37–42. doi: 10.1128/iai.51.1.37-42.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu P V. Exotoxins of Pseudomonas aeruginosa. I. Factors that influence the production of exotoxin A. J Infect Dis. 1973;128:506–513. doi: 10.1093/infdis/128.4.506. [DOI] [PubMed] [Google Scholar]
- 8.Miller J H. Experiments in molecular genetics. 1st ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 9.Ochsner U A, Johnson Z, Lamont I L, Cunliffe H E, Vasil M L. Exotoxin A production in Pseudomonas aeruginosa requires the iron-regulated alternative sigma factor PvdS. Mol Microbiol. 1996;21:1019–1028. doi: 10.1046/j.1365-2958.1996.481425.x. [DOI] [PubMed] [Google Scholar]
- 10.Ochsner U A, Vasil A I, Vasil M L. Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J Bacteriol. 1995;177:7194–7201. doi: 10.1128/jb.177.24.7194-7201.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ochsner U A, Vasil M L. Gene repression by the ferric uptake regulator (FUR) in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc Natl Acad Sci USA. 1996;93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ochsner, U. A., and M. L. Vasil. 1998. Unpublished observations.
- 13.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 14.Shortridge V D, Lazdunski A, Vasil Osmoprotectants and phosphate regulate expression of phospholipase C in Pseudomonas aeruginosa. Mol Microbiol. 1992;5:2823–2831. doi: 10.1111/j.1365-2958.1992.tb01537.x. [DOI] [PubMed] [Google Scholar]
- 15.Stintzi A, Cornelis P, Hohnadel D, Myer J M, Dean C, Poole K, Kourambas S, Krishnapillai V. Novel pyoverdine biosynthesis gene(s) of Pseudomonas aeruginosa. Microbiology. 1996;142:1181–1190. doi: 10.1099/13500872-142-5-1181. [DOI] [PubMed] [Google Scholar]
- 16.Storey D G, Frank D W, Farinha M A, Kropinski A M, Iglewski B H. Multiple promoters control the regulation of the Pseudomonas aeruginosa regA gene. Mol Microbiol. 1990;4:499–503. doi: 10.1111/j.1365-2958.1990.tb00616.x. [DOI] [PubMed] [Google Scholar]
- 17.Storey D G, Raivio T L, Frank D W, Wick M J, Kaye S, Iglewski B H. Effect of regB on expression from the P1 and P2 promoters of the Pseudomonas aeruginosa regAB operon. J Bacteriol. 1991;173:6088–6094. doi: 10.1128/jb.173.19.6088-6094.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasil M L, Prince R W, Shortridge V D. Exoproducts: Pseudomonas exotoxin A and phospholipase C. In: Fick R B, editor. Pseudomonas aeruginosa the opportunist: pathogenesis and disease. Boca Raton, Fla: CRC Press, Inc.; 1992. pp. 59–77. [Google Scholar]
- 19.West S, Sample A, Runyen-Janecky L. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J Bacteriol. 1994;176:7532–7542. doi: 10.1128/jb.176.24.7532-7542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woods D E, Vasil M L. Pathogenesis of Pseudomonas aeruginosa infections. Infect Dis Ther. 1994;12:21–50. [Google Scholar]