Abstract
The Erwinia amylovora rpoS gene, encoding the alternative sigma factor RpoS, has been cloned and characterized. Though highly sensitive to a number of environmental stresses, an E. amylovora rpoS mutant was not compromised in its ability to grow or cause disease symptoms within apple seedlings or in an overwintering model.
Erwinia amylovora is the causative organism of the disease fireblight, which affects members of the family Pomoideae (7). Studies on nonpathogenic mutants of E. amylovora have identified a number of virulence factors, including the extracellular polysaccharide (EPS) and the products of the hrp and dsp gene clusters (29). Little is known concerning how pathogens sense entry into the host; however, it has been proposed that bacteria respond to a generalized nutrient limitation which activates expression of the alternative sigma factor RpoS, promoting transcription of genes required for in vivo survival (12). The alternative sigma factor RpoS regulates a panel of genes which serve to maintain viability during periods of starvation and environmental stress (15). In addition, a direct role for RpoS in regulation of virulence factor expression has been established for a number of pathogens (11, 13). We describe the cloning and characterization of the E. amylovora rpoS gene and the effect of insertional inactivation of the chromosomal copy on E. amylovora virulence.
Cloning and expression of the E. amylovora rpoS gene.
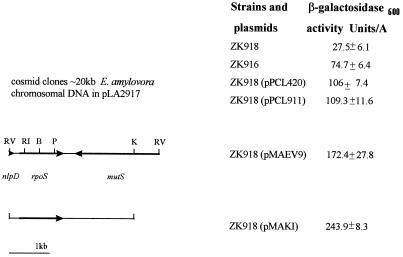
An E. amylovora cosmid library (2) was introduced into strain ZK918 (R. Kolter, Harvard Medical School, Boston, Mass.), which has a β-galactosidase reporter fusion in the bolZ gene and an insertional mutation in the chromosomal copy of rpoS. Transcription of bolZ is dependent on the rpoS gene product, and complementation of the mutation by E. amylovora rpoS was visualized as β-galactosidase activity (30). Of 1,200 E. amylovora cosmid clones, two, pPCL911 and pPCL420, showed high levels of β-galactosidase expression during stationary-phase growth (Fig. 1). Southern blot analysis of pPCL911 and pPCL420, using the Escherichia coli rpoS gene (24) as a probe, confirmed that both cosmids carried a homologue of E. coli rpoS (data not shown). A single 4-kb EcoRV fragment of pPCL911 that hybridized with the E. coli rpoS probe was subcloned into pBluescript SK+ (Stratagene) to generate plasmid pMAEV9, which retained the ability to complement ZK918. The rpoS gene was further localized to a 3.2-kb EcoRV/KpnI fragment on plasmid pMAKI (Fig. 1).
FIG. 1.
Restriction map of plasmid clones containing the E. amylovora rpoS gene and β-galactosidase activities of the plasmids in the reporter strain ZK918. ZK916 is the parent strain, with an intact chromosomal copy of rpoS.
Nucleotide sequence analysis of the E. amylovora rpoS gene.
The nucleotide sequence of the E. amylovora rpoS gene was determined by cycle sequencing (Applied Biosystems). Sequence analysis (1, 8) identified a single open reading frame (ORF) of 990 bp that was 81% homologous to the rpoS genes of E. coli and Salmonella species. Six bases upstream of the predicted start site is a putative Shine-Dalgarno sequence, AGGAG. The E. amylovora rpoS gene encoded a 330-amino-acid protein, the predicted sequence of which was 97% identical to the RpoS sequences of E. coli and Salmonella species. Fifty bases 5′ to the start site of the E. amylovora rpoS gene is an ORF with 64% identity to the E. coli nlpD gene (20). Further sequence analysis identified an incomplete ORF, 3′ to the E. amylovora rpoS gene, which was 76% identical to E. coli mutS. Therefore, the gene organization of nlpD, rpoS, and mutS would appear to be conserved in both E. amylovora and Erwinia carotovora.
Transcript mapping of the rpoS promoter region.
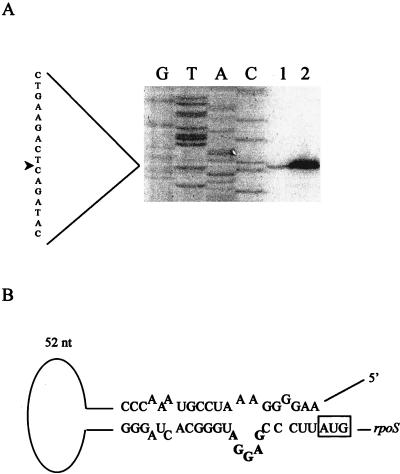
Transcript mapping of the E. amylovora rpoS gene (with the Promega RT primer extension system) identified a single transcriptional start site, within the nlpD gene, 536 bases 5′ of the start of the rpoS coding sequence (Fig. 2A). Putative −10 and −35 consensus sequences were identified 5′ of the transcriptional start. In E. coli the transcription start site for rpoS (P2) is also within the nlpD gene and is approximately 21 bp 5′ to that of E. amylovora (18, 32). No additional transcriptional start sites were detected.
FIG. 2.
(A) Promoter mapping of the E. amylovora rpoS gene. The first four lanes show the nucleotide sequence analysis of the promoter region. Lanes 1 and 2, reverse transcriptase primer extension of RNA from stationary-phase JM101 (pMAKI). The arrow indicates the transcriptional start site. (B) Model of the secondary structure of rpoS promoter region. The Shine-Dalgarno sequence is indicated in bold. nt, nucleotides.
Analysis of the secondary structure of the region containing the rpoS ribosome binding site demonstrated extensive folding (Fig. 2B), which could facilitate formation of a stem structure comparable to that seen in Salmonella typhimurium (3). Though only 66% identity is seen with the intergenic region of S. typhimurium nlpD and rpoS, the nucleotides postulated to be required for formation of the stem structure are conserved. This similarity in folding at the ribosome binding site suggests that the regulation of RpoS expression at the translational level may be comparable to that in other gram-negative bacteria (3, 19).
Characterization of an E. amylovora rpoS mutant.
The E. amylovora rpoS gene was mutated by insertional inactivation with a streptomycin resistance cassette. The EcoRV/PstI fragment from pMAKI was cloned into pRDH20 (9), and the streptomycin cassette from pHP45Ω (38) was ligated into the unique BamHI site in the rpoS gene. The resulting suicide vector, pMA16, was introduced into E. amylovora OT1 by filter mating (6), and the second crossover event was selected by using a sucrose-resistant phenotype. The E. amylovora rpoS mutant was termed OT1.Cm5.
RpoS has been shown to be important for the virulence of a number of animal pathogens. In Salmonella species, rpoS mutants are attenuated for virulence (28, 35) and RpoS protein is required for colonization and destruction of GALT tissue (25), persistence in the liver and spleen (16, 35), and survival in phagocytic cells (5). The ability of OT1.Cm5 to cause disease in the apple seedling assay (23) was determined. The virulence of OT1.Cm5 was not compromised: 61% of the plants developed symptoms compared to 57% of the plants inoculated with OT1 (Fig. 3A). In addition, the times required for the onset of disease symptoms caused by OT1 and OT1.Cm5 were comparable, with necrotic lesions becoming apparent 5 days after inoculation in both cases.
FIG. 3.
Phenotypic analysis of the E. amylovora rpoS mutant. (A) Disease incidence in apple seedlings inoculated with OT1 (solid line) and OT1.Cm5 (dashed line); (B) growth of E. amylovora in apple seedlings; (C) survival of exposure to 25 mM H2O2; (D) survival of exposure to 0.5 M NaCl; (E) survival post-stationary phase. All results are presented as means ± standard errors of the means.
The E. amylovora mutant showed in planta growth characteristics comparable to those of the wild type, with a maximal population of approximately 5 × 106 CFU per plant at day 7 (Fig. 3B). The ability of OT1.Cm5 to grow in planta indicates the absence of nutritional stress. The level of glucose in the apoplast has been estimated at 3 to 6 mM, a level likely to be sufficient to permit bacterial growth (10) and in excess of the 10−4 mM glucose concentration thought to induce the onset of stationary phase (26). As such, it is unclear how nutritionally limited the plant environment is to a pathogen and whether expression of the RpoS regulon would be activated. In our model the situation may be complicated by the release of cellular nutrients due to localized tissue damage at the inoculation site.
The entry of a pathogen into plant tissue evokes a defense response in the form of the generation of active oxygen species. Elevated levels of hydrogen peroxide in transgenic plants results in increased disease resistance (37). It has been proposed that for successful induction of disease symptoms plant pathogens require catalase activity to degrade hydrogen peroxide formed in response to infection (8, 17). Bacterial mechanisms for surviving oxidative stress have been well characterized; in E. coli a panel of protective genes, including katE and katG, are induced in an RpoS-dependent manner (4, 22). The ability of OT1.Cm5 to resist oxidative stress was determined. Strains OT1 and OT1.Cm5 were exposed to 25 mM H2O2 and assessed for viability at 30-s intervals over a 2-min period. Strain OT1 retained over 90% viability after 2 min of exposure, whereas OT1.Cm5 was highly sensitive, with no viable bacteria being recovered after as little as 30 s of exposure (Fig. 3C). Given the postulated role of hydrogen peroxide as a plant defense mechanism, it is surprising to find that, though sensitive to oxidative stress, OT1.Cm5 retains virulence. A likely explanation is that when infecting host plants E. amylovora evades recognition and does not trigger defense mechanisms (21), so levels of hydrogen peroxide in plant tissues remain low and the survival of the mutant strain is not compromised.
Although our results do not identify a role for RpoS in induction of disease symptoms in the apple seedling assay, we can readily propose a function for RpoS in the complex E. amylovora disease cycle. The most common route of fireblight infection follows colonization of the stigma and subsequent invasion via the nectaries (36). To colonize these sites, E. amylovora needs to resist the osmotic stress imposed as a consequence of the high sugar concentration in nectar and stigmal secretions (33). In an in vitro assay, OT1.Cm5 was more sensitive to osmotic shock than wild-type E. amylovora. Strain OT1 retained 70 to 80% viability after 2 min of exposure to 0.5 M NaCl, whereas OT1.Cm5 showed a rapid reduction over the first minute and exhibited only 45% survival (Fig. 3D). The sensitivity of OT1.Cm5 to osmotic stress suggests that RpoS may be essential for successful colonization, allowing the organism to survive the osmotic stress encountered during blossom infection.
During systemic infection the host may attempt to limit E. amylovora spread by walling off diseased tissue to form a canker. Cankers act as a reservoir in which E. amylovora overwinters (34). Canker formation may impose a nutritional stress on the infecting bacteria, inducing rpoS expression and promoting survival under the harsh environmental conditions associated with overwintering. To assess the role of rpoS in the starvation survival of E. amylovora, OT1 and OT1.Cm5 were grown to stationary phase and viable counts were determined every 24 h over a 10-day period. During the first 5 days there was a rapid loss of viability of both OT1 and OT1.Cm5 (Fig. 3E). However, after day 5 there was a slight increase in the numbers of viable OT1, with levels remaining constant for the rest of the experiment. In contrast, OT1.Cm5 continued to lose viability, until by day 7 no bacteria were recovered.
To determine whether this starvation sensitivity affects in planta survival of E. amylovora in mature tissue, OT1 and OT1.Cm5 (1,000 CFU/plant) were inoculated into apple bud sticks and incubated at 1°C for 10 weeks. The rpoS mutation had no effect on survival, with similar bacterial populations being recovered from both sets of bud sticks following overwintering (data not shown). In addition, the ability of OT1.Cm5 to replicate in mature tissue and induce water-soaking symptoms following temperature increase (1°C per day up to 25°C) was also unaffected (data not shown). These results suggest that RpoS plays no role in the survival of E. amylovora during overwintering in mature tissue. This result is surprising, since it is known that there is increased expression of RpoS at reduced temperatures during exponential growth, which leads to activation of a subset of RpoS-dependent genes (31). It has been suggested that under low-temperature conditions RpoS may act as a housekeeping sigma factor and that increased expression of members of the RpoS regulon facilitates efficient growth. The mechanisms by which E. amylovora is able to survive overwintering are currently being investigated in our laboratory.
These results identify an rpoS homologue in the plant pathogen E. amylovora and demonstrate a role for the gene in survival of stationary-phase, oxidative and osmotic stresses. In contrast to the situation seen with a number of animal pathogens, an rpoS mutation does not compromise the in planta growth or virulence of E. amylovora at different stages of the disease cycle. This implies that major differences exist in the natures of the bacterium-host interactions between plant and animal pathogens.
Nucleotide sequence accession number.
The GenBank accession number of the nucleotide sequence of the E. amylovora rpoS gene is AJ222716.
Acknowledgments
This work is supported by a grant from the Leverhulme Trust to J.A.E. and I.S.R. I.S.R. is a Lister Institute Fellow and gratefully acknowledges the support of The Lister Institute for Preventative Medicine. The work is authorized by the Ministry for Agriculture, Fisheries and Food, license no. PHF 1537/1033/44.
We thank M.-A. Barney, P. Loewen, R. Kolter, R. Haigh, and R. Burgess for the generous gifts of strains, plasmids, and antibodies and J. Easton for apple bud sticks.
REFERENCES
- 1.Altschul F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Barney M A, Guinebretiere M H, Marcais B, Coissac E, Paulin J P, Laurent J. Cloning of a large gene cluster involved in Erwinia amylovora CFBP1430 virulence. Mol Microbiol. 1990;4:777–786. doi: 10.1111/j.1365-2958.1990.tb00648.x. [DOI] [PubMed] [Google Scholar]
- 3.Brown L, Elliott T. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calcutt M J, Becker-Hapak M, Gaut M, Hoerter J, Eisenstark A. The rpoS gene of Erwinia carotovora: gene organisation and functional expression in E. coli. FEMS Microbiol Lett. 1998;159:275–281. doi: 10.1111/j.1574-6968.1998.tb12872.x. [DOI] [PubMed] [Google Scholar]
- 5.Chen C-Y, Eckmann L, Libby S J, Fang F C, Okamoto S, Kagnoff M F, Fierer J, Guiney D G. Expression of Salmonella typhimurium rpoS and rpoS-dependent genes in the intracellular environment of eukaryotic cells. Infect Immun. 1996;64:4739–4743. doi: 10.1128/iai.64.11.4739-4743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman M J, Milner J S, Cooper R M, Roberts I S. The use of TnPhoA in Erwinia amylovora to generate random fusions of alkaline phosphatase to extracytoplasmic proteins. FEMS Microbiol Lett. 1991;80:167–172. [Google Scholar]
- 7.Eden-Green S J, Billing E. Fireblight. Rev Plant Pathol. 1974;53:353–365. [Google Scholar]
- 8.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 9.Haigh, R. D. (University of Leicester). 1995. Personal communication.
- 10.Hancock J G, Huisman O C. Nutrient movement in host-pathogen systems. Annu Rev Phytopathol. 1981;19:309–331. [Google Scholar]
- 11.Heiskanen P, Taira S, Rhen M. Role of rpoS in the regulation of Salmonella plasmid virulence (spv) genes. FEMS Microbiol Lett. 1994;123:125–130. doi: 10.1111/j.1574-6968.1994.tb07211.x. [DOI] [PubMed] [Google Scholar]
- 12.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iriarte M, Stainier I, Cornelis G R. The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect Immun. 1995;63:1840–1847. doi: 10.1128/iai.63.5.1840-1847.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahn A Q, Zhao L, Hirose K, Miyake M, Li T, Hashimoto Y, Kawamura Y, Ezaki T. Salmonella typhi rpoS mutant is less cytotoxic than the parent strain but survives inside resting THP-1 macrophages. FEMS Microbiol Lett. 1998;161:201–208. doi: 10.1111/j.1574-6968.1998.tb12949.x. [DOI] [PubMed] [Google Scholar]
- 15.Kolter R, Siegele D A, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 16.Kowarz L, Coynault C, Robbe-Saule V, Norel F. The Salmonella typhimurium katF (rpoS) gene: cloning, nucleotide sequence, and regulation of spvR and spvABCD virulence plasmid genes. J Bacteriol. 1994;176:6852–6860. doi: 10.1128/jb.176.22.6852-6860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laby R J, Beer S V, Bonn W G. Identification of an Erwinia amylovora gene encoding catalase. Acta Hortic. 1995;411:227–228. [Google Scholar]
- 18.Lange R, Fischer D, Hengge-Aronis R. Identification of the transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the ςs subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1995;177:4676–4680. doi: 10.1128/jb.177.16.4676-4680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lange R, Hengge-Aronis R. The cellular concentration of the ςs subunit of RNA polymerase in E. coli is controlled at the levels of transcription, translation and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 20.Lange R, Hengge-Aronis R. The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol Microbiol. 1994;13:733–743. doi: 10.1111/j.1365-2958.1994.tb00466.x. [DOI] [PubMed] [Google Scholar]
- 21.Mansfield J W, Brown I R. The biology of interactions between plants and bacteria. In: Bailey J, editor. Biology and molecular biology of plant-pathogen interactions. Berlin, Germany: Springer-Verlag; 1986. pp. 71–98. [Google Scholar]
- 22.McCann M P, Kidwell J P, Matin A. The putative ς factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991;173:4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milner J S, Dymock D, Cooper R M, Roberts I S. Penicillin-binding-proteins from Erwinia amylovora: mutants defective in PBP2 are avirulent. J Bacteriol. 1993;175:6082–6088. doi: 10.1128/jb.175.19.6082-6088.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulvey M R, Loewen P C. Nucleotide sequence analysis of katF of Escherichia coli suggests KatF protein is a novel transcription factor. Nucleic Acids Res. 1989;17:9979–9990. doi: 10.1093/nar/17.23.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nickerson C A, Curtiss R., III Role of sigma factor RpoS in initial stages of Salmonella typhimurium infection. Infect Immun. 1997;65:1814–1823. doi: 10.1128/iai.65.5.1814-1823.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notely L, Ferenci T. Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli? J Bacteriol. 1996;178:1465–1468. doi: 10.1128/jb.178.5.1465-1468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 28.Robbe-Saule V, Coynault C, Norel F. The live oral typhoid vaccine Ty21a is a rpoS mutant and is susceptible to various environmental stresses. FEMS Microbiol Lett. 1995;126:171–176. doi: 10.1111/j.1574-6968.1995.tb07412.x. [DOI] [PubMed] [Google Scholar]
- 29.Roberts I S, Coleman M J. The virulence of Erwinia amylovora: molecular genetic perspectives. J Gen Microbiol. 1991;137:1453–1457. [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sledjeski D D, Gupta A, Gottesman S. The small RNA, DsrA is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 32.Takayanagi K, Tanaka K, Takahashi H. Structure of the 5′ upstream region and the regulation of the rpoS gene of Escherichia coli. Mol Gen Genet. 1994;243:525–531. doi: 10.1007/BF00284200. [DOI] [PubMed] [Google Scholar]
- 33.Thomas H E, Ark P A. Nectar and rain in relation to fireblight. Phytopathology. 1934;24:682–685. [Google Scholar]
- 34.Vanneste J L. Erwinia amylovora. In: Singh U S, Singh R P, Kohmoto K, editors. Pathogenesis and host specificity in plant diseases. 1. Prokaryotes. Oxford, United Kingdom: Pergamon Press; 1995. pp. 21–41. [Google Scholar]
- 35.Wilmes-Riesenberg M R, Foster J W, Curtiss R., III An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect Immun. 1997;65:203–210. doi: 10.1128/iai.65.1.203-210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson M, Sigee D C, Epton H A S. Erwinia amylovora infection of hawthorn blossom. III. The nectary. J Phytopathol. 1990;128:62–74. [Google Scholar]
- 37.Wu G, Shortt B J, Lawrence E B, Levine E B, Fitzsimmons K C, Shah D M. Disease resistance conferred by expression of a gene encoding H2O2-generating glucose oxidase in transgenic potato plants. Plant Cell. 1995;7:1357–1368. doi: 10.1105/tpc.7.9.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]