Abstract
Aspergillus fumigatus, an important opportunistic pathogen which commonly affects neutropenic patients, produces conidia with a bluish-green color. We identified a gene, alb1, which is required for conidial pigmentation. The alb1 gene encodes a putative polyketide synthase, and disruption of alb1 resulted in an albino conidial phenotype. Expression of alb1 is developmentally regulated, and the 7-kb transcript is detected only during the conidiation stage. The alb1 mutation was found to block 1,3,6,8-tetrahydroxynaphthalene production, indicating that alb1 is involved in dihydroxynaphthalene-melanin biosynthesis. Scanning electron microscopy studies showed that the alb1 disruptant exhibited a smooth conidial surface, whereas complementation of the alb1 deletion restored the echinulate wild-type surface. Disruption of alb1 resulted in a significant increase in C3 binding on conidial surfaces, and the conidia of the alb1 disruptant were ingested by human neutrophils at a higher rate than were those of the wild type. The alb1-complemented strain producing bluish-green conidia exhibited inefficient C3 binding and neutrophil-mediated phagocytosis quantitatively similar to those of the wild type. Importantly, the alb1 disruptant had a statistically significant loss of virulence compared to the wild-type and alb1-complemented strains in a murine model. These results suggest that disruption of alb1 causes pleiotropic effects on conidial morphology and fungal virulence.
Aspergillus fumigatus, a ubiquitous fungus, causes allergy, noninvasive colonization, or life-threatening invasive pulmonary aspergillosis (25). Recently, the incidence of invasive pulmonary aspergillosis caused by A. fumigatus has significantly increased secondary to increased use of immunosuppressive therapy and prolongation of life for neutropenic patients (7, 8, 13, 14). In nature, A. fumigatus survives as a saprophyte and propagates by highly dispersible conidia. The ability to adapt to broad ranges of environmental conditions may account for its worldwide distribution. Conidia, which are the first fungal structures encountered by the human host, are small (2 to 3.5 μm in diameter), extremely hydrophobic, and bluish green in color (32, 41). The inhaled conidia can cause invasive aspergillosis, a life-threatening disease, if the immune system is impaired, especially in patients with prolonged neutropenia (25, 56).
To establish an infection in the human host, inhaled conidia must protect themselves from the host defense system. At the initial stage of the A. fumigatus-host interaction, conidial surface components may play an important role in evasion of the host immune system. One of the visible conidial surface components is the bluish-green pigment. Fungal pigments have been shown to contribute to the survival and longevity of fungal propagules in the environment (61). Recently, we showed that complement component C3 binds more efficiently to conidia of a reddish-pink conidial color mutant of A. fumigatus than to bluish-green wild-type conidia (54). Analysis of the mutation led to the isolation of the alb1 gene, encoding a homolog of scytalone dehydratase, which is involved in the dihydroxynaphthalene (DHN)-melanin pathway. DHN-melanin has been shown to be a pathogenic factor in plant pathogenic fungi such as Magnaporthe grisea and Colletotrichum lagenarium (21, 23, 28, 37). Furthermore, melanin has been shown to be important for virulence in human pathogenic fungi, including Cryptococcus neoformans and Wangiella dermatitidis (10, 26, 27, 42, 55, 61).
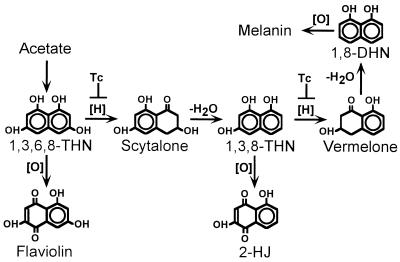
Most studies of pigment biosynthesis in fungi have focused on the brown and black fungi, since these fungi destroy many important agricultural crops (1). In the well-studied DHN-melanin pathway in the brown and black fungi, polyketides are synthesized by polyketide synthases. Using acetate as a precursor, 1,3,6,8-tetrahydroxynaphthalene (1,3,6,8-THN) is synthesized and subsequently reduced to form scytalone (Fig. 1) (5, 61). Scytalone is then dehydrated to produce 1,3,8-trihydroxynaphthalene (1,3,8-THN), which is converted to 1,8-dihydroxynaphthalene (1,8-DHN) after additional reduction and dehydration steps. Finally, 1,8-DHN is polymerized to form DHN-melanin. Both reduction steps are sensitive to tricyclazole, a fungicide which specifically inhibits THN reductases involved in the DHN-melanin pathway. In contrast to pigment biosynthesis in brown and black fungi, little is known about pigment biosynthesis in green and bluish-green fungi. In Aspergillus nidulans, a green-spored species, two conidial pigmentation genes, wA and yA, have been isolated (2, 29, 30, 34). The wA gene encodes a putative polyketide synthase, and yA encodes a laccase. Disruption of wA and yA individually resulted in white and yellow conidial-color mutants, respectively. Interestingly, although tricyclazole does not inhibit the conidial pigment biosynthesis of A. nidulans, it does inhibit conidial pigmentation in A. fumigatus (53, 54, 61, 62). That finding suggests that unlike A. nidulans, A. fumigatus uses a DHN-melanin pathway for conidial pigmentation.
FIG. 1.
Biosynthetic pathway of DHN-melanin in brown and black fungi. [H], a reduction step; [O], an oxidation step; −H2O, a dehydration step; Tc, a reduction step which can be inhibited by the fungicide tricyclazole.
During analysis of the alb1 gene, we found that alb1 is linked to a gene that likely encodes a polyketide synthase. The molecular genetics of polyketide biosynthesis has been most thoroughly analyzed in bacteria (15, 16). These studies showed that polyketide synthases can be divided into two types (18, 33). Type I enzymes are large, multifunctional polypeptides, while type II enzymes consist of several single-function polypeptides associated in a complex. To date, all the fungal polyketide synthases that have been characterized are type I enzymes (65) and are involved in the synthesis of pigments and/or mycotoxins.
Here, we report an additional dimension of fungal polyketide synthase which includes modulation of conidial morphology and virulence in A. fumigatus.
MATERIALS AND METHODS
Strains and media.
A. fumigatus B-5233 is a clinical isolate that produces conidia with a bluish-green color. B-5233/RGD12-8 is an alb1 disruptant which produces white conidia. RGD12-8/PKS33-3, obtained by transforming B-5233/RGD12-8 with pPKS33, produces bluish-green conidia. Aspergillus minimal medium contains 1% glucose, 10 mM NaNO3, and trace elements (20). Malt extract medium contains 2% glucose, 2% malt extract, and 0.1% peptone. Cultures were grown at 37°C. Asparagine-sucrose agar (ASA) medium is identical to a previously described alkaline medium (TM medium [64]). This medium was required for the growth of A. fumigatus under slightly alkaline conditions, near pH 7.5, to optimize flaviolin and 2-hydroxyjuglone (2-HJ) production. Scytalone and tricyclazole (Eli Lilly Research Laboratories, Greenfield, Ind.) were dissolved in absolute ethanol (EtOH). ASA medium was modified to contain 1% EtOH without tricyclazole or scytalone, with 30 μg of tricyclazole per ml, with 1 mM scytalone, or with a combination of 30 μg of tricyclazole per ml and 1 mM scytalone. Controls with 1% EtOH alone or no EtOH were compared to exclude the possibility that 1% EtOH affects morphology or conidial pigment production.
Preparation and analysis of nucleic acids.
Isolation of total DNA and RNA from Aspergillus cultures was performed as previously described (52). Cultures were harvested at 0 h (hyphal stage) and 14 h (conidiophores and conidial chains were observed) after induction of conidiation for RNA preparation (36). DNA sequencing was done with a Sequenase version 2.0 kit (U.S. Biochemical, Cleveland, Ohio) and an ABI automatic DNA sequencing system (Perkin-Elmer, Foster City, Calif.). A Geneclean II kit (Bio101, Vista, Calif.) was used to purify recovered DNA fragments. DNA cloning and Southern blot analyses were performed according to standard protocols (43). A Hybond-N nylon membrane (Amersham, Arlington Heights, Ill.) was used for blot analysis. DNA probes were labeled with [α-32P]dCTP (Amersham) by using a Prime It kit (Stratagene, La Jolla, Calif.).
Partial-length cDNA clones were obtained by screening a cDNA library of A. fumigatus B-5233, using the 5-kb AvrII DNA fragment as a probe (54). To confirm the presence of two introns at the 5′-end region of alb1, double-stranded cDNA was obtained by reverse transcription (RT)-PCR (Life Technologies, Gaithersburg, Md.), using poly(A)+ RNA of strain B-5233 harvested 14 h after induction of conidiation as the template and DW10 (5′-ACTCGGTGACTTTGTCCC-3′) and DW16N (5′-TTGCAGGCGAAGAACCAT-3′) as primers. The amplified double-stranded cDNA was cloned into pCR2.1TOPO (Invitrogen, Carlsbad, Calif.) and sequenced with primers DW16N and DW34 (5′-ATTGACTATCCACCTCGG-3′).
Plasmids.
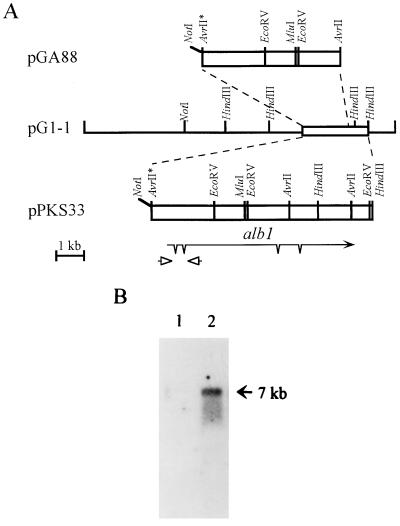
Cosmid pG1-1 (Fig. 2A) was rescued from a restored conidial-color mutant, RP3/G1-1 (54), and contained a 44-kb insert. Plasmid pGA88 was derived from pG1-1 and carried a 5-kb AvrII DNA fragment containing a truncated alb1 gene in pCosHX (obtained from J. Hamer, Purdue University, West Lafayette, Ind.). Plasmid pPKS33 carried a 8.2-kb AvrII-HindIII fragment which encompassed the alb1 gene on the pBC-phleo vector (44). To create an alb1 gene disruption construct, pRGD12, a 6.2-kb AvrII-HindIII alb1 DNA fragment was cloned into SpeI- and HindIII-digested pBC KS+ vector (Stratagene); subsequently, a 1.5-kb MluI-AvrII fragment was replaced with a 2.8-kb hph cassette from pAN7-1 (39) (see Fig. 4A).
FIG. 2.
Structure and expression of alb1. (A) Restriction enzyme map and structure of alb1. The asterisk indicates an AvrII site destroyed during cloning. The alb1 transcript is represented by an arrow, and introns are marked with spikes. Arrowheads represent the two oligonucleotides used to amplify cDNA in the RT-PCR. (B) Northern analysis of alb1 expression at different developmental stages. Twelve micrograms of total RNA from strain B-5233 harvested at 0 h (lane 1) or 14 h (lane 2) after induction of conidiation was fractionated on a 1% formaldehyde–agarose gel. The 5-kb AvrII DNA fragment was used as a probe. The size of the hybridizing signal is indicated by an arrow.
FIG. 4.
Disruption of alb1 in strain B-5233. (A) Diagram of gene replacement via double-crossover recombination. White boxes represent A. fumigatus DNA; black boxes represent hph genes. The alb1 region deleted during construction of pRGD12 is shown as a hatched box. Arrows aligned with boxes represent transcripts, with spikes indicating introns. Asterisks indicate the destroyer restriction enzyme sites. (B) Southern blot analysis. Total DNA from strain B-5233/RGD12-8 (lane 1) or strain B-5233 (lane 2) was digested with AvrII and fractionated on a 0.8% agarose gel. The membrane was hybridized with the 1.5-kb MluI-AvrII fragment probe (hatched box, panel A) (I) or pRGD12 (II). The sizes of hybridizing signals are indicated by arrows.
Transformation of A. fumigatus.
A. fumigatus protoplasts were prepared with mureinase (Amersham) and transformed by the polyethylene glycol method described by Yelton et al. (66). Transformants were selected on Aspergillus minimal medium containing hygromycin B (200 μg/ml) for pCosHX-based constructs or phleomycin (30 μg/ml) for pPKS33.
TLC analysis.
Technical-grade tricyclazole (5-methyl-1,2,4-triazole[3,4-b]benzothiazole) was obtained from Eli Lilly Research Laboratories. Flaviolin and 2-HJ were made synthetically as previously described (60, 63). Scytalone was obtained from a Brm-1 mutant strain of Verticillium dahliae (60).
For thin-layer chromatography (TLC) analysis, cultures were grown on ASA medium at 24°C. Inoculation of ASA medium was achieved by spreading 5 × 105 conidia in 0.25 ml of H2O containing 0.05% Tween 80 over the agar plate. Eight-day-old cultures of strains B-5233 and B-5233/RGD12-8 (grown on ASA medium with various modifications) were examined for the presence of flaviolin, 2-HJ, scytalone, 1,3,6,8-THN, 1,3,8-THN, and DHN by ethyl acetate extraction and TLC procedures as previously described (62). In these studies, the crude extracts were spotted on TLC plates coated with silica gel G/HR (J. T. Baker Inc., Phillipsburg, N.J.) containing 2.5% zinc silicate phosphor (Sigma Chemical Co., St. Louis, Mo.), a fluorescent indicator. The TLC plates were developed in diethyl ether-hexanes (boiling point, 68 to 70°C)-formic acid (70:29:1, vol/vol/vol). Standards of naphthoquinones, flaviolin, and 2-HJ were yellow to orange under visible light, and scytalone quenched 254-nm UV light and fluoresced yellow under 365-nm UV light. Flaviolin, 2-HJ, and scytalone also gave characteristic colors after the plates were sprayed with 1% FeCl3. These three compounds also were identified by high-performance liquid chromatography as described previously (62).
The inhibitory effects of tricyclazole on the metabolism of scytalone by strains B-5233 and B-5233/RGD12-8 were visually compared by TLC. For comparison, 60-μl samples of ethyl acetate extracts from the different cultures were spotted on TLC plates with a 50-μl syringe (Hamilton Co., Reno, Nev.). Each 60-μl sample was representative of material from 2.4 ml of ASA culture medium and was obtained from a larger, 2.5-ml ethyl acetate sample, obtained from five ASA culture plates, that collectively contained approximately 100 ml of medium and fungal material. The results were confirmed by additional repetitions.
SEM study.
The scanning electron microscopy (SEM) studies were performed by Tim Maugel at the University of Maryland, College Park. Conidia from 7-day-old cultures were fixed in 2% glutaraldehyde for 60 min. One milliliter of the fixative solution was collected onto a 0.6-μm-pore-size Nuclepore filter, and fixation was continued for an additional 60 min. The filters were subsequently washed three times in double-distilled water over a period of 10 min. Samples were then postfixed with 1% osmium tetroxide for 50 min and washed three times as described above. The filters were subsequently dehydrated, mounted on aluminum stubs, and coated with gold-palladium alloy. An Amray 1820D scanning electron microscope was operated at an accelerating potential of 25 kV to view the conidia.
Complement component C3 binding assay.
Opsonization of resting conidia was performed as described previously (54). Briefly, conidia were harvested from 10-day-old cultures in phosphate-buffered saline (PBS), filtered through sterile cheesecloth to remove mycelial fragments, washed three times with PBS, and finally resuspended in Hanks’ balanced salt solution (HBSS) with Ca2+ and Mg2+. Complement component C3 (Advanced Research Technologies, San Diego, Calif.) was labeled with 125I to a specific activity of 200,000 cpm/μg. C3 binding assays were performed with 2 × 107 resting conidia and an opsonization system containing 5% nonimmune human serum, 125I-labeled C3 (200,000 cpm), and HBSS with Ca2+ and Mg2+ (30 min, 37°C). Covalently bound 125I-C3 was measured with a gamma counter (Packard Instrument Co., Downer’s Grove, Ill.). Data were expressed as 125I-C3 bound to 2 × 107 conidia. Heat-inactivated serum served as a negative control.
Phagocytosis assay.
Phagocytosis was measured as described previously (58). Human neutrophils were purified by Ficoll-Hypaque density gradient centrifugation and mixed with resting conidia at an effector/target ratio of 1:1 (106 neutrophils: 106 conidia in 1 ml of HBSS with Ca2+ and Mg2+). Fresh nonimmune autologous serum served as a source of fresh complement (5% by volume). Incubation mixtures were tumbled for 30 min at 37°C; this was followed by cytospin preparation, Wright-Giemsa staining, and visual scoring of phagocytosis. The phagocytic index was defined as the mean number of intracellular conidia per 100 randomly visualized neutrophils. Neutrophil viability was consistently >95% as judged by trypan blue exclusion.
Animal study.
Conidia were harvested from 10-day-old cultures grown on Aspergillus minimal medium at 37°C with PBS containing 0.1% Tween 20. Seven-week-old BALB/c mice were injected with 1.5 × 105 conidia in 0.2 ml of PBS containing 0.002% Tween 20 via the tail vein. Mouse mortality was monitored daily for 21 days.
Nucleotide sequence accession number.
Sequence data reported in this paper have been submitted to the GenBank database under accession no. AF025541.
RESULTS
Cloning and analysis of the alb1 gene from A. fumigatus.
Previous analysis of a reddish-pink (RP3) conidial-color mutant of A. fumigatus led to identification of a complementing cosmid, pG1-1 (54). pGA88, a subclone of pG1-1 carrying a 5-kb AvrII DNA fragment (Fig. 2A), transformed strain RP3 to produce white conidia at a high frequency (49 of 58 transformants). A Blast search revealed that sequences of this 5-kb DNA were highly similar to the sequences of the wA gene of A. nidulans, which encodes a putative polyketide synthase (29). This 5-kb DNA contains a 3′-truncated gene which we designated alb1 (for albino). The complete alb1 gene was cloned from cosmid pG1-1 (Fig. 2A, pPKS33) and sequenced. Several cDNA clones were obtained after screening a cDNA library of wild-type strain B-5233, using the 5-kb AvrII DNA fragment as a probe. Comparison of genomic and cDNA sequences revealed the presence of two introns at the 3′-end region of the alb1 gene. Due to the large size (7 kb) of the alb1 transcript, a full-length cDNA clone was never obtained. Comparison of the alb1 genomic sequences with the A. nidulans wA cDNA sequence suggested the possible presence of two additional introns at the 5′-end region of the alb1 gene. The presence of these two introns at the 5′ end was confirmed by RT-PCR and DNA sequencing (see Materials and Methods) (Fig. 2A). The sizes of the four introns ranged from 47 to 73 nucleotides. The DNA sequences of alb1 and wA were well conserved downstream of the proposed ATG translation initiation codon (A as base 1) but not upstream of this codon. A stop codon was located 132 bp upstream of the proposed translation initiation codon. These observations render the ATG as the most likely initiation codon for alb1. The cDNA sequences of alb1 suggest that there is a poly(A) addition site at base 6840. The alb1 gene encodes a putative protein of 2,146 amino acids.
Northern blot analysis using the 5-kb AvrII DNA as a probe revealed that alb1 encodes a 7-kb transcript. This 7-kb transcript was observed only during conidiation and not in the hyphal stage (Fig. 2B).
alb1 encodes a putative polyketide synthase.
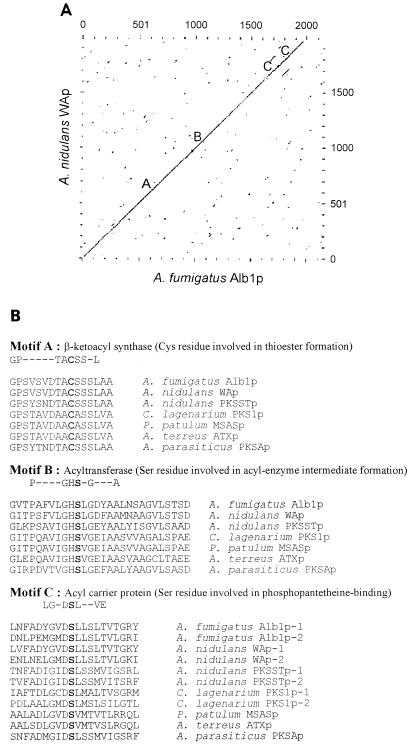
The dot plot matrix of the deduced Alb1 protein (Alb1p) and the putative polyketide synthase encoded by wA of A. nidulans showed a high degree of similarity existed throughout the complete open reading frame (Fig. 3A). Alb1p exhibits 66% identity and 79% similarity to the WA protein based on Gap analysis (9). Alb1p contains three motifs conserved among polyketide synthases: a β-ketoacyl synthase motif, an acetyltransferase motif, and an acyl carrier protein motif (Fig. 3B) (4, 6, 12, 29, 49, 67). Alb1p, however, lacks a β-ketoacyl reductase motif, which is conserved among fatty acid synthases in addition to the three previously described motifs. Collectively, that information suggests that Alb1p is a polyketide synthase rather than a fatty acid synthase (16, 18).
FIG. 3.
Comparison of Alb1p and other polyketide synthases. (A) Dot plot matrix of Alb1p and the A. nidulans WA protein (WAp) (EMBL accession no. X65866) (29). The locations of the conserved active sites among polyketide synthases are labeled as follows: A, β-ketoacyl synthase; B, acyltransferase; and C, acyl carrier protein. Parameters used for the comparison were a window size of 10 and a stringency of 16. (B) Alignment of Alb1p with active sites of other polyketide synthases. GenBank accession numbers for the corresponding genes are as follows: A. nidulans pksST, L39121 (67); Aspergillus terreus atx, D85860 (12); Aspergillus parasiticus pksA, Z47198 (6); C. lagenarium pks1, D83643 (49); and Penicillium patulum msas, X55776 (4). Conserved active-site residues important for enzyme function are in boldfaced letters, and their functions are indicated.
alb1 is required for conidial pigmentation.
Gene disruption was carried out to delineate the role of alb1 in conidial pigmentation of A. fumigatus. A gene disruption construct, pRGD12, was used to transform strain B-5233, and transformants were selected with hygromycin B. Transformants producing white conidia were observed at a frequency of 30% among 27 hygromycin B-resistant transformants. Southern analysis was used to determine whether the wild-type alb1 copy was deleted from these white transformants. Genomic DNA of B-5233 and a white transformant, B-5233/RGD12-8, was hybridized with the 1.5-kb MluI-AvrII DNA fragment which was replaced with the hph gene in the disruption cassette. B-5233 gave a hybridizing signal of 5.0 kb, while B-5233/RGD12-8 did not reveal any hybridizing fragment (Fig. 4B, panel I). The blot was stripped and rehybridized with the entire disruption cassette, pRGD12. B-5233 gave two hybridizing signals, of 2.5 and 5.0 kb, while B-5233/RGD12-8 also showed two hybridizing fragments, of 2.9 and 5.4 kb (Fig. 4B, panel II). These results indicate that the wild-type copy of alb1 in B-5233/RGD12-8 was replaced by the disrupted copy of alb1 from pRGD12 via a double-crossover event. Thus, the albino phenotype of strain B-5233/RGD12-8 was the result of alb1 disruption.
To confirm that the albino phenotype was not due to a secondary, hidden mutation, the albino strain, B-5233/RGD12-8, was transformed with pPKS33 (Fig. 2A) which lacks a portion of the 5′ flanking region of alb1. In the resulting analysis, 8 of 27 phleomycin-resistant transformants were restored to bluish-green pigment synthesis. One of these transformants, RGD12-8/PKS33-3, was analyzed by Southern hybridization to confirm that pPKS33 had integrated into the alb1 locus in B-5233/RGD12-8 and restored the functional copy of alb1 (data not shown).
Involvement of alb1 in the DHN-melanin pathway.
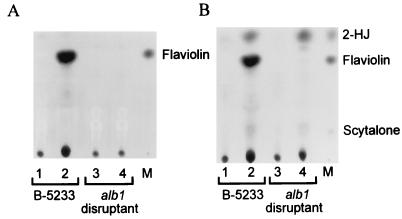
Previously we reported that A. fumigatus uses a DHN-melanin pathway to synthesize its conidial pigments; thus, alb1 is likely to be involved in DHN-melanin biosynthesis. Growth of strain B-5233 on ASA medium containing tricyclazole resulted in the accumulation of flaviolin, an autoxidative product of 1,3,6,8-THN, while controls grown without tricyclazole did not accumulate flaviolin (Fig. 5A). Accumulation of flaviolin is due to blockage of the first reduction step (conversion of 1,3,6,8-THN to scytalone [Fig. 1]). Simultaneous feeding of scytalone and tricyclazole to strain B-5233 led to accumulation of flaviolin and 2-HJ (Fig. 5B). 2-HJ is an autoxidative product derived from 1,3,8-THN, which is derived from scytalone (Fig. 1). Accumulation of 2-HJ is due to blockage of the reduction step of 1,3,8-THN. In contrast, the alb1 disruptant (B-5233/RGD12-8) showed no accumulation of flaviolin while growing on ASA medium containing tricyclazole. This indicates that the production of 1,3,6,8-THN was blocked and therefore no flaviolin was produced. Accumulation of 2-HJ was observed when the alb1 disruptant was fed scytalone and tricyclazole simultaneously, indicating that the alb1 disruptant retained the ability to convert scytalone to 1,3,8-THN. These results suggest that the alb1 disruptant had lost the ability to produce 1,3,6,8-THN without changing the machinery involved in the downstream DHN-melanin pathway.
FIG. 5.
TLC analysis of scytalone metabolism. A. fumigatus was grown on ASA medium without (A) or with (B) scytalone supplementation. The extract samples were obtained from a mixture of mycelia and agar medium by an ethyl acetate extraction procedure. Lanes: 1 and 3, extract samples from cultures without tricyclazole; 2 and 4, extract samples from cultures with tricyclazole; M, standards.
Disruption of alb1 resulted in altered conidial surface morphology.
Wild-type conidia are readily released from their conidiophores; conidia of cultures more than 5 days of age were often observed to be deposited on the inner side of the petri dish lids. In contrast, the alb1 disruptant culture rarely showed such a phenomenon, suggesting that disruption of alb1 altered the conidial surface properties which affect conidial dispersibility. Since we suspected that one of the changed properties was conidial hydrophobicity, the wettable phenotype of conidia was tested with water or various concentrations of sodium dodecyl sulfate, Tween 20, or Tween 80 with or without EDTA (47). We did not observe any difference among the wild-type, alb1 disruptant, and alb1-complemented strains. This suggests that conidial hydrophobicity was not affected by the disruption of alb1.
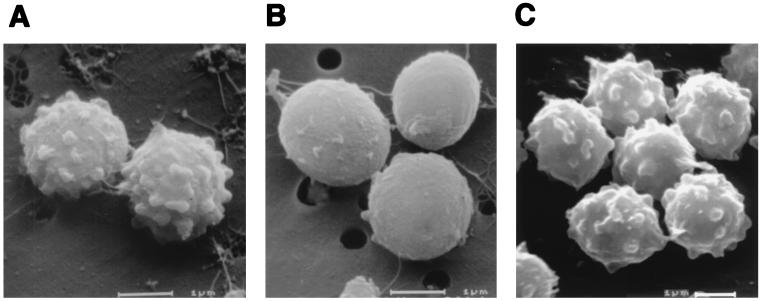
SEM studies, however, revealed a distinct change in the conidial surface morphology of the alb1 disruptant. While conidia of the wild type exhibited echinulate surfaces, the alb1 disruptant produced nearly smooth conidia (Fig. 6). Furthermore, the alb1-complemented strain, RGD12-8/PKS33-3, produced bluish-green conidia with the echinulate surface of wild-type conidia (Fig. 6). These findings indicate that mutation of alb1 had altered not only conidial pigmentation but also conidial surface morphology.
FIG. 6.
SEM study of conidial surface structure. Conidia were from 7-day-old cultures. (A) B-5233, the wild-type strain; (B) B-5233/RGD12-8, the alb1 gene disruptant; (C) RGD12-8/PKS33-3, the alb1-complemented strain. Bars, 1 μm.
Significant influence of alb1 on fungal virulence.
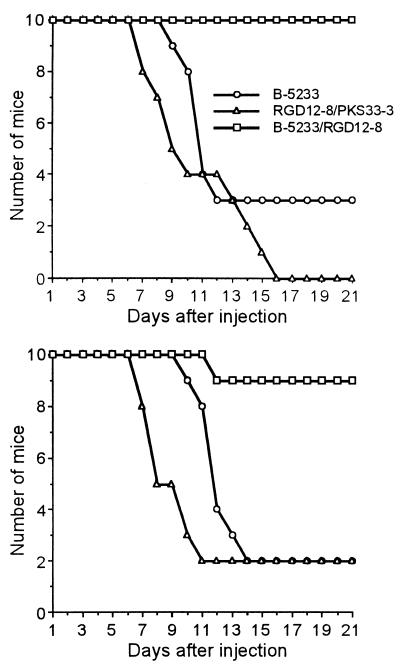
Two different sets of animal studies were carried out to compare the degrees of virulence of the wild-type, alb1 disruptant, and alb1-complemented strains. In these two experiments, mice injected with the alb1 disruptant showed a dramatic reduction in mortality compared to the rates of the wild-type and the alb1-complemented strains. Lethality in mice infected with strain B-5233 began on days 9 and 10, with mortality rates of 70 and 80% by the end of experiment, day 21 (Fig. 7). In contrast, mice injected with the alb1 disruptant showed only 0 and 10% mortality at day 21. However, the alb1-complemented strain recovered full virulence; mortality started on day 7 and reached 80 to 100% by the termination date. Statistical analysis showed a significantly reduced lethality for the alb1 disruptant compared to that for B-5233 or the alb1-complemented strain (P < 0.05, Kaplan-Meier analysis).
FIG. 7.
Virulence studies. Each study included 10 mice per strain in two independent experiments. Mice were injected intravenously with 1.5 × 105 conidia. Mouse mortality was monitored daily for 21 days. Strains: B-5233, the wild-type strain; B-5233/RGD12-8, the alb1 disruptant; RGD12-8/PKS33-3, the alb1-complemented strain. P < 0.05 for comparison of the alb1 disruptant to B-5233 or the alb1-complemented strain (Kaplan-Meier analysis).
alb1 modulates C3 deposition on conidia.
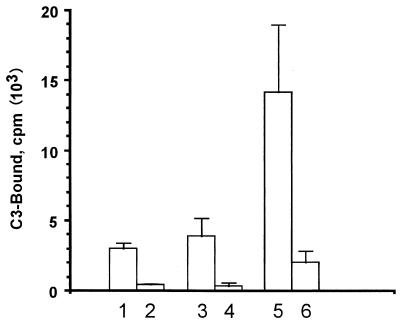
Our previous study showed that conidial pigmentation affects the deposition of complement component C3 on A. fumigatus conidia (54). Since disruption of alb1 resulted in an albino conidial phenotype, C3 binding assays were performed to delineate the effect of alb1 on C3 binding. The level of quantitative binding of complement component C3 to bluish-green conidia, produced by the wild-type and the alb1-complemented strains, was significantly lower than that to conidia of the alb1 disruptant (Fig. 8). These results suggest that, similar to the arp1 gene, alb1 modulates C3 deposition on conidia.
FIG. 8.
Complement component C3 binding analysis. Strains are as follows: lanes 1 and 2, wild-type strain B-5233; lanes 3 and 4, the alb1-complemented strain, RGD12-8/PKS33-3; and lanes 5 and 6, the alb1 disruptant strain, B-5233/RGD12-8. Lanes 1, 3, and 5 were with 5% fresh serum, while lanes 2, 4, and 6 were with heat-inactivated serum. Data represent means ± standard deviations (n = 4). P < 0.05 by Student’s t test for comparison of the alb1 disruptant to strain B-5233 or the alb1-complemented strain.
alb1 affects neutrophil-mediated phagocytosis.
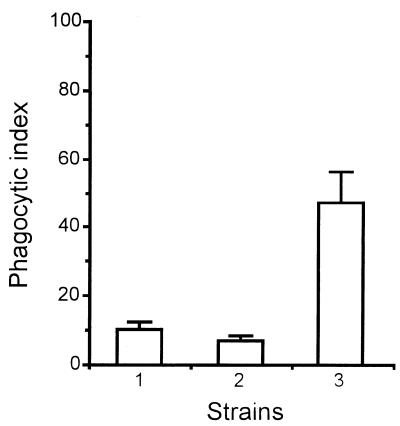
Results for neutrophil-mediated phagocytosis of resting conidia paralleled those for quantitative C3 deposition. As shown in Fig. 9, quantitative phagocytosis for the alb1 disruptant was significantly greater than that for the wild-type strain and the alb1-complemented strain. Neutrophils that were incubated with conidia of the alb1 disruptant became aggregated, suggesting activation. In contrast, neutrophils incubated with the wild-type or alb1-complemented strain remained individually suspended (data not shown). These results suggest that enhanced phagocytosis is an important mechanism accounting for the reduced virulence of the alb1 disruptant.
FIG. 9.
Phagocytosis assay. Strains are as follows: lane 1, wild-type strain B-5233; lane 2, the alb1-complemented strain, RGD12-8/PKS33-3; and lane 3, the alb1 disruptant strain, B-5233/RGD12-8. Data represent phagocytic index means ± standard deviations for three experiments performed in duplicate. P < 0.05 for comparison of the alb1 disruptant to strain B-5233 or the alb1-complemented strain (Student’s t test).
DISCUSSION
Only 16 of over 130 known Aspergillus spp. have been reported from clinical cases of infection in humans (25). Among these aspergilli, A. fumigatus is the leading cause of both noninvasive and invasive aspergillosis. This fungus is responsible for more than 60% of invasive aspergillosis cases in immunosuppressed patients. Identification of the determinants rendering A. fumigatus more virulent than the other aspergilli has been a research interest. Recently, several putative virulence factors have been analyzed using molecular genetic tools. These factors included alkaline proteases, elastase, catalase, chitin synthase, hydrophobin, cytotoxin ASPFI, and restrictocin. However, disruption of those genes individually or in various combinations in A. fumigatus failed to yield significant changes in fungal virulence (3, 31, 35, 40, 45, 46, 50, 51).
A. fumigatus propagates by conidia, which are encountered by the human host chiefly through inhalation. Inhaled conidia face challenges from the host immune system, and evasion of these challenges is a pivotal step in establishing an invasive infection. In this context, conidial surface components are likely to play important roles because of their initial contact with host defenses. We have focused our attention on molecular aspects of conidial pigment biosynthesis since the pigments are among the visible components of the conidial wall that protects the conidium.
The alb1 gene is developmentally regulated, being expressed only during sporulation. The gene is not essential in A. fumigatus, since disruption of the gene did not cause any visible change in fungal growth in vitro. The pleiotropic effects on conidial morphology caused by disruption of alb1 underscore its importance for A. fumigatus. The alb1-disrupted strain exhibited an albino conidial phenotype and did not accumulate flaviolin, a hallmark shunt product of the DHN-melanin pathway. These results support our previous hypothesis that A. fumigatus synthesizes its conidial pigments via a DHN-melanin pathway, based on the observations that arp1 encodes a scytalone dehydratase and that conidial pigmentation is sensitive to tricyclazole (54). Although alb1 is apparently a homolog of the wA gene of A. nidulans, we did not detect any arp1 homolog in A. nidulans even under low-stringency hybridization conditions (53a). In addition, tricyclazole did not inhibit conidial pigmentation of A. nidulans (62). Together, these observations suggest that the polyketide pathways for conidial pigmentation in A. fumigatus and A. nidulans may share the upstream steps but diverge at the downstream steps.
One of the phenotypes caused by disruption of the alb1 gene was the loss of echinulation on the conidial surface. Changes in conidial surface structure may have been secondary to the defect in conidial pigment biosynthesis, because conidial pigment is one of the main conidial surface components. Whether the pigment is independently formed and deposited on the outer surface or is codeposited with other molecules remains to be elucidated. If the protrusions formed on conidial surfaces were solely composed of pigments, the loss of the pigment layer could produce smooth conidial surfaces. Alternatively, if pigments are codeposited with other molecules (e.g., proteins or polysaccharides), pigment deposition may serve a crucial function for the formation of the echinulate surface.
The most intriguing phenotype of the alb1 disruptant was its significant loss of virulence. Reduced virulence in the murine model was also observed with a white conidial mutant obtained by UV mutagenesis (19). This genetically uncharacterized mutant also exhibited smooth conidial surfaces. It is likely that the reduced virulence in the present study was due to altered conidial pigmentation and surface morphology induced by the alb1 disruption. In the plant pathogenic fungi C. lagenarium and M. grisea, deficient DHN-melanin biosynthesis resulted in nonmelanized appressoria (24, 28, 37). Those mutants lost their ability to penetrate plant leaf tissue and became avirulent, possibly due to the fact that melanin is important for the rigidity of appressoria. These reports indicate an association between melanin biosynthesis and both cell wall rigidity and virulence. Similarly, melanin biosynthesis in A. fumigatus may be associated with conidial surface characteristics that protect these structures from environmental and immunological challenges, including C3 deposition and oxidative killing (19).
Disruption of alb1 not only altered conidial morphology but also led to efficient C3 binding on conidial surfaces. It is tempting to speculate that the alteration of the conidial surface topography is the major factor leading to enhanced C3 binding. Previous observations about complement activation by A. fumigatus include inefficient C3 deposition on resting conidia compared to that of swollen conidia (22) and identification of a 54- to 58-kDa C3 binding protein (48). Deposition of pigments on conidial surfaces may mask the C3 binding sites and reduce the C3 binding capacity, while loss of conidial pigments due to swelling or alb1 disruption may result in elevated C3 binding due to exposure of more binding sites. Alternatively, extracellular C3 binding inhibitors derived from the DHN-melanin pathway may directly downregulate C3 deposition on wild-type conidia. In fact, extracellular compounds which inhibit complement activation have been isolated from A. fumigatus and found to possess properties consistent with phenolic compounds (57, 59). Since the DHN-melanin pathway produces various phenolic compounds, it is possible that some of these compounds interfere with complement activation. The absence of these phenolic compounds due to defects in DHN-melanin biosynthesis may enhance C3 deposition on conidial surfaces. Finally, Alb1p may be involved in spore wall assembly, raising the possibility that alb1 disruption produces structural changes in conidial walls that enhance the surface exposure of C3 binding sites.
The present studies showed that the bluish-green pigmentation of wild-type conidia contributes to virulence in an animal model and modulates human complement-mediated opsonization and neutrophil-mediated phagocytosis. Thus, bluish-green pigmentation at least partially thwarts the complement-phagocyte axis of the mammalian host defense against invasive aspergillosis. To our knowledge, this represents the first demonstration of such an effect for A. fumigatus at the molecular genetic level. Of interest, these findings fit into a pattern of pigment-mediated virulence that has been emerging in recent literature for both mammalian and plant fungal pathogens, including C. neoformans and dematiaceous molds such as M. grisea and C. lagenarium (11, 17, 19, 38, 55). Several different virulence mechanisms have been proposed for those fungal pigments, but to our knowledge the present study provides the first definitive evidence that fungal pigmentation is a factor interfering with complement-mediated opsonization and phagocytosis. Thus, the bluish-green conidial pigmentation of A. fumigatus provides a novel mechanism of escape from mammalian defenses.
In summary, alb1 is an important virulence determinant for the human pathogenic fungus A. fumigatus. This novel discovery offers an impetus to further explore interactions between fungal pathogens and the human host. Since the alb1 gene plays an important role in modulating conidial morphology and fungal virulence in vivo, a strong association between conidial morphology and fungal virulence is apparent. Further investigation into the interactions between conidia and the host immune system will now lead to a better understanding of the mechanisms involved in fungal pathogenesis.
ACKNOWLEDGMENTS
We thank Herman Edskes for critical reviews and helpful suggestions, Tim Maugel for assistance in the SEM study, Lisa Penoyer for assistance in DNA sequencing, Amy Rye and Sandeep Kumar for technical assistance, and P. Silar for providing plasmids pBC-hygro and pBC-phleo.
R.G.W. was supported by NIH grant AI-01036 and Pfizer, U.S. Pharmaceuticals.
REFERENCES
- 1.Agrios G N. Plant pathology. 4th ed. London, United Kingdom: Academic Press; 1997. [Google Scholar]
- 2.Aramayo R, Timberlake W E. Sequence and molecular structure of the Aspergillus nidulans yA(laccase I) gene. Nucleic Acids Res. 1990;18:3415. doi: 10.1093/nar/18.11.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aufauvre-Brown A, Mellado E, Gow N A R, Holden D W. Aspergillus fumigatus chsE: a gene related to CHS3 of Saccharomyces cerevisiaeand important for hyphal growth and conidiophore development but not pathogenicity. Fungal Genet Biol. 1997;21:141–152. doi: 10.1006/fgbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 4.Beck J, Ripka S, Siegner A, Schiltz E, Schweizer E. The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum—its gene structure relative to that of other polyketide synthases. Eur J Biochem. 1990;192:487–498. doi: 10.1111/j.1432-1033.1990.tb19252.x. [DOI] [PubMed] [Google Scholar]
- 5.Bell A A, Wheeler M H. Biosynthesis and functions of fungal melanins. Annu Rev Phytopathol. 1986;24:411–451. [Google Scholar]
- 6.Chang P-K, Cary J W, Yu J, Bhatnagar D, Cleveland T E. The Aspergillus parasiticus polyketide synthase gene pksA, a homolog of Aspergillus nidulans wA, is required for aflatoxin B1biosynthesis. Mol Gen Genet. 1995;248:270–277. doi: 10.1007/BF02191593. [DOI] [PubMed] [Google Scholar]
- 7.Cohen M S, Isturiz R E, Malech H L, Root R K, Wilfert C M, Gutman L, Buckley R H. Fungal infection in chronic granulomatous disease: the importance of the phagocyte in defense against fungi. Am J Med. 1981;71:59–66. doi: 10.1016/0002-9343(81)90259-x. [DOI] [PubMed] [Google Scholar]
- 8.Denning D W. Therapeutic outcome in invasive aspergillosis. Clin Infect Dis. 1996;23:608–615. doi: 10.1093/clinids/23.3.608. [DOI] [PubMed] [Google Scholar]
- 9.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon D M, Polak A, Szaniszlo P J. Pathogenicity and virulence of wild-type and melanin-deficient Wangiella dermatitidis. J Med Vet Mycol. 1987;25:97–106. doi: 10.1080/02681218780000141. [DOI] [PubMed] [Google Scholar]
- 11.Emery H S, Shelburne C P, Bowman J P, Fallon P G, Schulz C A, Jacobson E S. Genetic study of oxygen resistance and melanization in Cryptococcus neoformans. Infect Immun. 1994;62:5694–5697. doi: 10.1128/iai.62.12.5694-5697.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii I, Ono Y, Tada H, Gomi K, Ebizuka Y, Sankawa U. Cloning of the polyketide synthase gene atX from Aspergillus terreusand its identification as the 6-methylsalicylic acid synthase gene by heterologous expression. Mol Gen Genet. 1996;253:1–10. doi: 10.1007/s004380050289. [DOI] [PubMed] [Google Scholar]
- 13.Gerson S T, Talbot G H, Hurwitz S, Strom B L, Lusk E J, Cassileth P A. Prolonged granulocytopenia: the major risk factor for invasive pulmonary aspergillosis in patients with acute leukemia. Ann Intern Med. 1984;100:345–351. doi: 10.7326/0003-4819-100-3-345. [DOI] [PubMed] [Google Scholar]
- 14.Gustafson T L, Schaffner W, Lavely G B, Stratton C W, Johnson H K, Hutcheson R H., Jr Invasive aspergillosis in renal transplant recipients: correlation with corticosteroid therapy. J Infect Dis. 1983;148:230–238. doi: 10.1093/infdis/148.2.230. [DOI] [PubMed] [Google Scholar]
- 15.Hopwood D A, Khosla C. Genes for polyketide secondary metabolic pathways in microorganisms and plants. Ciba Found Symp. 1992;171:88–112. doi: 10.1002/9780470514344.ch6. [DOI] [PubMed] [Google Scholar]
- 16.Hopwood D A, Sherman D H. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet. 1990;24:37–66. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- 17.Huffnagle G B, Chen G-H, Curtis J L, McDonald R A, Strieter R M, Toews G B. Down-regulation of the afferent phase of T cell-mediated pulmonary inflammation and immunity by a high melanin-producing strain of Cryptococcus neoformans. J Immunol. 1995;155:3507–3516. [PubMed] [Google Scholar]
- 18.Hutchinson C R, Fujii I. Polyketide synthase gene manipulation: a structure-function approach in engineering novel antibiotics. Annu Rev Microbiol. 1995;49:201–238. doi: 10.1146/annurev.mi.49.100195.001221. [DOI] [PubMed] [Google Scholar]
- 19.Jahn B, Koch A, Schmidt A, Wanner G, Gehringer H, Bhakdi S, Brakhage A A. Isolation and characterization of a pigmentless-conidium mutant of Aspergillus fumigatuswith altered conidial surface and reduced virulence. Infect Immun. 1997;65:5110–5117. doi: 10.1128/iai.65.12.5110-5117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaefer E. Meiotic and mitotic recombination in Aspergillusand its chromosomal aberrations. Adv Genet. 1977;19:33–131. doi: 10.1016/s0065-2660(08)60245-x. [DOI] [PubMed] [Google Scholar]
- 21.Keller N P, Hohn T M. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet Biol. 1997;21:17–29. [PubMed] [Google Scholar]
- 22.Kozel T R, Wilson M A, Farrell T P, Levitz S M. Activation of C3 and binding to Aspergillus fumigatusconidia and hyphae. Infect Immun. 1989;57:3412–3417. doi: 10.1128/iai.57.11.3412-3417.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubo Y, Takano Y, Endo N, Yasuda N, Tajima S, Furusawa I. Cloning and structural analysis of the melanin biosynthesis gene SCD1 encoding scytalone dehydratase in Colletotrichum lagenarium. Appl Environ Microbiol. 1996;62:4340–4344. doi: 10.1128/aem.62.12.4340-4344.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubo Y, Takano Y, Furusawa I. Molecular genetic analysis of melanin biosynthetic genes essential for appressorium function in Colletotrichum lagenarium. In: Mills D, Kunoh H, Keen N, Mayama S, editors. Molecular aspects of pathogenicity: requirements for signal transduction. St. Paul, Minn: APS Press; 1996. pp. 73–82. [Google Scholar]
- 25.Kwon-Chung K J, Bennett J E. Medical mycology. Baltimore, Md: Williams & Wilkins; 1992. Aspergillosis; pp. 201–247. [Google Scholar]
- 26.Kwon-Chung K J, Polacheck I, Popkin T J. Melanin-lacking mutants of Cryptococcus neoformansand their virulence for mice. J Bacteriol. 1982;150:1414–1421. doi: 10.1128/jb.150.3.1414-1421.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon-Chung K J, Rhodes J C. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect Immun. 1986;51:218–223. doi: 10.1128/iai.51.1.218-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundqvist T, Rice J, Hodge C N, Basarab G S, Pierce J, Lundqvist Y. Crystal structure of scytalone dehydratase—the disease determinant of a rice pathogen, Magnaporthe grisea. Structure. 1994;15:937–944. doi: 10.1016/s0969-2126(94)00095-6. [DOI] [PubMed] [Google Scholar]
- 29.Mayorga M E, Timberlake W E. The developmentally regulated Aspergillus nidulans wAgene encodes a polypeptide homologous to polyketide and fatty acid synthases. Mol Gen Genet. 1992;235:205–212. doi: 10.1007/BF00279362. [DOI] [PubMed] [Google Scholar]
- 30.Mayorga M E, Timberlake W E. Isolation and molecular characterization of the Aspergillus nidulans wAgene. Genetics. 1990;126:73–79. doi: 10.1093/genetics/126.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monod M, Paris S, Sarfati J, Jaton-Ogay K, Ave P, Latgé J-P. Virulence of alkaline protease-deficient mutants of Aspergillus fumigatus. FEMS Microbiol Lett. 1993;106:39–46. doi: 10.1111/j.1574-6968.1993.tb05932.x. [DOI] [PubMed] [Google Scholar]
- 32.Mullins J, Harvey R, Seaton A. Sources and incidences of airborne Aspergillus fumigatus. Clin Allergy. 1976;6:209–217. doi: 10.1111/j.1365-2222.1976.tb01899.x. [DOI] [PubMed] [Google Scholar]
- 33.O’Hagan D. Biosynthesis of polyketide metabolites. Nat Prod Rep. 1992;9:447–479. doi: 10.1039/np9920900447. [DOI] [PubMed] [Google Scholar]
- 34.O’Hara E B, Timberlake W E. Molecular characterization of the Aspergillus nidulans yAlocus. Genetics. 1989;121:249–254. doi: 10.1093/genetics/121.2.249. . (Erratum, 124:791, 1990.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paris S, Monod M, Diaquin M, Lamy B, Arruda L K, Punt P J, Latgé J-P. A transformant of Aspergillus fumigatusdeficient in the antigenic cytotoxin ASPFI. FEMS Microbiol Lett. 1993;111:31–36. doi: 10.1111/j.1574-6968.1993.tb06357.x. [DOI] [PubMed] [Google Scholar]
- 36.Parta M, Chang Y, Rulong S, Pinto-DaSilva P, Kwon-Chung K J. HYP1, a hydrophobin gene from Aspergillus fumigatus, complements the rodletless phenotype in Aspergillus nidulans. Infect Immun. 1994;62:4389–4395. doi: 10.1128/iai.62.10.4389-4395.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perpetua N S, Kubo Y, Yasuda N, Takano Y, Furusawa I. Cloning and characterization of a melanin biosynthetic THR1 reductase gene essential for appressorial penetration of Colletotrichum lagenarium. Mol Plant-Microbe Interact. 1996;9:323–329. doi: 10.1094/mpmi-9-0323. [DOI] [PubMed] [Google Scholar]
- 38.Polacheck I, Platt Y, Aronovitch J. Catecholamines and virulence of Cryptococcus neoformans. Infect Immun. 1990;58:2919–2922. doi: 10.1128/iai.58.9.2919-2922.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Punt P J, Oliver R P, Dingemase M A, Pouwel P H, van den Hondel C A. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;56:117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- 40.Ramesh M V, Sirakova T, Kolattukudy P E. Isolation, characterization, and cloning of cDNA and the gene for an elastinolytic serine proteinase from Aspergillus flavus. Infect Immun. 1994;62:79–85. doi: 10.1128/iai.62.1.79-85.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raper K B, Fennell D I. Aspergillus fumigatus group. In: Raper K B, Fennell D I, editors. The genus Aspergillus. Baltimore, Md: Williams & Wilkins; 1965. pp. 238–268. [Google Scholar]
- 42.Salas S D, Bennett J E, Kwon-Chung K J, Perfect J R, Williamson P R. Effect of the laccase gene, CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Silar P. Two new easy to use vectors for transformation. Fungal Genet Newsletter. 1995;42:73. [Google Scholar]
- 45.Smith J M, Davies J E, Holden D W. Construction and pathogenicity of Aspergillus fumigatusmutants that do not produce the ribotoxin restrictocin. Mol Microbiol. 1993;9:1071–1077. doi: 10.1111/j.1365-2958.1993.tb01236.x. [DOI] [PubMed] [Google Scholar]
- 46.Smith J M, Tang C M, Van Noorden S, Holden D W. Virulence of Aspergillus fumigatusdouble mutants lacking restrictocin and an alkaline protease in a low-dose model of invasive pulmonary aspergillosis. Infect Immun. 1994;62:5247–5254. doi: 10.1128/iai.62.12.5247-5254.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stringer M A, Timberlake W E. dewA encodes a fungal hydrophobin component of the Aspergillusspore wall. Mol Microbiol. 1995;16:33–44. doi: 10.1111/j.1365-2958.1995.tb02389.x. [DOI] [PubMed] [Google Scholar]
- 48.Sturtevant J E, Latgé J-P. Interactions between conidia of Aspergillus fumigatusand human complement component C3. Infect Immun. 1992;60:1913–1918. doi: 10.1128/iai.60.5.1913-1918.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takano Y, Kubo Y, Shimizu K, Mise K, Okuno T, Furusawa I. Structural analysis of PKS1, a polyketide synthase gene involved in melanin biosynthesis in Colletotrichum lagenarium. Mol Gen Genet. 1995;249:162–167. doi: 10.1007/BF00290362. [DOI] [PubMed] [Google Scholar]
- 50.Tang C M, Cohen J, Krausz T, Van Noorden S, Holden D W. The alkaline protease of Aspergillus fumigatusis not a virulence determinant in two murine models of invasive pulmonary aspergillosis. Infect Immun. 1993;61:1650–1656. doi: 10.1128/iai.61.5.1650-1656.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thau N, Monod M, Crestani B, Rolland C, Tronchin G, Latgé J-P, Paris S. rodletless mutants of Aspergillus fumigatus. Infect Immun. 1994;62:4380–4388. doi: 10.1128/iai.62.10.4380-4388.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Timberlake W E. Molecular genetics of Aspergillusdevelopment. Annu Rev Genet. 1986;24:5–36. doi: 10.1146/annurev.ge.24.120190.000253. [DOI] [PubMed] [Google Scholar]
- 53.Tokousbalides M C, Sisler H D. Site of inhibition by tricyclazole in the melanin biosynthetic pathway of Verticillium dahliae. Pestic Biochem Physiol. 1979;11:64–73. [Google Scholar]
- 53a.Tsai, H.-F. Unpublished data.
- 54.Tsai H-F, Washburn R G, Chang Y C, Kwon-Chung K J. Aspergillus fumigatus arp1modulates conidial pigmentation and complement deposition. Mol Microbiol. 1997;26:175–183. doi: 10.1046/j.1365-2958.1997.5681921.x. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Aisen P, Casadevall A. Cryptococcus neoformansmelanin and virulence: mechanism of action. Infect Immun. 1995;63:3131–3136. doi: 10.1128/iai.63.8.3131-3136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Washburn R G. Opportunistic mold infections. In: Esser L, Lemke P A, editors. The Mycota. VI. Heidelberg, Germany: Springer-Verlag; 1996. pp. 147–158. [Google Scholar]
- 57.Washburn R G, DeHart D J, Agwu D E, Bryant-Varela B J, Julian N C. Aspergillus fumigatuscomplement inhibitor: production, characterization, and purification by hydrophobic interaction and thin-layer chromatography. Infect Immun. 1990;58:3508–3515. doi: 10.1128/iai.58.11.3508-3515.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Washburn R G, Gallin J I, Bennett J E. Oxidative killing of Aspergillus fumigatusproceeds by parallel myeloperoxidase-dependent and -independent pathways. Infect Immun. 1987;55:2088–2092. doi: 10.1128/iai.55.9.2088-2092.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Washburn R G, Hammer C H, Bennett J E. Inhibition of complement by culture supernatants of Aspergillus fumigatus. J Infect Dis. 1986;154:944–951. doi: 10.1093/infdis/154.6.944. [DOI] [PubMed] [Google Scholar]
- 60.Wheeler M H. Melanin biosynthesis in Verticillium dahliae: dehydration and reduction reactions in cell-free homogenates. Exp Mycol. 1982;6:171–179. [Google Scholar]
- 61.Wheeler M H, Bell A A. Melanins and their importance in pathogenic fungi. In: McGinnis M R, editor. Current topics in medical mycology. Vol. 2. New York, N.Y: Springer-Verlag; 1988. pp. 338–387. [DOI] [PubMed] [Google Scholar]
- 62.Wheeler M H, Klich M A. The effects of tricyclazole, pyroquilon, phthalide, and related fungicides on the production of conidial wall pigments by Penicillium and Aspergillusspecies. Pestic Biochem Physiol. 1995;52:125–136. [Google Scholar]
- 63.Wheeler M H, Stipanovic R D. Melanin biosynthesis and the metabolism of flaviolin and 2-hydroxyjuglone in Wangiella dermatitidis. Arch Microbiol. 1985;142:234–241. doi: 10.1007/BF00693396. [DOI] [PubMed] [Google Scholar]
- 64.Wheeler M H, Stipanovic R D. Melanin biosynthesis in Thielaviopsis basicola. Exp Mycol. 1979;3:340–350. [Google Scholar]
- 65.Yang G, Rose M S, Turgeon B G, Yoder O C. A polyketide synthase is required for fungal virulence and production of the polyketide T-toxin. Plant Cell. 1996;8:2139–2150. doi: 10.1105/tpc.8.11.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yelton M M, Hamer J E, Timberlake W E. Transformation of Aspergillus nidulans by using a trpCplasmid. Proc Natl Acad Sci USA. 1984;81:1470–1474. doi: 10.1073/pnas.81.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu J-H, Leonard T J. Sterigmatocystin biosynthesis in Aspergillus nidulansrequires a novel type I polyketide synthase. J Bacteriol. 1995;177:4792–4800. doi: 10.1128/jb.177.16.4792-4800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]