Abstract
The four antibiotics produced by Streptomyces coelicolor are all affected by mutations in the absA and absB loci. The absA locus encodes a putative two-component signal transduction system, and the absB locus encodes a homolog of Escherichia coli RNase III. We assessed whether these loci control synthesis of the antibiotics actinorhodin and undecylprodigiosin by regulating transcript abundance from the biosynthetic and regulatory genes specific for each antibiotic. Strains that were Abs− (for antibiotic synthesis deficient) due to mutations in absA or absB were examined. In the Abs− absA mutant strain, transcripts for the actinorhodin biosynthetic genes actVI-ORF1 and actI, and for the pathway-specific regulatory gene actII-ORF4, were substantially lower in abundance than in the parent strain. The level of the transcript for the undecylprodigiosin pathway-specific regulatory gene redD was similarly reduced in this mutant. Additionally, a strain that exhibits precocious hyperproduction of antibiotics (Pha phenotype) due to disruption of the absA locus contained elevated levels of the actVI-ORF1, actII-ORF4, and redD transcripts. In the absB mutant strain, actVI-ORF1, actI, actII-ORF4, and redD transcript levels were also substantially lower than in the parent strain. These results establish that the abs genes affect production of antibiotics through regulation of expression of the antibiotic-specific regulatory genes in S. coelicolor.
Bacteria of the genus Streptomyces produce most of the antibiotics currently used in medicine. Bacterial cultures synthesize these compounds as secondary metabolites such that production follows the rapid growth phase of liquid-grown cultures or is coupled to sporulation of plate-grown cultures. Despite the many years of research on antibiotics driven by their commercial importance, the regulatory mechanisms responsible for antibiotics’ growth phase regulation are poorly understood.
Streptomyces coelicolor, the streptomycete best understood genetically, produces four biochemically and genetically distinct antibiotics: actinorhodin (Act [48]), undecylprodigiosin (Red [18, 48]), methylenomycin (Mmy [38, 56]), and calcium-dependent antibiotic (CDA [28]). The gene clusters responsible for Act (39), Red (18, 40), CDA (15a), and Mmy (13) synthesis have been cloned. The act cluster is the best characterized of these and consists of at least six transcripts and 20 open reading frames (ORFs). Transcription of the act biosynthetic genes is regulated by a cluster-linked regulatory gene, actII-ORF4 (19). The red cluster also contains a regulator, redD (42, 52), which is in turn regulated by another cluster-linked regulator, redZ (54). The sequences of ActII-ORF4 and RedD have considerable similarity at the amino acid level and appear to contain an OmpR-like DNA-binding fold (55). Both actII-ORF4 and redD are growth phase regulated; their transcripts accumulate at approximately the onset of stationary phase (25, 52). Shortly thereafter, accumulation of biosynthetic gene transcripts is seen (25, 52). ActII-ORF4 and RedD have been called “pathway-specific regulators,” and the evidence accumulated thus far indicates that they are positive regulators for the respective biosynthetic genes. For example, strains carrying mutant genes fail to accumulate biosynthetic gene transcripts and fail to cosynthesize Act (reviewed in reference 14) or Red (18, 49), respectively, with other act or red mutant classes. Moreover, overexpression of actII-ORF4 or redD early in a culture’s growth leads to early biosynthetic gene transcription and antibiotic production (25, 52). Thus, growth-phase-regulated transcription of antibiotic-specific regulators is one aspect of antibiotics’ temporal regulation.
A genetic analysis of S. coelicolor antibiotic regulation that sought to identify genes potentially involved in global temporal regulation of antibiotics defined the absA (2) and absB (1) loci. Both loci were defined by mutations that caused a phenotype of global loss of antibiotic synthesis (Abs− [for antibiotic synthesis deficient]). Hence these loci mutated to phenotypes that suggested that they encode global regulators of antibiotic synthesis.
The sequence of absA is predicted to encode a two-component signal transduction system composed of AbsA1, a putative sensor-transmitter, and AbsA2, a putative DNA-binding response regulator (5). The original collection of Abs− mutant strains (2), which was obtained from UV mutagenesis, carried mutations of the absA1 gene (5). In contrast, disruptions of the absA1A2 genes result in a phenotype of early-onset enhanced antibiotic production (Pha [for precocious hyperproduction of antibiotics]) (5). Hence, we hypothesize that the absA locus plays a negative regulatory role in antibiotic production and that the Abs− absA strains are mutationally locked into a negatively acting mode of regulation (5).
The sequence of absB is predicted to encode a homolog of Escherichia coli RNase III, a double-stranded-RNA-specific endonuclease (45). RNase III processes a number of mRNAs of E. coli and coliphage and thus regulates expression of numerous genes (17, 43). E. coli RNase III is also involved in rRNA processing, but this is a nonessential activity (17, 43).
A number of additional loci that are relevant to the regulation of S. coelicolor’s antibiotics have been identified. The serine-threonine-tyrosine phosphotransfer system-encoding afsRKS locus stimulates Act and Red synthesis when in high copy, while disruptions of genes encoded by the locus lead to medium-dependent reductions of Act, Red, and CDA (23, 27, 31–34, 53). The recently described cutRS putative two-component system appears to be a negative regulator of Act, since gene disruption causes overproduction of that antibiotic (11). The afsQ1Q2 genes, which encode another putative two-component system, stimulate Act and Red (36). Other, less-well-characterized loci influence one, two, or three of the antibiotics: afsB (affecting Act and Red [26]), abaA (Act, Red, and CDA [20]), and the abaB (Act and Red [51]) and “Romero” sequences (Act [47]). Synthesis of the compound (p)ppGpp by the relA gene product has been tied to the induction of antibiotic synthesis (reviewed in reference 4). RNA polymerase sigma factors are also potentially important to the regulation of antibiotic synthesis, but the sigma factor(s) recognizing antibiotic gene promoters has not been definitively specified. The sigma factor ςhrdD transcribes actII-ORF4 and redD in vitro but is dispensible in vivo. The essential vegetative sigma factor ςhrdB (7, 24) is a strong candidate for in vivo transcription. Finally, streptomycete antibiotic production is temporally coupled to sporulation, and numerous genetic loci named bld can mutate to a phenotype characterized by loss of both antibiotic production and sporulation (reviewed in references 10 and 30).
The mechanisms by which the above-mentioned genes influence antibiotic production have not been well defined, but some types of mutants blocked in Act and Red production have been evaluated for actII-ORF4 and redD transcription. bldA encodes the only tRNA of S. coelicolor that can efficiently translate the rare leucine codon UUA, which is found in the actII-ORF4 gene and the redZ-encoded regulator of redD. Hence, bldA mutants are Red− because of diminished redD transcription (54) but are Act− because of defective actII-ORF4 translation (19).
Some strains carrying mutations in the afsRK locus, including a deletion mutant of afsR (23), show a reduction in transcription of biosynthetic transcripts for Act but no effects on the transcripts for actII-ORF4 or redD. Finally, some strains with mutations in relA, which encodes (p)ppGpp synthetase, are affected in actinorhodin and undecylprodigiosin production with accompanying defects in actII-ORF4 and redD transcription (8, 41).
Here, we report a characterization of the regulation of act and red transcripts by the absA and absB loci. Using S1 nuclease protection assays, we show that both absA and absB are regulators of antibiotic biosynthetic gene expression and, moreover, are global regulators of expression of the antibiotic pathway-specific activators.
MATERIALS AND METHODS
Bacterial strains.
The following S. coelicolor A3(2) strains were used: J1501 (15) and its derivatives C542 (absA542 [2]), C120 (absB120 [1]), J1501/KC900 (actI::KC900 [5, 6]), C542/KC900, and C120/KC900.
XylE enzyme assays.
Growth conditions and assay techniques for the KC900 lysogens were as described previously (35) except that XylE enzyme activity was assayed on R5-thiostrepton plates. The KC900 phage (6) contains a fragment internal to the actI coding region and creates lysogens through homologous recombination with actI. Thus, it creates a single-copy transcriptional fusion of the actI promoter to the reporter gene xylE (6). Color development was visually evaluated 1 h after spraying with catechol.
Growth conditions and RNA isolation.
S. coelicolor strains were grown for RNA isolation on 8.5-cm-diameter cellophane disks (Cannings Packaging Ltd., Bristol, England) placed on plate media. Disks were washed by autoclaving twice for 15 min in 1 to 2 liters of distilled water. Two media were used for RNA isolation: a mannitol minimal medium (9) and a peptone-glucose (PGA) medium (0.5% peptone, 1.0% glucose, 2.2% agar [pH 7.1] [modified from reference 16]). The experiment shown in Fig. 1 used mannitol minimal medium, but other experiments used PGA medium because it allowed reproducible production of both Act and Red by cultures grown on cellophane disks. A variety of other medium formulations were surveyed, but they were not used, because only one antibiotic was produced. In some cases, certain media supported antibiotic production only in the absence of cellophane disks. Approximately 105 spores were spread onto each disk, followed by incubation at 30°C. RNA was harvested at time points postinoculation chosen to span the initiation of antibiotic synthesis; these are stated for each experiment. In a 4°C cold room, growth from each of 8 to 10 disks was scraped into a tube containing 5 ml of chilled modified Kirby mixture (29) and 14 g of 4-mm-diameter glass beads. Tube contents were alternately mixed vigorously for 30 s by vortex mixer and incubated on ice for 30 s, for four cycles. Samples were then processed as previously described for Northern blot RNA preparation (29). RNA quality was tested by gel electrophoresis and ethidium bromide staining; little or no degradation of rRNA was evident.
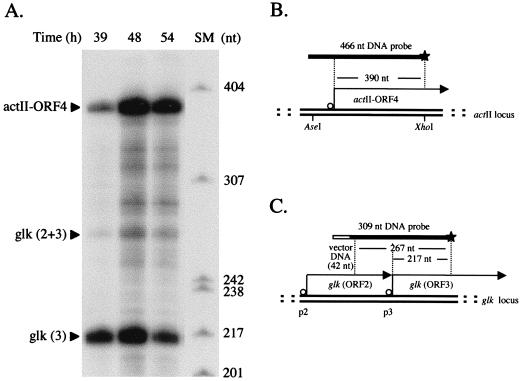
FIG. 1.
Growth-phase-dependent expression of actII-ORF4 mRNA in plate-grown J1501. (A) RNA isolated from mannitol minimal medium plates at the times indicated was used in S1 nuclease protection analyses. Size markers (SM), 32P-labeled MspI-digested pBR322. (B) The probe for the actII-ORF4 gene was end labeled at the XhoI site (indicated by a star; see Materials and Methods) and should yield a protected fragment of 390 nt (19, 25). (C) The uniquely end-labeled probe for glk was generated by PCR amplification (see Materials and Methods) and should yield two protected fragments of 267 and 217 nt that correspond to transcripts initiated at promoters p2 and p3, respectively (3).
S1 nuclease protection assays.
For each assay, 50 μg of RNA was dried down with the appropriate 32P-labeled DNA probes (30,000 or 100,000 cpm of restriction digest-generated or PCR-generated probes, respectively). Pellets were suspended in 20 μl of 80% formamide buffer (29) by pipetting and vortex mixing. Tubes were placed in a water bath that was kept at 85°C for 10 min then allowed to cool to 57°C overnight. Samples were processed as described previously (29) except as follows: S1 nuclease buffer was that described by Sambrook et al. (50); isopropanol precipitations were incubated on ice for 30 min; and final pellets were dissolved in standard sequencing gel-loading buffer (50), boiled for 2 min, and electrophoresed on denaturing 6% polyacrylamide sequencing gels (50). Size markers were purchased from New England Biolabs, Beverly, Mass. In concert with the experiments shown, control experiments in which samples of total RNA were hybridized with twice the probe concentration used in other lanes were performed; the control signals were not significantly different from corresponding experimental signals, confirming that probes were present in excess (data not shown). Control experiments in which 50 μg of yeast tRNA replaced experimental total RNA samples were performed with each set of S1 nuclease protection assays; no signals resulted (data not shown). Radioactivity on gels was quantitated with an AMBIS Radioanalytic Imaging System and AMBIS Quantprobe, version 3.0, software (AMBIS, Inc., San Diego, Calif.). Bands on autoradiograph films were quantitated by densitometry with a Molecular Dynamics (Sunnyvale, Calif.) computing densitometer and ImageQuant, version 3.0, software. A glk probe was generated by PCR amplification with the labeled 3′ primer 5′-GATGCCCACTGCGACGATCT-3′ and the unlabeled 5′ primer 5′-CCAGATCTGCAGCCAAGCTT-3′ to produce a 309-bp fragment that included the glk promoter region (3). The PCR template was pIJ2423, which carries the 1.2 kb SmaI-(BclI)-HindIII glk fragment from pIJ2420 (3) blunt-ended and cloned into the SmaI site of pIJ2925 (37). Two glk signals were seen; those labeled glk (3) represent transcript from the glk promoter, and those labeled glk (2 + 3) represent readthrough from an upstream promoter. In most time courses, the average glk band intensities were comparable for the J1501 and abs strains. In some time courses, e.g., that shown in Fig. 2, the glk band intensities dropped at very late times in all strains. The transcript abundances in abs mutant strains relative to the J1501 parent were determined by comparing the band intensities of the antibiotic gene transcripts in question to the glk band intensities. An actII-ORF4 probe was made from a 466-bp XhoI/AseI DNA fragment that included the promoter region of actII-ORF4 (25). The fragment was uniquely labeled at the 5′ end of the XhoI site with [32P]ATP by using T4 polynucleotide kinase. The actVI-ORF1 probe was generated by PCR amplification of a sequence that included the promoter region of that gene (22) by using the end-labeled 3′ oligonucleotide primer 5′-ACGTCCGGCTCGTACTCGATG-3′ and the unlabeled 5′ primer 5′-CTTGCGGTGGAAGTCCTCCAG-3′. The PCR template was a 4.5-kb BamHI act fragment (sites 1 to 3 in reference 22). A redD probe was made with the 3′ oligonucleotide primer 5′-ACAGTTCGTCCACCAGGTCCGCGA-3′ (end labeled before use) in the PCR with the unlabeled 5′ primer 5′-TGCTTCGTTTGCGTCGTTCAGTTC-3′ to generate a 497-bp fragment that included the redD promoter region (42, 52). The PCR template was the 2.1-kb redD fragment in pCLL38 (42) cloned into pUC18. The PCR mix contained 1× PCR buffer lacking MgCl2 (Perkin-Elmer), 2 mM MgCl2, 1% glycerol, 0.2 to 0.4 mM (each) deoxynucleoside triphosphate, 20 pmol of each primer, 100 ng of template DNA, and 2.5 U of Taq polymerase (Perkin-Elmer). Formamide at 2% (vol/vol) was included for redD amplification. Samples were subjected to 29 (actVI-ORF1 and glk) or 36 (redD) cycles of 3 min at 95°C, 2 min at 65°C, and 1 min at 72°C, and a final extension of 10 min at 72°C. The 497-nucleotide (nt) redD product was further purified from contaminating PCR products by agarose gel electrophoresis and isolation.
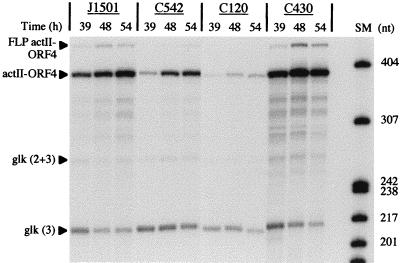
FIG. 2.
Expression of actII-ORF4 mRNA in J1501 (abs+), C542 (absA), C120 (absB), and C430 (absA disruption). RNA was isolated from PGA plate-grown cultures at the times indicated and used for S1 nuclease protection analyses. The size markers (SM) used were as described in the legend to Fig. 1. The probes used for actII-ORF4 and glk were the same as for Fig. 1. “FLP actII-ORF4” indicates the position of full-length probe; transcriptional readthrough from the upstream promoter for actII-ORF3 likely contributes to the signal as well (19, 25). This signal intensity varied from experiment to experiment but, when present, followed the same kinetics as the labeled actII-ORF4.
RESULTS
Regulation of the Act-specific regulator actII-ORF4 by the absA and absB loci.
Because the Abs− mutant phenotypes suggested that absA and absB were regulatory loci, transcription of antibiotic genes was assessed in absA and absB mutant strains and in the parental strain J1501 (abs+). The expression profile of act transcripts has been especially well characterized (25), and current evidence indicates that actII-ORF4 is the most directly acting regulator of the Act biosynthetic transcripts (19, 25, 44). The actII-ORF4 transcript is growth phase regulated in liquid-grown cultures of S. coelicolor, its level increasing greatly in the transition and stationary phases (25).
Figure 1A shows that plate-grown cultures of strain J1501 (Abs+) also exhibit temporal regulation of actII-ORF4 transcription, as assessed by S1 nuclease protection analysis of isolated RNA. In this experiment, Act was made visibly at 48 hours, approximately coordinated with sporulation.
The nuclease protection experiments included an assessment of the S. coelicolor glucose kinase transcript level. The glucose kinase gene (glk [ORF3] in Fig. 1C) is expressed throughout the growth of S. coelicolor from two promoters (3), referred to as p2 and p3 in Fig. 1C. The abundance of transcript from p2 is much lower than that from p3. Each S1 assay included an assay for glk mRNA, providing an internal control for in vivo RNA levels and the S1 procedure. The interpretations of relative transcript levels in the experiments discussed below reflect adjustments based on this control (see Materials and Methods).
Figure 2 shows a comparison of actII-ORF4 transcript levels in J1501, C542 (absA), and C120 (absB). In this time course, Act was produced by J1501 at about 42 h. For this experiment, and those following, PGA plate medium was used because J1501 reproducibly produced both Act and Red when grown with PGA on cellophane disks.
In comparison to J1501, actII-ORF4 transcript abundance in C542 was substantially reduced over the course of the experiment. The maximum level in C542, observed at 54 h in this experiment, was approximately 25% of the level in J1501. In four similar experiments involving two independent time courses of RNA isolation, the actII-ORF4 transcript levels were decreased approximately three- to sixfold in C542 (data not shown).
Figure 2 also shows that, in comparison to J1501, actII-ORF4 transcript abundance was reduced about 12-fold in the absB strain C120. In four similar experiments involving two independent time courses of RNA isolation, the actII-ORF4 transcript levels were decreased approximately two- to sixfold in C120 (data not shown).
Note that the abundance of the signal at the position of the actII-ORF4 probe generally follows the same pattern as that indicating probe protection due to transcription from the actII-ORF4 promoter. This most likely reflects coregulation of the upstream transcript (actII-ORF3 [19]) with actII-ORF4.
Regulation of Act biosynthetic genes by the absA and absB loci.
To assess the extent to which the reduced actII-ORF4 expression in strains C542 and C120 affected expression of act biosynthetic genes, two representative act transcripts were studied: actVI-ORF1, proposed to encode a dehydrogenase catalyzing an early reductive step in Act synthesis (22), and actI (ORF1 and ORF2), encoding components of the polyketide synthase that assembles the Act carbon backbone (21). The temporal expression of one of these, actVI-ORF1, has been well characterized in liquid culture, and the transcript is seen to accumulate as the culture enters stationary phase (25). In this work, transcript levels from actVI-ORF1 were monitored in the parental strain J1501 and in mutant strains C542 and C120. Expression of an actI transcriptional reporter gene fusion was also monitored.
In the experiment shown in Fig. 3A, the level of actVI-ORF1 transcript in the C542 (absA) mutant strain was reduced approximately sixfold over the time course of the experiment in comparison to J1501. Thus, the reduction in actVI-ORF1 transcript levels paralleled the reduction in actII-ORF4 transcript levels seen in the same time course of RNA isolation in Fig. 2A. In each of two similar assays of an independent RNA time course, actVI-ORF1 transcript levels were decreased more than eightfold in C542 (data not shown).
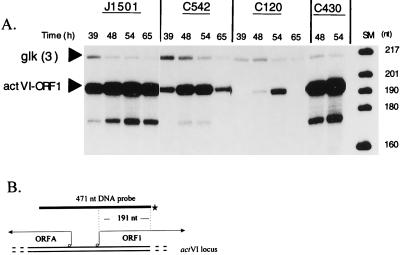
FIG. 3.
Expression of actVI-ORF1 mRNA in J1501 (abs+), C542 (absA), C120 (absB), and C430 (absA disruption). RNA was isolated from PGA plate-grown cultures at the times indicated and used for S1 nuclease protection analyses with a probe for the actVI-ORF1 transcript that should yield a protected fragment of 191 nt (22, 25). (A) The secondary bands below the actVI-ORF1 bands (approximately 167 nt), also obtained by other researchers using this probe (25), are of unknown origin; they may represent a secondary initiation site, a degradation product, or an artifact of the procedure. The glk probe was as described in the legend to Fig. 1. The size markers (SM) were as described for Fig. 1. (B) The uniquely end-labeled probe used for the actVI-ORF1 transcript is described in Materials and Methods. ORFA has been implicated in Act synthesis, but its role is unknown (22).
Figure 3A also shows that the levels of the actVI-ORF1 transcript were approximately 12-fold lower over the time course in the C120 (absB) strain than in J1501. In two similar assays of actVI-ORF1 transcript in an independent RNA time course, 12-fold- and 25-fold-lower amounts of actVI-ORF1 transcript were observed in C120 (data not shown).
To monitor transcription from another act promoter, actI, a xylE reporter gene fusion was used. The actinophage clone KC900 was used to produce a transcriptional fusion of xylE to the actI chromosomal promoter (see Materials and Methods). Fusions were constructed in the parental strain J1501 and in C542 (absA) and C120 (absB). J1501 strongly expressed the XylE product after 2 days of plate culture on R5 medium (5), while C542 and C120 did not visibly express XylE product during the 5-day time course (data not shown).
Regulation of expression of the Red-specific regulator redD by the absA and absB loci.
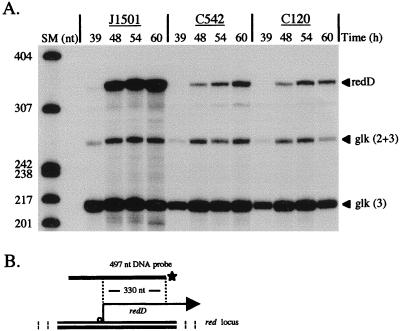
Like Act production, production of Red is growth phase regulated, initiating in the transition phase of liquid-grown cultures and continuing in stationary phase. The regulation of Red production depends, at least in part, on the growth-phase-regulated expression of the Red antibiotic-specific regulator redD, which is seen to appear in the transition phase of liquid-grown cultures of S. coelicolor M145 (52). Figure 4 shows redD expression in a time course in which Red was visibly detectable at 48 h. Figure 4A shows that plate-grown cultures of J1501 also exhibit temporal regulation of redD and that the C542 (absA) culture exhibited approximately sevenfold-reduced levels of redD transcript. Figure 4A also shows that redD transcript abundance was reduced approximately sevenfold in C120 (absB). Transcripts encoding biosynthetic genes have not been characterized, and red biosynthetic gene transcription was not assessed in this study.
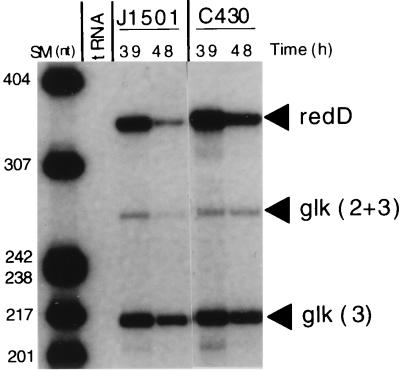
FIG. 4.
Expression of redD mRNA in J1501 (abs+), C542 (absA), and C120 (absB). RNA was isolated from PGA plate-grown cultures at the times indicated and used in S1 nuclease protection assays with a probe for the redD transcript that should yield a protected fragment of 330 nt (45, 52). (A) Bands labeled “redD” represent transcript from the redD gene. The glk probe was as described for Fig. 1. Size markers (SM) were as described for Fig. 1. (B) The uniquely end-labeled probe for the redD transcript is described in Materials and Methods.
Effect of a disruption mutation of the absA locus on expression of actII-ORF4, actVI-ORF1, and redD transcripts.
In contrast to the Abs− (antibiotic-deficient) phenotype of strain C542, another absA mutant strain, C430, displays a Pha phenotype (for precocious hyperproduction of antibiotics). Pha mutant strains such as C430 produce Act and Red 6 to 12 h earlier than J1501 and produce five- to eightfold-more Act and Red. This phenotype is associated with certain disruption mutations of the absA locus (5); strain C430 carries a disrupted absA2 gene due to insertion of phage φC31 DNA (5).
In previous work, use of an actI::xylE chromosomal fusion to assess the expression of the actI biosynthetic gene in an absA-disrupted Pha strain demonstrated significant XylE activity 6 to 12 h earlier than in J1501, and the activity reached a fourfold-higher peak (5). In Fig. 2A, 3A, and 5, all representing experiments using RNA from the same time course, strain C430 was evaluated for expression of the actII-ORF4, actVI-ORF1, and redD transcripts, respectively. C430 expressed all of these transcripts at approximately twofold-higher levels than J1501 at the times shown. In this time course, Red was produced earlier than in the experiment shown in Fig. 4A, appearing by 30 h in J1501. redD transcript abundance has been seen to drop at later times in previous experiments (23) and here does so in both the J1501 and C430 strains.
FIG. 5.
Expression of redD mRNA in J1501 (abs+) and C430 (absA disruption). RNA was isolated from PGA plate-grown cultures at the times indicated and used in S1 nuclease protection assays. The probes for redD and glk were as described for Fig. 4.
DISCUSSION
The results of this work indicate that the absA and absB gene products regulate expression of act and red antibiotic gene transcripts through regulation of the respective antibiotic-specific regulator genes. Previous work had established that S. coelicolor Act and Red production is limited by accumulation of sufficient quantities of the actII-ORF4- and redD-encoded regulators (25, 52). Hence, this work demonstrates that an important aspect of global regulation of antibiotic production is absA- and absB-mediated regulation of antibiotic pathway-specific regulators.
The visible phenotypes of both absA and absB Abs− strains were tight under the conditions of incubation used in these experiments, with little or no antibiotic produced after 5 days of incubation. However, the mutant effects exerted on the antibiotic gene transcripts were partial, with variability observed in different time courses, and so the amount of transcript accumulated in the Abs− strains was higher relative to that in J1501 than was the comparable amount of antibiotic detectably produced by these strains. One factor contributing to the amount of transcript seen in the C542 strain may be the presence in the cultures of a subpopulation of antibiotic-overproducing hyphae that result from sab (for suppressor of abs) suppressor mutations, which are spontaneously accumulating second-site suppressor mutations that restore antibiotic production. Such Act and Red pigment-overproducing absA sab mutants exist in a frequency high enough to cause extensive visible speckling of plate-grown cultures (46). It is not clear at this time whether the absA and absB mutations cause additional blocks to antibiotic production besides their effects on transcript levels as reported here, but the previous observation (10) that extra cloned copies of actII-ORF4 or redD are sufficient to restore Act or Red production, respectively, to the mutants suggests that reduced actII-ORF4 and redD expression is the critical limitation to antibiotic production.
Despite the Abs− mutants’ effects on antibiotic transcript levels, it is noteworthy that the temporal profile of mRNA expression is not perturbed in the mutants. This observation would be compatible with a primary role for the abs gene products in maximizing antibiotic gene expression rather than in determining its timing. An alternative view of the data involving the Abs− absA strain is that the signals evident at later times reflect, at least in part, antibiotic production in the cultures’ sab-suppressed subpopulation, as discussed above.
Although both Act and Red are subject to regulation by absA and absB, differences in the two antibiotics’ temporal profiles are evident. For example, in the course of these studies, Red was visibly produced about 12 h earlier than Act in all time courses. However, the timing of the increases in actII-ORF4 transcript levels were similar to the increases of the redD transcript levels. Thus, the visible accumulation of Act lagged well behind the detection of act transcripts, whereas Red was detectable within hours of redD accumulation. Some studies have seen even greater lags between transcription and production (23); clearly, more factors responsible for the temporal profiles of these antibiotics remain to be elucidated.
In addition to their effects on the antibiotics Act and Red, both the absA and absB mutations also abolish production of the antibiotics Mmy and CDA (1, 2). Determination of the effects of absA and absB on mmy and cda regulation awaits characterization of the mmy and cda transcripts.
This work does not determine whether the absA and absB mutants’ effects on message levels occur at the level of promoter usage or of transcript stability. Recent results (45) indicate that the S. coelicolor absB gene encodes a homolog of E. coli RNase III, a double-strand-specific RNase. It is tempting to speculate that an absB-encoded RNase III activity exerts control over antibiotic gene expression through posttranscriptional regulation of specific target genes. Definition of a structural or sequence motif recognized by E. coli RNase III has proven difficult (17, 43), so it is not currently feasible to use precedent from E. coli to predict whether potential RNase III targets are associated with the act and red genes studied here.
The absA locus encodes a putative negatively regulating two-component signal transduction system in which the absA1 gene encodes a protein with similarity to the sensor-transmitter class and the adjacent absA2 gene encodes a response regulator. It was the observation of an increase in antibiotics seen in absA-disrupted Pha strains that led to the hypothesis (5) that the absA1A2 genes exert negative control over antibiotic gene expression. The increase in message levels seen in the C430 strain in this study lends support to this hypothesis. However, this work does not address the issue of whether AbsA2 acts directly as a repressor of actII-ORF4 and redD or whether the observed negative regulation involves additional proteins. It is also not known what signal AbsA1 senses.
Current evidence does not determine whether the absA1A2 and absB genes function in the same or different pathways or whether they function in concert with or independently of other antibiotic regulators, such as afsRKS, afsQ1Q2, abaA, abaB, cutRS, and relA (4, 10, 12). Further analysis of the relationships of the absA- and absB-encoded products to these genes should provide significant information about the network of elements controlling antibiotic production.
ACKNOWLEDGMENTS
We thank M. J. Bibb, J. White, and E. Takano for plasmids and helpful discussions and K. Chater and J. Feitelson for plasmids and phage.
This work was supported by NSF grants MCB9206068, MCB9306676, and MCB9604055 to W.C.C. D.J.A. received support from NSF grant DEB9120006 to the Center for Microbial Ecology at Michigan State University.
REFERENCES
- 1.Adamidis T, Champness W. Genetic analysis of absB, a Streptomyces coelicolor locus involved in global antibiotic regulation. J Bacteriol. 1992;174:4622–4628. doi: 10.1128/jb.174.14.4622-4628.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamidis T, Riggle P, Champness W. Mutations in a new Streptomyces coelicolor locus which globally block antibiotic biosynthesis but not sporulation. J Bacteriol. 1990;172:2962–2969. doi: 10.1128/jb.172.6.2962-2969.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angell S, Schwarz E, Bibb M J. The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol Microbiol. 1992;6:2833–2844. doi: 10.1111/j.1365-2958.1992.tb01463.x. [DOI] [PubMed] [Google Scholar]
- 4.Bibb M. 1995 Colworth Prize Lecture. The regulation of antibiotic production in Streptomyces coelicolor A3(2) Microbiology. 1996;142:1335–1344. doi: 10.1099/13500872-142-6-1335. [DOI] [PubMed] [Google Scholar]
- 5.Brian P, Riggle P J, Santos R A, Champness W C. Global negative regulation of Streptomyces coelicolor antibiotic synthesis mediated by an absA-encoded putative signal transduction system. J Bacteriol. 1996;178:3221–3231. doi: 10.1128/jb.178.11.3221-3231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruton C J, Guthrie E P, Chater K F. Phage vectors that allow monitoring of transcription of secondary metabolism genes in Streptomyces. Bio/Technology. 1991;9:652–656. doi: 10.1038/nbt0791-652. [DOI] [PubMed] [Google Scholar]
- 7.Buttner M J, Lewis C G. Construction and characterization of Streptomyces coelicolor A3(2) mutants that are multiply deficient in the nonessential hrd-encoded RNA polymerase sigma factors. J Bacteriol. 1992;174:5165–5167. doi: 10.1128/jb.174.15.5165-5167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraburtty R, Bibb M. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J Bacteriol. 1997;179:5854–5861. doi: 10.1128/jb.179.18.5854-5861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Champness W C. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J Bacteriol. 1988;170:1168–1174. doi: 10.1128/jb.170.3.1168-1174.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Champness W C, Chater K F. The regulation and integration of antibiotic production and morphological differentiation in Streptomyces spp. In: Piggot P, Moran C P Jr, Youngman P, editors. Regulation of bacterial differentiation. Washington, D.C: American Society for Microbiology; 1994. pp. 61–94. [Google Scholar]
- 11.Chang H-M, Chen M-Y, Shieh Y-T, Bibb M J, Chen C W. The cutRS signal transduction system of Streptomyces lividans represses the biosynthesis of the polyketide antibiotic actinorhodin. Mol Microbiol. 1996;21:1075–1085. [PubMed] [Google Scholar]
- 12.Chater K F, Bibb M J. Regulation of bacterial antibiotic production. In: Kleinkauf H, von Dohren H, editors. Products of secondary metabolism. Biotechnology. Vol. 6. Weinheim, Germany: VCH; 1997. pp. 57–105. [Google Scholar]
- 13.Chater K F, Bruton C J. Mutational cloning in Streptomyces and the isolation of antibiotic production genes. Gene. 1983;26:67–78. doi: 10.1016/0378-1119(83)90037-9. [DOI] [PubMed] [Google Scholar]
- 14.Chater K F, Hopwood D A. Antibiotic biosynthesis in Streptomyces. In: Hopwood D A, Chater K F, editors. Genetics of bacterial diversity. London, England: Academic Press; 1989. pp. 129–150. [Google Scholar]
- 15.Chater K F, Bruton C J, King A A, Suarez J E. The expression of Streptomyces and Escherichia drug resistance determinants cloned into the Streptomyces φC31. Gene. 1982;19:21–32. doi: 10.1016/0378-1119(82)90185-8. [DOI] [PubMed] [Google Scholar]
- 15a.Chong P D, Podmore S M, Kieser H M, Redenbach M, Turgay K, Marahiel M, Hopwood D A, Smith C P. Physical identification of a chromosomal locus encoding biosynthetic genes for the lipopeptide calcium-dependent antibiotic (CDA) of Streptomyces coelicolor A3(2) Microbiology. 1998;144:193–199. doi: 10.1099/00221287-144-1-193. [DOI] [PubMed] [Google Scholar]
- 16.Coco E A, Narva K E, Feitelson J S. New classes of Streptomyces coelicolor A3(2) mutants blocked in undecylprodigiosin (Red) biosynthesis. Mol Gen Genet. 1991;227:28–32. doi: 10.1007/BF00260702. [DOI] [PubMed] [Google Scholar]
- 17.Court D. RNA processing and degradation by RNAseIII. In: Belasco J G, Brawerman G, editors. Control of messenger RNA stability. San Diego, Calif: Academic Press, Inc.; 1993. pp. 71–116. [Google Scholar]
- 18.Feitelson J S, Malpartida F, Hopwood D A. Genetic and biochemical characterization of the red gene cluster of Streptomyces coelicolor A3(2) J Gen Microbiol. 1985;131:2431–2441. doi: 10.1099/00221287-131-9-2431. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Moreno M A, Caballero J L, Hopwood D A, Malpartida F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell. 1991;66:769–780. doi: 10.1016/0092-8674(91)90120-n. [DOI] [PubMed] [Google Scholar]
- 20.Fernández-Moreno M A, Martín-Triana A J, Martínez E, Niemi J, Kieser H M, Hopwood D A, Malpartida F. abaA, a new pleiotropic regulatory locus for antibiotic production in Streptomyces coelicolor. J Bacteriol. 1992;174:2958–2967. doi: 10.1128/jb.174.9.2958-2967.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez-Moreno M A, Martinez E, Boto L, Hopwood D A, Malpartida F. Nucleotide sequence and deduced functions of a set of cotranscribed genes of Streptomyces coelicolor A3(2) including the polyketide synthase for the antibiotic actinorhodin. J Biol Chem. 1992;267:19278–19290. [PubMed] [Google Scholar]
- 22.Fernandez-Moreno M A, Martinez E, Caballero J L, Ichinose K, Hopwood D A, Malpartida F. DNA sequence and functions of the actVI region of the actinorhodin biosynthetic gene cluster of Streptomyces coelicolor A3(2) J Biol Chem. 1994;269:24854–24863. [PubMed] [Google Scholar]
- 23.Floriano B, Bibb M. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2) Mol Microbiol. 1996;21:385–396. doi: 10.1046/j.1365-2958.1996.6491364.x. [DOI] [PubMed] [Google Scholar]
- 24.Fujii T, Gramajo H C, Takano E, Bibb M J. redD and actII-ORF4, pathway-specific regulatory genes for antibiotic production in Streptomyces coelicolor A3(2), are transcribed in vitro by an RNA polymerase holoenzyme containing ςhrdD. J Bacteriol. 1996;178:3402–3405. doi: 10.1128/jb.178.11.3402-3405.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gramajo H C, Takano E, Bibb M J. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol Microbiol. 1993;7:837–845. doi: 10.1111/j.1365-2958.1993.tb01174.x. [DOI] [PubMed] [Google Scholar]
- 26.Hara O, Horinouchi S, Uozumi T, Beppu T. Genetic analysis of A-factor synthesis in Streptomyces coelicolor A3(2) and Streptomyces griseus. J Gen Microbiol. 1983;129:2939–2944. doi: 10.1099/00221287-129-9-2939. [DOI] [PubMed] [Google Scholar]
- 27.Hong S-K, Kito M, Beppu T, Horinouchi S. Phosphorylation of the AfsR product, a global regulatory protein for secondary-metabolite formation in Streptomyces coelicolor A3(2) J Bacteriol. 1991;173:2311–2318. doi: 10.1128/jb.173.7.2311-2318.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopwood D A, Wright H M. CDA is a new chromosomally-determined antibiotic from Streptomyces coelicolor A3(2) J Gen Microbiol. 1983;129:3575–3579. doi: 10.1099/00221287-129-12-3575. [DOI] [PubMed] [Google Scholar]
- 29.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 30.Hopwood D A, Chater K F, Bibb M J. Genetics of antibiotic production in Streptomyces coelicolor A3(2) In: Vining L C, Stuttard C, editors. Regulation and biochemistry of antibiotic production. Newton, Mass: Butterworth-Heinemann; 1995. pp. 71–108. [DOI] [PubMed] [Google Scholar]
- 31.Horinouchi S, Beppu T. Production in large quantities of actinorhodin and undecylprodigiosin induced by afsB in Streptomyces lividans. Agric Biol Chem. 1984;48:2131–2133. [Google Scholar]
- 32.Horinouchi S, Beppu T. Regulation of secondary metabolism and cell differentiation in Streptomyces: A-factor as a microbial hormone and the AfsR protein as a component of a two-component regulatory system. Gene. 1992;115:167–172. doi: 10.1016/0378-1119(92)90555-4. [DOI] [PubMed] [Google Scholar]
- 33.Horinouchi S, Hara O, Beppu T. Cloning of a pleiotropic gene that positively controls biosynthesis of A-factor, actinorhodin, and prodigiosin in Streptomyces coelicolor A3(2) and Streptomyces lividans. J Bacteriol. 1983;155:1238–1248. doi: 10.1128/jb.155.3.1238-1248.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horinouchi S, Kito M, Nishiyama M, Furuya K, Hong S-K, Miyake K, Beppu T. Primary structure of AfsR, a global regulatory protein for secondary metabolite formation in Streptomyces coelicolor A3(2) Gene. 1990;95:49–56. doi: 10.1016/0378-1119(90)90412-k. [DOI] [PubMed] [Google Scholar]
- 35.Ingram C, Brawner M, Youngman P, Westpheling J. xylE functions as an efficient reporter gene in Streptomyces spp.: use for the study of galP1, a catabolite-controlled promoter. J Bacteriol. 1989;171:6617–6624. doi: 10.1128/jb.171.12.6617-6624.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishizuka H, Horinouchi S, Kieser H M, Hopwood D A, Beppu T. A putative two-component regulatory system involved in secondary metabolism in Streptomyces spp. J Bacteriol. 1992;174:7585–7594. doi: 10.1128/jb.174.23.7585-7594.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janssen G R, Bibb M J. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene. 1993;124:133–134. doi: 10.1016/0378-1119(93)90774-w. [DOI] [PubMed] [Google Scholar]
- 38.Kirby R, Hopwood D A. Genetic determination of methylenomycin synthesis by the SCP1 plasmid of Streptomyces coelicolor A3(2) J Gen Microbiol. 1977;98:239–252. doi: 10.1099/00221287-98-1-239. [DOI] [PubMed] [Google Scholar]
- 39.Malpartida F, Hopwood D A. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. Nature. 1984;309:462–464. doi: 10.1038/309462a0. [DOI] [PubMed] [Google Scholar]
- 40.Malpartida F, Niemi J, Navarrete R, Hopwood D A. Cloning and expression in a heterologous host of the complete set of genes for biosynthesis of the Streptomyces coelicolor antibiotic undecylprodigiosin. Gene. 1990;93:91–99. doi: 10.1016/0378-1119(90)90141-d. [DOI] [PubMed] [Google Scholar]
- 41.Martinez-Costa O H, Arias P, Romero N M, Parro V, Mellado R P, Malpartida F. A relA/spoT homologous gene from Streptomyces coelicolor A3(2) controls antibiotic biosynthetic genes. J Biol Chem. 1996;271:10627–10634. doi: 10.1074/jbc.271.18.10627. [DOI] [PubMed] [Google Scholar]
- 42.Narva K E, Feitelson J S. Nucleotide sequence and transcriptional analysis of the redD locus of Streptomyces coelicolor A3(2) J Bacteriol. 1990;172:326–333. doi: 10.1128/jb.172.1.326-333.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicholson A W. Escherichia coli ribonucleases: paradigms for understanding cellular RNA metabolism and regulation. In: D’Alessio G, Riordan J F, editors. Ribonucleases: structures and functions. San Diego, Calif: Academic Press, Inc.; 1997. pp. 1–49. [Google Scholar]
- 44.Passantino R, Puglia A-M, Chater K. Additional copies of the actII regulatory gene induce actinorhodin production in pleiotropic bld mutants of Streptomyces coelicolor A3(3) J Gen Microbiol. 1991;137:2059–2064. [Google Scholar]
- 45.Price, B., and W. Champness. Unpublished data.
- 46.Riggle, P., and W. Champness. Unpublished data.
- 47.Romero N M, Parro V, Malpartida F, Mellado R P. Heterologous activation of the actinorhodin biosynthetic pathway in Streptomyces lividans. Nucleic Acids Res. 1992;20:2767–2772. doi: 10.1093/nar/20.11.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rudd B A M, Hopwood D A. Genetics of actinorhodin biosynthesis by Streptomyces coelicolor A3(2) J Gen Microbiol. 1979;114:35–43. doi: 10.1099/00221287-114-1-35. [DOI] [PubMed] [Google Scholar]
- 49.Rudd B A M, Hopwood D A. A pigmented mycelial antibiotic in Streptomyces coelicolor: control by a chromosomal gene cluster. J Gen Microbiol. 1980;119:333–340. doi: 10.1099/00221287-119-2-333. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Scheu A-K, Martinez E, Soliveri J, Malpartida F. abaB, a putative regulator for secondary metabolism in Streptomyces. FEMS Microbiol Lett. 1997;147:29–36. doi: 10.1111/j.1574-6968.1997.tb10216.x. [DOI] [PubMed] [Google Scholar]
- 52.Takano E, Gramajo H C, Strauch E, Andres N, White J, Bibb M J. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2) Mol Microbiol. 1992;6:2797–2804. doi: 10.1111/j.1365-2958.1992.tb01459.x. [DOI] [PubMed] [Google Scholar]
- 53.Vogtli M, Chang P-C, Cohen S N. afsR2: a previously undetected gene encoding a 63-amino-acid protein that stimulates antibiotic production in Streptomyces lividans. Mol Microbiol. 1994;14:643–654. doi: 10.1111/j.1365-2958.1994.tb01303.x. [DOI] [PubMed] [Google Scholar]
- 54.White J, Bibb M. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J Bacteriol. 1997;179:627–633. doi: 10.1128/jb.179.3.627-633.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wietzorrek A, Bibb M. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol. 1997;25:1181–1185. doi: 10.1046/j.1365-2958.1997.5421903.x. [DOI] [PubMed] [Google Scholar]
- 56.Wright L F, Hopwood D A. Identification of the antibiotic determined by the SCP1 plasmid of Streptomyces coelicolor A3(2) J Gen Microbiol. 1976;95:96–106. doi: 10.1099/00221287-95-1-96. [DOI] [PubMed] [Google Scholar]