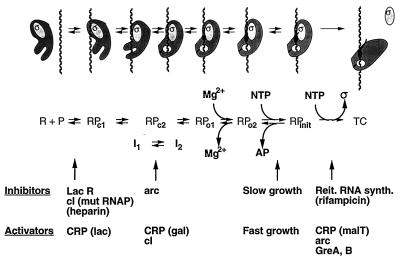
Initiation of transcription is a complicated process involving several different phases: promoter location by RNA polymerase, formation of a competent initiation complex, synthesis of the initial phosphodiester bonds, and movement of RNA polymerase from the promoter as it enters the elongation phase; as shown below, further subdivisions may be warranted. The large number of different steps has afforded a multitude of focal points at which control of the process could be exerted. Many of these have been exploited by the bacterial cell, as exemplified by the rich variety of regulatory mechanisms that have been uncovered (Fig. 1). To arrive at a better understanding of both the sequence of events in the initiation of RNA synthesis and the control of the process, it is important to identify the intermediate species involved. As detailed in a recent review (68), a major challenge is not only to correlate kinetically observed intermediates with those that can be trapped and characterized but also to put all intermediates in the context of structural information as it becomes available.
FIG. 1.
Summary of the intermediates in the process of initiation of RNA synthesis for which structural and/or kinetic evidence has been obtained; see the text for details. The complexes are shown in cartoon form at the top with a descriptive notation below. In the designation of structurally characterized complexes, R stands for RNA polymerase, P stands for promoter DNA, and c and o indicate closed and open complexes, respectively (where strand separation has not or has occurred, respectively). I1 and I2 are the kinetically significant intermediates, shown by DNA footprinting to be RPc2-like complexes. AP stands for abortive RNA product, and TC stands for the transcribing complex in elongation mode. Also indicated are examples of agents or conditions that can affect (inhibit or activate) the interconversion of intermediates; some of these (CRP, cI, and arc) have different effects, depending on the promoter or RNA polymerase. Some are E. coli proteins: the lac repressor (LacR [78]), CRP (which activates different processes, depending on the promoter [18, 34, 52, 55, 62]), and elongation factors GreA and GreB (which can affect promoter escape in vitro [36]). Phage-encoded proteins cI (lambda) and arc (P22) display polymerase- or promoter-dependent effects: cI inhibits the binding of a mutant RNA polymerase (mut RNAP) (49) but affects I1-I2 isomerization with wild-type RNA polymerase (31); arc represses wild-type promoters by slowing I1-I2 isomerization but activates a consensus promoter mutant by accelerating clearance (85). Rifampin is an antibiotic which targets the β subunit of RNA polymerase; heparin binds and inactivates free RNA polymerase. The latter two agents have been useful tools in the study of RNA polymerase-promoter interactions. Growth rate control (20) and reiterative (Reit.) RNA synthesis (synth.) (50) are responsive to NTP levels (see text).
Here we will emphasize mechanistic aspects of the interaction of RNA polymerase containing the initiation factor ς70 (which is responsible for the vast majority of initiation events in Escherichia coli) with promoters to form initiation-competent complexes and on the dissolution of these complexes when the RNA polymerase leaves the promoter in the course of chain initiation. Possible intermediates for open-complex formation at various promoters have been identified as well (68; cf. Fig. 1). Here we focus on the PR promoter of bacteriophage λ. It is likely that open-complex formation at other promoters involves a similar order of events, so that the principles which are emerging for PR should be generally valid. Following a brief overview of RNA polymerase and its promoters, we review the following sequential events in the initiation of transcription: promoter location, initial reversible binding of RNA polymerase, conformational changes in RNA polymerase, conformational changes in DNA, binding of nucleoside triphosphate (NTP) to the functional RNA polymerase-promoter complex, and nonproductive and productive initiation of RNA synthesis.
RNA POLYMERASE AND PROMOTERS
The initiating RNA polymerase holoenzyme (Eς70) has a molecular weight of about 4.5 × 105 and contains five subunits with the stoichiometry α2ββ′ς (see Fig. 2). The ς subunit is bound relatively weakly to the rest of the enzyme (the core polymerase, E); ς is responsible for specific promoter recognition by RNA polymerase but is released during the initiation process. The core polymerase then continues to catalyze phosphodiester bond formation between the growing RNA chain and the next NTP, whose identity is specified by the sequence of the template strand. After chain termination and dissociation of the ternary complex of the core RNA polymerase, RNA, and DNA, the released core can again bind a ς subunit and start a new cycle of RNA synthesis (8, 93). The Escherichia coli genome encodes multiple ς factors, of which ς70 (named for its molecular weight of 70,000; it is also referred to as ςD) is by far the most abundant. The ς subunit is an important determinant of the sequence-specific recognition of promoter DNA. Through the use of different ς factors, RNA polymerase can be targeted to promoters with different sequences (15, 33, 47, 51, 68, 87, 97, 101).
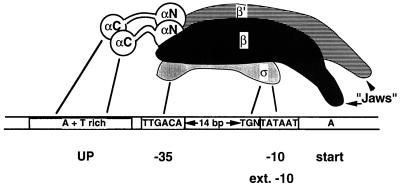
FIG. 2.
Summary of RNA polymerase-promoter interactions. A promoter with consensus sequences for the −10 and −35 regions (boxed) is shown; the sequences of actual promoters deviate from those shown here. In general, the more closely a promoter resembles the one shown, the stronger (more active) the promoter will be. Also shown is the TGN sequence just upstream of the −10 region; together, they comprise an “extended” −10 region which has promoter activity even in the absence of a −35 sequence. The model of RNA polymerase shown is based on recently available evidence (27, 60, 98); however, it is intended to show contacts between the subunits and promoter DNA and not necessarily to reflect the precise orientation of the subunits. The “jaws” of RNA polymerase are shown on the right of the molecule. This region of the RNA polymerase would grasp the DNA downstream of the catalytic site. Contacts between RNA polymerase and promoter DNA are shown by the solid lines. Not all contacts occur in every RNA polymerase-promoter interaction, but in all known cases (including promoters activated by regulator proteins), at a minimum, some contacts between ς and the −10 region appear to be required.
We will consider almost exclusively those promoters recognized by the Eς70 holoenzyme and will therefore use the term RNA polymerase for this particular form. Four important promoter elements can be distinguished: two hexamers centered at or near positions −10 and −35 upstream from the transcription start site (designated by their locations as the −10 and −35 regions), the spacer DNA separating them, and a region between −40 and −60 (the UP element) (see Fig. 2). Various compilations of promoter sequences recognized by Eς70 have established the importance of the −35 and −10 regions (29, 32). The consensus sequences of these two regions as read on the nontemplate strand are, respectively, TTGACA and TATAAT (Fig. 2). With a few exceptions (19, 42), the general rule holds that the greater the similarity of the −10 and −35 regions to their consensus sequence, the better the promoter functions in vitro, as well as in vivo (19, 29, 32, 42, 68); in this regard, well-conserved base pairs may be of greater importance than the less-conserved ones (57). The ability of RNA polymerase to discriminate between promoters with different sequences at the −10 and −35 regions has allowed the unregulated (or basal) rate of transcription of various genes to be set at different levels. A consensus length of 17 bp has been established for the spacer between the −10 and −35 regions. Promoters with such a spacer length have been found to be more active in vitro, as well as in vivo, than those with shorter or longer spacers (2, 58, 86). Quite recently, a very A+T-rich region between positions −40 and −60 (the UP element) was recognized in some promoters as an additional important determinant of promoter activity. This region is contacted by the α subunit (5, 22, 48, 67, 74). In an initiation-competent complex between RNA polymerase and promoter DNA, the two strands of a 12- to 15-bp region from about the middle of the −10 element to just past the start site (+2 or +3) become more reactive to footprinting agents (43, 68, 76, 82, 90, 100); therefore, such a complex is referred to as the “open” promoter complex.
A subclass of E. coli promoters function quite well without a recognizable −35 region or the involvement of any activating proteins. Such promoters have been found to have an “extended −10 region” with the sequence TGNTATAAT (15, 56, 75, 96) (Fig. 2). Open-complex formation at these promoters proceeds at rates comparable to those for promoters with consensus −10 and −35 regions (14a) and appears to be less temperature dependent than open-complex formation at promoters with −35 regions (56). It was recently shown that a region of ς70 not previously identified as conserved is implicated in recognition of the upstream TG sequence (4).
PROMOTER LOCATION
There has been much speculation concerning the kinetic pathway along which RNA polymerase locates promoter sequences embedded in nonspecific DNA. The RNA polymerase holoenzyme has a measurable affinity for nonspecific sites (14), but whether this mode of binding plays a role in facilitating promoter location (41, 83, 84) is still uncertain. For several other proteins, including the lac repressor and the EcoRI restriction enzyme (3, 92, 95), considerable evidence has been obtained for acceleration of target site location through facilitated diffusion mediated by nonspecific binding. Possible mechanisms by which this is accomplished include one-dimensional diffusion along the DNA, “hopping” (successive association and dissociation within the domain of the DNA molecule), and intersegment transfer between regions of DNA that are close in space but separated by long DNA sequence stretches (reviewed in reference 94). Direct evidence for translocation of RNA polymerase along DNA fragments has been obtained (41), but it is unclear whether this was by one-dimensional diffusion or hopping. In other reports, linear diffusion of RNA polymerase along DNA was inferred, but it was not clearly established that the diffusion played a role in the acceleration of promoter location of the enzyme (70, 83, 84). Intersegment transfer would require the availability of two separate DNA binding sites which transiently are simultaneously occupied during the transfer process. The existence of two such sites has not been demonstrated for the RNA polymerase protomer.
At the strongest known promoters (those for the synthesis of rRNA), in vivo chain initiation occurs at the rate of about 1/s (68), which is thus the lower limit for the rate of binding of RNA polymerase to the promoter. As the free RNA polymerase concentration in the cell has been estimated to be about 30 nM (54), a crude estimate of the association rate constant is the reciprocal of this, or about 3 × 107 M−1 s−1. Rates of up to 3 × 108 M−1 s−1 have been observed in vitro (6, 67) for promoters contained on restriction fragments (typically, 200 to 1,000 bp, long enough to benefit from facilitated diffusion). These rates do not exceed reasonable estimates for the rate of a reaction limited by three-dimensional diffusion, i.e., direct interaction of the reactants without nonspecific binding: 108 to 109 M−1 s−1 (68). In addition, comparable or faster rates have been observed by using fragments of synthetic DNA too short (90 bp) for facilitated diffusion to significantly accelerate the target search (12a). Finally, accelerated diffusion is not expected to lead to a rate enhancement for the majority of promoters, where the initial (closed) promoter complex is in rapid equilibrium with free promoter and RNA polymerase prior to proceeding down the pathway shown in Fig. 1. Therefore, McClure’s 1985 conclusion (54) for RNA polymerase is still valid today: “… the experiments that show the enzyme can slide do not establish that it must do so as a rate enhancement mechanism.”
INITIAL REVERSIBLE BINDING OF RNA POLYMERASE TO PROMOTER SITES
Some time ago, it was proposed (24) that sequences in the −35 region would affect the initial binding of RNA polymerase to the promoter and that those in the −10 region would affect the isomerization to the open complex. Thus, mutations in the two promoter elements would affect different steps (30, 80, 81). The model fell out of favor when kinetic studies failed to find consistent differences in the way base changes in either region affected the kinetics of open-complex formation (e.g., see references 54 and 91). Current experimental evidence indicates that the initial contacts between RNA polymerase and the promoter to form the first double-stranded, or closed, intermediate (RPc1; Fig. 1) probably do involve the −35 region, but the role of the −10 region remains unclear. The −35 region remains double stranded throughout the process of open-complex formation. It has been demonstrated that polypeptides of ς70 can recognize both −35 and −10 sequences as double-helical DNA, although only the upstream half of the −10 region appears to be important in this respect (15–17). Finally, the RNA polymerase holoenzyme has been observed to bind single-stranded oligodeoxynucleotides bearing the −10 sequence of the nontemplate strand (37, 53, 71, and see below) with greater affinity than double-stranded DNA spanning the same region (37). Perhaps initial recognition of −10 sequences involves the upstream half of the region, approximately coinciding with the part of the region remaining double helical in an open complex.
From studies of several promoters at low temperatures (10°C and below), the equilibrium constant (K1) for the initial binding of RNA polymerase to form a closed complex has been estimated to be about 107 to 108 M−1 in buffer containing 0.1 M monovalent cation and 0.01 M MgCl2. DNase I footprinting experiments with this low-temperature complex (RPc1) at some promoters indicate that RNA polymerase contacts the DNA from −55 to −5. At somewhat higher temperatures, a second closed RNA polymerase-promoter complex is experimentally detectable by virtue of a downstream extension in the footprint; this intermediate has been designated RPc2 (reviewed in reference 68). These and other complexes which have been structurally characterized by probing with DNase I and KMnO4 are indicated in the first line below the cartoons in Fig. 1.
REVERSIBLE CONFORMATIONAL CHANGES IN RNA POLYMERASE
Two intermediates (designated I1 and I2 in Fig. 1) are kinetically significant; their interconversion is the rate-limiting step in open-complex formation at all temperatures, in both the association and dissociation directions (78a). A key question in recent years has involved the correlation of the kinetic and structural intermediates. To address this question, I1 and I2 were trapped by using thermodynamic conditions under which each was predicted to accumulate. It was found that both intermediates are closed complexes with extended footprints; in this regard, they are both RPc2-like complexes (11a).
Analysis of the thermodynamics of the conversion of I1 to I2 indicates that a very large amount of the nonpolar surface is buried, leading to the proposal that this transition involves a major conformational change in RNA polymerase (72, 73, 78a). Although the nature of this conformational change has yet to be elucidated, one possibility is that the conversion of I1 to I2 is the step at which a jawlike structure in RNA polymerase closes around the DNA (Fig. 1). This structure is believed to contain the active site of the polymerase and has been shown to be open in the holoenzyme (and by inference in the early steps of promoter recognition) (12) but closed in the core polymerase (and, by inference, at the end of the long chain of events that results in initiation of RNA synthesis) (66). A similar hypothesis has been advanced by Heumann’s group (77), although the case has also been made for jaw closing at a later stage, in response to RNA synthesis (64).
CONFORMATIONAL CHANGES IN THE DNA
In the process of open-complex formation, conformational changes also take place in promoter DNA; of these, the most salient is the strand separation extending from the −10 region to past the start site (7, 10, 11, 43, 76, 82, 90). The process takes place under conditions in which the double-stranded form of DNA is resistant to denaturation by about 1 kcal/mol bp, yet no external source of energy is required to drive the strand separation process. Open complexes generally are very stable, and their formation is quite fast (see above). Thus, it seems plausible that RNA polymerase would not only stabilize the open complex once it was formed but also lower the activation energy for the strand separation process. It is envisaged that this is accomplished by RNA polymerase-induced DNA distortions that destabilize the double-helical form. Such distortions might include torqueing of the DNA to introduce an unwinding twist and DNA bending across the promoter DNA. The most compelling evidence for the former (recently reviewed [13]) is derived from studies of the effects of DNA supercoiling on promoter utilization. The results can be interpreted to indicate that RNA polymerase unwinds promoter DNA by as much as half a turn early in the process of open-complex formation (1, 88). It seems likely that the untwisting torque would be applied directly across the region of strand separation. While this possibility has previously been explicitly suggested (88), to the best of our knowledge, there is no direct experimental data to support it; some studies suggest a rotational distortion across the spacer DNA separating the −10 and −35 regions (13). Strong evidence in support of promoter DNA bending by RNA polymerase has been obtained from the visualization of the complexes by atomic force microscopy (69), as well as from gel shift experiments (35). The results of footprinting and chemical probing experiments suggest that the DNA is wrapped around RNA polymerase over a region of 70 to 80 bp (11, 46, 61); reviewed in references 23 and 68). Whether the wrapping or bending is required to lower the stability of the helix in the region where strand opening occurs has not been established experimentally.
Once formed, the open complex is presumably stabilized by interactions between single-stranded DNA and the RNA polymerase. Indeed, Roberts and coworkers (53, 71; see also reference 37) have recently described the sequence-specific recognition by the holoenzyme of nontemplate strand −10 sequences contained on small single-stranded oligodeoxynucleotides. The rate of binding of such molecules is an order of magnitude slower than that for open-complex formation at a promoter with a similar −10 region and a consensus −35 (12a). This observation is consistent with the notion that for the single-stranded DNA to interact with the RNA polymerase, the latter has to undergo a change in conformation, which would be facilitated by the binding of double-helical promoter DNA but not of a short, single-strand oligomer.
Completion of the process of RNA polymerase-induced strand opening is dependent on Mg2+ (89, 90, 100). The complex formed in the absence of Mg2+ has been designated RPo1 (Fig. 1); it shows base pair opening extending from the middle of the −10 region to bp −1, just upstream of the start site (90). It is thought to be an intermediate in the formation of the functional open complex RPo2 (Fig. 1), formed in the presence of Mg2+, in which opening extends from −12 to +2. An interesting question involves the order in which DNA bases become unpaired to form RPo2 (10, 13): is the process cooperative, or can partially melted intermediates be identified from which the progression of strand opening can be deduced? Current experimental evidence points to initiation of strand opening in the −10 region and then propagation downstream toward the start site. This conclusion is based primarily on chemical probing experiments carried out on complexes detected at different temperatures (10), but recent fast-kinetic studies also are consistent with such a model (12a).
NTP BINDING TO DRIVE FORMATION OF TERNARY COMPLEXES
After strand opening, the template strand becomes accessible to the NTPs so that it can (by Watson-Crick base pairing) specify the sequence of the RNA to be synthesized. The kinetic stability of the open complex can be enhanced (i.e., the rate of dissociation is diminished) by the binding of the initiating NTP without requiring its hydrolysis or formation of a phosphodiester bond (20). The rrnB P1 promoter for the synthesis of rRNA forms an unstable open complex even at 37°C (25, 65); most likely, this is due to both the nonconsensus spacer length between the −10 and −35 regions (16 instead of 17 bp) and the presence of an abundance of G+C base pairs in the melted region between −10 and +1. Kinetic stabilization of the open complex by a high concentration (in the millimolar range) of ATP (the initiating NTP) at this promoter is observed in vitro (20). In agreement with this result, it was found that the activity of rrnB P1 in vivo is controlled by the intracellular concentration of ATP, the level of which was shown to vary with the growth rate of the cell. High levels of ATP (as found in fast-growing cells) result in greatly enhanced synthesis of rRNA, presumably by kinetically stabilizing the open complexes at the promoter and allowing enough time for formation of the first phosphodiester bond to occur (20). These observations go a long way toward solving the long-standing problem of growth rate control of rRNA synthesis (21).
NONPRODUCTIVE AND PRODUCTIVE INITIATION OF RNA SYNTHESIS
Ternary complexes of promoter, RNA polymerase, and NTP(s) are poised to initiate productive RNA synthesis, but after formation of the first phosphodiester bond, several hurdles still need to be overcome before the complex is committed to productive RNA synthesis. It has been known since the early 1980s that a significant fraction of the complexes engages in abortive RNA synthesis in which short (<10-nt) RNA transcripts are made and released in a repetitive manner (9, 26, 59). As a result of these studies, abortive initiation has been regarded as a phase in the process of productive chain initiation. After each catalytic step, a stochastic event would determine whether the nascent RNA was released or further elongated, until a critical length (about 10 nt) was reached, whereupon the complex became committed to productive RNA synthesis. Based on recent results obtained with the PR promoter, an alternative possibility has been suggested. From a homogeneous population of RNA polymerases, each molecule has a certain probability of becoming irreversibly trapped in carrying out abortive synthesis or of escaping this fate and entering the elongation mode (45). It is unclear how the two different models can be reconciled.
Another nonproductive mode of initiation involves a process termed “reiterative RNA synthesis,” which was found to play a role in the UTP concentration-dependent regulation at the pyrBI operon (40). Here, the first six nucleotide residues of the nascent RNA are AAUUUG. At high UTP concentrations, RNA polymerase reiteratively transcribes bases 3 to 5 in the template strand to synthesize RNA products containing 30 or more U residues. While engaged in this process, productive initiation at the promoter is blocked and expression of the operon is inhibited. At low UTP concentrations, extension by G successfully competes with the reiterative addition of U and this mode is bypassed. This is an additional example of the regulation of productive initiation of RNA synthesis by levels of NTP. Two important differences from the rrnB P1 promoter are that the regulation is by a noninitiating NTP and that at high levels it promotes a decrease in initiation of transcription. Several other cases of reiterative RNA synthesis have been described (39, 50), including some at promoters bearing mutations which introduce a string of A residues in the template strand near the position of initiation of RNA synthesis (38, 99).
Once an RNA chain of eight or nine nucleotide residues has been synthesized, ς factor is released (28, 44); at a similar length of the nascent RNA chain (10 nucleotides [44]), RNA polymerase leaves the promoter and becomes committed to productive chain elongation. The physical movement of the RNA polymerase away from the promoter (as assayed, for instance, by the reclosure of the region of strand separation between −10 and +2) is referred to as “promoter clearance.” The rates of clearance at various promoters vary significantly (19, 79, 85). It is thought that there is a direct relationship between the strengths of the RNA polymerase-promoter contacts and the rate with which promoter clearance is achieved (19, 42). The fact that ς factor is responsible for many of the RNA polymerase-promoter contacts may then explain the coincidence of ς factor release and the commitment to elongation, both occurring when the length of the nascent RNA is about 10 nucleotides.
Clearly, the RNA polymerase-promoter complex is held together by contacts in addition to those involving ς factor, as the complex does not fall apart upon ς factor release. Promoter clearance is dependent upon the sequence of the region downstream of the start site (19, 26, 42). It is unknown whether these differences in sequence affect the downstream contacts between the RNA polymerase and DNA or whether they are exerted at the level of RNA synthesis or structure. Based on available evidence, either of these possibilities could be envisaged. Contacts between the β′ subunit and the downstream DNA have been found to stabilize the elongating complex (63). However, whether similar contacts may be formed during initiation and whether they would be sequence dependent has not been established.
CONCLUDING REMARKS
Substantial progress has been made in understanding the interaction of RNA polymerase with promoters in prokaryotes. The various intermediates which have been identified kinetically or structurally, as well as some agents or conditions which can affect their interconversion, are shown in Fig. 1 (details concerning the effectors can be found in the legend). Several aspects of the scheme in Fig. 1 remain unclear, however, and require further experimentation for their elucidation. The nature of the conformational change that occurs during the transition from I1 to I2 is a subject of great importance. The hypothesis that this step involves the closing of the jaw on RNA polymerase needs to be tested. How RNA polymerase weakens the base pairing interactions of DNA in order to be able to orchestrate the separation of the strands also remains to be established, as does the precise site of initiation of strand separation and the involvement of specific amino acid residues of RNA polymerase in this process. It is necessary to pinpoint the precise amino acid residue-nucleotide interactions which occur in the various intermediate initiation complexes to develop a clearer picture of the early steps of transcription. Also, it needs to be determined what triggers the release of ς factor from the initiating RNA polymerase. Finally, the role of NTPs in open-complex stabilization, start site selection, and the transition from initiation to elongation needs to be further explored.
ACKNOWLEDGMENTS
We thank P. de Boer, P. Rather, and D. Setzer, as well as members of the deHaseth and Record groups, for their comments and suggestions.
Our research is supported by N.I.H. grants GM 31808 (P.L.H.) and GM 23467 (M.T.R.).
REFERENCES
- 1.Amouyal M, Buc H. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J Mol Biol. 1987;195:795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
- 2.Ayers D G, Auble D T, deHaseth P L. Promoter recognition by Escherichia coli RNA polymerase. Role of the spacer DNA in functional complex formation. J Mol Biol. 1989;207:749–756. doi: 10.1016/0022-2836(89)90241-6. [DOI] [PubMed] [Google Scholar]
- 3.Barkley M D. Salt dependence of the kinetics of the lac repressor-operator interaction: role of nonoperator deoxyribonucleic acid in the association reaction. Biochemistry. 1981;20:3833–3842. doi: 10.1021/bi00516a026. [DOI] [PubMed] [Google Scholar]
- 4.Barne K A, Bown J A, Busby S J W, Minchin S D. Region 2.5 of the Escherichia coli RNA polymerase ς70 subunit is responsible for the recognition of the “extended −10” motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blatter E E, Ross W, Tang H, Gourse R L, Ebright R H. Domain organization of RNA polymerase α subunit: C-terminal 85 amino acids constitute an independently folded domain capable of dimerization and DNA binding. Cell. 1994;78:889–896. doi: 10.1016/s0092-8674(94)90682-3. [DOI] [PubMed] [Google Scholar]
- 6.Brunner M, Bujard H. Promoter recognition and promoter strength in the Escherichia coli system. EMBO J. 1987;6:3139–3144. doi: 10.1002/j.1460-2075.1987.tb02624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckle M, Buc H. Fine mapping of DNA single stranded regions using base-specific chemical probes: study of an open complex formed between RNA polymerase and the lac UV5 promoter. Biochemistry. 1989;28:4388–4396. doi: 10.1021/bi00436a040. [DOI] [PubMed] [Google Scholar]
- 8.Burgess R, Travers A, Dunn J J, Bautz E K F. Factor stimulating transcription by RNA polymerase. Nature. 1969;221:43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- 9.Carpousis A J, Gralla J D. Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry. 1980;19:3245–3253. doi: 10.1021/bi00555a023. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y-F, Helmann J D. DNA melting at the Bacillus subtilis flagellin promoter nucleates near −10 and expands unidirectionally. J Mol Biol. 1997;267:47–59. doi: 10.1006/jmbi.1996.0853. [DOI] [PubMed] [Google Scholar]
- 11.Craig M L, Suh W-C, Record M T., Jr HO· and DNase I probing of Eς70 RNA polymerase-λPR promoter open complexes: Mg2+ binding and its structural consequences at the transcription start site. Biochemistry. 1995;34:15624–15632. doi: 10.1021/bi00048a004. [DOI] [PubMed] [Google Scholar]
- 11a.Craig, M. L., •. •. Tsodikov, •. •. Saecker, and M. T. Record, Jr. Unpublished data.
- 12.Darst S A, Kubalek E W, Kornberg R D. Three-dimensional structure of Escherichia coli RNA polymerase holoenzyme determined by electron crystallography. Nature. 1989;340:730–732. doi: 10.1038/340730a0. [DOI] [PubMed] [Google Scholar]
- 12a.deHaseth, P. L., et al. Unpublished data.
- 13.deHaseth P L, Helmann J D. Open complex formation by Escherichia coli RNA polymerase: the mechanism of polymerase-induced strand separation of double helical DNA. Mol Microbiol. 1995;16:817–824. doi: 10.1111/j.1365-2958.1995.tb02309.x. [DOI] [PubMed] [Google Scholar]
- 14.deHaseth P L, Lohman T M, Burgess R R, Record M T., Jr Nonspecific interactions of Escherichia coli RNA polymerase with native and denatured DNA: differences in the binding behavior of core and holoenzyme. Biochemistry. 1978;17:1612–1622. doi: 10.1021/bi00602a006. [DOI] [PubMed] [Google Scholar]
- 15.Dombroski A J. Recognition of the −10 promoter sequence by a partial polypeptide of ς70in vitro. J Biol Chem. 1997;272:3487–3494. [PubMed] [Google Scholar]
- 16.Dombroski A J, Johnson B D, Lonetto M, Gross C A. The sigma subunit of Escherichia coli RNA polymerase senses promoter spacing. Proc Natl Acad Sci USA. 1996;93:8858–8862. doi: 10.1073/pnas.93.17.8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dombroski A J, Walter W A, Record M T, Jr, Siegele D A, Gross C A. Polypeptides containing highly conserved regions of transcription factor ς70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 18.Eichenberger P, Dethiolliaz S, Buc H, Geiselmann J. Structural kinetics of transcription activation at the malT promoter of Escherichia coli by UV laser footprinting. Proc Natl Acad Sci USA. 1997;94:9022–9027. doi: 10.1073/pnas.94.17.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellinger T, Behnke D, Bujard H, Gralla J D. Stalling of Escherichia coli RNA polymerase in the +6 to +12 region in vivo is associated with tight binding to consensus promoter elements. J Mol Biol. 1994;239:455–465. doi: 10.1006/jmbi.1994.1388. [DOI] [PubMed] [Google Scholar]
- 20.Gaal T, Bartlett M S, Ross W, Turnbough C L, Jr, Gourse R L. NTP concentration as a regulator of transcription initiation: control of rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 21.Gaal T, Gourse R L. Guanosine 3′-diphosphate 5′-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:5533–5537. doi: 10.1073/pnas.87.14.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaal T, Ross W, Blatter E E, Tang H, Jia X, Krishnan V V, Assa-Munt N, Ebright R H, Gourse R L. DNA-binding determinants of the α subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev. 1996;10:16–26. doi: 10.1101/gad.10.1.16. [DOI] [PubMed] [Google Scholar]
- 23.Geiselmann J. The role of DNA conformation in transcriptional initiation and activation in Escherichia coli. Biol Chem. 1997;378:599–607. [PubMed] [Google Scholar]
- 24.Gilbert W. Starting and stopping sequences for the RNA polymerase. In: Losick R, Chamberlin M, editors. RNA polymerase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1976. pp. 193–205. [Google Scholar]
- 25.Gourse R L. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 1988;16:9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gralla J D, Carpousis A J, Stefano J E. Productive and abortive initiation of transcription in vitro at the lac UV5 promoter. Biochemistry. 1980;19:5864–5869. doi: 10.1021/bi00566a031. [DOI] [PubMed] [Google Scholar]
- 27.Greiner D P, Hughes K A, Gunasekera A H, Meares C F. Binding of the ς70 protein to the core subunits of Escherichia coli RNA polymerase, studied by iron-EDTA protein footprinting. Proc Natl Acad Sci USA. 1996;93:71–75. doi: 10.1073/pnas.93.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen U M, McClure W R. Role of the sigma subunit of Escherichia coli RNA polymerase in initiation. II. Release of sigma from ternary complexes. J Biol Chem. 1980;255:9564–9570. [PubMed] [Google Scholar]
- 29.Harley C B, Reynolds R P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hawley D, McClure W R. In vitro comparison of initiation properties of bacteriophage lambda wild-type PR and x3 mutant promoters. Proc Natl Acad Sci USA. 1980;77:6381–6385. doi: 10.1073/pnas.77.11.6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawley D, McClure W R. Mechanism of activation of transcription initiation from the lambda PRM promoter. J Mol Biol. 1982;157:493–525. doi: 10.1016/0022-2836(82)90473-9. [DOI] [PubMed] [Google Scholar]
- 32.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helmann J D. Bacterial sigma factors. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 1–17. [Google Scholar]
- 34.Herbert M, Kolb A, Buc H. Overlapping promoters and their control in Escherichia coli: the gal case. Proc Natl Acad Sci USA. 1986;83:2807–2811. doi: 10.1073/pnas.83.9.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heumann H, Ricchetti M, Werel W. DNA-dependent RNA polymerase of Escherichia coli induces bending or an increased flexibility of DNA by specific complex formation. EMBO J. 1988;7:4379–4381. doi: 10.1002/j.1460-2075.1988.tb03336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu L M, Vo N M, Chamberlin M C. Escherichia coli transcript cleavage factors GreA and GreB stimulate promoter escape and gene expression in vivo and in vitro. Proc Natl Acad Sci USA. 1995;92:11588–11592. doi: 10.1073/pnas.92.25.11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang X, Lopez de Saro F J, Helmann J D. Sigma factor mutations affecting the sequence-selective interaction of RNA polymerase with −10 region single-stranded DNA. Nucleic Acids Res. 1997;25:2603–2609. doi: 10.1093/nar/25.13.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacques J P, Susskind M M. Pseudo-templated transcription by Escherichia coli RNA polymerase at a mutant promoter. Genes Dev. 1990;4:1801–1810. doi: 10.1101/gad.4.10.1801. [DOI] [PubMed] [Google Scholar]
- 39.Jin D J. A mutant RNA polymerase reveals a kinetic mechanism for the switch between nonproductive stuttering synthesis and productive initiation during promoter clearance. J Biol Chem. 1996;271:11659–11667. [PubMed] [Google Scholar]
- 40.Jin D J, Turnbough C L. An Escherichia coli RNA polymerase defective in transcription due to its overproduction of abortive initiation products. J Mol Biol. 1994;236:72–80. doi: 10.1006/jmbi.1994.1119. [DOI] [PubMed] [Google Scholar]
- 41.Kabata H, Kurosawa O, Arai I, Washizu M, Margarson S A, Glass R E, Shimamoto N. Visualization of single molecules of RNA polymerase sliding along DNA. Science. 1993;262:1561–1563. doi: 10.1126/science.8248804. [DOI] [PubMed] [Google Scholar]
- 42.Kammerer W, Deuschle U, Gentz R, Bujard H. Functional dissection of Escherichia coli promoters: information in the transcribed region is involved in the late steps of the overall process. EMBO J. 1986;5:2995–3000. doi: 10.1002/j.1460-2075.1986.tb04597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirkegaard K, Buc H, Spassky A, Wang J. Mapping of single-stranded regions in duplex DNA at the sequence level: Single-strand-specific cytosine methylation in RNA polymerase-promoter complexes. Proc Natl Acad Sci USA. 1983;80:2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krummel B, Chamberlin M J. RNA chain initiation by Escherichia coli RNA polymerase. Structural transitions of the enzyme in early ternary complexes. Biochemistry. 1989;28:7829–7842. doi: 10.1021/bi00445a045. [DOI] [PubMed] [Google Scholar]
- 45.Kubori T, Shimamoto N. A branched pathway in the early stage of transcription by Escherichia coli RNA polymerase. J Mol Biol. 1996;256:449–457. doi: 10.1006/jmbi.1996.0100. [DOI] [PubMed] [Google Scholar]
- 46.Kuhnke G, Fritz H-J, Ehring R. Unusual properties of promoter-up mutations in the Escherichia coli galactose operon and evidence suggesting RNA polymerase-induced DNA bending. EMBO J. 1987;6:507–513. doi: 10.1002/j.1460-2075.1987.tb04782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar A, Malloch R A, Fujita N, Smillie D A, Ishihama A, Hayward R S. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J Mol Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 48.Landini P, Volkert M. RNA polymerase α subunit binding site in positively controlled promoters: a new model for RNA polymerase-promoter interaction and transcriptional activation in the Escherichia coli ada and aidB genes. EMBO J. 1995;14:4329–4335. doi: 10.1002/j.1460-2075.1995.tb00107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li M, McClure W R, Susskind M M. Changing the mechanism of transcriptional activation by phage λ repressor. Proc Natl Acad Sci USA. 1997;94:3691–3696. doi: 10.1073/pnas.94.8.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J, Turnbough C L., Jr Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J Bacteriol. 1994;176:2938–2945. doi: 10.1128/jb.176.10.2938-2945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lonetto M, Gribskov M, Gross C A. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malan P, McClure W R. Dual promoter control of the Escherichia coli lactose operon. Cell. 1984;39:173–180. doi: 10.1016/0092-8674(84)90203-4. [DOI] [PubMed] [Google Scholar]
- 53.Marr M T, Roberts J W. Promoter recognition as measured by binding of RNA polymerase to nontemplate strand oligonucleotides. Science. 1997;276:1258–1260. doi: 10.1126/science.276.5316.1258. [DOI] [PubMed] [Google Scholar]
- 54.McClure W R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 55.Menendez M, Kolb A, Bus H. A new target for CRP action at the MalT promoter. EMBO J. 1987;6:4227–4234. doi: 10.1002/j.1460-2075.1987.tb02771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Minchin S, Busby S. Location of close contacts between Escherichia coli RNA polymerase and guanine residues at promoters either with or without consensus −35 region sequences. Biochem J. 1993;289:771–775. doi: 10.1042/bj2890771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moyle H, Waldburger C, Susskind M M. Hierarchies of base pair preferences in the P22 ant promoter. J Bacteriol. 1991;173:1944–1950. doi: 10.1128/jb.173.6.1944-1950.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mulligan M E, Brosius J, McClure W R. Characterization of the effect of spacer length on the activity of Escherichia coli RNA polymerase at the TAC promoter. J Biol Chem. 1985;260:3529–3538. [PubMed] [Google Scholar]
- 59.Munson L M, Reznikoff W S. Abortive initiation and long ribonucleic acid synthesis. Biochemistry. 1981;20:2081–2085. doi: 10.1021/bi00511a003. [DOI] [PubMed] [Google Scholar]
- 60.Murakami K, Kimura M, Owens J T, Meares C F, Ishihama A. The two α subunits of Escherichia coli RNA polymerase are asymmetrically arranged and contact different halves of the DNA upstream element. Proc Natl Acad Sci USA. 1997;94:1709–1714. doi: 10.1073/pnas.94.5.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nickerson C A, Achberger E C. Role of curved DNA in binding of Escherichia coli RNA polymerase to promoters. J Bacteriol. 1995;177:5756–5761. doi: 10.1128/jb.177.20.5756-5761.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niu W, Kim Y, Tau G, Heyduk T, Ebright R. Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Transcription processivity: protein-DNA interactions holding together the elongation complex. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 64.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 65.Ohlsen K L, Gralla J D. DNA melting within stable closed complexes at the Escherichia coli rrnB P1 promoter. J Biol Chem. 1992;267:19813–19818. [PubMed] [Google Scholar]
- 66.Polyakov A, Severinova E, Darst S A. Three-dimensional structure of E. coli RNA polymerase: promoter binding and elongation conformations of the enzyme. Cell. 1995;83:365–373. doi: 10.1016/0092-8674(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 67.Rao L, Ross W, Appleman J A, Gaal T, Leirmo S, Schlax P J, Record M T, Jr, Gourse R L. Factor independent activation of rrnB P1. An “extended” promoter with an upstream element that dramatically increases promoter strength. J Mol Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 68.Record M T, Jr, Reznikoff W S, Craig M L, McQuade K L, Schlaz P J. Escherichia coli RNA polymerase (Eς70), promoters, and the kinetics of the steps of transcription initiation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K R, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 792–820. [Google Scholar]
- 69.Rees W A, Keller W R, Vesenka J P, Yang G, Bustamante C. Evidence for DNA bending in transcription complexes imaged by scanning force microscopy. Science. 1993;260:1646–1649. doi: 10.1126/science.8503010. [DOI] [PubMed] [Google Scholar]
- 70.Ricchetti M, Netzger W, Heumann H. One-dimensional diffusion of Escherichia coli DNA-dependent RNA polymerase: a mechanism to facilitate promoter location. Proc Natl Acad Sci USA. 1988;85:4610–4614. doi: 10.1073/pnas.85.13.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roberts C W, Roberts J W. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 72.Roe J H, Burgess R R, Record M T. Kinetics and mechanism of the interaction of E. coli RNA polymerase with the λPR promoter. J Mol Biol. 1984;176:495–521. doi: 10.1016/0022-2836(84)90174-8. [DOI] [PubMed] [Google Scholar]
- 73.Roe J H, Burgess R R, Record M T. Temperature dependence of the rate constants of the Escherichia coli RNA polymerase lambda PR promoter interaction. Assignment of the kinetic steps corresponding to protein conformational change and DNA opening. J Mol Biol. 1985;184:441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- 74.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 75.Sabelnikov A G, Greenberg B, Lacks S A. An extended −10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J Mol Biol. 1995;250:144–155. doi: 10.1006/jmbi.1995.0366. [DOI] [PubMed] [Google Scholar]
- 76.Sasse-Dwight S, Gralla J D. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989;264:8074–8081. [PubMed] [Google Scholar]
- 77.Schickor P, Metzger W, Werel W, Lederer H, Heumann H. Topography of intermediates in transcription initiation of E. coli. EMBO J. 1990;9:2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schlax P J, Capp M W, Record M T., Jr Inhibition of transcription initiation by lac repressor. J Mol Biol. 1995;245:331–350. doi: 10.1006/jmbi.1994.0028. [DOI] [PubMed] [Google Scholar]
- 78a.Schlax, P. E., Jr., K. L. McQuade, O. V. Tsodikov, M. L. Craig, and M. T. Record, Jr. Unpublished data.
- 79.Schmitt B, Reiss C. Kinetic study in vitro of Escherichia coli promoter closure during transcription initiation. Biochem J. 1995;306:123–128. doi: 10.1042/bj3060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shih M-C, Gussin G N. Differential effects of mutations on discrete steps in transcription initiation at the λ PR E promoter. Cell. 1983;34:941–949. doi: 10.1016/0092-8674(83)90551-2. [DOI] [PubMed] [Google Scholar]
- 81.Shih M-C, Gussin G N. Mutations affecting two different steps in transcription initiation at the phage λ PRM promoter. Proc Natl Acad Sci USA. 1983;80:496–500. doi: 10.1073/pnas.80.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Siebenlist U, Simpson R B, Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980;20:269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- 83.Singer P, Wu C-W. Promoter search by Escherichia coli RNA polymerase on a circular DNA template. J Biol Chem. 1987;262:14178–14189. [PubMed] [Google Scholar]
- 84.Singer P T, Wu C-W. Kinetics of promoter search by Escherichia coli RNA polymerase. Effects of monovalent and divalent cations and temperature. J Biol Chem. 1988;263:4208–4214. [PubMed] [Google Scholar]
- 85.Smith T L, Sauer R T. Dual regulation of open-complex formation and promoter clearance by Arc explains a novel repressor to activator switch. Proc Natl Acad Sci USA. 1996;93:8868–8872. doi: 10.1073/pnas.93.17.8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stefano J E, Gralla J D. Spacer mutations in the lacPs promoter. Proc Natl Acad Sci USA. 1982;79:1069–1072. doi: 10.1073/pnas.79.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Strickland M, Thompson N, Burgess R. Structure and function of the sigma 70 subunit of Escherichia coli RNA polymerase. Monoclonal antibodies: localization of epitopes by peptide mapping and effects on transcription. Biochemistry. 1988;27:5755–5765. doi: 10.1021/bi00415a054. [DOI] [PubMed] [Google Scholar]
- 88.Su T T, McClure W R. Selective binding of Escherichia coli RNA polymerase to topoisomers of minicircles carrying the TAC16 and TAC17 promoters. J Biol Chem. 1994;269:13511–13521. [PubMed] [Google Scholar]
- 89.Suh W-C, Leirmo S, Record M T J. Roles of Mg2+ in the mechanism of formation and dissociation of open complexes between Escherichia coli RNA polymerase and the lambda PR promoter: kinetic evidence for a second open complex requiring Mg2+ Biochemistry. 1992;31:7815–7825. doi: 10.1021/bi00149a011. [DOI] [PubMed] [Google Scholar]
- 90.Suh W-C, Ross W, Record M T J. Two open complexes and a requirement for Mg2+ to open the λPR transcription start site. Science. 1993;259:358–361. doi: 10.1126/science.8420002. [DOI] [PubMed] [Google Scholar]
- 91.Szoke P A, Allen T A, deHaseth P L. Promoter recognition by Escherichia coli RNA polymerase. Effects of base substitution in the −10 and −35 regions. Biochemistry. 1987;26:6188–6194. doi: 10.1021/bi00393a035. [DOI] [PubMed] [Google Scholar]
- 92.Terry B J, Jack W E, Modrich P. Facilitated diffusion during catalysis by Eco RI endonuclease. Nonspecific interactions in Eco RI catalysis. J Biol Chem. 1985;260:13130–13137. [PubMed] [Google Scholar]
- 93.Travers A, Burgess R R. Cyclic re-use of the RNA polymerase sigma factor. Nature. 1969;222:537–540. doi: 10.1038/222537a0. [DOI] [PubMed] [Google Scholar]
- 94.von Hippel P H, Bear D G, Morgan W D, McSwiggen J A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]
- 95.von Hippel P H, Berg O G. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 96.Voskuil M I, Voepel K, Chambliss G H. The −16 region, a vital sequence for the utilization of a promoter in B. subtilis and Escherichia coli. Mol Microbiol. 1995;17:271–279. doi: 10.1111/j.1365-2958.1995.mmi_17020271.x. [DOI] [PubMed] [Google Scholar]
- 97.Waldburger C T G R W, Susskind M M. Changes in conserved region 2 of Escherichia coli sigma 70 affecting promoter recognition. J Mol Biol. 1990;215:1–10. doi: 10.1016/s0022-2836(05)80345-6. [DOI] [PubMed] [Google Scholar]
- 98.Wang Y, Severinov K, Loizos N, Fenyo D, Heyduk E, Heyduk T, Chait B T, Darst S A. Determinants for Escherichia coli RNA polymerase assembly within the β subunit. J Mol Biol. 1997;270:648–662. doi: 10.1006/jmbi.1997.1139. [DOI] [PubMed] [Google Scholar]
- 99.Xiong X F, Reznikoff W S. Transcriptional slippage during the transcription initiation process at a mutant lac promoter in vivo. J Mol Biol. 1993;231:569–580. doi: 10.1006/jmbi.1993.1310. [DOI] [PubMed] [Google Scholar]
- 100.Zaychikov E, Denissova L, Meier T, Gotte M, Heumann H. Influence of Mg2+ and temperature on formation of the transcription bubble. J Biol Chem. 1997;272:2259–2267. doi: 10.1074/jbc.272.4.2259. [DOI] [PubMed] [Google Scholar]
- 101.Zuber P, Healy J, Carter III H L, Cutting S, Moran C P, Losick R. Mutation changing the specificity of an RNA polymerase sigma factor. J Mol Biol. 1989;206:605–614. doi: 10.1016/0022-2836(89)90569-x. [DOI] [PubMed] [Google Scholar]