Abstract
The present investigation reports the efficient multistep synthesis of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f) in good yields. All the 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f) were characterized by spectroscopic techniques. Five among the six compounds were tested against 56 cancer cell lines at 10 µM as per the standard protocol. 1-(4-Bromophenyl)-3-(1,3-dioxoisoindolin-2-yl)urea (7c) exhibited moderate but significant anticancer activity against EKVX, CAKI-1, UACC-62, MCF7, LOX IMVI, and ACHN with percentage growth inhibitions (PGIs) of 75.46, 78.52, 80.81, 83.48, 84.52, and 89.61, respectively. Compound 7c was found to exhibit better anticancer activity than thalidomide against non-small cell lung, CNS, melanoma, renal, prostate, and breast cancer cell lines. It was also found to exhibit superior anticancer activity against melanoma cancer compared to imatinib. Among the tested compounds, the 4-bromosubstitution (7c) on the phenyl ring demonstrated good anticancer activity. Docking scores ranging from −6.363 to −7.565 kcal/mol were observed in the docking studies against the molecular target EGFR. The ligand 7c displayed an efficient binding against the EGFR with a docking score of −7.558 kcal/mol and displayed an H-bond interaction with Lys745 and the carbonyl functional group. Compound 7c demonstrated a moderate inhibition of EGFR with an IC50 of 42.91 ± 0.80 nM, in comparison to erlotinib (IC50 = 26.85 ± 0.72 nM), the standard drug. The antioxidant potential was also calculated for the compounds (7a–f), which exhibited good to low activity. 1-(2-Methoxyphenyl)-3-(1,3-dioxoisoindolin-2-yl)urea (7f) and 1-(4-Methoxyphenyl)-3-(1,3-dioxoisoindolin-2-yl)urea (7d) demonstrated significant antioxidant activity with IC50 values of 15.99 ± 0.10 and 16.05 ± 0.15 µM, respectively. The 2- and 4-methoxysubstitutions on the N-phenyl ring showed good antioxidant activity among the series of compounds (7a–f). An in silico ADMET prediction studies showed the compounds’ adherence to Lipinski’s rule of five: they were free from toxicities, including mutagenicity, cytotoxicity, and immunotoxicity, but not for hepatotoxicity. The toxicity prediction demonstrated LD50 values between 1000 and 5000 mg/Kg, putting the compounds either in class IV or class V toxicity classes. Our findings might create opportunities for more advancements in cancer therapeutics.
Keywords: ADMET, anticancer, antioxidant, cell lines, phthalimide, molecular docking
1. Introduction
With nearly 10 million fatalities and nearly 19.3 million new cases of cancer worldwide in 2020, cancer is currently the leading cause of death. The number of cancer cases is expected to increase to 28.4 million by 2040 [1]. Cancer’s etiology is complex, involving the uncontrolled growth of cells invading neighboring tissues or organs. Adults over 50 are typically prone to developing cancer, but incidence rates of early-onset cancer (those diagnosed before 50) have increased globally [2,3]. In 2020, the global lifetime risk of cancer from birth to death was estimated to be 25.10% [4]. The United States is expected to have 1,958,310 new cancer cases and 609,820 cancer deaths in 2023 [5]. Similarly, the European Union (EU-27) is expected to have 1,261,990 cancer deaths in 2023, while the estimated number of cancer incidents in India in 2022 was 1,461,427 [6,7]. For many years, patients with cancer had only a limited number of treatment options, which included surgery, radiation therapy, and chemotherapy either individually or in combination [8]. Surgery offers the best chance of a cure for tumors that can be removed, especially in cases of early-stage disease [9]. Despite its known harmful effects on the patient’s physical as well as psychological health, chemotherapy is still a frequently chosen therapeutic option used either alone or in combination with radiotherapy [8,10]. One can avoid early-onset cancers by altering their diet, giving up alcohol, and quitting smoking [2]. Medicinal chemistry involves the invention, discovery, identification, synthesis of organic compounds with medicinal importance, in silico studies, target identification, and structure–activity relationship studies [11]. Medicinal chemists are crucial players in the process of discovering new, more effective drugs, and they are intensely engaged in cancer research worldwide to find a more pleasant cancer treatment option.
Heterocyclic compounds have drawn the attention of scientists due to their ever-increasing need for therapeutic compounds. Phthalimide is a fused heterocyclic ring with a variety of therapeutic uses, including antibacterial [12], antiviral [13], anticancer [14,15,16], anticonvulsant [17], anti-EGFR [15], anti-inflammatory [18], inflammatory and neuropathic pain [19], antimalarial [20], schistosomiasis [21], and many more [22,23]. Some of phthalimide-based anticancer compounds include cantharidin, CC-11006, lenalidomide, pomalidomide, and thalidomide (Figure 1) [24,25]. Inspired by the enormous therapeutic potential of phthalimide, we report herein the synthesis of a series of six phthalimide-based 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f) and their anticancer and antioxidant activities. Recent years have seen incredible progress in the expansion of anticancer drugs, and a number of molecular targets have been explored [26]. DNA topoisomerase I, epidermal growth factor receptor, focal-adhesion kinase, glycogen synthase kinase-3, histone deacetylase, methionine aminopeptidase, nuclear factor, poly(ADP-ribose) polymerase-1, tubulin, telomerase, thymidine phosphorylase, thymidylate synthase, and vascular endothelial growth factor receptor are some of the important targets reported in the literature [15,27]. 1-(1,3-Dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f) were subjected to molecular docking studies against three different molecular targets including DNA topoisomerase I, tubulin, and the EGFR. Phthalimide analogs were also reported as an inhibitor of the EGFR in previous studies; hence, we explored the binding affinity of compounds 7a–f with the EGFR [15]. Many anticancer drugs, including gefitinib, laptinib, orantinib, nintedanib, semaxanibs, sunitinib, etc., have the EGFR as one of their primary molecular targets.
Figure 1.
Phthalimide-based anticancer agents.
The body constantly produces free radicals, which can become toxic in high concentrations or when the body’s natural antioxidant defenses are not working properly [28]. High concentrations of free radicals cause lipids, proteins, and DNA damage in cells and tissues, which lead to serious illnesses like cancer, inflammatory diseases, CVS disorders, immunosuppression, and neurodegenerative disorders [29]. Cancer may develop as a result of free radicals’ formation brought on by oxidative stress. Antioxidants are a class of compounds that aid in the capture and neutralization of free radicals, reducing the harm that free radicals can cause to the body. In addition, antioxidants help prevent oxidative-stress-related illnesses like cancer and inflammation [30,31,32]. The antioxidant activity of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogues (7a–f) was also assessed in the current study because phthalimide analogues have also been reported to be antioxidants [33,34,35]. Earlier a few 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogues were prepared and reported by researchers by two different routes. One research group prepared 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogues by refluxing substituted phenyl semicarbazide and phthalic anhydride in glacial acetic acid for 5 h with very low yields ranging from 41 to 61 percent [36]. Another group of researchers also prepared these compounds by refluxing N-aminophthalimide and p-substituted phenylcarbamate in ethanol for 1–2 h with yields varying from 35.9 to 86.2 percent [37]. We have opted for an alternative synthetic pathway for the synthesis, as it was discovered to be highly effective in terms of its yield. Earlier these compounds were reported as anticonvulsant and antiviral agents [36,37]. Since some of the compounds were reported to be non-toxic (cytotoxicity ≥ 400 µg/mL), it made sense to investigate their anticancer potential using cell line experiments [37].
Modern therapeutic development is aided by in silico studies. During the COVID-19 pandemic, bioinformatics and in silico techniques were valued [38]. In silico screening methods based on algorithms rapidly screen numerous molecules to identify potential drugs [39]. Thanks to cutting-edge in silico and artificial intelligence techniques, research is no longer limited to laboratory settings. This has resulted in the development of INS018_055, a first-in-class AI-generated anti-fibrotic medication that is undergoing a multiregional Phase II clinical trial in both China and the United States [40,41,42]. The current study aimed to synthesize a few 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogues (7a–f), evaluate their biological potentials, and investigate in silico docking and ADMET prediction.
2. Results
2.1. Chemistry
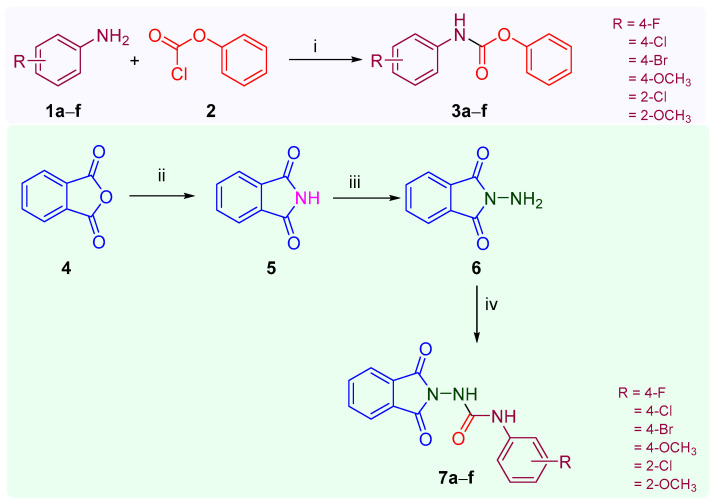
The protocol for the synthesis of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f) is summarized in Scheme 1. In the initial step, phenyl(substituted phenyl)carbamate (3a–f) was prepared by adding a solution of substituted aniline (1a–f) (2.5 mmol) in triethylamine to a solution of phenyl chloroformate (2) in chloroform [34]. In the second step, phthalimide (5) was prepared through the fusion of phthalic anhydride (4) and urea at 140 °C, and in the subsequent step, phthalimide (5) was allowed to react with hydrazine hydrate (NH2NH2.H2O) in ethanol at 85 °C for 5 min to obtain 2-aminoisoindoline-1,3-dione (6) [43]. In the concluding step, an equimolar quantity of phenyl(substituted phenyl)carbamate (3a–f) and aminoisoindoline-1,3-dione (6) in dichloromethane (DCM) was continuously stirred for 6–8 h at room temperature to obtain the final product, 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f) [43]. Preparatory thin-layer chromatography (TLC Silica gel 60 F254) using toluene/ethylacetate/formic acid (5:4:1) as the mobile phase was used to continuously track the reaction’s progress. The compounds (7a–f) were identified and characterized based on the spectral data as well as their melting points reported in the previous literature. Yogeeswari et al. reported the synthesis of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs from phthalic anhydride and substituted phenyl semicarbazide in glacial acetic acid with very low yields ranging from 41 to 61 percent. Similarly, Verma et al. reported their synthesis with low to good yields ranging from 35.9 to 86.2 percent [36,37]. The title compounds (7a–f) were successfully synthesized with excellent yields ranging from 81 to 92 percent in the current investigation using a productive synthetic scheme (Scheme 1). The physical constant and NSC code of compounds (7a–f) are given in Table 1. The title compounds (7a–f) were identified using spectroscopic techniques, such as mass and NMR spectral data. The 1H NMR of the title compounds (7a–f) showed a singlet at δ ppm 3.74 corresponding to the three protons of the OCH3 group present in the compounds 7d and 7f; a multiplet at δ ppm 7.88–8.15 corresponding to the four aromatic protons of phthalimide; two multiplets at δ ppm 7.33–7.42 (H3 and H5) and 7.48–7.60 (H2 and H6) corresponding to each of the two protons of the 4-flurophenyl ring of compound 7a; two doublets at δ ppm ranging from 6.95 to 7.43 (H3 and H5) and 7.31 to 7.74 (H2 and H6) corresponding to each of the two aromatic protons of the substituted phenyl ring of compound 7b–d; and two multiplets at δ ppm 7.05–7.52 and 7.53–7.98 corresponding to each of four the aromatic protons of the N-phenyl ring and phenyl ring of phthalimide, respectively, for the compounds 7e and 7f. Similarly, a singlet ranging from δ ppm 9.59 to 9.69 corresponding to the secondary amine (ArNH) was observed, while a singlet ranging from δ ppm 10.03 to 10.65 was observed, corresponding to the amide (CONH) group present in the compounds (7a–f). The 13C NMR spectroscopy identified the nature and number of carbons present in any of the compounds (7a–f). The mass spectra (ESI-MS) of the compounds (7a–f) revealed (M + 1)+ (7a–f) and (M + 3)+ (7b,c, and 7e) peaks corresponding to their molecular formula. One of the representative compounds, 7c, had a purity of 98.74 percent in HPLC.
Scheme 1.
Synthetic protocol of 1-(1,3-dioxoisoindolin-2-yl)-3-(substituted phenyl)urea analogs (7a–f): Reagents and conditions: (i) CHCl3, Et2N, rt, 4–5 h, 78–80% yield%; (ii) NH2CONH2, 140 °C, 5 min, 86%, yield; (iii) EtOH, NH2NH2.H2O; 85 °C, 5 min, 72 yield; (iv) 3a–f, DCM, rt, 6–8 h, 81–92% yield.
Table 1.
The comparative yields, melting point, and NSC code of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f).

| ||||||||
|---|---|---|---|---|---|---|---|---|
| S. No. | Compound | R | NSC Code | Rf * | % Yield | Mp (°C) | ||
| Found | Reported | Found | Reported | |||||
| 1 | 7a | 4-Fluoro | 838925 | 0.69 | 89 | 67.7 a | 248–250 | >245 a |
| 2 | 7b | 4-Chloro | 838926 | 0.71 | 90 | 86.2 a | 252–254 | >250 a |
| 3 | 7c | 4-Bromo | 838927 | 0.72 | 92 | 73.1 a | 250–252 | 250 a |
| 4 | 7d | 4-Methoxy | - | 0.67 | 83 | 35.9 a | >260 | >260 a |
| 5 | 7e | 2-Chloro | 838928 | 0.68 | 81 | 46 b | 140–142 | 139 b |
| 6 | 7f | 2-Methoxy | 838934 | 0.73 | 85 | - | 156–158 | - |
2.2. Anticancer Activity
According to the reported protocol, the anticancer activity of the compounds was evaluated against the nine diverse panels of 56 NCI cancer cell lines at 10 µM in terms of their growth control [44,45,46,47]. When tested against the breast cancer cell line MCF7, compound 7e showed anticancer activity with a growth percentage (GP) of 87.43%, while compound 7f showed anticancer activity when tested against the renal cancer cell line CAKI-1 with a GP of 89.58%. Compound 7a exhibited anticancer activity against the MCF7 (breast cancer), HL-60(TB) (leukemia), and EKVX (non-small cell lung cancer) cancer cell lines with GPs of 84.87, 86.23, and 89.44 percent, respectively. Compound 7b exhibited moderate anticancer activity against the EKVX (non-small cell lung cancer), CAKI-1 (renal cancer), HL-60(TB) (leukemia), UACC-62 (melanoma), and LOX IMVI (melanoma) cancer cell lines with GPs of 80.73, 82.3, 83.07, 85.6, and 87.19 percent, respectively. Compound 7c exhibited significant anticancer activity against the EKVX (non-small cell lung cancer), CAKI-1 (renal cancer), UACC-62 (renal cancer), MCF7 (breast cancer), LOX IMVI (melanoma), and ACHN (renal cancer) cancer cell lines with GPs of 75.46, 78.52, 80.81, 83.48, 84.52, and 89.61, respectively, among the series of compounds. The anticancer data of the compounds at 10 µM are given in Table 2. The five most sensitive cell lines are summarized in Table 3. The compounds’ average percentage growth inhibitions (PGIs) against each panel were calculated and compared to the reference drug, imatinib (Table 4). The compounds 7a–c and 7e,f demonstrated good inhibition against melanoma and ovarian cancer compared to imatinib. Compound 7c displayed better anticancer activity than imatinib (PGI = −0.87%) against melanoma with a percentage growth inhibition (PGI) of 5.03 percent. The compounds 7a–c and 7e,f demonstrated good inhibition against CNS, melanoma, and prostate cancer compared to thalidomide. Compound 7c displayed better anticancer activity than thalidomide against the non-small cell lung, CNS, melanoma, renal, prostate, and breast cancer cell lines. The anticancer activity of the compounds 7a–c and 7e,f was found to be less than that of gefitinib. The 4-bromo substitution on the N-phenyl ring demonstrated promising results. The structure–activity relationship using the mean PGI was measured for the following substitutions: N-phenyl 4-bromo > 4-fluoro > 4-chloro > 2-chloro > 2-methoxy. Similarly, the structure–activity relationship against the EKVX and MCF7 cell lines are shown in Figure 2. The replacement of the NH function with CH2 increased the anticancer activity against PC3 and HT29 as evidenced from previous reported work [48]. Previous reports have demonstrated a decrease in anticancer activity against PC3 and HT29 when the NH function was replaced with CH2 together with the replacement of the phenyl ring with the bulky benzylthio-1,3,4-thiadiazole moiety [49]. Further research is required to determine whether the substitution of the 1,3-dioxoisoindole moiety affects the anticancer activity, as in some cases, the substitution of the 1,3-dioxoisoindole moiety alters the anticancer activity [24,25]. Other changes, such as replacing NH with CH2 or C=O with C=S, may be possible and could be the focus of future studies on SAR establishment. The anticancer data of the compounds against 56 NCI cancer cell lines can be found within the Supplementary Material (Figures S1–S5).
Table 2.
The anticancer activity of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f) at 10 µM as GP.
| Panel | Cell Line | 7a | 7b | 7c | 7e | 7f |
|---|---|---|---|---|---|---|
| Leukemia | CCRF-CEM | 103.9 | 104.8 | 101.2 | 103.37 | 95.13 |
| HL-60(TB) | 86.23 | 83.07 | 92.99 | 101.97 | 101.22 | |
| K-562 | 100.3 | 91.98 | 92.8 | 110.7 | 92.54 | |
| MOLT-4 | 98.29 | 101.1 | 99.67 | 97.49 | 106.2 | |
| RPMI-8226 | 105.9 | 106.5 | 108.3 | 109.46 | 103.93 | |
| SR | 94.86 | 96.2 | 94.2 | 92.66 | 93.35 | |
| Non-small cell lung cancer | A549/ATCC | 99.08 | 102.7 | 101.2 | 103.02 | 103.59 |
| EKVX | 89.44 | 80.73 | 75.46 | 90.83 | 91.2 | |
| HOP-62 | 107.5 | 107.1 | 108.4 | 99.95 | 115.38 | |
| HOP-92 | 120.9 | 109.3 | 107.7 | 110.12 | 115.4 | |
| NCI-H226 | 99.82 | 94.57 | 91.94 | 96.96 | 100.71 | |
| NCI-H23 | 102.4 | 93.59 | 93.98 | 100.32 | 97.03 | |
| NCI-H322M | 94 | 96.1 | 98.49 | 101.21 | 95.89 | |
| NCI-H460 | 116.7 | 121.1 | 107.8 | 106.44 | 118.73 | |
| NCI-H522 | 96.78 | 94.82 | 96.41 | 97.3 | 96.7 | |
| Colon cancer | COLO 205 | 111.6 | 111.5 | 108.9 | 100.1 | 113.79 |
| HCC-2998 | 105.2 | 107.6 | 104.5 | 101.52 | 104.81 | |
| HCT-116 | 97.82 | 100.6 | 97.06 | 104.92 | 110.15 | |
| HCT-15 | 99.39 | 94.99 | 94.12 | 101.91 | 100.49 | |
| HT29 | 103.4 | 109.3 | 107.1 | 104.06 | 113.18 | |
| KM12 | 100.2 | 99.16 | 98.57 | 99.5 | 100.71 | |
| SW-620 | 103.7 | 108.3 | 99.5 | 96.91 | 101.88 | |
| CNS cancer | SF-268 | 97.06 | 97.16 | 96.17 | 94.28 | 104.11 |
| SF-295 | 94.85 | 98.09 | 99.52 | 96.86 | 101.54 | |
| SF-539 | 95.37 | 92.59 | 92.35 | 94.08 | 95.62 | |
| SNB-19 | 97.85 | 96.23 | 96.31 | 98.24 | 97.73 | |
| U251 | 103.1 | 107.2 | 105.5 | 104.47 | 104.72 | |
| Melanoma | LOX IMVI | 96.84 | 87.19 | 84.52 | 92.19 | 96.42 |
| MALME-3M | 93.65 | 94.22 | 96.35 | 94.53 | 94.69 | |
| M14 | 101.2 | 101.9 | 101.8 | 101.76 | 104.17 | |
| MDA-MB-435 | 97.22 | 96.66 | 96.81 | 100.53 | 99.08 | |
| SK-MEL-28 | 99.56 | 102.6 | 104.4 | 104.47 | 101.02 | |
| SK-MEL-5 | 92.01 | 86.59 | 90.69 | 92.19 | 93.38 | |
| UACC-257 | 102 | 105.8 | 104.4 | 94.53 | 103.72 | |
| UACC-62 | 90.53 | 85.6 | 80.81 | 101.76 | 93.41 | |
| Ovarian cancer | IGROV1 | 105 | 106.3 | 104.3 | 100.53 | 110.36 |
| OVCAR-3 | 109.2 | 113.9 | 111.8 | 104.47 | 106.37 | |
| OVCAR-4 | 97.78 | 96.67 | 93.93 | 94.18 | 99.61 | |
| OVCAR-5 | 104.8 | 102.9 | 101.1 | 105.8 | 102.63 | |
| OVCAR-8 | 99.87 | 103.1 | 102.2 | 92.98 | 97.77 | |
| NCI/ADR-RES | 102.4 | 102.1 | 103.1 | 108.87 | 102.13 | |
| SK-OV-3 | 102.6 | 105.6 | 99.28 | 107.11 | 104.8 | |
| Renal cancer | 786-0 | 98.32 | 100.2 | 100.1 | 100.59 | 104.72 |
| A498 | 103.9 | 117 | 114.4 | 108.88 | 115.67 | |
| ACHN | 98.77 | 92.38 | 89.61 | 99.89 | 99.4 | |
| CAKI-1 | 92.91 | 82.3 | 78.52 | 99.75 | 89.58 | |
| RXF 393 | 96.91 | 102 | 105.8 | 114.5 | 105.46 | |
| SN 12C | 93.94 | 93.88 | 94.56 | 93.07 | 99.81 | |
| TK-10 | 96.11 | 109.1 | 113.5 | 102.14 | 111.49 | |
| Prostate cancer | PC-3 | 99.99 | 102.2 | 99.48 | 96.67 | 104.34 |
| DU-145 | 104.9 | 104.6 | 103.3 | 105.71 | 104.85 | |
| Breast cancer | MCF7 | 84.87 | 90.11 | 83.48 | 87.43 | 91.63 |
| HS 578T | 98.14 | 96.97 | 92.76 | 93.19 | 100.05 | |
| BT-549 | 103.5 | 117.7 | 112.2 | 117.25 | 115.87 | |
| T-47D | 103.8 | 102.3 | 98.97 | 103.41 | 105.35 | |
| MDA-MB-468 | 99.41 | 97.99 | 100 | 98.35 | 97.8 | |
| Mean GP | 99.92 | 100.11 | 98.79 | 100.64 | 102.35 | |
Table 3.
The anticancer activity (GP and PGI) of the five most sensitive cell lines at 10 µM.
| Compound | Anticancer Activity in One Dose (10 µM) | ||
|---|---|---|---|
| The Most Sensitive Cell Lines | GP | PGI | |
| 7a | MCF7 (Breast cancer) | 84.87 | 15.13 |
| HL-60(TB) (Leukemia) | 86.23 | 13.77 | |
| EKVX (Non-small cell lung cancer) | 89.44 | 10.56 | |
| SK-MEL-5 (Melanoma) | 92.01 | 7.99 | |
| CAKI-1 (Renal cancer) | 92.91 | 7.09 | |
| 7b | EKVX (Non-small cell lung cancer) | 80.73 | 19.27 |
| CAKI-1 (Renal cancer) | 82.3 | 17.7 | |
| HL-60(TB) (Leukemia) | 83.07 | 16.93 | |
| UACC-62 (Melanoma) | 85.6 | 14.4 | |
| LOX IMVI (Melanoma) | 87.19 | 12.81 | |
| 7c | EKVX (Non-small cell lung cancer) | 75.46 | 24.54 |
| CAKI-1 (Renal cancer) | 78.52 | 21.48 | |
| UACC-62 (Melanoma) | 80.81 | 19.19 | |
| MCF7 (Breast cancer) | 83.48 | 16.52 | |
| LOX IMVI (Melanoma) | 84.52 | 15.48 | |
| 7e | MCF7 (Breast cancer) | 87.43 | 12.57 |
| EKVX (Non-small cell lung cancer) | 90.83 | 9.17 | |
| LOX IMVI (Melanoma) | 92.19 | 7.81 | |
| SK-MEL-5 (Melanoma) | 92.19 | 7.81 | |
| SR (Leukemia) | 92.66 | 7.34 | |
| 7f | CAKI-1 (Renal cancer) | 89.58 | 10.42 |
| EKVX (Non-small cell lung cancer) | 91.2 | 8.8 | |
| MCF7 (Breast cancer) | 91.63 | 8.37 | |
| K-562 (Leukemia) | 92.54 | 7.46 | |
| SR (Leukemia) | 93.35 | 6.47 | |
Table 4.
The average PGI of the 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f) and imatinib at 10 µM.
| Panels | 7a | 7b | 7c | 7e | 7f | Imatinib * | Gefitinib * | Thalidomide * |
|---|---|---|---|---|---|---|---|---|
| Leukemia | 1.76 | 2.72 | 1.82 | −2.6 | 1.27 | 9 | 79.68 | 15.28 |
| Non-small cell lung cancer | −2.94 | 0.01 | 2.07 | −0.7 | −3.85 | 15.68 | 63.97 | 0.06 |
| Colon cancer | −3.05 | −4.5 | −1.39 | −1.3 | −6.43 | 5.34 | 52.19 | −0.27 |
| CNS cancer | 2.36 | 1.74 | 2.02 | 2.41 | −0.74 | 5.8 | 46.13 | −4.95 |
| Melanoma | 3.38 | 4.93 | 5.03 | 2.26 | 1.76 | −0.87 | 44.99 | −0.39 |
| Ovarian cancer | −3.08 | −4.38 | −2.23 | −1.99 | −3.38 | −7.16 | 60.93 | 1.90 |
| Renal cancer | 2.74 | 0.45 | 0.50 | −2.69 | −3.733 | 3.25 | 77.89 | −2.37 |
| Prostate cancer | −2.46 | −3.40 | −1.41 | −1.19 | −4.59 | 12.5 | 59.60 | −8.60 |
| Breast cancer | 2.06 | −0.99 | 2.25 | 0.07 | −2.35 | 12.15 | 52.88 | −2.04 |
The best outcome is indicated by bold font; * The anticancer data of gefitinib and thalidomide with NSC Codes 759854 and 66847, respectively, were retrieved from official website of NCI, USA [44].
Figure 2.
The structure–inhibition relationship for anticancer activity of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs.
2.3. In Vitro EGFR Kinase Inhibition Assay
In order to investigate the EGFR inhibition potential of the most active anticancer compound (7c), an EFGR kinase assay kit (BPS Bioscience, San Diego, CA, USA) was used as per the manufacturer’s instructions and the protocol is reported [50]. The IC50 values for the tested compound (7c) and the reference drug (erlotinib) were determined in a molar concentration using the GraphPad prism 9 software. The inhibition assays were performed in triplicate and their results showed that the compound displayed an inhibition of the EGFR with an IC50 value of 42.91 ± 0.80 nM, while the standard drug erlotinib displayed an IC50 value of 26.85 ± 0.72 nM. Compound 7c displayed less EGFR inhibitory activity than erlotinib.
2.4. Molecular Docking Studies
The molecular docking of the ligands (7a–f) was studied against the three important molecular targets of anticancer drugs: tubulin, DNA topoisomerase I, and the epidermal growth factor receptor (EGFR). Higher eukaryotes depend on DNA topoisomerase I enzymes to relax DNA supercoils caused by transcription, replication, and chromatin remodeling [51]. It is the target of the commonly used anticancer medications topotecan and irinotecan, which are made from the alkaloid camptothecin [52,53]. In vitro and in vivo studies of a fluoroglycosyl-3,9-difluoroindolocarbazole (BMS-250749) containing the phthalimide moiety demonstrated DNA topoisomerase I’s inhibitory activity; therefore, we selected DNA topoisomerase I for our molecular docking studies [54,55]. Microtubules are an important target for cancer therapy because they are essential for many cellular processes, such as mitosis, cell signaling, and organelle trafficking [56]. The development of novel tubulin inhibitors (TIs) that specifically target the colchicine binding site seems to be a promising avenue for tubulin inhibitor advancement [57]. Paclitaxel, docetaxel, epothilones, vincristine, vinblastine, vinflunine, dolastatin 10, colchicine, podophyllotoxin, and noscapine are some of the reported tubulin inhibitors [58]. Phthalimide analogues were also reported as a tubulin inhibitor [59]. From the protein data bank, the X-ray crystallographic structure of DNA topoisomerase I (PDB ID: 1SC7) and tubulin (PDB ID: 1SA0) was retrieved for docking studies [60,61]. Docking scores ranging from −5.830 to −7.292 kcal/mol were observed in the docking studies of ligands 7a–f against tubulin as a molecular target (PDB ID: 1SA0). The binding site of colchicine in tubulin enzyme (PDB ID: 1SA0) consists of a hydrophobic cavity, lined with a few active residues, like Lys254, Cys241, Asn248, Lys352, Ala316, Val315, Met259, Leu248, Ala250, and Ala317. It contains glycine 717, the polar residues Thr718 and Asn722, the hydrophobic residue Leu721, and the positively charged residue Arg364. Similarly, docking scores of ranging from −6.363 to −7.565 kcal/mol were observed in the docking studies of ligands 7a–f against human DNA topoisomerase I (PDB ID: 1SC7). An efficient binding was observed with the EGFR as a putative target with docking scores ranging from −7.702 to −8.644 kcal/mol. The active site of DNA topoisomerase I (PDB ID: 1SC7) contains glycine 717, the polar residues Thr718 and Asn722, the hydrophobic residue Leu721, and the positively charged residue Arg364. The EGFR is an important molecular target for many anticancer drugs, including gefitinib, laptinib, orantinib, nintedanib, semaxanibs, sunitinib, etc. [62]. From the protein data bank, the X-ray crystallographic structure of the EGFR (PDB ID: 3W2R) was retrieved for the docking studies [63]. The ligands 7a–f were saved as mol files and were prepared for docking using Ligprep, and the docking was carried out in accordance with the method outlined by Sogabe et al. (2013) [64]. The active site contains the hydrophobic residues Phe723, Leu747, Ile759, Met766, Cys775, Leu788, Met790, and Phe856; the positively charged residues Lys745 and Arg858; the negatively charged residues Glu762 and Asp855; and the polar residue Thr854. The docking studies with two types of electrostatic interactions, H-bond and π–π stacking, showed docking scores ranging from −6.363 to −7.565 kcal/mol. The ligands 7d and 7c showed a similar type of binding interactions with the residue Lys745 through an H-bond, whereas the ligands 7a and 7d showed identical interactions with the residue Lys745 through an H-bond and π–π stacking with the residue Phe856. The ligand 7e showed two H-bond interactions with the residues Lys745 and Asp855, while the ligand 7f displayed an H-bond interaction with Thr854 via the water molecule. The results of the molecular docking studies are compiled in Table 5, and Figure 3 displays the interaction of the ligands three-dimensionally (7a–f). The two- and three-dimensional interactions of ligand 7c are shown in Figure 4, while the two- and three-dimensional interactions of ligand 7a are shown in Figure 5. The electrostatic interaction in the molecular docking studies of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a−f) against tubulin (PDB ID: 1SA0) and DNA topoisomerase I (PDB ID: 1SC7) are given in Table S1 (Supplementary Materials).
Table 5.
The molecular docking studies of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f).
| S. No. | Compound | PDB: 1SA0 | PDB ID: 1SC7 | PDB ID: 3W2R | ||||
|---|---|---|---|---|---|---|---|---|
| Docking Score | Emodel Score | Docking Score | Emodel Score | Docking Score | Emodel Score | Interaction | ||
| 1 | 7a | −6.469 | −54.331 | −7.169 | −59.500 | −7.934 | −66.771 | H-bond (Lys745; 2.05 Å), π-π-stacking (Phe856; 5.21 Å) |
| 2 | 7b | −6.426 | −56.002 | −7.473 | −63.130 | −7.816 | −64.552 | H-bond (Lys745; 1.97 Å) |
| 3 | 7c | −6.480 | −57.825 | −7.318 | −64.802 | −7.558 | −64.096 | H-bond (Lys745; 2.04 Å) |
| 4 | 7d | −7.069 | −57.606 | −7.565 | −66.006 | −7.702 | −65.761 | H-bond (Lys745; 2.04 Å), π-π-stacking (Phe856; 5.48 Å) |
| 5 | 7e | −7.292 | −62.624 | −6.363 | −65.436 | −7.119 | −65.715 | H-bond (Lys745; 1.64 Å), H-bond (Asp855; 2.13 Å) |
| 6 | 7f | −5.830 | −54.813 | −7.306 | −63.183 | −8.644 | −65.599 | H-bond (Thr854 via H2O; 1.76 and 2.39 Å) |
Figure 3.
The 3D interaction of ligands (7a–f) with EGFR (PBD: 3W2R).
Figure 4.
The 2D and 3D interactions of ligand 7c with EGFR (PBD: 3W2R).
Figure 5.
The 2D and 3D interactions of ligand 7a with EGFR (PBD: 3W2R).
2.5. Antioxidant Activity
Oxidative stress causes inflammation and transforms normal cells into cancerous ones [65,66,67]. Free radical formation as a result of oxidative stress plays an important role in the progress of many chronic illnesses, including CVS disorders, ageing, anaemia, cancer, and inflammation. Antioxidants provide protection against these disease by scavenging available free radicals [68,69]. The antioxidant activity was calculated as the IC50 in µM, using the DPPH free radicals scavenging activity as described by Koleva et al., 2002, and is given in Table 6 [70]. The compounds displayed antioxidant activity with IC50 values ranging between 15.99 ± 0.10 and 54.27 ± 0.49 µM. The compounds 7f and 7d demonstrated significant antioxidant activity with IC50 values of 15.99 ± 0.10 and 16.05 ± 0.15 µM, respectively, while rest of the compounds displayed moderate to low antioxidant activity in comparison to the standard ascorbic acid. The compound 7c demonstrated moderate antioxidant activity with IC50 values of 24.02 ± 0.29. The 2- and 4-methoxy substitution on the 3-aryl ring displayed promising results, while rest of the compounds containing electronegative group substitutions (4-fluoro, 4-chloro, 4-bromo, and 2-chloro) at the 2 or 4 positions displayed moderate to low antioxidant activity (Figure 6). As demonstrated in earlier, adding an ethylene linkage between an amide and an aryl NH enhanced the antioxidant potential for the 4-methoxy substitution on the phenyl ring [71]. Additionally, studies are needed to ascertain whether the substitution of the 1,3-dioxoisoindole moiety influences the antioxidant activity. Other modifications, such as replacing NH with CH2 or C=O with C=S, may be feasible and will be the focus of future research to establish SAR.
Table 6.
The DPPH free radical scavenging activity of compounds 7a–f.
| S. No. | Compound | Free Radical Scavenging Activity IC50 (μM) |
|---|---|---|
| 1 | 7a | 36.18 ± 0.31 |
| 2 | 7b | 39.22 ± 0.12 |
| 3 | 7c | 24.02 ± 0.29 |
| 4 | 7d | 16.05 ± 0.15 |
| 5 | 7e | 54.27 ± 0.49 |
| 6 | 7f | 15.99 ± 0.10 |
| 7 | Ascorbic acid | 14.02 ± 0.15 |
Figure 6.
The structure–activity relationship for antioxidant activity of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs.
2.6. ADMET Prediction
ADMET predictions are indispensable in drug discovery programs to reduce the possibility of pharmacokinetics-related clinical failures [72]. Pharmaceutical companies suffer significant financial losses as a result of ADMET-related post-marketing failures of drug candidates [73]. ADMET properties are crucial variables that must be researched during clinical trials for drug development. Since evaluating every compound in in vivo ADMET studies is a laborious and time-consuming process, in silico studies could be of significant help in reducing such clinical failures. All the compounds were subjected to ADMET predictions using the software programs swissADME and ProTox II, which are available for free [74,75]. The compounds (7a–f) demonstrated that they adhered to Lipinski’s rule of five and had molecular weights between 299 and 360.16 (MW < 500), log p values between 1.74 and 2.05 (log P ≤ 5), and a range of three to four hydrogen bond acceptors (HBAs) (<HBA 10) and two hydrogen bond donors (HBD < 5), among other characteristics [76]. The percent absorption (% ABS), which was calculated as , was found to be 78.73 and 81.91 percent [77]. The in silico ADMET prediction results are summarized in Table 7. The compounds were also predicted to be toxicity-free, with the exception of hepatotoxicity (HEP). The toxicity prediction demonstrated LD50 values between 1000 and 5000 mg/Kg, putting the compounds in class IV (300 < LD50 ≤ 2000) and class V (2000 < LD50 ≤ 5000) toxicity classes [75]. The in silico toxicity prediction results are summarized in Table 8. Some of the compounds (7a–d) were reported to be non-toxic (cytotoxicity > 400 µg/mL) in a previous report [37].
Table 7.
In silico ADME prediction of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f).
| S. No. | Compound | % ABS | TPSA | HBA (<10) | HBD (<5) | NROTB | Log p (<5) | MW (500) | GI Absorption | Lipinski’s Violation (≤1) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7a | 81.91 | 78.51 | 4 | 2 | 4 | 1.74 | 299.26 | High | 0 |
| 2 | 7b | 81.51 | 78.51 | 3 | 2 | 4 | 1.92 | 315.71 | High | 0 |
| 3 | 7c | 81.51 | 78.51 | 3 | 2 | 4 | 2.03 | 360.16 | High | 0 |
| 4 | 7d | 78.73 | 87.74 | 4 | 2 | 5 | 1.93 | 311.29 | High | 0 |
| 5 | 7e | 81.51 | 78.51 | 3 | 2 | 4 | 1.94 | 315.71 | High | 0 |
| 6 | 7f | 78.73 | 87.74 | 4 | 2 | 5 | 2.05 | 311.29 | High | 0 |
% ABS = Percent absorption; TPSA = Total polar surface area; HBAs = Hydrogen bond acceptors; HBDs = Hydrogen bond donors; NROTB = Number of rotatable bonds; Log p = Lipophilicity; MW = Molecular weight; GI = Gastrointestinal.
Table 8.
In silico toxicity prediction of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f).
| S. No. | Compound | HEP | IMMUNO | MUTAGEN | CYTOTOX | LD50 (mg/Kg) | Toxicity Class |
|---|---|---|---|---|---|---|---|
| 1 | 7a | + | − | − | − | 1000 | IV |
| 2 | 7b | + | − | − | − | 5000 | V |
| 3 | 7c | + | − | − | − | 1000 | IV |
| 4 | 7d | + | − | − | − | 5000 | V |
| 5 | 7e | + | − | − | − | 1500 | IV |
| 6 | 7f | + | − | − | − | 5000 | V |
HEP = Hepatotoxicity; IMMUNO = Immunotoxicity; MUTAGEN = Mutagenicity; CYTOTOX = Cytotoxicity; + = active; − = inactive.
3. Discussion
Phthalimide analogs have been reported to be biologically active compounds, and a number of biological activities have been reported to date. Inspired by their biological activities we report herein the synthesis of as well as anticancer, antioxidant, and in silico studies on 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f). The synthesis of the compounds (7a–f) were accomplished in multiple steps as per the reported protocols [36,37,43]. Yogeeswari et al. synthesized 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs from phthalic anhydride and substituted phenyl semicarbazide in glacial acetic acid with very low yields ranging from 41 to 61 percent. Similarly, Verma et al. synthesized (1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs with low to good yields ranging from 35.9 to 86.2 percent [36,37]. Phenyl(substituted phenyl)carbamate (3a–f) was prepared from substituted aniline (1a–f) and phenyl chloroformate (2) in the first step, while aminoisoindoline-1,3-dione (6) was prepared in two steps starting from phthalic anhydride (4). In the final step, aminoisoindoline-1,3-dione (6) and phenyl(substituted phenyl)carbamate (3a–f) was stirred in DCM at room temperature for 6–8 h to obtain 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f); however, in a previous work, 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs were prepared from aminoisoindoline-1,3-dione and ethyl N-p-substituted phenyl carbamate but the yield was low, with some exceptions (a few compounds were obtained in good yields) [37]. Overall, the synthetic protocol reported herein was found to be efficient with good yields. Five compounds were assessed for their anticancer activity against 56 cancer cell lines in single dose assay at 10 µM. Among the series, compound 7c demonstrated a moderate but significant anticancer activity against EKVX, CAKI-1, UACC-62, MCF7, LOX IMVI, and ACHN with GP of 75.46, 78.52, 80.81, 83.48, 84.52, and 89.61, respectively. Compound 7c displayed better anticancer activity than imatinib against melanoma with a PGI of 5.03 percent. The EGFR is an important target for many anticancer drugs; hence, the compounds were subjected to docking studies against the EGFR. Efficient binding with two types of electrostatic interactions, H-bond and π–π stacking interactions, and docking scores ranging from −6.363 to −7.565 kcal/mol was observed in the docking studies. The ligand 7c displayed an H-bond interaction with Lys745 with a docking score of −7.558 kcal/mol. Free radical formation as a result of oxidative stress could be another reason to develop cancer; hence, the in vitro antioxidant potentials of the compounds 7a–f was also studied. The compounds displayed an antioxidant activity with IC50 values ranging between 15.99 ± 0.10 and 54.27 ± 0.49 µM. The compounds 7f and 7d demonstrated significant antioxidant activity with IC50 values of 15.99 ± 0.10 and 16.05 ± 0.15 µM, respectively, while rest of the compounds displayed moderate to low antioxidant activity in comparison to the standard ascorbic acid. The in silico ADMET prediction displayed the compounds’ adherence to Lipinski’s rule of five and they were predicted to be free from toxicities, except for hepatotoxicity (HEP). The toxicity prediction demonstrated LD50 values between 1000 and 5000 mg/Kg, putting the compounds either in class IV (300 < LD50 ≤ 2000) or class V (2000 < LD50 ≤ 5000) toxicity classes. Overall, in the present investigation, an efficient synthesis of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f) was accomplished adopting different synthetic routes, followed by studies on their biological activities and in silico ADMET predictions.
4. Materials and Methods
4.1. General Procedure for the Synthesis of Phenyl(substituted phenyl)carbamate Analogs (3a–f)
A solution of 2- and 4-substituted aniline (1a–f) (1 mmol) in triethylamine (1 mmol; ~2.8 mL) was added dropwise to a solution of phenyl chloroformate (1 mmol; 1.30 mL) in chloroform (5 mL), and reaction mixture was continuously stirred with magnetic stirrer at room temperature (rt) for 4–5 h. The reaction mixture was then concentrated under vacuum and cooled to room temperature. Petroleum ether (20 mL) was added to obtain precipitate, which was filtered, washed with water, and dried at room temperature to obtain phenyl (substituted phenyl)carbamate (3a–f).
4.2. Procedure for the Synthesis of Phthalimide (5)
Equimolar amounts of phthalic anhydride (25 mmol; 3.70 g) (4) and urea (25 mmol; 1.5 g) were fused at high temperature (140 °C) for 5 min, then the reaction mixture was cooled to obtain white precipitate. The solid was triturated with hydroalcoholic (H2O-EtOH, 2:1) solution to obtain phthalimide (5).
4.3. Procedure for the Synthesis of 2-Aminoisoindoline-1,3-dione (6)
Equimolar amounts of phthalimide (5) (20 mmol; 2.94 g) and hydrazine hydrate (20 mmol; 0.65 mL) in absolute ethanol (50 mL) were shaken at room temperature for 2 min followed by quickly heating them for 3 min in water bath. The reaction mixture was then poured into crushed ice to obtain a precipitate of 2-aminoisoindoline-1,3-dione (6).
4.4. General Procedure for the Synthesis of 1-(1,3-Dioxoisoindolin-2-yl)-3-aryl Urea Analogs (7a–f)
An equimolar mixture of phenyl(substituted phenyl)carbamate (3a–f) (1 mmol) and aminoisoindoline-1,3-dione (6) (1 mmol; 162 mg) in dichloromethane (DCM) (10 mL) was continuously stirred for 6–8 h at room temperature. The precipitate of the final 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f) was separated out, filtered, washed with DCM, and dried at room temperature, followed by recrystallization from ethanol.
1-(4-Fluorophenyl)-3-(1,3-dioxoisoindolin-2-yl)urea (7a): 1H NMR (300 MHz, DMSO-d6): δ ppm: 7.35–7.42 (m, 2H, ArH), 7.48–7.60 (m, 2H, ArH), 7.88–8.15 (m, 4H, ArH), 9.69 (s, 1H, ArNH), 10.65 (s, 1H, CONH); 13C NMR (75 MHz, DMSO-d6): δ ppm: 167.2, 163.1, 154.4, 135.1, 132.5, 132.1, 124.1, 119.9, 116.2; Anal. Calc. for C15H10FN3O3: C, 60.20; H, 3.37; N, 14.04 found: C, 60.01; H, 3.35; N, 14.05%. ESI-MS m/z = 300 (M + 1)+, 301 (M + 2)+.
1-(4-Chlorophenyl)-3-(1,3-dioxoisoindolin-2-yl)urea (7b): 1H NMR (300 MHz, DMSO-d6): δ ppm: 7.33 (d, 1H, J = 6.9 Hz, ArH), 7.49 (d, 1H, J = 9.0 Hz, ArH), 7.61 (d, 2H, J = 6.1 Hz, ArH), 7.88–8.15 (m, 4H, ArH), 9.69 (s, 1H, ArNH), 10.03 (s, 1H, CONH); 13C NMR (75 MHz, DMSO-d6): δ ppm: 166.2, 154.9, 137.5, 133.9, 132.6, 132.1, 129.3, 123.9, 120.9; Anal. Calc. for C15H10ClN3O3: C, 57.07; H, 3.19; N, 13.31 found: C, 57.00; H, 3.20; N, 13.35%. ESI-MS m/z = 316 (M + 1)+, 318 (M + 3)+.
1-(4-Bromophenyl)-3-(1,3-dioxoisoindolin-2-yl)urea (7c): 1H NMR (300 MHz, DMSO-d6): δ ppm: 7.43 (d, J = 8.7 Hz, 2H, ArH), 7.74 (d, J = 8.4 Hz, 2H, ArH), 7.89–8.14 (m, 4H, ArH), 9.62 (s, 1H, ArNH), 10.09 (s, 1H, CONH); 13C NMR (75 MHz, DMSO-d6): δ ppm: 164.9, 154.7, 139.1, 132.9, 132.1, 131.9, 123.9, 122.7, 121.9; Anal. Calc. for C15H10BrN3O3: C, 50.02; H, 2.80; N, 11.67 found: C, 49.98; H, 2.78; N, 11.70%. ESI-MS m/z = 360 (M + 1)+, 362 (M + 3)+. HPLC purity: 98.74%.
1-(1,3-Dioxoisoindolin-2-yl)-3-(4-methoxyphenyl)urea (7d): 1H NMR (300 MHz, DMSO-d6): δ ppm: 3.74 (s, 3H, OCH3), 6.95 (d, J = 6.9 Hz, 2H, ArH), 7.31 (d, J = 6.6 Hz, 2H, ArH), 7.88–8.02 (m, 4H, ArH); 9.59 (s, 1H, ArNH), 10.53 (s, 1H, CONH); 13C NMR (75 MHz, DMSO-d6): δ ppm: 164.9, 159.1, 154.5, 139.9, 132.1, 131.9, 124.1, 120.1, 114.9, 56.2; Anal. Calc. for C16H13N3O4: C, 61.73; H, 4.21; N, 13.50 found: C, 61.69; H, 4.19; N, 13.53%. ESI-MS m/z = 312 (M + 1)+.
1-(2-Chlorophenyl)-3-(1,3-dioxoisoindolin-2-yl)urea (7e): 1H NMR (300 MHz, DMSO-d6): δ ppm: 7.53–7.72 (m, 3H, ArH), 7.93–7.98 (m, 1H, ArH), 8.00–8.15 (m, 4H, ArH), 9.69 (s, 1H, ArNH), 10.03 (s, 1H, CONH); 13C NMR (75 MHz, DMSO-d6): δ ppm: 164.9, 154.3, 134.9, 132.7, 132.1, 131.4, 130.5, 129.3, 125.3, 124.6, 123.8; Anal. Calc. for C15H10ClN3O3: C, 57.07; H, 3.19; N, 13.31 found: C, 57.02; H, 3.21; N, 13.34%. ESI-MS m/z = 316 (M + 1)+, 318 (M + 3)+.
1-(1,3-Dioxoisoindolin-2-yl)-3-(2-methoxyphenyl)urea (7f): 1H NMR (300 MHz, DMSO-d6): δ ppm: 3.74 (s, 3H, OCH3), 7.05–7.52 (m, 4H, ArH), 7.82–8.07 (m, 4H, ArH); 9.63 (s, 1H, ArNH), 10.61 (s, 1H, CONH); 13C NMR (75 MHz, DMSO-d6): δ ppm: 164.9, 153.9, 149.7, 132.9, 132.1, 128.7, 124.8, 123.9, 121.6, 120.5, 112.9, 56.2; Anal. Calc. for C16H13N3O4: C, 61.73; H, 4.21; N, 13.50 found: C, 61.68; H, 4.19; N, 13.54%. ESI-MS m/z = 312 (M + 1)+.
4.5. Anticancer Activity
According to the reported protocol, 56 cancer cell lines were taken to test the compounds’ anticancer in one-dose assay at 10 µM [44,45,46,47]. The previous report contains a comprehensive procedure for anticancer screening [78].
4.6. In Vitro EGFR Kinase Inhibition Assay
In order to investigate the EGFR inhibition potential of most active anticancer compound (7c), EFGR kinase assay kit (BPS Bioscience, San Diego, CA, USA) was used as per the manufacturer’s instructions and the reported protocol [50]. To each well, the master mixture (25 µL) containing 5× Kinase Buffer 1 (6 µL), ATP (1 µL; 500 µM), PTK substrate Poly (Glu:Tyr 4:1) (1 μL; 10 mg/mL), and water (17 μL) was added. After pipetting master mixture, 5 µL solutions (in aqueous DMSO) of varying concentrations of 7c and erlotinib, were added to the test inhibitor (7c and Erlotinib) wells. A total of 5 μL of inhibitor’s buffer (containing no inhibitors) was added to positive control and blank wells. Finally, 20 μL EGFR (1 ng/μL) was added to the positive control and test inhibitor wells, while 20 μL of 1× kinase buffer was added to the blank wells. Each well then contained a total of 50 μL solution and was incubated at 30 °C for 40 min. A total of 50 μL of Kinase-Glo MAX detection reagent (Promega, Madison, WI, USA) was then added and the wells were further incubated at 30 °C for 15 min under light protection. Luminescence intensity was measured using ELISA reader at 450 nm. IC50 values for the tested compound 7c and the reference drug (erlotinib) were determined in µM using GraphPad prism 9 software at five different concentrations. The inhibition assays were performed in triplicate.
4.7. Molecular Docking Studies
From the protein data bank, the X-ray crystallographic structure with a resolution of 2.05 Å and R-value of 0.220 (observed) of the EGFR with PDB ID: 3w2r was retrieved for docking studies [63]. The ligands 7a–f saved as mol files were prepared for docking using Ligprep, and protein was minimized. Afterwards, the grid was prepared and ligand docking was carried out as per the docking protocol reported by Sogabe et al., 2013 [64]. From the protein data bank, the X-ray crystallographic structure with a resolution of 3.00 Å and R-value of 0.233 of DNA topoisomerase I with PDB ID: 1sc7 was retrieved for docking studies, and ligand docking was carried out as per the docking protocol in the reported methods of [27,60]. From the protein data bank, the X-ray crystallographic structure with a resolution of 3.58 Å and R-value of 0.233 (observed) of the tubulin with PDB ID: 1as0 was retrieved for docking studies. Ligand docking was carried out as per the docking protocol reported in the methods of [59,79].
4.8. Antioxidant Activity
The compounds (7a–f) were tested for their antioxidant potentials using the DPPH free radical scavenging activity as described by Koleva et al., 2002 [68]. The IC50 is the concentration of test compounds (in µM) that showed 50% scavenging activity of free radicals, and this value was used to calculate the antioxidant activity.
4.9. ADMET Prediction
All the compounds were subjected to ADMET prediction using the software programs swissADME and ProTox II, which are available for free [74,75].
5. Conclusions
The efficient multistep synthesis of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f) was reported herein. Their anticancer activity against some cancer cell lines was found to be significant. Against a few cancer cell lines, including EKVX, CAKI-1, UACC-62, MCF7, LOX IMVI, and ACHN, compound 7c showed notable anticancer activity. In order to assess the compounds’ affinity towards the active site of the EGFR for potential inhibition, molecular docking studies were also conducted against the putative target EGFR. As the formation of free radicals as a result of oxidative stress may be another factor in the transformation of normal cells to cancerous ones, the antioxidant potentials of compounds 7a–f were also investigated. Only the compounds 7f and 7d demonstrated notable antioxidant activity when compared to the antioxidant activity of ascorbic acid. Our in silico ADMET predictions showed the compounds’ adherence to the Lipinski’s rule of five and they were free from toxicities, except for hepatotoxicity. The toxicity prediction demonstrated LD50 values between 1000 and 5000 mg/Kg, putting the compounds either in class IV or class V toxicity classes. The present investigation may potentiate future drug discovery programs for anticancer drugs.
Acknowledgments
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (IF2/PSAU/2022/03/21467).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules29010067/s1, Figures S1–S5: anticancer data of compound 7a–c, 7e,f against 56 cancer cell lines at 10 µM; Figures S6–S16: characterization data of compounds 7a–f. Table S1: the molecular docking studies of 1-(1,3-dioxoisoindolin-2-yl)-3-aryl urea analogs (7a–f).
Author Contributions
Conceptualization, O.A. and M.J.A.; methodology, O.A. and M.J.A.; software, O.A. and M.J.A.; validation, M.J.A. and O.A.; formal analysis, O.A.; investigation, M.J.A. and O.A.; data curation, M.J.A. and O.A.; writing—original draft preparation, M.J.A. and O.A.; writing—review and editing, M.J.A.; visualization, M.J.A.; supervision, M.J.A.; project administration, O.A.; funding acquisition, O.A. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The Supplementary Material to this article contains the information that supports the findings of this study.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research work is funded by the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia through the project number (IF2/PSAU/2022/03/21467).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Sung H., Ferley J., Siegel R.L., Laversanne M., Soerjomartaram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Zhao J., Xu L., Sun J., Song M., Wang L., Yuan S., Zhu Y., Wan Z., Larsson S., Tsilidis K., et al. Global trends in incidence, death, burden and risk factors of early-onset cancer from 1990 to 2019. BMJ Oncol. 2023;2:e000049. doi: 10.1136/bmjonc-2023-000049. [DOI] [Google Scholar]
- 3.Ugai T., Sasamoto N., Lee H.-Y., Ando M., Song M., Tamimi R.M., Kawachi I., Campbell P.T., Giovannucci E.L., Weiderpass E., et al. Is early-onset cancer an emerging global epidemic? Current evidence and future implications. Nat. Rev. Clin. Oncol. 2022;19:656–673. doi: 10.1038/s41571-022-00672-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng R., Wanga S., Zhang S., Zeng H., Chen R., Sun K., Li L., Bray F., Wei W. Global, regional, and national lifetime probabilities of developing cancer in 2020. Sci. Bull. 2023;68:2620–2628. doi: 10.1016/j.scib.2023.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics, 2023. CA Cancer J. Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 6.Malvezzi M., Santucci C., Boffetta P., Collatuzzo G., Levi F., Vecchia C.L., Negri E. European cancer mortality predictions for the year 2023 with focus on lung cancer. Ann. Oncol. 2023;34:410–419. doi: 10.1016/j.annonc.2023.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Sathishkumar K., Chaturvedi M., Das P., Stephen S., Mathur P. Cancer incidence estimates for 2022 & projection for 2025: Result from National Cancer Registry Programme, India. Indian J. Med. Res. 2022;156:598–607. doi: 10.4103/ijmr.ijmr_1821_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Debela D.T., Muzazu S.G.Y., Heraro K.D., Ndalama M.T., Mesele B.W., Haile D.C., Kitui S.K., Manyazewal T. New approaches and procedures for cancer treatment: Current perspectives. SAGE Open Med. 2021;9:20503121211034366. doi: 10.1177/20503121211034366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.GlobalSurg Collaborative. National Institute for Health Research Global Health Research Unit on Global Surgery Global variation in postoperative mortality and complications after cancer surgery: A multicentre, prospective cohort study in 82 countries. Lancet. 2021;397:387–397. doi: 10.1016/S0140-6736(21)00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anand U., Dey A., Chandel A.K.S., Sanyal R., Mishra A., Pandey D.K., De Falco V., Upadhyay A., Kandimalla R., Chaudhary A., et al. Cancer chemotherapy and beyond: Current status, drug candidates, associated risks and progress in targeted therapeutics. Genes. Dis. 2023;10:1367–1401. doi: 10.1016/j.gendis.2022.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandes J.P.S. The Importance of Medicinal Chemistry Knowledge in the Clinical Pharmacist’s Education. Am. J. Pharm. Educ. 2018;82:6083. doi: 10.5688/ajpe6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed H.E.A., Abdel-Salam H.A., Shaker M.A. Synthesis, characterization, molecular modeling, and potential antimicrobial and anticancer activities of novel 2-aminoisoindoline-1,3-dione derivatives. Bioorg. Chem. 2016;66:1–11. doi: 10.1016/j.bioorg.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Al-Masoudi N.A., Abood E., Al-Maliki Z.T., Al-Masoudi W.A., Pannecouque C. Amino acid derivatives. Part 6. Synthesis, in vitro antiviral activity and molecular docking study of new N-α-amino acid derivatives conjugated spacer phthalimide backbone. Med. Chem. Res. 2016;25:2578–2588. doi: 10.1007/s00044-016-1693-9. [DOI] [Google Scholar]
- 14.Kok S.H., Gambari R., Chui C.H., Yuen M.C., Lin E., Wong R.S., Lau F.Y., Cheng G.Y., Lam W.S., Chan S.H., et al. Synthesis and anti-cancer activity of benzothiazole containing phthalimide on human carcinoma cell lines. Bioorg. Med. Chem. 2008;16:3626–3631. doi: 10.1016/j.bmc.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Ihmaid S.K., Alraqa S.Y., Aouad M.R., Aljuhani A., Elbadawy H.M., Salama S.A., Rezki N., Ahmed H.E.A. Design of molecular hybrids of phthalimide-triazole agents with potent selective MCF-7/HepG2 cytotoxicity: Synthesis, EGFR inhibitory effect, and metabolic stability. Bioorg. Med. Chem. 2021;111:104835. doi: 10.1016/j.bioorg.2021.104835. [DOI] [PubMed] [Google Scholar]
- 16.Gunkara O.T., Ocal N. Synthesis of New N-Phthalimide Substituted Tricyclic Imide Containing Isoxazoline and Bispiro Functional Group as Possible Anti-cancer Agents. J. Turk. Chem. Soc. Sec. A Chem. 2018;5:73–84. doi: 10.18596/jotcsa.338218. [DOI] [Google Scholar]
- 17.Davood A., Iman M., Pouriaiee H., Shafaroodi H., Akhbari S., Azimidoost L., Imani E., Rahmatpour S. Novel derivatives of phthalimide with potent anticonvulsant activity in PTZ and MES seizure models. Iran. J. Basic. Med. Sci. 2017;20:430–437. doi: 10.22038/IJBMS.2017.8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bach D.-H., Liu J.-Y., Kim W.K., Hong J.-Y., Park S.H., Kim D., Qin S.-N., Luu T.-T.-T., Park H.J., Xu Y.-N., et al. Synthesis and biological activity of new phthalimides as potential anti-inflammatory agents. Bioorg. Med. Chem. 2017;25:3396–3405. doi: 10.1016/j.bmc.2017.04.027. [DOI] [PubMed] [Google Scholar]
- 19.Batista C.R.A., Godin A.M., Melo I.S.F., Coura G.M.E., Matsui T.C., Dutra M.M.G.B., Brito A.M.S., Canhestro W.G., Alves R.J., Araujo D.P., et al. The phthalimide analogues N-3-hydroxypropylphthalimide and N-carboxymethyl-3-nitrophthalimide exhibit activity in experimental models of inflammatory and neuropathic pain. Pharmacol. Rep. 2019;71:1177–1183. doi: 10.1016/j.pharep.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Bansal M., Upadhyay C., Ponam, Kumar S., Rathi B. Phthalimide analogs for antimalarial drug discovery. RSC Med. Chem. 2021;12:1854–1867. doi: 10.1039/D1MD00244A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh S., El-Sakkary N., Skinner D.E., Sharma P.P., Ottilie S., Antonova-Koch Y., Kumar P., Winzeler E., Poonam, Caffrey C.R., et al. Synthesis and Bioactivity of Phthalimide Analogs as Potential Drugs to Treat Schistosomiasis, a Neglected Disease of Poverty. Pharmaceuticals. 2020;13:25. doi: 10.3390/ph13020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma U., Kumar P., Kumar N., Singh B. Recent Advances in the Chemistry of Phthalimide Analogues and their Therapeutic Potential. Mini-Rev. Med. Chem. 2010;10:678–704. doi: 10.2174/138955710791572442. [DOI] [PubMed] [Google Scholar]
- 23.Fernandes G.F.S., Lopes J.R., Santos J.L.D., Scarim C.B. Phthalimide as a versatile pharmacophore scaffold: Unlocking its diverse biological activities. Drug Dev. Res. 2023;87:1346–1375. doi: 10.1002/ddr.22094. [DOI] [PubMed] [Google Scholar]
- 24.Jacques V., Czarnik A.W., Judge T.M., der Ploeg L.H.T.V., DeWitt S.H. Differentiation of anti-inflammatory and antitumorigenic properties of stabilized enantiomers of thalidomide analogs. Proc. Natl. Acad. Sci. USA. 2015;112:E1471–E1479. doi: 10.1073/pnas.1417832112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotla V., Goel S., Nischal S., Heuck C., Vivek K., Das B., Verma A. Mechanism of action of lenalidomide in hematological malignancies. J. Hematol. Oncol. 2009;2:36. doi: 10.1186/1756-8722-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahsan M.J. 1,3,4-Oxadiazole containing compounds as therapeutic targets for cancer therapy. Mini-Rev. Med. Chem. 2022;22:144–197. doi: 10.2174/1389557521666210226145837. [DOI] [PubMed] [Google Scholar]
- 27.Staker B.L., Feese M.D., Cushman M., Pommier Y., Zembower D., Stewart L., Burgin A.B. Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J. Med. Chem. 2005;48:2336–2345. doi: 10.1021/jm049146p. [DOI] [PubMed] [Google Scholar]
- 28.AlNeyadi S.S., Amer N., Thomas T.G., Al Ajeil R., Breitener P., Munawar N. Synthesis, Characterization, and Antioxidant Activity of Some 2-Methoxyphenols derivatives. Heterocycl. Commun. 2020;26:112–122. doi: 10.1515/hc-2020-0112. [DOI] [Google Scholar]
- 29.Braughler J.M., Duncan L.A., Chase R.L. The involvement of iron in lipid peroxidation. Importance of ferric to ferrous ratios in initiation. J. Biol. Chem. 1986;261:10282–10289. doi: 10.1016/S0021-9258(18)67521-0. [DOI] [PubMed] [Google Scholar]
- 30.Bouyahya A., El Menyiy N., Oumeslakht L., El Allam A., Balahbib A., Rauf A., Muhammad N., Kuznetsova E., Derkho M., Thiruvengadam M., et al. Preclinical and Clinical Antioxidant Effects of Natural Compounds against Oxidative Stress-Induced Epigenetic Instability in Tumor Cells. Antioxidants. 2021;10:1553. doi: 10.3390/antiox10101553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mineo S., Takahashi N., Yamada-Hara M., Tsuzuno T., Aoki-Nonaka Y., Tabeta K. Rice bran-derived protein fractions enhance sulforaphane-induced anti-oxidative activity in gingival epithelial cells. Arch. Oral. Biol. 2021;129:105215. doi: 10.1016/j.archoralbio.2021.105215. [DOI] [PubMed] [Google Scholar]
- 32.Mo Y.-N., Cheng F., Yang Z., Shang X.-F., Liang J.-P., Shang R.-F., Hao B.-C., Wang X.-H., Zhang H.-J., Wali A., et al. Antioxidant Activity and the Potential Mechanism of the Fruit From Ailanthus altissima Swingle. Front. Vet. Sci. 2021;8:784898. doi: 10.3389/fvets.2021.784898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bădiceanu C.D., Mares C., Nuță D.C., Avram S., Drăghici C., Udrea A.-M., Zarafu I., Chiriță C., Hovaneț M.V., Limban C. N-Substituted (Hexahydro)-1H-isoindole-1,3(2H)-dione Derivatives: New Insights into Synthesis and Characterization. Processes. 2023;11:1616. doi: 10.3390/pr11061616. [DOI] [Google Scholar]
- 34.Perveen S., Orfali R. L-Proline-Catalyzed Synthesis of Phthalimide Derivatives and Evaluation of Their Antioxidant, Anti-Inflammatory, and Lipoxygenase Inhibition Activities. J. Chem. 2018;2018:5198325. doi: 10.1155/2018/5198325. [DOI] [Google Scholar]
- 35.Karthik C.S., Mallesha L., Mallu P. Investigation of antioxidant properties of phthalimide derivatives. Can. Chem. Trans. 2015;3:199–206. [Google Scholar]
- 36.Yogeeswari P., Sriram D., Saraswat V., Ragavendran J.V., Kumar M.M., Murugesan S., Thirumurugan R., Stables J.P. Synthesis and anticonvulsant and neurotoxicity evaluation of N4-phthalimido phenyl (thio) semicarbazides. Eur. J. Pharm. Sci. 2003;20:341–346. doi: 10.1016/j.ejps.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Verma M., Singh K.N., Clercq E.D. Synthesis of some heterocycle containing urea derivatives and their antiviral activity. Heterocycles. 2006;68:11–22. [Google Scholar]
- 38.Moradi M., Golmohammadi R., Najafi A., Moghaddam M.M., Fasihi-Ramandi M., Mirnejad R. A contemporary review on the important role of in silico approaches for managing different aspects of COVID-19 crisis. Inform. Med. Unlocked. 2022;28:100862. doi: 10.1016/j.imu.2022.100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puttaswamy H., Gowtham H.G., Ojha M.D., Yadav A., Choudhir G., Raguraman V., Kongkham B., Selvaraju K., Shareef S., Gehlot P., et al. In silico studies evidenced the role of structurally diverse plant secondary metabolites in reducing SARS-CoV-2 pathogenesis. Sci. Rep. 2020;10:20584. doi: 10.1038/s41598-020-77602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huanca P.I.J., Veras B.A.F., de Sousa Oliveira I., Ferreira S.B. In Silico Analysis Applied to the Study of Cytotoxicity in Natural Products. Chem. Proc. 2022;12:69. doi: 10.3390/ecsoc-26-13557. [DOI] [Google Scholar]
- 41.A Phase 1, Evaluate the Safety, Tolerability, and Pharmacokinetics of INS018_055 in Healthy Subjects. [(accessed on 2 December 2023)]; Available online: https://clinicaltrials.gov/study/NCT05154240.
- 42.First Drug Discovered and Designed with Generative AI Enters Phase II Trials, with First Patients Dosed. [(accessed on 2 December 2023)]. Available online: https://www.eurekalert.org/news-releases/993844.
- 43.Yogeeswari P., Sriram D., Thirumurugan R., Saxena A., Stables J., Vaigunda Raghuvendran J., Suddan K., Pavana R.K. Discovery of N-(2,6-Dimethyphenyl)-substituted semicarbazones as Anticonvulsants: Hybrid Pharmacophore-Based Design. J. Med. Chem. 2005;48:6202–6211. doi: 10.1021/jm050283b. [DOI] [PubMed] [Google Scholar]
- 44.DTP Developmental Therapeutic Programs. [(accessed on 10 October 2023)]; Available online: http://dtp.nci.nih.gov.
- 45.Monks A., Scudiero D., Skehan P., Shoemaker R., Paull K., Vistica D., Hose C., Langley J., Cronise P., Vaigro-Wolff A., et al. Feasibility of a highflux anticancer drug screening using a diverse panel of cultured human tumor cell lines. J. Nat. Cancer Inst. 1991;83:757–766. doi: 10.1093/jnci/83.11.757. [DOI] [PubMed] [Google Scholar]
- 46.Boyd M.R., Paull K.D. Some practical considerations and applications of the National Cancer Institute in vitro anticancer drug discovery screen. Drug Dev. Res. 1995;34:91–109. doi: 10.1002/ddr.430340203. [DOI] [Google Scholar]
- 47.Shoemaker R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 48.Aliabadi A., Mohammadi-Farani A., Seydi-Kangarshahi S., Ahmadi F. Discovery of 2-(1,3-dioxoisoindolin-2-yl)-n-phenylacetamide derivatives as probable 15-lipoxygenase-1 Inhibitors with potential anticancer effects. Farmacia. 2017;65:268–274. [Google Scholar]
- 49.Aliabadi A., Mohammadi-Farani A., Hosseinzadeh Z., Nadri H., Moradi A., Ahmadi F. Phthalimide analogs as probable 15-lipoxygenase-1 inhibitors: Synthesis, biological evaluation and docking studies. DARU J. Pharm. Sci. 2015;23:36. doi: 10.1186/s40199-015-0118-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mourad A.A., Farouk N.A., El-Sayed E.S.H., Mahdy A.R. EGFR/VEGFR-2 dual inhibitor and apoptotic inducer: Design, synthesis, anticancer activity and docking study of new 2-thioxoimidazolidin-4one derivatives. Life Sci. 2021;277:119531. doi: 10.1016/j.lfs.2021.119531. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi D.T., Gadelle D., Agama K., Kiselev E., Zhang H., Yab E., Petrella S., Forterre P., Pommier Y., Mayer C. Topoisomerase I (TOP1) dynamics: Conformational transition from open to closed states. Nat. Comm. 2022;13:59. doi: 10.1038/s41467-021-27686-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Capranico G., Marinello J., Chillemi G. Type I DNA Topoisomerases. J. Med. Chem. 2017;60:2169–2192. doi: 10.1021/acs.jmedchem.6b00966. [DOI] [PubMed] [Google Scholar]
- 53.Pommier Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 54.Saulnier M.G., Balasubramanian B.N., Long B.H., Frennesson D.B., Ruediger E., Zimmermann K., Eummer J.T., St. Laurent D.R., Stoffan K.M., Naidu B.N., et al. Discovery of a Fluoroindolo[2,3-a]carbazole Clinical Candidate with Broad Spectrum Antitumor Activity in Preclinical Tumor Models Superior to the Marketed Oncology Drug, CPT-11. J. Med. Chem. 2005;48:2258–2261. doi: 10.1021/jm049090z. [DOI] [PubMed] [Google Scholar]
- 55.Pommier Y. DNA Topoisomerase I Inhibitors: Chemistry, Biology, and Interfacial Inhibition. Chem. Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shuai W., Wang G., Zhang Y., Bu F., Zhang S., Miller D.D., Li W., Ouyang L., Wang Y. Recent Progress on Tubulin Inhibitors with Dual Targeting Capabilities for Cancer Therapy. J. Med. Chem. 2021;64:7963–7990. doi: 10.1021/acs.jmedchem.1c00100. [DOI] [PubMed] [Google Scholar]
- 57.Hawash M. Recent Advances of Tubulin Inhibitors Targeting the Colchicine Binding Site for Cancer Therapy. Biomolecules. 2022;12:1843. doi: 10.3390/biom12121843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng Z., Lu X., Feng B. A review of research progress of antitumor drugs based on tubulin targets. Transl. Cancer Res. 2020;9:4020–4027. doi: 10.21037/tcr-20-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yanagawa T., Noguchi T., Miyachi H., Kobayashi H., Hashimoto Y. Tubulin polymerization inhibitors with a fluorinated phthalimide skeleton derived from thalidomide. Bioorg. Med. Chem. Lett. 2006;16:4748–4751. doi: 10.1016/j.bmcl.2006.06.091. [DOI] [PubMed] [Google Scholar]
- 60.X-ray Crystal Structure of DNA Topoisomerase I. [(accessed on 10 October 2023)]. Available online: https://www.rcsb.org/structure/1sc7.
- 61.X-ray Crystal Structure of Tubulin. [(accessed on 10 October 2023)]. Available online: https://www.rcsb.org/structure/1sa0.
- 62.Zubair T., Bandyopadhyay D. Small Molecule EGFR Inhibitors as Anti-Cancer Agents: Discovery, Mechanisms of Action, and Opportunities. Int. J. Mol. Sci. 2023;24:2651. doi: 10.3390/ijms24032651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.X-ray Crystal Structure of EGFR. [(accessed on 10 October 2023)]. Available online: https://www.rcsb.org/structure/3W2R.
- 64.Sogabe S., Kawakita Y., Igaki S., Iwata H., Miki H., Cary D.R., Takagi T., Takagi S., Ohta Y., Ishikawa T. Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors. ACS Med. Chem. Lett. 2013;4:201–205. doi: 10.1021/ml300327z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Birben E., Sahiner U.M., Sackesen C., Erzurum S., Kalayci O. Oxidative stress and antioxidant defence. World Allergy Organ J. 2012;5:9–19. doi: 10.1097/WOX.0b013e3182439613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reuter S., Gupta S.C., Chaturvedi M.M., Aggarwal B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010;49:1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pham-Huy L.A., He H., Pham-Huy C. Free radicals, antioxidants in disease and health. Int. J. Biomed. Sci. 2008;4:89–96. doi: 10.59566/IJBS.2008.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zehiroglu C., Ozturk Sarikaya S.B. The importance of antioxidants and place in today’s scientific and technological studies. J. Food Sci. Technol. 2019;56:4757–4774. doi: 10.1007/s13197-019-03952-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vaibhav D.A., Arunkumar W., Abhijit M.P., Arvind S. Antioxidants as an immunomodulator. Int. J. Curr. Pharm. Res. 2011;1:8–10. [Google Scholar]
- 70.Koleva I.I., Van-Beek T.A., Linssen J.P., de-Groot A., Evstatieva L.N. Screening of plant extracts for antioxidant activity: A comparative study on three testing methods. Phytochem. Anal. 2002;13:8–17. doi: 10.1002/pca.611. [DOI] [PubMed] [Google Scholar]
- 71.Tumosienė I., Kantminienė K., Klevinskas A., Petrikaitė V., Jonuškienė I., Mickevičius V. Antioxidant and Anticancer Activity of Novel Derivatives of 3-[(4-Methoxyphenyl)amino]propanehydrazide. Molecules. 2020;25:2980. doi: 10.3390/molecules25132980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Daina A., Michielin O., Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, druglikeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daina A., Zoete V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem. 2016;11:1117–1121. doi: 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.ADME Prediction. [(accessed on 12 October 2023)]. Available online: http://www.swissadme.ch/
- 75.Toxicity Prediction. [(accessed on 12 October 2023)]. Available online: https://tox-new.charite.de/protox_II/
- 76.Lipinski C.A., Lombardo L., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 2001;46:3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 77.Ertl P., Rohde B., Selzer P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 2000;43:3714–3717. doi: 10.1021/jm000942e. [DOI] [PubMed] [Google Scholar]
- 78.Ali A., Ali A., Tahir A., Bakht M.A., Salahuddin, Ahsan M.J. Molecular Engineering of Curcumin, an Active Constituent of Curcuma longa L. (Turmeric) of the Family Zingiberaceae with Improved Antiproliferative Activity. Plants. 2021;10:1559. doi: 10.3390/plants10081559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ravelli R.B., Gigant B., Curmi P.A., Jourdain I., Lachkar S., Sobel A., Knossow M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Supplementary Material to this article contains the information that supports the findings of this study.