Abstract
Agrobacterium tumefaciens 1D1609, which was originally isolated from alfalfa (Medicago sativa L.), contains genes that increase competitive root colonization on that plant by reducing the accumulation of alfalfa isoflavonoids in the bacterial cells. Mutant strain I-1 was isolated by its isoflavonoid-inducible neomycin resistance following mutagenesis with the transposable promoter probe Tn5-B30. Nucleotide sequence analysis showed the transposon had inserted in the first open reading frame, ifeA, of a three-gene locus (ifeA, ifeB, and ifeR), which shows high homology to bacterial efflux pump operons. Assays on alfalfa showed that mutant strain I-1 colonized roots normally in single-strain tests but was impaired significantly (P ≤ 0.01) in competition against wild-type strain 1D1609. Site-directed mutagenesis experiments, which produced strains I-4 (ifeA::gusA) and I-6 (ifeA::Ω-Tc), confirmed the importance of ifeA for competitive root colonization. Exposure to the isoflavonoid coumestrol increased β-glucuronidase activity in strain I-4 21-fold during the period when coumestrol accumulation in wild-type cells declined. In the same test, coumestrol accumulation in mutant strain I-6 did not decline. Expression of the ifeA-gusA reporter was also induced by the alfalfa root isoflavonoids formononetin and medicarpin but not by two triterpenoids present in alfalfa. These results show that an efflux pump can confer measurable ecological benefits on A. tumefaciens in an environment where the inducing molecules are known to be present.
Agrobacterium tumefaciens causes crown gall tumors on a wide range of dicotyledonous plants by colonizing wounded tissues and transferring oncogenes into the plant genome (15). Successful interaction between A. tumefaciens and target cells of the host plant depends on the capacity of the bacteria to elude deleterious plant defense compounds that can slow growth. In the rhizosphere, A. tumefaciens also must compete effectively with other microorganisms as it colonizes the root (38).
Alfalfa roots release a wide variety of molecules, including many flavonoids (29). Isoflavonoids, a subgroup of flavonoids, have been studied both for their negative effects on microorganisms and for their role as inducers of nodulation genes in symbiotic Rhizobium, Bradyrhizobium, and Sinorhizobium spp. (8). Alfalfa roots are specifically known to store glucosides of the isoflavonoids formononetin, coumestrol, and medicarpin (42), as well as saponins of the hydrophobic triterpenoids hederagenin and medicagenic acid (21). Root exudates from this species have been shown to contain various forms of coumestrol, formononetin, and medicarpin (7, 19). One can therefore ask whether bacteria which colonize alfalfa roots have evolved any particular mechanism either to use or to avoid these compounds.
The first A. tumefaciens naturally infective on alfalfa was isolated recently from a crown gall. This strain, designated 1D1609, is exceptionally virulent on alfalfa (27). Studies with strain 1D1609 showed that virulence on alfalfa depended not only on the Ti plasmid but also on undefined chromosomal loci that were absent in other A. tumefaciens strains. These loci could play many roles, but it is reasonable to postulate their involvement with factors known to be present in the alfalfa rhizosphere. We hypothesized these loci might confer some beneficial interaction with isoflavonoids exuded from alfalfa roots. To test this hypothesis, a mutant bank of A. tumefaciens 1D1609 was generated with the transposable promoter probe Tn5-B30, which carries a promoterless nptII gene (40). Screening for flavonoid- and isoflavonoid-inducible neomycin resistance found one particularly responsive locus which is identified here as an isoflavonoid efflux pump involved in the competitive colonization of alfalfa roots.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
Strains and plasmids used in this study are listed in Table 1. A. tumefaciens strains were grown in AB mineral medium (5) or LB medium (37) at 30°C with shaking (200 rpm). D1M agar, a selective medium for A. tumefaciens, contained (per liter) 5 g of cellobiose, 3 g of K2HPO4, 1 g of NaH2PO4, 1 g of NH4Cl, 0.3 g of MgSO4 · 7H2O, 10 mg of malachite green, and 15 g of agar. Escherichia coli strains were grown in LB medium. When appropriate, media were supplemented with tetracycline (2 μg/ml for A. tumefaciens and 10 μg/ml for E. coli), neomycin (80 μg/ml), ampicillin (50 μg/ml), or gentamicin (25 μg/ml). Standard chemicals, reagents, and antibiotics were purchased from Fisher Scientific (Santa Clara, Calif.) or Sigma Chemical Co. (St. Louis, Mo.).
TABLE 1.
Strains, plasmids, and transposons
| Strain, plasmid, or transposon | Characteristics | Source or reference |
|---|---|---|
| A. tumefaciens | ||
| 1D1609 | Wild-type alfalfa isolate, Kmr Spr | 27 |
| I-1 | ifeA::Tn5-B30 mutant of 1D1609 | This study |
| I-4 | ifeA::gusA mutant of 1D1609 | This study |
| I-6 | ifeA::Ω-Tc mutant of 1D1609 | This study |
| E. coli | ||
| DH5α | recA1 ΔlacZ | Gibco BRL |
| HB101 | pRK2013 helper strain for matings | Stratagene |
| S17-1 | Modified RP4 integrated into genome | 39 |
| VCS257 | Host strain for cosmid libraries | Stratagene |
| Plasmids and transposons | ||
| pUC19 | Multicopy cloning vector | Gibco BRL |
| pBSK+ | Multicopy cloning vector | Stratagene |
| pJQ200mp18 | pACYC184ori, mob sacB Gmr | 33 |
| pRK2013 | pRK212.2 derivative for matings | 11 |
| pSUP205 | pBR325 derivative, mob λcos Cmr Tcr | 39 |
| Tn5-B30 | pSUP102 with transposable nptII promoter probe, Tcr | 40 |
| Ω-Tc | pUT::mini-Tn5Tc, Tcr | 9 |
| pCAM140 | mini-Tn5SSgusA40, promoterless gusA | 44 |
| pAC107 | pSUP205 with 1D1609 ifeABR locus | This study |
| pMC11 | pSUP205 with I-1 ifeA::Tn5-B30 locus | This study |
| pMC115 | pUC19 with SalI fragment upstream of Tn5-B30 from pMC11 | This study |
| pMC113 | pUC19 with SalI fragment downstream of Tn5-B30 from pMC11 | This study |
| pAC1073+ | pBSK+ with 5.2-kb EcoRI ifeABR′ fragment of pAC107 | This study |
| pAC1073− | pBSK+ with 5.2-kb EcoRI ifeABR′ fragment from pAC107 | This study |
| pAC1074 | pJQ200mp18 with 0.9-kb EcoRI-SalI fragment of pAC1073+ | This study |
| pAC1075tet | pAC1074::Ω-Tc | This study |
| pAC1075gus | pAC1074::gusA | This study |
| pAC1079 | pUC19 with 7.5-kb PstI ifeABR fragment of pAC107 | This study |
| p18Not | NotI-EcoRI-SalI-HindIII-NotI as MCS in pUC18 | 13 |
Unless noted otherwise, flavonoids were purchased from Spectrum Chemical MFG Corp. (Gardena, Calif.). Coumestrol was purchased from Eastman Kodak Co. (Rochester, N.Y.). The 4,4′-dihydroxy-2′-methoxychalcone was synthesized (6). Medicarpin and the triterpenoids hederagenin and medicagenic acid were isolated as glycosides from mature alfalfa roots, hydrolyzed to aglycones, and purified by high-pressure liquid chromatography (21, 22, 26, 36). All compounds obtained by purification or synthesis were confirmed by nuclear magnetic resonance and mass spectrometry against published values. Flavonoids, isoflavonoids, and triterpenoids were prepared as 2.5 mM stock solutions in 100% methanol.
Transposon mutagenesis and screening.
Mutants were constructed by conjugating pSUP102::Tn5-B30 from E. coli S17-1 into A. tumefaciens 1D1609 under standard conditions (39). Transposon mutants were selected on D1M agar containing 2 μg of tetracycline per ml and maintained in microtiter plates in AB medium with the same antibiotic. Inducibility of the nptII promoter probe from Tn5-B30 was screened in mutants by testing for differential neomycin resistance in the presence or absence of isoflavonoids and flavonoids. Mutants were replicated from microtiter plates onto AB agar containing 2 μg of tetracycline per ml, 80 μg of neomycin per ml, and either 0.2% methanol (negative control)–1 μM coumestrol or a mixture of 10 μM each of formononetin, quercetin, luteolin, 4′,7-dihydroxyflavone, and 4,4′-dihydroxy-2′-methoxychalcone. Mutants which grew better on medium containing the flavonoid-isoflavonoid mixture or coumestrol than on medium containing methanol were retested for induced neomycin resistance on agar-containing individual compounds.
Rhizosphere tests.
Assays for bacterial colonization of alfalfa (Medicago sativa L. cv. CUF101) roots were performed in vermiculite as described previously (41) with the following modifications. Plant nutrient solution was supplemented with 8 mM NH4NO3. Inocula were prepared by growing cultures of strains 1D1609, I-1, and I-6 overnight in AB medium, washing once in sterile dilution buffer (25 mM NaH2PO4, 25 mM Na2HPO4, 0.01% Tween 20; pH 7.0), and resuspension in dilution buffer to an optical density of 0.5. In single-strain inoculations, suspensions of each strain were diluted to 1 × 103 to 3 × 103 CFU/ml; for dual-strain inoculations, suspensions of strain 1D1609 and either strain I-1 or strain I-6 were mixed 1:1 and diluted to 1 × 103 to 3 × 103 CFU/ml. At each sampling point, bacteria were recovered as described previously (41) from entire plant roots of uniform size. Serial dilutions of bacterial suspensions were plated on AB agar with and without tetracycline (2 μg/ml) for enumeration. Data for each sampling point consisted of bacterial counts recovered from 7 to 10 plants. Each root colonization experiment was repeated at least three times.
Molecular analysis of mutant strain I-1.
Total DNA from strain I-1 was isolated from cells grown overnight in LB medium and collected by centrifugation (3 min at 10,000 × g). Cell pellets were resuspended in 200 μl of TEN buffer (10 mM Tris, 1 mM EDTA, 10 mM NaCl; pH 8.0). Cells were lysed by adding 100 μl of proteinase K solution (1 mg/ml in TEN buffer) and 100 μl of sodium dodecyl sulfate (5% in TEN buffer) and incubating 1 h at 37°C. NaCl was then added to a final concentration of 0.5 M, and lysates were incubated at 68°C for 30 min before extracting twice with buffer-saturated phenol-chloroform and twice with chloroform. DNA was precipitated with an equal volume of cold isopropanol and washed twice with 70% ethanol. Precipitated DNA was dissolved in TE buffer (10 mM Tris, 1 mM EDTA; pH 8.0). Restriction digests (37) were performed with commercial enzymes (Promega, Madison, Wis.).
Cosmid clones of strain I-1 DNA were prepared by standard protocols (14), and clones containing the DNA fragment with the Tn5-B30 insertion were isolated by selecting for tetracycline resistance conferred by Tn5-B30. Clone pMC11 was selected on LB agar with tetracycline, and a physical map was constructed from restriction digests and hybridization tests with Tn5-B30 DNA which was labeled randomly with a Genius digoxigenin kit (Boehringer, Mannheim, Germany). Hybridizations were performed at 68°C and washed under high-stringency conditions according to the manufacturer’s instructions. SalI fragments of pMC11 corresponding to chromosomal DNA flanking Tn5-B30 were subcloned into SalI-digested pUC19 to construct pMC115 and pMC113 containing the SalI fragments upstream and downstream of the Tn5-B30 insertion in pMC11, respectively. A DNA probe specific to the mutated locus in I-1 was prepared by PCR amplification of the subcloned SalI fragment in pMC113 with primers Tn5out (5′ GAA AGG TTC CGT TCA GGA CGC TAC 3′) and M13R (5′ TCA CAC AGG AAA CAG CTA TGA C 3′), followed by random labeling of this product with digoxigenin.
A genomic cosmid library was prepared in pSUP205 by using wild-type DNA from strain 1D1609, and clone pAC107 containing DNA corresponding to the locus mutated in strain I-1 was identified by colony hybridization by using the probe prepared from pMC113. The 5.2-kb EcoRI fragment from clone pAC107 corresponding to the mutated locus in strain I-1 was subcloned into pBSK+ in both orientations to create pAC1073+ and pAC1073−. Nested deletions for sequencing were prepared from XbaI- and SacI-digested pAC1073+ and pAC1073− DNA with a double-stranded nested deletion kit (Pharmacia Biotech, Piscataway, N.J.). Both DNA strands were sequenced automatically (ABI 377; Applied Biosystems Perkin-Elmer, Foster City, Calif.) with standard M13 primers at the Division of Biological Sciences DNA Sequencing Facility (University of California, Davis, Calif.). Assembled sequences were analyzed for similarities to known sequences by using the BLAST (1) internet site (http://www.ncbi.nlm.nih.gov/BLAST).
Site-directed mutations of ifeA.
Plasmids for gene insertion within ifeA in strain 1D1609 were constructed in the sacB positive-selection suicide vector pJQ200mp18 (33). A 0.9-kb EcoRI-SalI fragment of pAC1073+ containing the first 639 bp of the ifeA open reading frame was subcloned into pJQ200mp18 to form pAC1074. A 2.0-kb EcoRI-generated DNA fragment containing the tetracycline resistance interposon Ω-Tc (10) was excised from mini-Tn5Tc (9) and cloned into p18Not (13). The resulting Ω-Tc NotI fragment was cloned into the unique NotI site within the ifeA fragment in pAC1074 to form pAC1075tet. A 2.0-kb NotI fragment containing a promoterless gusA gene was excised from pCAM140 (44) and cloned into the unique NotI site within the ifeA fragment in pAC1074 to form pAC1075gus. Plasmids pAC1075tet and pAC1075gus were conjugated into A. tumefaciens 1D1609 by triparental mating by using pRK2013 (11).
Single-recombinant clones resulting from chromosomal integration of pAC1075tet were selected on AB agar containing tetracycline and gentamicin. Double-recombinant clones resulting from a second homologous recombination event, which deleted the vector portion of the introduced plasmid along with the wild-type ifeA fragment, were selected on AB agar containing tetracycline and 5% sucrose. Likewise, single-recombinant clones containing chromosomally integrated pAC1074gus were selected on AB agar containing gentamicin. Double-recombinant clones containing only ifeA::gusA were selected on AB agar containing 5% sucrose and screened for coumestrol-inducible β-glucuronidase (GUS) activity on AB medium containing 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (25 μg/ml).
Insertion of the ifeA::gusA gene fusion in mutant strain I-4 was confirmed by PCR by using primers gusu (5′ AGA CTG AAT GCC CAC AGG CCG TCG 3′) and uncd (5′ CAT GTC GTC CAT CCA TGT AGA TAG 3′) to amplify the fragment upstream of the insertion and primers gusd (5′ GCG TTG GCG GTA ACA AGA AAG G 3′) and dclu (5′ CTT CAC ACG ATC CAG ACG GAG 3′) to amplify the downstream fragment. Insertion of Ω-Tc within ifeA in mutant strain I-6 was confirmed by PCR by using primers Ωout (5′ CCG GTG GAT GAC CTT TTG AAT GA 3′) and uncd to amplify the upstream fragment and primers Ωout and dclu to amplify the downstream fragment.
Isoflavonoid accumulation tests.
Coumestrol was added to a final concentration of 50 μM to cultures of wild-type strain 1D1609 and mutant strain I-6 grown to late exponential phase (optical density at 600 nm = 0.8 to 1.0). Triplicate samples (1 ml) of each culture were removed at selected time points. Cells were pelleted by centrifugation (3 min at 10,000 × g) and resuspended in methanol (1 ml) for 4 h at room temperature with shaking to extract coumestrol. The extraction was terminated by removing cells (3 min at 10,000 × g), and coumestrol in the supernatant was quantified by measuring the A342 value in a Lambda 6 dual-beam spectrophotometer (Perkin-Elmer, Norwalk, Conn.) relative to a standard curve (A342 versus concentration). The coumestrol content of culture samples was normalized to cell number and expressed as nmol/109 CFU. Each experiment was repeated twice.
Induction of ifeA::gusA expression.
Coumestrol induction of ifeA expression was measured as GUS activity in cultures of strain I-4 which were treated identically and run in parallel to wild-type 1D1609 and mutant I-6 cells during the isoflavonoid accumulation tests. After cells were centrifuged (3 min at 10,000 × g) out of the coumestrol-containing medium, pellets were washed once in carbon substrate-free AB medium containing 100 μg of chloramphenicol per ml and resuspended in 1 ml of the same medium. GUS assays measured the hydrolysis of p-nitrophenylglucuronide (43), and GUS activity was normalized to cell number as nanomoles of p-nitrophenyglucuronide hydrolyzed/min/109 CFU. Each experiment was repeated twice.
Induction of ifeA was assayed as GUS activity in cultures of strain I-4 which were grown overnight in the presence of the test compound. Exponentially growing cultures of strain I-4 in AB medium were diluted 100-fold into AB medium containing different concentrations of either coumestrol, formononetin, medicarpin, genistein, daidzein, biochanin-A, quercetin, 4,4′-dihydroxy-2′-methoxychalcone, hederagenin, or medicagenic acid and were grown overnight. GUS assays were performed and quantified as described above, with triplicate samples (1 ml) of each treatment. Each treatment was repeated twice.
RESULTS
Isolation of flavonoid-inducible mutants.
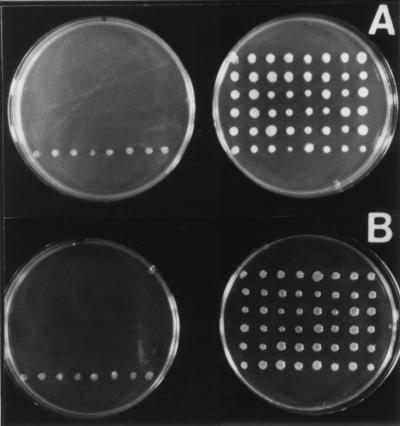
Approximately 5,000 Tn5-B30 mutants of strain 1D1609 were screened for inducible neomycin resistance in the presence of a mixture of flavonoids and isoflavonoids. These tests identified mutant strain I-1 as showing a reproducible induction of neomycin resistance, and further experiments with individual flavonoid and isoflavonoid compounds indicated that the promoterless nptII insertion in strain I-1 was induced by 1 μM coumestrol and 10 μM formononetin (Fig. 1).
FIG. 1.
Identification of an A. tumefaciens strain mutated in an isoflavonoid-inducible locus by the promoterless reporter gene nptII which confers neomycin resistance. (A) Growth of mutant strain I-1 on neomycin-containing medium with (right) or without (left) 10 μM formononetin. (B) Growth of mutant strain I-1 on neomycin-containing medium with (right) or without (left) 1 μM coumestrol. AB medium was supplemented with 0.2% methanol as a control (left) for the isoflavonoid solutions (right). All plates were inoculated with 40 replicate colonies of mutant strain I-1 (top five rows) and eight replicate colonies of a constitutively neomycin-resistant mutant (bottom rows).
Rhizosphere competence of mutant strain I-1.
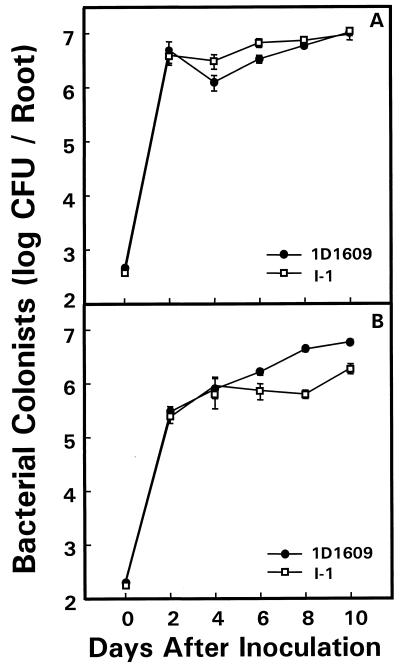
Wild-type strain 1D1609 and mutant strain I-1 achieved identical colonization densities on alfalfa roots 10 days after they were inoculated in single-strain tests (Fig. 2A), but the mutant competed poorly with wild-type cells when a 1:1 mixture of the two strains was introduced (Fig. 2B). On day 10 in the competitive assay, the ratio of mutant I-1 to wild-type 1D1609 cells was 0.33. Both strains grew rapidly in the first 2 days after inoculation when cell densities were low, and no effect of the mutation was detected at that time. These genotypic effects on alfalfa root colonization were seen in three independent experiments.
FIG. 2.
Alfalfa root colonization by wild-type A. tumefaciens 1D1609 and mutant strain I-1. Strains were inoculated separately (A) or as a 1:1 mixture (B) on sterile alfalfa seedlings at the time of germination. Root-colonizing bacteria were recovered and counted by dilution plating at the times indicated. Bacterial counts are reported as means ± standard errors from 7 to 10 replicate plants. Standard error bars are obscured by symbols in some cases.
Molecular analysis of mutant strain I-1.
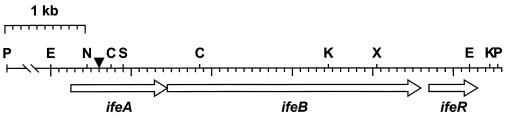
Hybridization tests of total DNA from strain I-1 with a Tn5-B30-specific DNA probe indicated that strain I-1 contained a single transposon insertion within a 5.2-kb EcoRI chromosomal restriction fragment. Colony hybridization tests of a wild-type 1D1609 DNA library with a probe specific for the flanking region downstream of the Tn5 insertion in mutant I-1 located four overlapping cosmid clones. These clones contained the same restriction pattern as that of the mutated locus in strain I-1 (Fig. 3). The 5.2-kb EcoRI fragment in pAC107 was sequenced and revealed a 5,227-bp fragment with two complete open reading frames and part of a third. The sequence of this third open reading frame was extended and completed by using sequence-specific primers and template DNA from pAC1079, which contains the PstI restriction fragment from pAC107 (Fig. 3). Sequence data for all three open reading frames were submitted to the GenBank (National Center for Biotechnology Information [NCBI]) database as accession number AF039653.
FIG. 3.
Restriction map of the 7.5-kb PstI fragment containing the Tn5-B30 insertion (▾) in A. tumefaciens mutant strain I-1. Restriction sites are represented as C, ClaI; E, EcoRI; K, KpnI; N, NotI; P, PstI; S, SalI; or X, XhoI. Arrows below the map indicate open reading frames predicted from nucleotide sequence analysis.
Sequence similarity searches with the NCBI BLAST database tool indicated that the predicted proteins encoded by these open reading frames are highly similar to proteins encoded by efflux system genes in other bacteria (Table 2). The first open reading frame, ifeA for isoflavonoid efflux, encodes a predicted protein of 384 amino acids with amino acid similarity (23 to 34% identical residues and 37 to 50% conserved residues) to members of a family of membrane-fusion proteins. The second open reading frame, ifeB, encodes a predicted protein of 1,046 amino acids with amino acid similarity (37 to 49% identical residues and 54 to 65% conserved residues) to members of a family of transmembrane efflux pump proteins. The third open reading frame, ifeR, encodes a predicted protein of 208 amino acids with amino acid similarity (27 to 49% identical residues and 52 to 63% conserved residues) to regulatory proteins of homologous efflux pump operons over the N-terminal regions. Sequence analysis of chromosomal DNA flanking the Tn5-B30 insertion in strain I-1 showed that the transposon inserted 320 bp downstream from the start codon for ifeA. Characteristic of Tn5 transposition, the insertion of Tn5-B30 in strain I-1 resulted in the duplication of 9 bp (5′ GGCCAATGT 3′) flanking the site of transposition.
TABLE 2.
Similarity of ifeA, ifeB, and ifeR open reading frames to efflux pump proteins in other bacteria
| Putative homologous protein (no. of amino acids) | % Identical residues | % Conserved residues | Reference |
|---|---|---|---|
| IfeA (384) | |||
| E. coli | |||
| AcrA (397) | 33 | 49 | 20 |
| AcrE (385) | 34 | 48 | 16 |
| EnvC (384) | 31 | 46 | 17 |
| P. aeruginosa | |||
| MexA (383) | 30 | 41 | 32 |
| MexC (387) | 29 | 43 | 31 |
| MexE (414) | 23 | 37 | 18 |
| N. gonorrhoeae MtrC (412) | 34 | 50 | 28 |
| IfeB (1,046) | |||
| E. coli | |||
| AcrB (1,049) | 49 | 65 | 20 |
| AcrF (1,034) | 48 | 63 | 16 |
| AcrD (1,034) | 46 | 60 | 4 |
| P. aeruginosa | |||
| MexB (1,046) | 48 | 64 | 32 |
| MexD (1,043) | 40 | 55 | 31 |
| MexF (1,062) | 37 | 54 | 18 |
| N. gonorrhoeae MtrD (1,067) | 43 | 62 | 12 |
| IfeR (208) | |||
| E. coli | |||
| AcrR (215) | 32a | 52a | 20 |
| EnvR (220) | 27a | 53a | 16 |
| N. gonorrhoeae MtrR (210) | 49b | 63b | 28 |
Sequence similarity to N-terminal 113 amino acids.
Sequence similarity to N-terminal 61 amino acids.
Verification of the ifeA mutant phenotype.
Mutant strain I-6 was constructed to contain an insertion of Ω-Tc, creating a polar mutation within ifeA. Successful construction was confirmed by PCR amplification of fragments flanking the site of Ω-Tc insertion and by DNA hybridization of total I-6 DNA to the pAC1074-derived ifeA probe, indicating an insertion of ∼2.0 kb within ifeA (data not shown).
Competitive root colonization tests established that the insertion in ifeA in strain I-6 impaired competitiveness against strain 1D1609 in a manner similar to the insertion in ifeA in strain I-1 (Table 3). In three separate experiments, strain I-6 was significantly (P ≤ 0.01) less competitive than wild-type 1D1609 when measured in populations recovered from alfalfa roots 10 days after inoculation. The absolute values measured for competition in strains I-1 and I-6 (i.e., the recovery ratio of mutant to wild type) of 0.33 and 0.27 were not significantly different. These data indicate that the insertion of Ω-Tc in ifeA in strain I-6 was phenotypically equivalent to the insertion of Tn5-B30 in ifeA in strain I-1.
TABLE 3.
Competitive alfalfa root colonization by wild-type and mutant I-6 (ifeA::Ω-Tc) cells of A. tumefaciens 1D1609
| Expt | Coinoculated cellsa | Recovered titer (106 CFU/plant)b | Recovered ratio (I-6/1D1609)b |
|---|---|---|---|
| 1 | I-6 | 1.28 ± 0.28 | 0.28 ± 0.08 |
| Wild type | 5.19 ± 0.53 | ||
| 2 | I-6 | 1.10 ± 0.15 | 0.27 ± 0.05 |
| Wild type | 4.74 ± 0.71 | ||
| 3 | I-6 | 2.30 ± 0.91 | 0.27 ± 0.09 |
| Wild type | 8.19 ± 1.06 |
Cells were coinoculated at a 1:1 ratio on day 0 with a total of 1 × 102 to 3 × 102 CFU/plant.
Mean titers and ratios ± standard error of bacteria recovered from 7 to 10 plants 10 days after inoculation.
Isoflavonoid accumulation in A. tumefaciens cells.
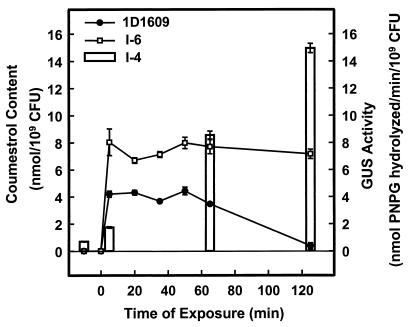
Within 5 min after its addition, coumestrol accumulated to a significantly (P ≤ 0.01) higher level in mutant strain I-6 than in the wild-type cells (Fig. 4). Coumestrol levels in mutant cells remained nearly twice as high as in wild-type cells for nearly 1 h, and then the difference increased markedly as the coumestrol content in wild-type cells decreased. At the end of the 120-min experiment, the coumestrol content of mutant cells was about 20-fold higher than that of the wild-type cells because coumestrol remained at high, unchanged levels in mutant strain I-6. Reduction of coumestrol in wild-type cells was associated with a 21-fold increase in expression of the ifeA-gusA reporter fusion in strain I-4 exposed to the same conditions (Fig. 4). We interpret these data as showing that an efflux pump associated with ifeA expression was responsible for the dramatic decline in the coumestrol content of wild-type cells during the second hour of this experiment. Similar results were obtained in two separate experiments.
FIG. 4.
Coumestrol accumulation and ifeA expression in A. tumefaciens. Coumestrol accumulation in wild-type strain 1D1609 (•) and mutant strain I-6 (□) was measured before and after the addition of 50 μM coumestrol (left axis). Corresponding expression of ifeA was measured as GUS activity (bar graph) in mutant strain I-4 (right axis). Data represent means ± standard errors from three replicates.
Induction of ifeA in A. tumefaciens.
Expression of the ifeA::gusA fusion in strain I-4 was induced by several flavonoids but not by two triterpenoids after overnight growth in media containing the test compounds (Table 4). GUS expression was significantly higher (P ≤ 0.05) in strain I-4 cells grown in the presence of the alfalfa isoflavonoids coumestrol, medicarpin, and formononetin or the soybean isoflavonoids genistein, daidzein, and biochanin-A. The alfalfa chalcone 4,4′-dihydroxy-2′-methoxychalcone also strongly induced expression of ifeA, but the flavonol quercetin, which is released in large amounts by germinating seeds of many Medicago species, including alfalfa (30), gave a variable response which was not significant. These results indicate that at least six isoflavonoids and one chalcone can serve as natural inducers of ifeA.
TABLE 4.
Induction of ifeA expression by flavonoids and triterpenoids
| Inducer | Inducer concna
|
|
|---|---|---|
| 10 μM | 50 μM | |
| Coumestrol | 9.8 ± 0.8 | 9.4 ± 1.6 |
| Formononetin | 1.7 ± 0.1 | 2.7 ± 0.2 |
| Medicarpin | 3.2 ± 0.2 | 17 ± 0.6 |
| Genistein | 4.8 ± 0.4 | 15 ± 1.6 |
| Daidzein | 4.1 ± 0.1 | 16 ± 1.8 |
| Biochanin-A | 1.4 ± 0.1b | 4.0 ± 0.3 |
| 4,4′-Dihydroxy-2′-methoxychalcone | 5.9 ± 0.8 | 9.9 ± 0.2 |
| Quercetin | 1.2 ± 0.1b | 2.9 ± 1.0b |
| Hederagenin | 1.0 ± 0.1b | 1.4 ± 0.1b |
| Medicagenic acid | 1.1 ± 0.1b | 1.4 ± 0.2b |
Fold induced GUS activity after overnight induction, presented as the mean ± standard error relative to methanol control.
Not significantly different from uninduced levels at P ≤ 0.05.
DISCUSSION
Evidence provided here shows that a new genetic locus, ifeABR, contributes significantly to the ecological competence of A. tumefaciens 1D1609 in its normal habitat by reducing cellular accumulation of isoflavonoids. The ifeA locus is induced by various isoflavonoids and a chalcone (Fig. 1 and 4; Table 4), which are present in alfalfa root exudate (7, 19, 24). Both random (strain I-1) and site-directed (strain I-6) mutations in ifeA impaired competitive root colonization significantly (Fig. 2B; Table 3). As the first microbial pumping system described for isoflavonoids, these observations indicate that the well-established phenomenon of hydrophobic efflux pumps (3) confers ecological benefits in the rhizosphere ecosystem. Interestingly, this locus was absent from A. tumefaciens C58 and Ach5 and Sinorhizobium meliloti 1021 as determined by DNA hybridization to ifeA probes (data not shown).
Functional interpretation of ifeABR as an isoflavonoid efflux pump operon in A. tumefaciens is based on its capacity to reduce accumulation of coumestrol (Fig. 4). This result was consistent with an efflux pumping of coumestrol after it accumulated to a level which induced expression of ifeA. Coumestrol was used as a substrate in these experiments because it was identified as an inducer of ifeA expression (Fig. 1 and 4; Table 4). The polar mutation in ifeA in strain I-6 presumably disrupted expression of both ifeA and ifeB and resulted in the absence of an active efflux pump system. Additional work is required to define the physiological functioning of this putative isoflavonoid pump, including an investigation of the effect of proton gradient uncouplers on coumestrol accumulation, but the rhizosphere phenotype associated with its absence (Fig. 2B) establishes its ecological significance. Wild-type strain 1D1609 shows an unusual, strong resistance to kanamycin (27), but that trait was not affected by mutations in the ifeA gene.
Structural interpretation of the ifeABR locus as an efflux pump is based on sequence analysis relative to reported proteins. The open reading frames found in this study are predicted to code for proteins that are quite similar to known membrane-fusion proteins, transmembrane transporter proteins, and regulatory proteins (Table 2). These proteins appear to belong to the “resistance-nodulation-division” family of efflux pump operons in gram-negative bacteria (25, 35). In cells expressing them, these operons confer resistance to a broad spectrum of hydrophobic and amphiphilic agents, including antibiotics, dyes, and nonionic detergents. The proposed mechanism of action of these pumps is that the transporter pump protein captures hydrophobic or amphiphilic substrate molecules from the cytoplasmic membrane and pumps them, via proton antiport, through a channel between the inner and outer membranes created by the membrane-fusion protein and possibly an outer-membrane porin channel (3, 25, 35). The generally hydrophobic nature of isoflavonoids makes them good candidates for such an efflux mechanism. However, since the growth of neither wild-type nor ifeA mutant cultures was inhibited by alfalfa isoflavonoids at concentrations of up to 50 μM (data not shown), ifeABR may be only one of several mechanisms conferring isoflavonoid resistance. This suggestion was supported by high-pressure liquid chromatography analyses, which showed that both mutant and wild-type cultures modified coumestrol (data not shown). It is also possible that the apparent ife efflux pump protects cells from rhizosphere compound(s) other than isoflavonoids.
Expression of efflux systems in other bacteria is regulated by signals of environmental stress (20, 32) and by efflux pump substrates (3). Induction of ifeA in A. tumefaciens by coumestrol (Fig. 4) may reflect the role of coumestrol as both an environmental signal and a pump substrate signal in the expression of the putative ifeABR efflux system. The broad specificity of known efflux pumping systems was mirrored to some extent by the tests for the specificity of ifeA expression (Table 4). In those experiments, the major isoflavonoids known to be present in alfalfa root exudate, as well as structurally related molecules such as genistein and daidzein, which are associated with soybean roots (8), were active inducers of ifeA. Whether the ifeABR locus confers any competitive advantage on A. tumefaciens 1D1609 in the rhizosphere of other legumes or nonlegume plants is not known.
Additional studies of the ifeABR locus are required to develop the depth of information already reported for many bacterial efflux pumps. For example, the transcriptional regulation of ifeB and the functional role of ifeR are not known. However, the identification here of an isoflavonoid-regulated locus which shows functional and structural similarities to known efflux pumps in bacteria offers a new direction for basic studies in rhizosphere ecology. The significant contribution of ifeA to competitive root colonization (Fig. 2B; Table 3) suggests that at least one rhizosphere bacterium reduces its exposure to isoflavonoids by a mechanism that differs from the widely recognized capacity of microorganisms to catabolize flavonoids (34) and isoflavonoids (2, 23).
ACKNOWLEDGMENTS
This work was supported in part by grants from the U.S. National Science Foundation (IBN-92-18567), from the U.S.-Israel Binational Agricultural Research and Development Fund, BARD (IS-2388-94), and from the National Institutes of Health (GM-45550).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Barz W. Isolation of rhizosphere bacterium capable of degrading flavonoids. Phytochemistry. 1970;9:1745–1749. [Google Scholar]
- 3.Bolhuis H, van Veen H W, Poolman B, Driessen A J M, Konings W N. Mechanisms of multidrug transporters. FEMS Microbiol Rev. 1997;21:55–84. doi: 10.1111/j.1574-6976.1997.tb00345.x. [DOI] [PubMed] [Google Scholar]
- 4.Bouvier J, Richaud C, Higgins W, Bogler O, Stragier P. Cloning, characterization, and expression of the dapE gene of Escherichia coli. J Bacteriol. 1992;174:5265–5271. doi: 10.1128/jb.174.16.5265-5271.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cangelosi G A, Best E A, Marinetti G, Nester E W. Genetic analysis of Agrobacterium. Methods Enzymol. 1991;204:384–397. doi: 10.1016/0076-6879(91)04020-o. [DOI] [PubMed] [Google Scholar]
- 6.Carlson R E, Dolphin D H. Pisum sativum stress metabolites: two cinnamylphenols and a 2′-methoxychalcone. Phytochemistry. 1982;21:1733–1736. [Google Scholar]
- 7.Dakora F D, Joseph C M, Phillips D A. Alfalfa (Medicago sativa L.) root exudates contain isoflavonoids in the presence of Rhizobium meliloti. Plant Physiol. 1993;101:819–824. doi: 10.1104/pp.101.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dakora F D, Phillips D A. Diverse functions of isoflavonoids in legumes transcend anti-microbial definitions of phytoalexins. Physiol Mol Plant Pathol. 1996;49:1–20. [Google Scholar]
- 9.De Lornezo V, Herrero M, Jakubzik U, Timms K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 11.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagman K E, Lucas C E, Balthazar J T, Snyder L, Nilles M, Judd R C, Shafer W M. The MtrD protein of Neisseria gonorrhoeae is a member of the resistance/nodulation/division protein family constituting part of an efflux system. Microbiology. 1997;143:2117–2125. doi: 10.1099/00221287-143-7-2117. [DOI] [PubMed] [Google Scholar]
- 13.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ish-Horowicz D, Burke J F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981;9:2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kado C I. Molecular mechanisms of crown gall tumorigenesis. Crit Rev Plant Sci. 1991;10:1–32. [Google Scholar]
- 16.Klein J R, Henrich B, Plapp R. Molecular analysis and nucleotide sequence of the envCD operon of Escherichia coli. Mol Gen Genet. 1991;230:230–240. doi: 10.1007/BF00290673. [DOI] [PubMed] [Google Scholar]
- 17.Klein J R, Henrich B, Plapp R. Molecular cloning of the envC gene of Escherichia coli. Curr Microbiol. 1990;21:341–347. [Google Scholar]
- 18.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 19.Koshino H, Masaoka Y, Ichihara A. A benzofuran derivative released by Fe-deficient Medicago sativa. Phytochemistry. 1993;33:1075–1077. [Google Scholar]
- 20.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 21.Massiot G, Lavaud C, Guillaume D, Le Men-Olivier L. Reinvestigation of the sapogenins and prosapogenins from alfalfa (Medicago sativa) J Agric Food Chem. 1988;36:902–909. [Google Scholar]
- 22.Massiot G, Lavaud C, Le Men-Olivier L, van Binst G, Miller S P F, Fales H M. Structural elucidation of alfalfa root saponins by mass spectrometry and nuclear magnetic resonance analysis. J Chem Soc Perkin Trans I. 1988;1988:3071–3079. [Google Scholar]
- 23.Matthews D E, Weiner E J, Matthews P S, VanEtten H D. Role of oxygenases in pisatin biosynthesis and in fungal degradation of maackiain. Plant Physiol. 1987;83:365–370. doi: 10.1104/pp.83.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maxwell C A, Hartwig U A, Joseph C M, Phillips D A. A chalcone and two related flavonoids released from alfalfa roots induce nod genes of Rhizobium meliloti. Plant Physiol. 1989;91:842–847. doi: 10.1104/pp.91.3.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oleszek W, Price K R, Colquhoun I J, Jurzysta M, Ploszynski M, Fenwick G R. Isolation and identification of alfalfa (Medicago sativa L.) root saponins: their activity in relation to a fungal bioassay. J Agric Food Chem. 1990;38:1810–1817. [Google Scholar]
- 27.Palumbo, J. D., D. A. Phillips, and C. I. Kado. Characterization of a new Agrobacterium tumefaciens strain from alfalfa (Medicago sativa L.). Arch. Microbiol., in press. [DOI] [PubMed]
- 28.Pan W, Spratt B G. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol Microbiol. 1994;11:769–775. doi: 10.1111/j.1365-2958.1994.tb00354.x. [DOI] [PubMed] [Google Scholar]
- 29.Phillips D A, Streit W R. Applying plant-microbe signalling concepts to alfalfa: roles for secondary metabolites. In: McKersie B D, Brown D C W, editors. Biotechnology and the improvement of forage legumes. Wallingford, England: CAB International; 1997. pp. 319–342. [Google Scholar]
- 30.Phillips D A, Wery J, Joseph C M, Jones A D, Teuber L R. Release of flavonoids and betaines from seeds of seven Medicago species. Crop Sci. 1995;35:805–808. [Google Scholar]
- 31.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 32.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7365–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 34.Rao J R, Cooper J E. Rhizobia catabolize nod gene-inducing flavonoids via C-ring fission mechanisms. J Bacteriol. 1994;176:5409–5413. doi: 10.1128/jb.176.17.5409-5413.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saier M H, Tam R, Reizer A, Reizer J. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol Microbiol. 1994;11:841–847. doi: 10.1111/j.1365-2958.1994.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 36.Sakagami Y, Kumai S, Suzuki A. Isolation and structure of medicarpin-β-d-glucoside in alfalfa. Agric Biol Chem. 1974;38:1031–1034. [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schmidt E L. Initiation of plant root-microbe interactions. Annu Rev Microbiol. 1979;33:355–376. doi: 10.1146/annurev.mi.33.100179.002035. [DOI] [PubMed] [Google Scholar]
- 39.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 40.Simon R, Quandt J, Klipp W. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in Gram-negative bacteria. Gene. 1989;80:161–169. doi: 10.1016/0378-1119(89)90262-x. [DOI] [PubMed] [Google Scholar]
- 41.Streit W R, Joseph C M, Phillips D A. Biotin and other water-soluble vitamins are key growth factors for alfalfa rhizosphere colonization by Rhizobium meliloti 1021. Mol Plant-Microbe Interact. 1996;9:330–338. doi: 10.1094/mpmi-9-0330. [DOI] [PubMed] [Google Scholar]
- 42.Tiller S A, Parry A D, Edwards R. Changes in the accumulation of flavonoid and isoflavonoid conjugates associated with plant age and nodulation in alfalfa (Medicago sativa) Physiol Plant. 1994;91:27–36. [Google Scholar]
- 43.Wilson K J, Hughes S G, Jefferson R A. The Escherichia coli gus operon, induction and expression of the gus operon in E. coli and the occurrence and use of GUS in other bacteria. In: Gallagher S R, editor. GUS protocols: using the GUS gene as a reporter of gene expression. New York, N.Y: Academic Press; 1992. pp. 7–23. [Google Scholar]
- 44.Wilson K J, Sessitsch A, Corbo J C, Giller K E, Akkermans A D L, Jefferson R A. β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other Gram-negative bacteria. Microbiology. 1995;141:1691–1705. doi: 10.1099/13500872-141-7-1691. [DOI] [PubMed] [Google Scholar]