Abstract
We have examined the targeting preference of the bacterial insertion element IS903 by determining the sites of insertion of a large number of transposition events into the 55-kb conjugative plasmid pOX38. Despite the large target size, all the insertions were clustered in four small distinct regions associated with conjugal DNA transfer. Within these regions, many different sites were used for insertion; however, there were a few sites that IS903 inserted into more than once. Alignment of the insertion sites showed that there was no consensus sequence within the 9-bp target duplication but that there were preferred sequences located symmetrically on either side of the target. This is consistent with target recognition by a dimer or multimer of transposase, with either sequence-specific or structure-specific interactions on both sides of the target. We show further that when one of these preferred regions was cloned into a second conjugative plasmid, pUB307, it was still a preferred target, implying that all the sequences necessary for target selection are contained within this DNA segment. Also, we observed a very strong preference for insertion in a single orientation in pUB307. We examined the possibility that either DNA replication from the origin of vegetative replication, oriV, or the origin of transfer, oriT, might determine this orientation effect. We find that reversing the direction of vegetative replication had no effect on the orientation of transposon insertions; however, reversing the direction of DNA transfer abolished the orientation effect. This supports the idea that conjugal DNA transfer imparts a polarity on the target that is sensed by the transposon.
Bacterial transposases are an interesting class of multifunctional DNA-binding proteins that catalyze the process of transposition. They bind specifically to sequences at the transposon ends and catalyze the DNA cleavage events that are the first step of transposition. The transposase also recognizes the target DNA into which the transposon integrates. The recognition and interaction properties of the transposase with the target DNA differ from those with the transposon ends in two ways. First, the transposase binds specifically to the ends of the transposon; in contrast, for most transposable elements, transposition is rarely targeted to a specific sequence, thus implying involvement of nonspecific interactions between the transposase and target DNA. The most notable exception to this is Tn7, which inserts site specifically into a unique site on the Escherichia coli chromosome, attTn7 (7). Second, the cleavage event at the target site is different from the precise strand cleavages that must occur for transposon excision. The 3′ ends of the transposon are joined to the target in a staggered fashion, and the resulting gaps are filled in to generate the characteristic target site duplications found on either side of the transposon. The length of the duplication is characteristic for each transposon and can vary from 2 to 13 bp (13). For example, IS903 and IS10 generate 9-bp duplications, while Mu and Tn7 generate 5-bp target duplications. The variation in the size of target duplication indicates that the sites of DNA cleavage for each strand, and the transposase residues that mediate the cleavage, are in very different positions along the DNA backbone and suggests that each transposase interacts with the DNA with a precise, but different, geometry.
Selection of a target site is an important part of the transposition process (reviewed in reference 6). Although it is chronologically perceived to be one of the final steps in transposition, it is clear that for some transposons the target DNA is an important and integral component of the earlier steps in transposition. Intermolecular transposition of Mu is enhanced by the addition of MuB protein and target DNA before any cleavage steps have occurred (25, 28), and for Tn7, it has been demonstrated that cleavage at the transposon ends, considered the first full commitment to transposition, will occur only in the presence of target DNA (1). The requirement for a target interaction prior to initiation of transposition is thought to coordinate and regulate transposition and thereby prevent the formation of aberrant transposition products. This is in contrast to the situation observed with IS10, in which transposon excision must occur before target interaction can take place (20, 31). The results obtained with IS10 suggest that the flanking donor DNA must be released to allow binding of target DNA into the same “binding pocket” of the transposase end complex.
The choice of target can have important biological and evolutionary consequences for both the host and the transposon (6, 33). Transposition can be a mutagenic process resulting in insertion into essential genes and deletion or inversion of genomic segments. Therefore, the ability to direct transposition events to certain regions or segments of DNA will increase the fitness of both the host and the transposon. By inserting into a unique site on the chromosome, Tn7 ensures that it does not inactivate essential host genes. In addition, intramolecular rearrangements can destroy the integrity of the transposon if a target is chosen within the element itself. Certain classes of transposons avoid self-insertion by a process called immunity, which favors insertion into distal sites or intermolecular targets (24). For Tn10, in vivo experiments suggest that the interaction of the transposon end with the target can be channeled in a precise manner to promote intramolecular inversions, which result in the formation of new mobile elements (20, 35).
In the few examples for which target selection has been systematically examined, it is clear that there are a variety of ways a target is selected. IS10 has a preferred target site, the selection of which clearly involves transposase-target interactions. Sequence analysis of multiple IS10 insertions has identified a symmetrical 6-bp consensus sequence that is found within the 9-bp target duplication (4, 16). In addition, the context of the flanking DNA has been shown to have a dramatic influence on the use of these preferred target sites, suggesting that there is a structural component to site selection (4). In retroviral integration (a process mechanistically equivalent to transposition), in vitro experiments have shown that there is a preference for insertion into nucleosomal DNA and that it is the bend induced in the DNA by the nucleosome that favors integration (27).
Protein-protein interactions can also target transposon insertion. Insertion of the Saccharomyces cerevisiae retrotransposon Ty3 is targeted immediately upstream of genes transcribed by polymerase III. In vitro experiments have demonstrated that this targeting involves protein-protein interactions between the integrase and the transcription factor TFIIIB or TFIIIC (19). Tn7 is directly targeted to its unique chromosomal site by the binding of the transposon-encoded TnsD protein to a site adjacent to attTn7 (1, 38). TnsD then recruits Tn7 to the target site by interacting directly with TnsC, which in turn interacts with the transposase proteins that are bound to the transposon ends (7). Tn7 can also transpose via an alternative pathway mediated by the TnsE protein. In this case, transposition is not directed to attTn7 but occurs at a low frequency to other, unrelated sites (7). Recently, it has been shown that the TnsE pathway prefers conjugative plasmids as targets (40). Through a series of elegant genetic experiments, it was shown that conjugal functions were required for this targeting, implying that transposition occurred during transfer. Furthermore, transposition into the conjugative plasmids occurred with a preferred orientation and with a distinct preference for the leading region of transfer. It was proposed that conjugal DNA synthesis in the recipient was a preferred target for transposition, and the process of replication provided the polarity to dictate the orientation of insertion.
Little is known about the target selection of most transposable elements. Either a limited number of insertions have been examined or a relatively small plasmid has been used as a target, which restricts both the availability of a preferred site and the size of the target that can be used and still give rise to a viable insertion. This paper describes an analysis of the insertion sites selected by IS903. We undertook this analysis for two reasons. First, there has been no comprehensive analysis of insertion preference for this element. Indeed, data accumulated over the years suggested that IS903 inserts randomly (9). Second, we wished to identify a target that had been used in vivo for our in vitro transposition studies. We reasoned that a DNA substrate containing a target site used by IS903 in vivo would be more likely to work in vitro. In addition, other information obtained concerning target selection might also help optimize such an in vitro system.
We show that IS903 inserts into discrete regions of the F plasmid derivative pOX38 and that within these regions there are a few sites that are used multiple times, thus allowing us to identify preferred in vivo hot spots for IS903 transposition. Alignment of the insertion sites showed that there are no preferred sequences within the target duplication but that there are distinct symmetrically located nucleotide preferences in the DNA flanking the target, implying that the transposase interacts with the target in a symmetrical fashion. IS903 also shows a very strong orientation bias for insertion into a second conjugative plasmid, pUB307. We have examined the plasmid features that are responsible for this and have shown that reversing the direction of plasmid transfer abolishes this orientation bias, suggesting that the process of conjugation directly influences IS903 transposition.
MATERIALS AND METHODS
Media and standard procedures.
Luria-Bertani (LB) medium was used for growth of all E. coli strains. The following antibiotics were used at the indicated concentrations: chloramphenicol, 20 μg/ml; kanamycin, 50 μg/ml; nalidixic acid, 40 μg/ml; ampicillin, 100 μg/ml; tetracycline, 10 μg/ml; streptomycin, 200 μg/ml. Restriction enzymes and DNA-modifying enzymes were purchased from New England Biolabs and Boehringer Mannheim and used as recommended. Oligonucleotide synthesis and DNA sequencing were performed at the molecular genetics core facilities of the Wadsworth Center. Plasmid DNA manipulations were carried out by using standard procedures (32). PCR colony screening was performed as described previously (37).
Bacterial strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. Segments of pOX38 used to assay target selection in pUB307 were generated by PCR, using the oligonucleotide primers (oKD94 to oKD97) listed in Table 2. These PCR fragments were cloned into the HindIII site of pUC118 before being recloned into the HindIII site of pUB307. Derivatives of pUB307 were isolated with each pOX38 DNA segment inserted in both orientations. The constructs were confirmed by restriction enzyme analysis, PCR analysis using appropriate pairs of fragment- and pUB307-specific primers, and in many cases DNA sequencing. A 71-bp deletion of the traY promoter (traYp) in region I (traY to traL) was made by digesting the pUC118 derivative pUCF1 with MluI and BstEII, end filling, and religating to generate pUCF2. These two sites are unique in that plasmid and flank the traYp. The 1.0-kb HindIII fragment of pUCF2 was subcloned into the HindIII site of pUB307 to generate pUBF36. To eliminate traYp activity without affecting the length of region I, point mutations were introduced in the −35 and −10 regions of the promoter in pUCF1 to generate pUCF9. The primer oKD151 was used to introduce multiple point mutations in the −35 and −10 hexamer sequences, which were predicted to eliminate transcription and to create BamHI and SphI recognition sites, respectively. The mutations were confirmed by restriction analysis and DNA sequencing before the HindIII fragment was subcloned into pUB307 to generate pUBF38. pUBF38Ω1 was generated from pUBF38 by introducing the omega fragment of pHP45Ω (30) into the HindIII site closest to the Kmr promoter of pUBF38. This is a 2-kb fragment that encodes resistance to spectinomycin and is flanked by transcriptional terminators.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference |
|---|---|---|
| Strains | ||
| DH1 | F−recA1 gyrA96 | 17 |
| RR1023 | pOX38 recA56 | 10 |
| NG135 | F−recA56 rpsL | 10 |
| Plasmids | ||
| pKD100 | pBR322 derivative carrying a model IS903 transposon; Cmr Apr Kmr | 10 |
| pKD1 | pBR322 derivative carrying a Kmr IS903 derivative | 39 |
| pOX38 | A 55-kb F derivative that is conjugative | 14 |
| pUCF1 | 1.1-kb PCR fragment (pOX38 tra region from nt 1700–2846)a inserted into pUC118 HindIII site; fragment amplified with primers oKD94 and oKD95 | This work |
| pUCF2 | pUCF1 with MluI-BstEII (end-filled) deletion of the traY promoter | This work |
| pUCF9 | Point mutations at the −35 (BamHI site created) and −10 (SphI site created) hexamers of traY promoter in pUCF1 | This work |
| RK2 | 60-kb naturally occurring conjugative plasmid; Tcr Apr Kmr | 29 |
| pRK21761 | RK2 derivative with E. coli lac operon in tetA gene and four point mutations in oriT which creates an XbaI site; Apr Kmr | 34 |
| pUB307 | RK2 derivative with ∼5.5 kb deletion of Apr Tn1; Tcr Kmr | 5 |
| pUBF4 | 0.4-kb PCR fragment (pOX38 tra region from nt 2056–2471)a cloned into the HindIII site of pUB307; fragment amplified with primers oKD96 and oKD97 | This work |
| pUBF12 | 1.1-kb HindIII fragment from pUCF1 inserted into HindIII site of pUB307 | This work |
| pUBF20 | Same as for pUBF4 except the fragment is cloned in the opposite orientation | This work |
| pUBF28 | 0.8-kb PCR fragment (pOX38 tra region from nt 1700–2471)a cloned into the HindIII site of pUB307; fragment amplified with primers oKD94 and oKD97 | This work |
| pUBF30 | Same as for pUBF28 except the fragment is cloned in the opposite orientation | This work |
| pUBF31 | 0.8-kb PCR fragment (pOX38 tra region from nt 2056–2846)a cloned into the HindIII site of pUB307; fragment amplified with primers oKD95 and oKD96 | This work |
| pUBF33 | Same as for pUBF31 except the fragment is cloned in the opposite orientation | This work |
| pUBF36 | 1.0-kb HindIII fragment from pUCF2 inserted into HindIII site of pUB307 | This work |
| pUBF38 | 1.1-kb HindIII fragment from pUCF9 inserted into HindIII site of pUB307 | This work |
| pUBF42 | Same as for pUBF12 except the fragment is cloned in the opposite orientation | This work |
| pUBF46 | 1.1-kb HindIII fragment from pUCF1 inserted into HindIII site of pUBT10 | This work |
| pUBF51 | 1.1-kb HindIII fragment from pUCF1 inserted into HindIII site of pUBT13 | This work |
| pRKF2 | Replacement of the 7.1-kb AseI-BglII fragment of RK2 with a 947-bp PCR fragment (nt 12044–12990)b containing oriV; the direction of DNA replication is opposite that of RK2 and pUB307 | This work |
| pRKF6 | Same as for pRKF2 except the oriV fragment is inserted in the native orientation | This work |
| pUBT2 | 16-kb EcoRI-AvrII fragment of pRK21761 replaced with the EcoRI-AvrII fragment of pUB307; Tcr Kmr | This work |
| pUBT10 | 273-bp PCR fragment (RK2 oriT region from nt 51123–51395)b cloned into the XbaI site of pUBT2; fragment amplified with primers oKD156 and oKD157 | This work |
| pUBT13 | Same as for pUBT10 except the oriT fragment is cloned in the opposite orientation | This work |
| pHP45Ω | Ω fragment encoding Smr/Spr gene flanked by transcription and translation termination signals in pBR322 | 30 |
| pUBF38Ω1 | HindIII Ω fragment from pHP45Ω is inserted at the HindIII site of pUBF38 between the Kmr promoter and the pOX38 fragment | This work |
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| oKD20 | GATATATTTTTATCTTGTGC |
| oKD21 | TTACGCTGACTTGACGGG |
| oKD78 | GACGAAAGCAGGTATGCCTGAAAGC |
| oKD79 | GCCCACCGGAAGGAGCTGACTGGG |
| oKD94 | CGGAATTCAAGCTTTGTGTATCATAAATACGCGT |
| oKD95 | CGGAATTCAAGCTTAACCAGGAACTGCCCCGCCC |
| oKD96 | CGGAATTCAAGCTTACCAGCTACTTATTGCAGCC |
| oKD97 | CGGAATTCAAGCTTAGTCCTTACCGAAGGTCGCC |
| oKD125 | GATGAAATGGTGAGATTGCG |
| oKD126 | CCGATTTCAGCACTTGATAT |
| oKD151 | ATCTTCTGGTTACCACTTGCATGCGCAGAATAATCAGAAAAGGATCCTAGACGGAAAG |
| oKD156 | GCTCTAGACGGTAGCCGGCCAGCCTCGC |
| oKD157 | GCTCTAGAACAAAACAGCAGGGAAGCAG |
| oKD158 | GCTCTTCGTTCGTCTGGAAGGCAG |
| oKD159 | ACTCGGCCAGCTCGTCGGTGTAGC |
| oRK2 | TAGCGCAGATTAATCGGATCCGTAATTGAGCATTTCC |
| oRK3 | TTGCGCAGATTAATTAGATCTAGCGTGGACTCAAGGC |
pRKF2 and pRKF6 contain region I (traY to traL) of pOX38 in the HindIII site of pUB307 but differ in the relative orientation of oriV. They were constructed by replacing the 7.1-kb AseI-BglII fragment of RK2 with a 947-bp PCR fragment containing the RK2 oriV region (RK2 coordinates 12044 to 12990) (36) generated by primers oRK2 and oRK3. Vegetative DNA replication in pRKF6 occurs in the same direction as that in RK2 and pUB307, while DNA replication in pRKF2 is in the opposite direction.
Plasmids pUBF46 and pUBF51 differ with respect to the orientation of oriT and were constructed in several steps. pUBT2 is a derivative of pUB307 that contains a defective oriT site subcloned from pRK21761 (34). The oriT site contains four point mutations at the nick site that inactivate oriT and introduce an XbaI site. PCR primers oKD156 and oKD157 were used to generate a 273-bp fragment containing the wild-type RK2 oriT. The fragment was cloned into the unique XbaI site of pUBT2 in both orientations to generate pUBT10 and pUBT13. The orientation of these insertions was confirmed by restriction enzyme analysis and DNA sequencing using primers oKD158 and oKD159. Region I of pOX38 was subcloned into the unique HindIII site of these derivatives to generate pUBF46 and pUBF51. The oriT of pUBF46 is in the native orientation.
Transposition and conjugation assays.
Transposon insertions into the target plasmids were obtained by a mating-out assay using E. coli NG135 as the recipient (10). To ensure that all insertion events were unique, each mating was carried out with a single colony from independent transformations of the donor strain. RR1023 was used as a donor to isolate independent insertions into pOX38 as previously described (10). In the transposition analysis using pUB307 and its derivatives as a target, DH1 was used as the donor (17). Donors and recipients were grown overnight with selection for resident plasmids and inoculated in fresh medium at a 1:20 dilution. The cells were grown to mid-log phase, and donor (0.1 ml) and recipient (1 ml) strains were mixed and resuspended in 20 μl of broth and then deposited on an LB plate for 2 h at 37°C. The mating mixture was scraped from the plate and resuspended in 0.5 ml of LB medium. Appropriate amounts of mating mixture were spread on media selective for transposition events. Conjugation and transposition frequencies were determined from at least three matings and are expressed as the number of transconjugants per donor.
RESULTS
IS903 prefers to insert into distinct regions of plasmid pOX38.
To determine whether IS903 exhibits a target preference, we have mapped the locations of independent insertions into the plasmid pOX38, which is a transfer-proficient deletion derivative of the F plasmid (14). It provides an ideal target for transposition since insertions can be selected by use of a mating-out assay, it lacks insertion sequences, it provides a large target (55 kb) that includes many nonessential genes, and the majority of its sequence is known (12, 18, 23). We used pKD100 (10) to deliver an IS903 derivative that includes genes encoding resistance to chloramphenicol (Cmr) and ampicillin (Apr) and IS903 transposase and carries the pBR322 origin of replication (Fig. 1A).
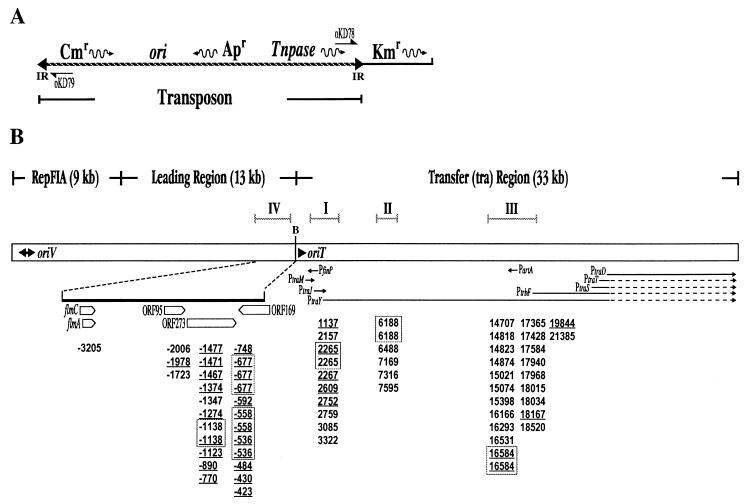
FIG. 1.
(A) Linear diagram of the transposon donor plasmid pKD100 (7.0 kb). The inverted repeats (IR) are represented by triangles. The direction of transcription of genes is indicated by wavy arrows. The transposon in pKD100 carries transposase (Tnpase), genes encoding resistance to ampicillin (Apr) and chloramphenicol (Cmr), and the pBR322 origin of replication (ori). The gene for kanamycin resistance (Kmr) is present on the plasmid backbone. Primers oKD78 and oKD79, indicated by half arrows, were used to sequence the target duplications and to identify the orientation of insertion by PCR analysis. (B) Transposon insertion sites in pOX38. The open box represents a linear map (to scale) of 55-kb pOX38, showing three regions: the RepFIA replication region containing the origin of vegetative replication (oriV), the leading region, and the transfer (tra) region. The arrowheads indicate the direction of vegetative DNA replication from oriV and the direction of conjugal DNA transfer from oriT. Arrowed lines in the polycistronic tra region indicate transcription initiating at the indicated promoters; dotted segments signify uncertainty in the extent of the transcript (12). Transposon insertion sites were determined by DNA sequencing using both primers oKD78 and oKD79. The numbers below the physical map of pOX38 represent the first nucleotide of each IS903 insertion. The target sites used more than once are boxed, and an underline indicates an insertion in the opposite orientation. Nucleotide coordinates are designated with respect to the published sequence of the tra operon which begins at the BglII site (marked B) (accession no. U01159, 33.6 kb). The leading region (accession no. M97768) upstream of oriT has been assigned negative numbers beginning from the BglII site. The thick line represents an enlargement of region IV. The arrowed boxes represent coding regions and directions of transcription of the genes in region IV.
The sites of insertion of 69 independent transposition events were determined by DNA sequence analysis of both inverted repeat-target junctions using primers that anneal within the ends of the transposon (Fig. 1A). A 9-bp target duplication was observed for all insertions. Despite the large size of pOX38, the majority of insertions (i.e., 66) mapped to four small regions of the plasmid (Fig. 1B). These were all located in the transfer region (tra) or the leading region immediately proximal to the origin of transfer (oriT). Not only was there a regional preference for insertion, but a few sites were used multiple times (Fig. 1B), implying a distinct sequence preference for insertion.
Thirty-nine insertions (57%) were clustered in three regions of the large polycistronic tra operon: region I, traJ to traE, with two insertions at position 2265; region II, traP to traR with two insertions at position 6188; region III, traN to traH with two insertions at position 16584. Twenty-six insertions (38%) mapped to a 1.6-kb segment in the leading region, adjacent to oriT. Three of the 69 insertions did not map to the four regions defined above and were located in a segment of pOX38 associated with plasmid replication (data not shown). The insertions had a noticeable orientation bias depending on their location. Those within the transfer region were inserted predominantly in one orientation, while those in the leading region (on the opposite side of oriT) were mainly inserted in the opposite orientation (Fig. 1B). The bias was not exclusive, thus ruling out inviability of one particular insertion orientation or influences of transposon sequences.
Alignment of target sites.
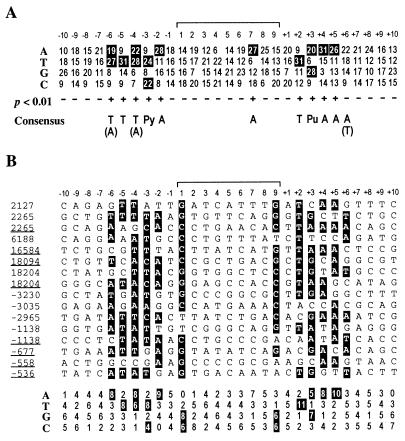
IS903 generates a 9-bp duplication, which is likely a result of a 9-bp staggered cleavage made by the transposase during integration of the transposon. The 63 unique duplications sequenced in this study were aligned to determine if there was a preferred consensus sequence for insertion. As can be seen in Fig. 2A, there are no preferred sequences found within the target duplication; however, certain nucleotides are not favored at several positions, implying that they negatively influence the selection of that target. In contrast, in the 10 bp flanking the target site duplication, there are distinct nucleotide preferences symmetrically located about the target site. The consensus for this region of symmetry is 5′-T/A-T-T/A-Py-A-3′ and extends from positions 2 to 6 on either side of the duplication. This preference is highly significant (greater than 99% confidence level) as determined by the chi-square test based on an AT content of 48% for the pOX38 plasmid. The least significant position in this consensus is at position +6, which occurred only within confidence limits of 95%. The symmetrical location of these sequences suggests either that the transposase makes base-specific symmetrical contacts with these nucleotides or that this sequence forms a unique structure on either side of the target which predisposes it to recognition, cleavage, and integration.
FIG. 2.
Alignment of target sites used by IS903. (A) Sixty-three target sites and the flanking DNA sequences have been aligned to generate the matrix shown. The sequences compiled are those adjacent to the left end of IS903 (−1 to −10) and those adjacent to the right end of IS903 (+1 to +10). The central 9 nucleotides are those that would be duplicated on insertion. The consensus sequence derived from the outlined numbers is shown below the matrix. Positions at which a nucleotide bias occurs with greater than 99% confidence levels as determined by a chi-square test are indicated by a “+” symbol. An A(T) at position +6 in the consensus occurs with 95% confidence limits. The parentheses around a nucleotide designation indicate that the preference for that base is not as strong. (B) Target and flanking sequences of sites used by IS903 more than once are shown. The data are compiled from insertions mapped in Fig. 1 and 3. Boxed nucleotides are those that match the consensus sequence in panel A and also include the preference for a G/C nucleotide at positions 1 and 9 of the target duplication. Note that three sites contained insertions in both orientations and so each orientation has been included. Numbering at the top is as for panel A.
The 13 sites that were used multiple times also match the consensus sequence (Fig. 2B). Again there is a preference for consensus bases on both sides of the insertion, reflecting a preference for symmetrical contacts. There are two differences between the general consensus (Fig. 2A) and that generated from the multiply used sites (Fig. 2B). First, it is clear that there is a strong preference for G/C nucleotides at positions 1 and 9 in the target site. Second, the preference for T/A nucleotides at position +6 in the overall consensus sequence is lost. More extensive searches on either side of the target failed to detect any significant correlation with specific host factor binding sites (e.g., IHF, Chi, DnaA, or dam methylation sites) that may have influenced target site selection.
Insertion preference is reproducible with a second IS903 derivative.
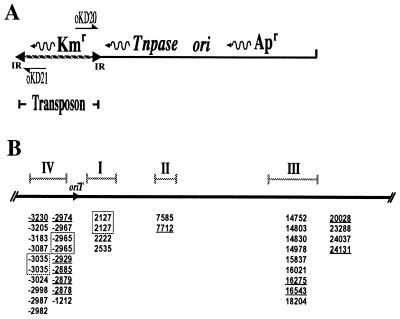
To rule out the possibility that target preference was influenced by internal sequences of the IS903 derivative in pKD100, we used a second transposon derivative carried on pKD1 (39). This transposon carries a Kmr gene flanked by 180 bp and 113 bp of DNA from the left and right ends of IS903, respectively (Fig. 3A). Thirty-eight independent transposition events were isolated by the mating-out assay, and all but four insertions mapped to the same four clusters seen previously (Fig. 3B).
FIG. 3.
(A) Linear diagram of the transposon donor plasmid pKD1. The transposon carries the Kmr gene, and the transposase gene (Tnpase) is located immediately adjacent to one end of the transposon. The direction of transcription of genes is indicated by wavy arrows. The annealing sites of oligonucleotides oKD20 and oKD21 are indicated by half arrows; they were used to determine the site of transposon insertion. IR, inverted repeat. (B) Insertion sites of IS903 from pKD1 into pOX38. Only the tra region and leading region of pOX38 are shown. Thirty-eight transposon insertion sites were determined. The majority of insertions were mapped to the same four regions shown in Fig. 1; these are indicated above the map. The numbers below the physical map of pOX38 represent the first nucleotide of each IS903 insertion. The target sites used more than once are boxed, and an underline indicates an insertion in the opposite orientation.
Transposition onto pOX38 does not occur at high frequencies during vegetative growth.
All of the experiments described above used a mating-out assay to select for transposition events. This assay prevented us from determining whether the process of conjugal transfer or transfer functions played a direct role in targeting. Therefore, we developed a second transposon delivery system that made use of a temperature-sensitive RK2-based plasmid that carries the Kmr transposon from pKD1. Transposition events could be directly selected by plating cells on media containing kanamycin at the nonpermissive temperature for the transposon donor plasmid. By using this transposon delivery system, we have monitored IS903 transposition in the presence and absence of pOX38. Transposition was not stimulated by the presence of pOX38 in the cell (data not shown). Furthermore, of 20 Kmr colonies screened, none contained insertions in pOX38, suggesting that pOX38 was not used efficiently as a target under these growth conditions. We have also been unable to detect efficient transposition onto a pUC118 derivative containing a wild-type oriT. Thus, at present, we are unable to directly test the role of conjugation in target selection by using transfer-deficient F derivatives.
IS903 insertion into pUB307 is not targeted but does occur with a unique orientation.
To determine if oriT sites in general, or proteins associated with them, were functionally “attractive” to IS903, we have examined insertion preference into a second conjugative plasmid, pUB307. pUB307 is a deletion derivative of RP1, a member of the IncP family of plasmids, and is unrelated to F (5, 29). This plasmid was chosen because conjugation could be used to select for transposition events, the entire nucleotide sequence is known (29), the conjugal transfer system is well characterized and is different from that of pOX38 (22), and the presence of a defined oriT and transfer region would allow us to make comparisons with the regional targeting seen in pOX38.
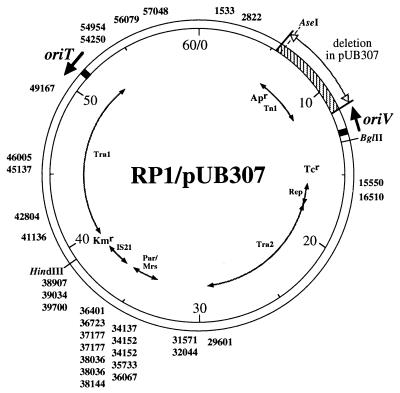
By using pKD100 as the transposon donor, 31 independent insertions in pUB307 were isolated and mapped by sequencing. The insertions were broadly distributed around the entire plasmid with the exception of a clustering in a 4-kb region around IS21 (Fig. 4). No preference for insertions around oriT was found. Thus, the very strong preference for insertion into localized regions, especially that adjacent to oriT, appears to be specific to pOX38. Notably, all the insertions seen in pUB307 were in the same orientation.
FIG. 4.
IS903 insertion sites in pUB307. Physical and genetic maps of RP1/pUB307 and the insertion sites of IS903 transposition are shown. The first nucleotides of each insertion site with respect to the RP1 sequence (Genbank database accession no. L27758) are marked around the map. The region of RP1 deleted in pUB307 (including the ampicillin-resistant transposon Tn1) is indicated by vertical bars. Genetic elements are indicated in the inner circle and include the following: the two transfer regions (Tra); genes encoding resistance to tetracycline (Tcr) and kanamycin (Kmr); the partition locus par; and the insertion sequence IS21. The unique HindIII site used for cloning pOX38 fragments is shown within the Kmr gene. The AseI and BglII sites used for inverting the oriV region are also indicated. Black boxes represent oriV and oriT; the directions of DNA replication from oriV and conjugal transfer from oriT are indicated by arrows.
All signals for targeting are present on a 1.1-kb traY-to-traL pOX38 DNA fragment.
Since IS903 insertions in pUB307 were well distributed, the plasmid was used as a vector to test the regional preference for pOX38 hot spots in a different DNA context. PCR-amplified segments of pOX38 DNA were subcloned into the unique HindIII site in the Kmr gene of pUB307 (Fig. 4). Segments of pOX38 DNA that included sites that had been used multiple times as a target were amplified from each of the preferred regions. Independent transposition events into the pUB307 derivatives containing the different pOX38 segments were generated by using a mating-out assay. Insertions into the pOX38 fragment were screened by a PCR assay that allowed insertions into the cloned fragment to be detected and their orientation to be determined. This was achieved with combinations of primers that were specific to the Kmr gene and each end of the transposon.
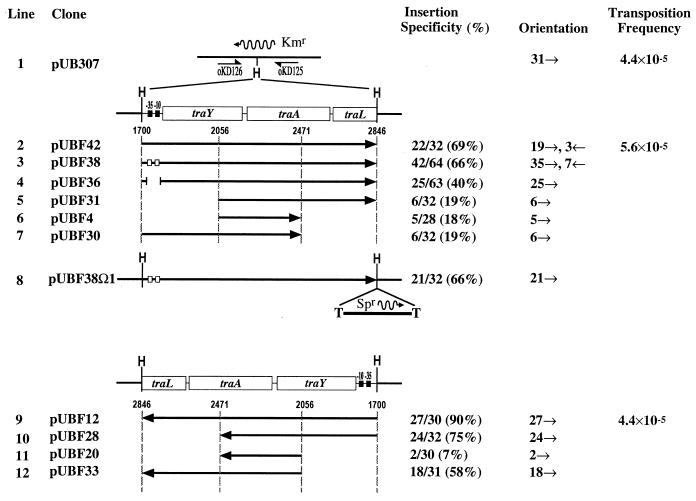
The pOX38 fragments were used as targets with varying efficiency. The most significant result was seen with a segment of pOX38 DNA spanning positions 1700 to 2846, which included most of region I (Fig. 5). Depending on the orientation of the cloned fragment, 69% (22 of 32) or 90% (27 of 30) of the insertions mapped to this segment of pOX38, indicating that it is a highly preferred target for IS903 transposition (Fig. 5, lines 2 and 9). The magnitude of this preference is emphasized by the fact that this 1.1-kb pOX38 segment represents only 2% of the available target. The other DNA fragments were used less efficiently. A segment from region III (positions 16062 to 17080) was used with varying efficiency depending on its orientation within pUB307. It was used 12 of 30 times in one orientation and not at all in the reverse orientation (0 of 30 times). pUB307 carrying a segment of DNA from the pOX38 leading region (positions −996 to −305) was not used efficiently as a target, with IS903 inserted 4 of 30 and 2 of 29 times for each orientation. The reduced targeting to these regions in pUB307 also implies that the preference for region I is not due to its being more AT rich than the vector DNA since all of these regions from pOX38 have similar AT contents. Finally, the presence of region I in pUB307 did not enhance the transposition frequency (Fig. 5); it simply changed the distribution of insertion sites. Further analysis was focused on region I in an attempt to define the important features of targeting (see below).
FIG. 5.
IS903 insertion specificity and orientation preference in region I from pOX38 when cloned into pUB307. A simple map of the Kmr gene in pUB307 is indicated in line 1. The direction of transcription of the Kmr gene is designated by the wavy arrow. Oligonucleotides oKD125 and oKD126 were used in a PCR analysis to identify IS903 insertions into the region. To determine the orientation of those insertions by PCR, oKD125 and oKD126 were used in combination with the transposon-specific primers oKD78 and oKD79 (Fig. 1). Region I and derivatives of it were cloned into the unique HindIII (H) site of pUB307 in both orientations (lines 2 to 12). The DNA present in each plasmid is shown by the solid arrow. The tra genes and their coordinates with respect to the BglII site in the pOX38 transfer region are indicated. The vertical dashed lines indicate the end points of subclones made from the traY-to-traL fragment. Filled and open boxes represent the wild-type and mutant −35 and −10 hexamers of the traY promoter, respectively. pUBF36 contains a 71-bp deletion of the traY promoter. The Ω fragment present in pUBF38Ω1 contains a gene encoding spectinomycin resistance (Spr) and is flanked by transcriptional terminators (T). Insertion specificity is defined as the number of insertions in region I divided by the total number of insertions. The orientation of IS903 insertion is indicated by small arrows (→ and ←). Line 1 shows that 31 insertions were mapped in the same orientation in pUB307 (data from Fig. 4). Transposition frequency was determined from at least three independent mating-out assays.
In these experiments and others described below (Fig. 5), the majority of insertions occurred in the same orientation. This was true for insertions within the pOX38 fragment (Fig. 5) and those in the vector backbone (data not shown). Furthermore, the orientation was not influenced by flipping the direction of the pOX38 target segment (compare upper and lower panels in Fig. 5), implying that it is an inherent property of the plasmid that imparts the bias. The ability to isolate insertions in both orientations demonstrates that viability of the flipped insertion product is not the reason for this bias.
Dissection of the traY-to-traL gene fragment.
The 1.1-kb fragment containing the first part of the tra operon (traY to traL) is clearly a preferred target for IS903, irrespective of its orientation when cloned into pUB307. In addition, target sites used within this region have been used before. For example, insertions at position 2265 of the tra region were isolated both in pOX38 and in pUB307, indicating that this is a true hot spot for IS903 insertion. To further define the critical regions required for insertion, this DNA fragment was divided into three almost equal segments. Primers were designed to amplify individual segments as well as combinations of them, and these were then subcloned into the HindIII site of pUB307. Independent transposition events into each pUB307 derivative were generated and then screened by the PCR assay for insertions into the pOX38 fragment. The PCR assay was also used to show that the majority of insertions had inserted in the same relative orientation.
In Fig. 5 we have compared the effect of deletions within the 1.1-kb traY-to-traL target fragment. In both orientations, deletion of the outer segments results in decreased targeting efficiency (Fig. 5, compare line 2 with lines 5 and 7 and line 9 with lines 10 and 12). It is also clear that although the middle segment is not as efficiently targeted as when the plasmid carries the entire region, targeting is still above that predicted for a 415-bp segment in a 55-kb plasmid. Thus, all three regions contribute to targeting. This region of pOX38 also includes the traY promoter (traYp). We examined a possible role for the traYp in targeting by constructing two derivatives of pUBF42 that eliminated transcriptional activity from this promoter as determined by a poison primer experiment (reference 21 and data not shown). Point mutations in both the −35 and −10 regions of the promoter had no significant effect on targeting (Fig. 5, line 3), and a 71-bp deletion that removed the entire promoter region reduced targeting to this region only to 40% (line 4). These derivatives thus rule out a direct role for the traYp in targeting. To rule out the additional effect of readthrough transcription from the Kmr gene promoter, a transcriptional terminator cassette carrying spectinomycin resistance (30) was cloned into the HindIII site most proximal to the Kmr promoter in pUBF38. Targeting to this transcriptionally silenced segment was unaffected (Fig. 5, line 8), and therefore a major role for transcription in targeting to this particular preferred region in pUB307 is ruled out.
Orientation of insertion is influenced by oriT.
One of the more striking results found was that IS903 insertion has a strong orientation bias (Fig. 4 and 5). This bias must result from the transposase sensing a strong polarity in the target. For pUB307, DNA replication from both oriV and oriT is unidirectional, and we reasoned that either one or both of these processes might explain the strict orientation of IS903 insertion. A simple test of this model would be to flip the orientation of oriT or oriV to see whether it affected the orientation of insertion. The ability to clone and manipulate DNA in pUB307 also made it feasible to carry out these flips in the native vector rather than with a high-copy or miniplasmid derivative. In addition, we could take advantage of the high targeting preference for the pOX38 hot spot to use a PCR-based assay to simplify the determination of the direction of transposon insertion.
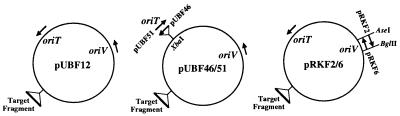
Two sets of pUB307 derivatives that differed simply in the orientation of oriV or oriT were made (Fig. 6). pUBF46 and pUBF51 contain the oriT region of RP1 inserted into an XbaI site and differ only in the relative orientation of oriT. Point mutations were introduced at the native nick site of RP1 that generated an XbaI site and reduced transfer 105-fold (reference 34 and data not shown). pRKF2 and pRKF6 differ by the relative orientation of oriV. A 0.9-kb fragment encompassing the minimal oriV region of RP1, as defined by Thomas et al. (36), was cloned in both orientations between the unique AseI and BglII sites of RP1 (Fig. 4 and 6). This cloning replaces the native oriV with the amplified fragment and, in addition, deletes the adjacent Tn1 transposon to generate a product that is essentially identical to pUB307. These plasmid derivatives were transferred and maintained at levels similar to those of pUB307, showing that we have not significantly affected plasmid replication or DNA transfer (Table 3 and data not shown).
FIG. 6.
A schematic representation of the pairs of plasmids used to monitor the effect of reversing the direction of either oriV or oriT. The native orientations of oriV and oriT are shown in pUBF12. All plasmids contain the 1.1-kb pOX38 fragment inserted in the HindIII site; this is the target for IS903 insertion.
TABLE 3.
Orientation preference of IS903 insertions in pUB307 derivatives with a flipped oriT or oriV
| Plasmida | Orientation of oriT/oriVb | Transfer frequencyc | Transposition frequencyc | Insertion specificityd | Orientation of insertionse |
|---|---|---|---|---|---|
| pUBF12 (native) | ←/← | 1.00 | 1.00 | 27/30 (90) | 0:27 |
| pUBF46 | ←/← | 0.10 | 0.10 | 52/54 (96) | 0:52 |
| pUBF51 | →/← | 0.16 | 0.27 | 55/63 (87) | 27:28 |
| pRKF2 | ←/→ | 1.15 | 0.67 | 36/39 (92) | 4:32 |
| pRKF6 | ←/← | 0.83 | 0.37 | 39/40 (98) | 2:37 |
All plasmids listed in this table are pUB307 derivatives containing the F traY-to-traL segment in the HindIII site in the same orientation (see Fig. 6).
The directions of vegetative DNA replication, oriV, and conjugal DNA transfer, oriT, are represented by arrows. The native orientation of oriV and oriT is that in pUBF12.
The transfer frequency of the control plasmid, pUBF12, is 3.51/donor cell in a 2-h mating, and transposition into this target occurs at 4.4 × 10−5/donor cell. All mating data were determined from at least three independent experiments.
Insertion specificity is defined as the number of insertions into the F segment relative to the total number of insertions. Percentage equivalents are shown in parentheses.
The numbers preceding the colons indicate insertions in one orientation which were identified by primers OKD78 and OKD126. The numbers following the colons indicate insertions in the opposite orientation, identified by primers OKD79 and OKD126 (Fig. 5).
Independent insertions into these derivatives were obtained, and the orientation of IS903 insertion was determined by PCR analysis (Table 3). Inverting the orientation of oriV and hence the direction of plasmid replication did not change the orientation bias (compare data for pRKF2 with pRKF6 in Table 3). However, changing the orientation of oriT and therefore reversing the direction of conjugal transfer had a dramatic effect on the orientation of insertions: IS903 now inserted without an orientation bias (compare data for pUBF46 with pUBF51 in Table 3). Target preference was unaffected, as shown by the fact that the segment of pOX38 in these derivatives was used as a target about 90% of the time.
DISCUSSION
Target preference.
Despite the availability of a large target (>55 kb) for transposon insertion, we have shown that IS903 prefers to insert into very discrete regions within pOX38. About half of the insertions were found in three small regions in the tra operon. This was a surprising result, in that our selection for transposition relied on transfer proficiency, and suggests that many of the events occurred during, or immediately prior to, DNA transfer since many of the insertions result in a transfer-deficient phenotype (data not shown). The remaining insertions were all located in the leading region of pOX38 transfer, immediately adjacent to oriT. These same regions were used as targets in multiple experiments using different IS903 derivatives. This rules out influences of transposon sequences and implies that these regions are intrinsically attractive as IS903 targets. In particular, region I was used efficiently as a target in both pOX38 and pUB307, implying that it contains all the information necessary for targeting. The fact that a precise region for targeting could not be defined by a simple deletion analysis (Fig. 5) suggests that there are multiple sequence components throughout the region that all contribute to targeting. The lack of an obvious association of host-factor binding sites with insertion sites in this region and the insensitivity to plasmid backbone context suggest that region I is capable of forming a structure or domain that makes it more accessible to transposition. In addition, by eliminating transcription through this region in pUB307, we demonstrated that it does not play a major role in targeting to region I.
Regions II to IV were not used as efficiently as targets when subcloned into pUB307, suggesting that their use in pOX38 was context dependent. As for region I, regions II to IV may adopt unusual structural conformations or domains that favor transposon integration, but their formation may require more extensive regions of pOX38 that were not included when subcloned into pUB307. Regional targeting has been observed with other transposable elements. Mu inserts into preferred regions which are binding sites for the MuB protein (26). Superimposed on this regional targeting, the MuA protein is thought to direct insertion into sites that contain the degenerate consensus Y-G/C-R. Also, IS1 has been observed to insert primarily into a relatively AT-rich sequence of pBR322 that contains multiple binding sites for IHF (41). Thus, it was suggested that a combination of IHF-induced bends in an AT-rich region of DNA may enhance the use of this region as a target. Coincidentally, a strong orientation bias was observed in these experiments but was not investigated further.
When pOX38 was used as a target for IS903, almost 50% of the insertions were located in the leading region of transfer (Fig. 1). However, when this region was subcloned into pUB307, it was not used as a preferred target, suggesting that it is the specific context of pOX38 that makes the leading region a preferred target. The close proximity to the oriT site and the influence of oriT on orientation (Table 3) make it tempting to speculate that oriT or a transfer-related process plays a role in this targeting. The leading region of pOX38 is a preferred target for Tn7 when transposing via the TnsE pathway, and conjugation is required for this targeting (40). In fact, the targeting of IS903 insertions to the leading region bears a striking resemblance to that seen with Tn7 when transposing via the TnsE pathway. In that study, 8 of 18 Tn7 insertions were found in the same two open reading frames (ORFs) of the leading region, ORF 273 and ORF 169. The other Tn7 insertions mapped in pOX38 were distributed throughout the transfer region with a slight preference for a region at the distal end of the tra operon. Unfortunately, in the absence of a mating assay, we could not detect transposition onto pOX38 or an oriT-containing plasmid. This precluded us from using transfer-deficient mutants of pOX38 to determine genetically the role conjugation plays in targeting IS903 to this region.
A preferred target site.
The availability of a large collection of insertions allowed us to examine the target sites for a consensus sequence. Although no consensus sequence could be found for the 9-bp target duplication, assessment of the flanking sequences showed that there is a preferred consensus sequence found on either side of the target duplication (5′-T/A-T-T/A-Py-A-3′). A similar consensus detected on both sides of the duplication was maintained when sites that had been used more than once were aligned (Fig. 2B). The most significant difference observed when the multiply used sites were compared with all the target sites was the very strong preference for G/C nucleotides at the first and last positions of the target duplication. The arrangement of symmetrical sequences is consistent with a dimer or multimer of transposase, in a complex with the ends of the transposon, recognizing a target by making specific, symmetrical contacts outside the target duplication. These flanking sequences could be recognized in two ways: (i) by base-specific contacts with the transposase or (ii) as a specific structural conformation in the DNA favored by those sequences. We note that there are two potential matches to the consensus sequence within the inverted repeat of IS903 (8), which, if significant, would provide a rational explanation for the presence of this sequence around insertion sites. In preliminary band shift experiments, we have been unable to detect binding of transposase to a segment containing a preferred target (data not shown). However, this might simply be because it is only recognized by the transposase when in the form of an active transpositional integration complex. Clarification of the significance of these sequences will require more detailed genetic and especially biochemical analyses using an in vitro transposition system.
Although this consensus represents an average of many targets, and no site matches the consensus perfectly, these sequences may be predisposed for use as targets by causing the transpososome to pause as it tracks along the DNA searching for a suitable site for integration. Similar contacts to a symmetrically located sequence on the other side of the potential 9-bp target by a second transposase molecule would stabilize the transposon-target interactions further and would increase the probability of integration before continuing the search. Clearly, a preferred insertion site alone is not sufficient for targeting as shown by our deletion analysis of region I (Fig. 5). Presumably, the initial search is influenced by the regional preference and once the transposon has gained access to the DNA, it would scan for consensus sites.
IS10 has a clearly defined consensus target sequence, found within the target duplication, to which the transposase is assumed to make base-specific contacts in the major groove (16). However, mutagenic studies have shown that flanking sequences can dramatically influence use of the target site (4). No consensus sequence or symmetry was detected in the DNA flanking the IS10 target sites, and so it was proposed that the helical structure of the flanking DNA influenced the use of the consensus target site. IS231 also has a preferred target site, which is influenced by the ability of flanking sequences to adopt an unusual S-shaped structure (15).
Orientation of insertion.
One of the more striking observations made in this work was that there was a distinct orientation preference for insertions in pUB307. A similar effect has been seen with Tn7 insertions into both pOX38 (40) and the parent of pUB307, RP1 (2, 3). To generate such a profound bias, there has to be an asymmetry in both the transposon donor and target; otherwise, insertion would occur equally in both orientations. For IS903, the asymmetry could arise if one end was presented in a more-or-less active form. For example, transcription across one end may reduce its activity, or delivery of the cis-acting transposase (or host factor) to one end may place that end in a more active conformation relative to the other end (11).
This asymmetric transposase end complex must then sense a polarity in the target DNA. Two obvious candidates for generating this polarity are transcription and replication, since they both would have influences on the entire plasmid. Several observations argue against its being transcription. First, we see insertions in both orientations within the same transcriptional unit. Second, the orientation of insertion is unaffected by flipping the pOX38 fragment in pUB307. Therefore, a likely candidate is DNA replication. The orientation bias of Tn7 insertion has been attributed to the polarity conferred by conjugal DNA replication. Our inability to detect IS903 insertions into pOX38 in the absence of transfer selection precludes us from directly testing whether the same is true for IS903. So, we have tested the influence of replication in a different way, by creating pUB307 derivatives that carry either an inverted oriT or oriV region. A prediction of this experiment is that if transposition is sensing either one of these replication processes, there should be a direct correlation between orientation and the direction of DNA replication (vegetative or conjugal).
As shown in Table 3, reversing the direction of DNA replication initiated from oriV had no effect on the orientation of transposon insertions. However, flipping the direction of conjugal transfer abolished the orientation bias completely and provides strong evidence for the process of conjugation playing a distinct role in IS903 integration. One surprising aspect of the experiment was that the orientation bias was abolished rather than reversed. This suggests that conjugal DNA replication per se is not the only determining factor influencing orientation of insertion. It is possible that by inverting oriT, we have uncoupled its communication with a second cis-acting sequence on the plasmid that together with oriT (or oriT-associated functions) imparted a polarity to the plasmid that was recognized by the transpososome. A second possibility is that changing the direction of DNA transfer results in a global disruption of the genomic topological and structural organization of the plasmid such that the polarity of the target is not reversed but disrupted. This could result from interference with other plasmid processes such as transcription and DNA replication or the disruption of supercoiling domains in the plasmid. Clearly, these and other possible explanations will require a more detailed experimental approach.
Many of the insertions into pOX38 result in transfer-defective plasmids (data not shown) and thus must have occurred just prior to, or during, conjugation. The absence of transpositional jackpots in a mating experiment also implies that transposition into a conjugative plasmid occurs at a late stage of the assay. These two indirect observations, combined with the sensitivity to the orientation of conjugal transfer, imply that there is a close connection between conjugation and IS903 transposition. Directing transposition to a conjugative plasmid would provide IS903 with the ideal opportunity to spread horizontally through a population.
ACKNOWLEDGMENTS
We thank David Figurski for pRK21761 and advice concerning the minimal regions required for oriT and oriV function in pUB307. We gratefully acknowledge the use of the Wadsworth Center’s molecular genetics core facilities. Finally, we thank Joan Curcio, Vicky Derbyshire, and members of the Derbyshire laboratory for their critical comments on the manuscript.
This work was supported by grant GM50699 from the National Institutes of Health.
REFERENCES
- 1.Bainton R J, Kubo K M, Feng J N, Craig N L. Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell. 1993;72:931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- 2.Barth P T, Grinter N J. Map of plasmid RP4 derived by insertion of transposon C. J Mol Biol. 1977;113:455–474. doi: 10.1016/0022-2836(77)90233-9. [DOI] [PubMed] [Google Scholar]
- 3.Barth P T, Grinter N J, Bradley D E. Conjugal transfer system of plasmid RP4: analysis by transposon 7 insertion. J Bacteriol. 1978;133:43–52. doi: 10.1128/jb.133.1.43-52.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender J, Kleckner N. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. Proc Natl Acad Sci USA. 1992;89:7996–8000. doi: 10.1073/pnas.89.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett P M, Grinsted J, Richmond M H. Transposition of TnA does generate deletions. Mol Gen Genet. 1977;154:205–211. doi: 10.1007/BF00330839. [DOI] [PubMed] [Google Scholar]
- 6.Craig N L. Target site selection in transposition. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 7.Craig N L. Transposon Tn7. Curr Top Microbiol Immunol. 1996;204:27–48. doi: 10.1007/978-3-642-79795-8_2. [DOI] [PubMed] [Google Scholar]
- 8.Derbyshire K M, Grindley N D. Binding of the IS903 transposase to its inverted repeat in vitro. EMBO J. 1992;11:3449–3455. doi: 10.1002/j.1460-2075.1992.tb05424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derbyshire, K. M., and N. D. F. Grindley. Unpublished data.
- 10.Derbyshire K M, Hwang L, Grindley N D. Genetic analysis of the interaction of the insertion sequence IS903 transposase with its terminal inverted repeats. Proc Natl Acad Sci USA. 1987;84:8049–8053. doi: 10.1073/pnas.84.22.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derbyshire K M, Kramer M, Grindley N D. Role of instability in the cis action of the insertion sequence IS903 transposase. Proc Natl Acad Sci USA. 1990;87:4048–4052. doi: 10.1073/pnas.87.11.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frost L S, Ippen-Ihler K, Skurray R A. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev. 1994;58:162–210. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galas D J, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 14.Guyer M S, Reed R R, Steitz J A, Low K B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harbor Symp Quant Biol. 1981;45:135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- 15.Hallet B, Rezsohazy R, Mahillon J, Delcour J. IS231A insertion specificity: consensus sequence and DNA bending at the target site. Mol Microbiol. 1994;14:131–139. doi: 10.1111/j.1365-2958.1994.tb01273.x. [DOI] [PubMed] [Google Scholar]
- 16.Halling S M, Kleckner N. A symmetrical six-basepair target site sequence determines Tn10 insertion specificity. Cell. 1982;28:155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 18.Helsberg M, Eichenlaub R. Twelve 43-base-pair repeats map in a cis-acting region essential for partition of plasmid mini-F. J Bacteriol. 1986;165:1043–1045. doi: 10.1128/jb.165.3.1043-1045.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirchner J, Connolly C M, Sandmeyer S B. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science. 1995;267:1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- 20.Kleckner N, Chalmers R M, Kwon D, Sakai J, Bolland S. Tn10 and IS10 transposition and chromosome rearrangements: mechanism and regulation in vivo and in vitro. Curr Top Microbiol Immunol. 1996;204:49–82. doi: 10.1007/978-3-642-79795-8_3. [DOI] [PubMed] [Google Scholar]
- 21.Kohrer K, Kutchan T M, Domdey H. Specific oligodeoxynucleotide probes obtained through RNA sequencing. DNA. 1989;8:143–147. doi: 10.1089/dna.1.1989.8.143. [DOI] [PubMed] [Google Scholar]
- 22.Lanka E, Wilkins B M. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 23.Loh S, Cram D, Skurray R. Nucleotide sequence of the leading region adjacent to the origin of transfer on plasmid F and its conservation among conjugative plasmids. Mol Gen Genet. 1989;219:177–186. doi: 10.1007/BF00261174. [DOI] [PubMed] [Google Scholar]
- 24.Mizuuchi K. Transpositional recombination: mechanistic insights from studies of Mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- 25.Mizuuchi M, Baker T A, Mizuuchi K. Assembly of the active form of the transposase-Mu DNA complex: a critical control point in Mu transposition. Cell. 1992;70:303–311. doi: 10.1016/0092-8674(92)90104-k. [DOI] [PubMed] [Google Scholar]
- 26.Mizuuchi M, Mizuuchi K. Target site selection in transposition of phage Mu. Cold Spring Harbor Symp Quant Biol. 1993;58:515–523. doi: 10.1101/sqb.1993.058.01.058. [DOI] [PubMed] [Google Scholar]
- 27.Muller H P, Varmus H E. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994;13:4704–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naigamwalla D Z, Chaconas G C. A new set of Mu DNA transposition intermediates: alternate pathways of target capture preceding strand transfer. EMBO J. 1997;16:5227–5234. doi: 10.1093/emboj/16.17.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pansegrau W, Lanka E, Barth P T, Figurski D H, Guiney D G, Haas D, Helinski D R, Schwab H, Stanisich V A, Thomas C M. Complete nucleotide sequence of Birmingham IncPα plasmids: compilation and comparative analysis. J Mol Biol. 1994;239:623–663. doi: 10.1006/jmbi.1994.1404. [DOI] [PubMed] [Google Scholar]
- 30.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 31.Sakai J, Kleckner N. The Tn10 synaptic complex can capture a target DNA only after transposon excision. Cell. 1997;89:205–214. doi: 10.1016/s0092-8674(00)80200-7. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. pp. 1.1–1.110. [Google Scholar]
- 33.Sandmeyer S B, Hansen L J, Chalker D L. Integration specificity of retrotransposons and retroviruses. Annu Rev Genet. 1990;24:491–518. doi: 10.1146/annurev.ge.24.120190.002423. [DOI] [PubMed] [Google Scholar]
- 34.Sia E A, Kuehner D M, Figurski D H. Mechanism of retrotransfer in conjugation: prior transfer of the conjugative plasmid is required. J Bacteriol. 1996;178:1457–1464. doi: 10.1128/jb.178.5.1457-1464.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Signon L, Kleckner N. Negative and positive regulation of Tn10/IS10-promoted recombination by IHF: two distinguishable processes inhibit transposition off of multicopy plasmid replicons and activate chromosomal events that favor evolution of new transposons. Genes Dev. 1995;9:1123–1136. doi: 10.1101/gad.9.9.1123. [DOI] [PubMed] [Google Scholar]
- 36.Thomas C M, Stalker D M, Helinski D R. Replication and incompatibility properties of segments of the origin region of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181:1–7. doi: 10.1007/BF00338996. [DOI] [PubMed] [Google Scholar]
- 37.Trower M K. A rapid PCR-based colony screening protocol for cloned inserts. Methods Mol Biol. 1996;58:329–333. doi: 10.1385/0-89603-402-X:329. [DOI] [PubMed] [Google Scholar]
- 38.Waddell C S, Craig N L. Tn7 transposition: recognition of the attTn7 target sequence. Proc Natl Acad Sci USA. 1989;86:3958–3962. doi: 10.1073/pnas.86.11.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinert T A, Derbyshire K M, Hughson F M, Grindley N D. Replicative and conservative transpositional recombination of insertion sequences. Cold Spring Harbor Symp Quant Biol. 1984;49:251–260. doi: 10.1101/sqb.1984.049.01.029. [DOI] [PubMed] [Google Scholar]
- 40.Wolkow C A, DeBoy R T, Craig N L. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 1996;10:2145–2157. doi: 10.1101/gad.10.17.2145. [DOI] [PubMed] [Google Scholar]
- 41.Zerbib D, Gamas P, Chandler M, Prentki P, Bass S, Galas D. Specificity of insertion of IS1. J Mol Biol. 1985;185:517–524. doi: 10.1016/0022-2836(85)90068-3. [DOI] [PubMed] [Google Scholar]