Abstract
The activity of ribosomes from a clinical isolate of Escherichia coli, exposed to starvation for 7 days in sea salts medium, was investigated by measuring the kinetic parameters of ribosomal peptidyltransferase, by using the puromycin reaction as a model reaction. No alterations in the extent of peptide bond formation were observed during starvation. In contrast, a 50% reduction in the kmax/Ks ratio could be seen after 24 h of starvation; an additional 6 days of starvation resulted in a progressive but less abrupt decline in the kmax/Ks value. {kmax is the apparent catalytic rate constant of peptidyl transferase, and Ks is the dissociation constant of the encounter complex between acetyl (Ac)[3H]Phe-tRNA-poly(U)-ribosome and puromycin.} Although the distribution of ribosomal particles remained constant, a substantial decrease in the number of ribosomes per starved cell and a clear decline in the ability of ribosomes to bind AcPhe-tRNA were observed, particularly during the first day of starvation. Further analysis indicated that rRNA in general, but especially 23S rRNA, was rapidly degraded during the starvation period. In addition, the L12/L7 molar ratio decreased from 1.5 to 1 during the initial phase of starvation (up to 24 h) but remained constant during the subsequent starvation period. Ribosomes isolated from 24-h-starved cells, when artificially depleted of L7/L12 protein and reconstituted with L7/L12 protein from mid-logarithmic-phase cells, regenerated an L12/L7 molar ratio of 1.5 and restored the peptidyltransferase activity to a substantial level. An analogous effect of reconstitution on the efficiency of ribosomes in binding AcPhe-tRNA was evident not only during the initial phase but throughout the starvation period.
Most of our understanding of bacterial metabolism has been obtained from cells undergoing exponential growth (22, 40). In contrast, our knowledge of bacterial metabolism in natural environments is still limited. Most ecosystems are grossly oligotrophic and characterized by drastically reduced concentrations of available carbon compounds. Growth of heterotrophic bacterial populations in such environments is limited, because the cost of maintenance for the organism is detrimentally high if the cell remains in a high metabolic state during nutritional deprivation.
Adaptation to a nongrowing state requires many physiological changes. A key control point in the regulation of cell growth is the protein-synthesizing capacity, which does not remain constant but is precisely adjusted to the growth demand (22, 40). During carbon starvation, a rapid degradation of ribosomes, which is essential for cell maintenance and survival, occurs (6, 15, 19, 20). However, the level of functional ribosomes cannot fall below a critical point, because some metabolic processes operate only during starvation (11, 20, 31, 39, 41). In parallel, ribosome synthesis is negatively regulated by various mechanisms. These mechanisms include ribosome feedback inhibition, stringent control, and growth-rate-dependent control (22, 40). For a long time, it was believed that the reduced rate of protein synthesis in starved cells is a direct effect of the accumulation of signal molecules, such as ppGpp (stringent control) (16, 25, 34, 42). This mode of ppGpp action has been refuted by recent studies (13, 32, 37), suggesting that the function of ppGpp is to maintain a tight coupling of translation and transcription by modulating the RNA polymerization rate or by a direct effect on RNA polymerase promoter selection.
During carbon starvation, the expression of a specific set of genes makes the cells more viable (11, 20, 23, 29, 31, 38, 39, 41). Among them, the rmf gene encodes a ribosome modulation factor (RMF), which associates with 70S ribosomes and converts them to 100S particles (38, 39, 41); disruption of the rmf gene results in a significant decrease of the viability of mutant Escherichia coli only at the stationary phase, suggesting that the dimerization of ribosomes is essential for stationary survival (41). Conformational changes in bacterial polysomes induced by amino acid starvation (2, 8, 24) also suggest that a possible correlation between starvation and ribosomal structure may exist. With regard to ribosomal proteins, it has been found that the L12/L7 molar ratio changes concomitantly with the activity of ribosomal peptidyltransferase in exponentially growing E. coli cells (14). Although both the amounts of L12 and L7 proteins and their molar ratio decrease progressively during the stationary phase (26), the physiological significance of these alterations has never been investigated.
The main objective of the present study was to elucidate whether alterations in the peptidyltransferase activity are related to specific structural changes occurring in ribosomes upon cell starvation. Given that the fate of certain pathogenic bacteria in marine environments is important in both fundamental and applied microbiology, and since various physicochemical parameters in marine environments may change due to seasonal and local conditions, we followed the efficiency of ribosomes from a clinical isolate of E. coli during prolonged culture in a commercially available sea salts solution.
MATERIALS AND METHODS
Materials.
GTP (disodium salt), ATP (disodium salt), poly(U), phenylalanine, spermine tetrahydrochloride, puromycin dihydrochloride, heterogenous tRNA from E. coli W, and artificial sea salts mixture (sea salts) were purchased from Sigma. l-Phenyl[2,3-3H]alanine was obtained from Amersham (Buckinghamshire, United Kingdom). The 16S and 23S rRNAs from E. coli MRE600 were obtained from Boehringer. Cellulose nitrate filters (type HA, 24-mm diameter, 0.45-μm pore size) were from Millipore. Bacto Peptone, Bacto yeast, and Casamino Acids were purchased from Difco.
Bacterial strains and growth.
In most of the following experiments we used an E. coli clinical isolate (EC 138 collection; biotype, S144573) (14). As a reference, we used E. coli B cells.
Cells were grown aerobically at 37°C with shaking in Erlenmeyer flasks containing M9 medium (18) supplemented with 0.03 mM FeCl3, 0.1 mM CaCl2, 1 mM MgSO4, 0.01 mM vitamin B1, and 0.6% glucose plus 0.2% Casamino Acids. Steady-state cultures were collected by pouring over crushed ice at an optical density at 540 nm (OD540) close to 0.5 (mid-logarithmic phase), centrifuged, and washed twice with sea salts solution (40 g/liter). A suitable aliquot (20 g [wet weight] of cells) was resuspended at a density of 7 × 106 CFU/ml in sea salts solution and incubated for 7 days, with vigorous aeration at 37°C. At the onset and after selected times of starvation, cells from the starved culture were harvested by centrifugation, washed once in cold 0.9 M KCl solution, frozen rapidly in a dry ice-acetone bath, and stored at −70°C.
The colony-forming ability of the cells (culturability) was assessed by spreading an appropriate aliquot of culture on agar plates (0.8% Bacto agar in rich medium [14]). The plates were incubated at 37°C for 24 h, and the colonies were counted.
Biochemical preparations.
Salt-washed ribosomes and crude acetyl (Ac)[3H]Phe-tRNA, charged with 29.8 pmol of [3H]Phe (160,000 cpm total) per A260 unit, were prepared as described elsewhere (14). The method used for polysome preparation was adopted from the freeze-thaw-lysozyme lysis procedure described by Ron et al. (27). Complex C, i.e., the Ac[3H]Phe-tRNA-poly(U)-ribosome complex adsorbed on cellulose nitrate filters, was prepared as reported previously (14). The adsorbed radioactivity was measured in a liquid scintillation spectrometer. Controls without poly(U) were included in each experiment, and the values obtained were subtracted.
rRNA was extracted from E. coli ribosomes as described before (18). The rRNA was precipitated with ethanol, dissolved in water, and analyzed by electrophoresis in a 1% denaturing agarose gel (18) or in an 8% polyacrylamide gel in the presence of 6 M urea (4). Ribosomal proteins were isolated from E. coli ribosomes by precipitation with acetone from acetic acid extracts (3) and redissolved in 20 mM bis-Tris-MES (morpholineethanesulfonic acid) (pH 7.0)–6 M urea–6 mM β-mercaptoethanol. The separation of L7 and L12 ribosomal proteins was carried out by electrophoresis in 4% polyacrylamide gels in the presence of 6 M urea (17). L7 and L12 ribosomal proteins were isolated from E. coli B cells according to Hamel et al. (12) and used as reference standards. For quantitation, gels stained with Coomassie or methylene blue were scanned at 550 nm in a BT 511 densitometer (Biotechnica Instruments, Rome, Italy). The identification of RMF in ribosomal samples was carried out by exposing the ribosomal particles to 1 M ammonium acetate (38) and analyzing the protein extract by ultrafiltration and Tricine-sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (28).
Ribosome sedimentation.
The ribosome preparations were loaded on 10 to 30% linear sucrose gradients in 10 mM Tris-HCl, pH 7.5–6 mM MgCl2–30 mM NH4Cl–6 mM β-mercaptoethanol and centrifuged at 85,000 × g for 6 h at 4°C in an SW41 Ti rotor (Beckman, Palo Alto, Calif.). Fractions were collected from the gradients and analyzed by optical scanning at 260 nm.
Peptide bond formation assay.
The peptidyltransferase activity of ribosomes was assessed by the puromycin reaction carried out at 25°C in the presence of 6 mM Mg2+ and 100 μM spermine (7). Under these conditions, the reaction between complex C and excess puromycin (S) proceeds as an irreversible pseudo-first-order reaction:
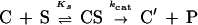 |
where C′ is a modified species of complex C not participating in reforming complex C, and P is the product (AcPhe-puromycin).
The percent (x) of the bound Ac[3H]Phe-tRNA that was converted to product was calculated by dividing the amount of product by the amount of Ac[3H]Phe-tRNA and multiplying the result by 100. The intervention of other species, except that of complex C, was erased by dividing the x values by the extent factor α. Additional details on α are given elsewhere (35).
At fixed initial concentrations of puromycin, the first-order rate constant (kobs) was determined by fitting the corrected values of x/α = x′ into the equation:
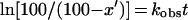 |
1 |
for each time point (t) and calculating the slope of this straight line. The relationship between kobs and [S] is given by the equation:
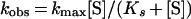 |
2 |
where kmax expresses the apparent catalytic rate constant of peptidyltransferase and Ks is the dissociation constant of the encounter complex CS (7). The values of kmax and Ks were determined from double-reciprocal plots of equation 2. All data used in the kinetic analysis were obtained with ribosomes isolated from four independently starved cultures. The standard errors of the means (Table 1) were calculated according to Daniel (5).
TABLE 1.
Kinetic parameters of ribosomal peptidyltransferase during incubation of the E. coli isolate in sea salts medium, as evaluated by the puromycin reactiona
| Ribosome source | kmax (min−1) | Ks (mM) | kmax/Ks (min−1/mM) |
|---|---|---|---|
| Supplemented M9 medium (mid-log-phase cells) | 1.54 ± 0.04 | 0.670 ± 0.009 | 2.30 ± 0.13 |
| Sea salts medium (24 h) | 1.00 ± 0.05 | 0.870 ± 0.012 | 1.15 ± 0.12 |
| Sea salts medium (72 h) | 1.05 ± 0.04 | 1.333 ± 0.018 | 0.79 ± 0.06 |
| Sea salts medium (168 h) | 1.05 ± 0.04 | 2.000 ± 0.025 | 0.53 ± 0.04 |
| Sea salts medium (24 h) (ribosomes − L7/L12b) | 1.02 ± 0.05 | 2.430 ± 0.020 | 0.42 ± 0.04 |
| Reconstituted ribosomesc | 1.21 ± 0.03 | 0.690 ± 0.008 | 1.75 ± 0.09 |
All data used in the kinetic analysis were obtained with ribosomes isolated from four independently starved cultures.
Ribosomes isolated from 24-h-starved cells and artificially depleted of L7/L12 protein.
Ribosomes isolated from 24-h-starved cells, artificially depleted of L7/L12 protein and then reconstituted with L7/L12 protein extracted from mid-logarithmic-phase cells.
Reconstitution experiments.
Ribosomal particles depleted of L7/L12 protein were prepared from 70S ribosomes by treatment with 0.5 M NH4Cl-ethanol at 0°C and centrifugation at 30,000 × g for 15 min at 4°C in an SS34 rotor (Sorvall, Newtown, Conn.) (12). By this procedure, proteins L7 and L12 were selectively and completely removed from 70S ribosomes. In reconstitution experiments, ribosomal particles depleted of L7/L12 protein were preincubated at 37°C for 20 min in 100 mM Tris-HCl (pH 7.2)–100 mM NH4Cl (pH 7.2)–6 mM Mg2+ (acetate)–100 μM spermine–6 mM β-mercaptoethanol, mixed with 8 molar equivalents of L7/L12 protein isolated from mid-logarithmic-phase ribosomes, and further incubated at 25°C for 20 min. Aliquots of this mixture were used for the preparation of complex C adsorbed on cellulose nitrate filters and, subsequently, for the puromycin reaction. To determine the protein composition of the reconstituted particles, these were isolated by centrifugation at 100,000 × g for 4 h at 4°C in a 75 Ti rotor (Beckman) and then analyzed by gel electrophoresis.
RESULTS
Growth characteristics and culturability of E. coli cells incubated in sea salts medium.
The exponential growth rate of the E. coli clinical isolate was 56% that of E. coli B cells, which grew with doubling times close to 30 min in supplemented M9 medium. Exponentially growing cells of the E. coli clinical isolate were collected by centrifugation and starved by resuspension in sea salts medium. Biomass was determined by measuring the OD540: no biomass increase was observed after the onset of starvation. The clinical isolate’s number of CFU was constant for at least 3 days and thereafter reduced slowly, not exceeding a 50% decrease at the end of 7 days of starvation. In contrast, E. coli B cells, when exposed in sea salts medium, exhibited a rapid decline in the number of CFU; 7 days after the onset of starvation, the concentration of culturable cells decreased more than 99%, dropping to values as low as 100 CFU/ml.
Distribution of ribosomal particles and alterations in the binding of AcPhe-tRNA to ribosomes after prolonged incubation of E. coli cells in sea salts medium.
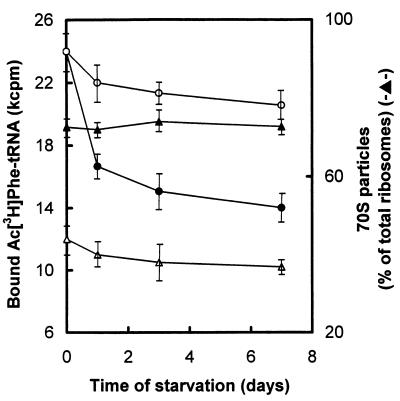
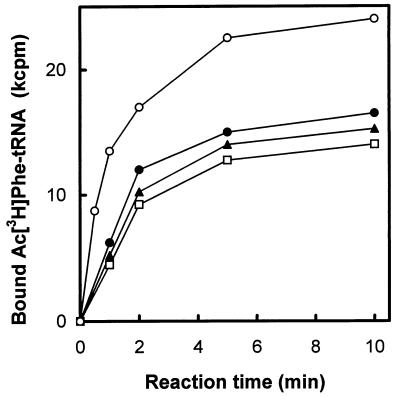
Polyribosomes were not detected in cells that were starved for either 1 or 7 days. Runoff ribosomes from cells harvested at various times of incubation were also subjected to sucrose gradient centrifugation under association conditions (6 mM MgCl2, 30 mM NH4Cl). The major species of ribosomes was found to be 70S monomers. In addition, native 50S and 30S subunits were detected in about 27% of the total ribosomes. The percentage of 70S monomers remained constant throughout the incubation period (Fig. 1). However, statistically significant changes in the efficiency of ribosomes to bind AcPhe-tRNA were observed (Fig. 1). The extent of binding declined, particularly during the first 24 h of starvation. Moreover, the initial rate of AcPhe-tRNA binding steeply decreased by 24 h of starvation and then remained almost unchanged (Fig. 2).
FIG. 1.
Levels of 70S ribosomal particles and AcPhe-tRNA binding to poly(U)-programmed ribosomes isolated from a clinical isolate of E. coli during starvation in sea salts medium. The level of 70S ribosomes was calculated by measuring at 260 nm the corresponding peak area of ribosomal profile after sucrose gradient centrifugation. Ribosomes were assayed for AcPhe-tRNA binding by the filter-binding technique. The values of radioactivity shown on the vertical axis represent the extent of Ac[3H]Phe-tRNA binding to 2.5 A260 units of intact ribosomes (•), ribosomes depleted of L7/L12 protein (▵), and ribosomes depleted of L7/L12 protein and reconstituted with L7/L12 protein from mid-logarithmic-phase cells (○). Controls without poly(U) were included in each experiment, and the values obtained were subtracted. Bars represent standard deviations as calculated from four independently starved cultures.
FIG. 2.
Time course of AcPhe-tRNA binding to E. coli ribosomes. The ribosomes were prepared from the clinical isolate cells harvested at time zero (○) and at 1 day (•), 3 days (▴), and 7 days (□) after the onset of starvation.
Analysis of rRNA.
Analysis of equivalent numbers of cells indicated that 35% ± 4% of ribosomes existing at time zero were completely disintegrated after 7 days of starvation. The distribution of the surviving ribosomal particles remained constant during this time (Fig. 1), which may indicate that, in addition to the ribosomal subunits, 70S particles were also subjected to degradation in a synchronized fashion.
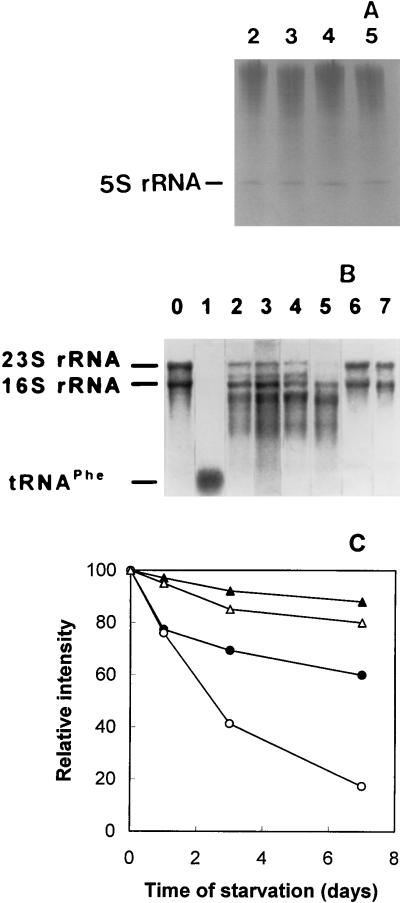
rRNA was isolated from the surviving ribosomes of starved cells and analyzed by gel electrophoresis. Low-molecular-mass rRNA species were examined by electrophoresis in 8% polyacrylamide gels in the presence of 6 M urea. Only one rRNA species was found, showing a mobility equivalent to that of 5S rRNA and exhibiting high stability throughout the starvation period (Fig. 3A). A different picture emerged with regard to high-molecular-mass rRNA. In this case, electrophoresis in 1% denaturing agarose gels showed that both the clinical isolate’s ribosomes and ribosomes from exponentially growing E. coli B cells contained fragmented rRNA even before the start of starvation, the rRNA fragmentation of the former being higher (Fig. 3B, lanes 2 and 6). After the onset of starvation, both 23S and 16S rRNA were further fragmented. However, the rRNA fragmentation of the clinical isolate (Fig. 3B, lanes 3 to 5, and C) was much more pronounced than that of the E. coli B cells (Fig. 3B, lane 7, and C).
FIG. 3.
Fragmentation of rRNA after incubation of E. coli cells in sea salts medium. rRNA was extracted from E. coli ribosomes and analyzed by 8% polyacrylamide gel electrophoresis in the presence of 6 M urea (A) or by 1% agarose denaturing gel electrophoresis (B). Lanes: 0, standards of 16S and 23S rRNA from E. coli W; 1, tRNAPhe from E. coli W; 2 to 5, rRNAs (5 μg) prepared from the clinical isolate cells harvested at the mid-logarithmic phase of growth (lane 2) or after 1 day (lane 3), 3 days (lane 4), or 7 days (lane 5) of starvation; 6, rRNA from mid-logarithmic-phase cells of E. coli B strain; 7, rRNA from E. coli B cells harvested at the end of the starvation period. (C) Quantitation of 16S and 23S rRNA fragmentation during starvation. Equivalent aliquots (5 μg) of total rRNA from each sample were analyzed by 1% agarose denaturing gel electrophoresis, such as that shown in panel B. After staining with methylene blue, the gels were scanned at 550 nm in a densitometer. The relative intensity represents the absorbance of 16S (solid symbols) or 23S (open symbols) rRNA bands at several points of the starvation period, given as the percentage of the rRNA band absorbance corresponding to 5 μg of rRNA isolated from surviving E. coli B ribosomes (triangles) or from surviving ribosomes of the clinical isolate (circles), prepared from cultures harvested at time zero.
Analysis of ribosomal proteins.
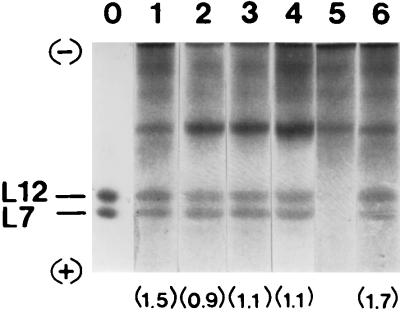
Analysis of ribosomal proteins by SDS-polyacrylamide gel electrophoresis showed no major differences in the patterns of proteins between the exponentially growing cells and those incubated in sea salts medium, with the exception of L7 and L12 proteins (data not shown). The changes in L7 and L12 proteins were further examined by electrophoresis in 6 M urea–4% polyacrylamide gels (Fig. 4). A small amount (15% ± 5%) of L7 and L12 proteins, found in mid-logarithmic-phase ribosomes, was lost within 24 h of incubation of cells in sea salts medium. However, the L12/L7 molar ratio changed from 1.5 to 1 (Fig. 4). Further incubation of cultures did not essentially alter the content of L7 plus L12 per ribosome or the L12/L7 molar ratio (Fig. 4, compare lanes 2, 3, and 4).
FIG. 4.
Gel electrophoresis of total ribosomal proteins (TP70) isolated from the E. coli clinical isolate during starvation. Proteins were separated by 6 M urea–4% polyacrylamide gel electrophoresis. Lanes: 0, L7 and L12 reference standards; 1 to 4, TP70 from cells harvested at time zero (lane 1) or at 1 day (lane 2), 3 days (lane 3), or 7 days (lane 4) after the onset of starvation; 5 and 6, TP70 from ribosomal particles isolated from 24-h-starved cells and depleted of L7/L12 protein before (lane 5) or after (lane 6) reconstitution with L7/L12 protein from exponentially growing cells. The numbers in parentheses are the corresponding mean L12/L7 molar ratios, obtained from four independently starved cultures. The standard error of these values was found to be less than 0.05.
Another point of interest is that no bands corresponding to RMF protein could be observed throughout the starvation period. In accordance with this finding, we failed to detect 100S particles (dimers of 70S ribosomal monomers). Similar results were also obtained when exponentially growing cells with low doubling time (0.4 doublings/h) were used instead of starved cells. Since salt washing of cells or ribosomes interferes with the state of ribosomes to various extents (38), this step of purification was omitted when cells or ribosomes were prepared in order to be used in such experiments.
Peptidyltransferase activity of E. coli cells after prolonged incubation in sea salts medium.
By using the puromycin reaction as a model reaction (7), changes in peptidyltransferase activity were followed throughout the incubation period (up to 7 days) in sea salts medium. Two distinct parameters of the peptide bond formation were examined: first, the extent (or final degree) of the puromycin reaction which determines the percentage of the bound AcPhe-tRNA that is converted to product at infinity (35) and, second, the ratio kmax/Ks, which provides an estimation of enzyme activity status (9).
The final degree value was constant and equal to 74% ± 5%. In contrast, a 50% reduction in the kmax/Ks ratio could be seen after 24 h of starvation, due to a decrease of kmax and a parallel increase of Ks (Table 1). To investigate whether this effect is related to the shift in the L12/L7 molar ratio, a series of reconstitution experiments was performed. Ribosomes prepared from 24-h-starved cells were artificially depleted of L7/L12 protein and then incubated with an eightfold molar excess of L7/L12 protein isolated from mid-logarithmic-phase ribosomes. Purification of reconstituted ribosomes by centrifugation showed that both the L12/L7 molar ratio (Fig. 4, lane 6) and the L7/L12 content per ribosome were similar to those corresponding to mid-logarithmic-phase ribosomes. Reconstituted ribosomes prepared as described above were further examined for their kinetic properties during peptide bond formation. As shown in Table 1, replacement of starved by exponential L7/L12 protein significantly improved the catalytic properties of ribosomes isolated from 24-h-starved cells; the Ks value was reduced nearly to the same value as that obtained from mid-logarithmic-phase ribosomes, while the kmax value showed a 78% recovery. Similar treatment of ribosomes isolated from 72-h- or 168-h-starved cells did not cause significant improvement in the catalytic properties of reconstituted ribosomes (data not shown). However, the efficiency of reconstituted ribosomes to bind AcPhe-tRNA was greatly restored, independent of whether the L7/L12-depleted ribosomes were prepared from 24-h- or from 168-h-starved cells (Fig. 1).
DISCUSSION
Despite intensive research of the regulation of protein synthesis in starved cells, the mechanisms involved in the adaptation to the nongrowing state are largely unknown. In the present study, we have focused on alterations in function and structure relationships regarding ribosomes prepared from an E. coli clinical isolate cultured for 7 days in sea salts medium.
In accordance with previous reports (6, 15, 19, 20), our experiments showed that the ribosomal content decreases when cells enter the nongrowing phase. In starved cultures, 35% ± 4% of rRNA material extracted from exponential cultures of the same turbidity is completely disintegrated. We found that this rRNA breakdown is lower than that observed in other studies (6, 19). Although this difference is probably related to the metabolic properties of the E. coli clinical isolate per se, it has been mentioned (19) that the degradation rate during complete starvation is much less than that observed during incomplete starvation. This may suggest that some level of energy is required to cause exhaustive degradation of rRNA.
Polyribosomes were not detected in starved cells of the E. coli clinical isolate. This finding is in accordance with observations made with E. coli during glucose starvation (8) or in a marine Vibrio sp. strain (10). However, a low level of protein synthesis cannot be precluded. Indeed, previous studies have demonstrated that, while the bulk of protein synthesis is largely restricted in E. coli, synthesis of 30 to 50 new proteins is induced in response to starvation (31). It was, therefore, attractive to reexamine the structural and functional characteristics of ribosomes remaining in the starved cells with more sensitive methods. In the present study, we used a kinetic procedure (35), which is the best method available for evaluating changes in the activity of ribosomal peptidyltransferase in vitro. Having separated all steps involved in Ac[3H]Phe-tRNA-poly(U)-ribosome formation (step 1) from the reaction between the donor (AcPhe-tRNA) and puromycin (step 2), we were able to examine the effect of starvation on each of these steps.
With respect to the influence of starvation on the binding of AcPhe-tRNA to ribosomes, our results show that ribosomes in starved cells have a reduced capacity to bind the initial aminoacyl-tRNA. Since the assembly state of ribosomes does not essentially change upon starvation, it seems that the 70S particles bind AcPhe-tRNA with altered efficiency, depending on the age of the culture. It could be proposed that starvation might activate a modifier of initiation factors, in analogy with the effect of amino acid starvation on regulation of polypeptide chain initiation in Ehrlich ascites tumor cells (25). Alternatively, it could be suggested that the translation initiation factors become less stable during carbon starvation, as previously detected by phosphate starvation studies (6). Although both suggestions may explain alterations in the initial rate of AcPhe-tRNA binding, they cannot interpret observed changes in the extent of binding (Fig. 1). Therefore, it might be supposed that some of the ribosomal particles have fully lost their efficiency for AcPhe-tRNA binding.
It is of particular interest that changes in peptidyltransferase activity occur when bacteria switch from the exponential to the nongrowing state (Table 1). The second-order rate constant kmax/Ks is reduced after 24 h of incubation or more in sea salts medium. While the catalytic rate constant kmax and the dissociation constant Ks clearly change, the extent of the puromycin reaction remains constant throughout the starvation period. This observation implies that the critical step, at which the peptide bond formation is inhibited, is the kinetic phase of the reaction. Furthermore, this finding suggests that the distribution of AcPhe-tRNA among different binding states (P/P or A/P ribosomal state [14]) is not influenced by starvation. Albertson and Nystrom (1) have proposed that the repression of protein chain elongation factors during starvation may play an important role in the reduction of the elongation rate. However, we did not observe any significant repression of protein chain elongation factors, at least during the first 24 h of starvation (data not shown). Furthermore, we failed to detect 100S ribosomal particles or electrophoretic bands corresponding to RMF protein in extracts from starved cells or cells growing at a low growth rate. It is obvious that the rmf gene, encoding RMF (39), is either mutated or negatively regulated in the E. coli clinical isolate.
Experiments on the constitution of isolated ribosomes provide several interesting observations on ribosomal metabolism during the time course of starvation. The 23S rRNA and to a lesser degree 16S rRNA undergo a cumulative fragmentation that is initiated even before the culture enters the incubation in sea salts medium. As we have recently reported (14), this clinical isolate is the first E. coli strain to exhibit such a high degree of rRNA fragmentation before growth has ceased. After the onset of starvation, ribosomes start disintegrating; after 7 days of incubation in sea salts medium, 35% ± 4% of ribosomes have been completely lost. Furthermore, in the surviving ribosomes neither 23S rRNA nor 16S rRNA remains intact. The corresponding fragmentation of rRNA from E. coli B cells is much lower, although the corresponding loss of cell culturability is remarkably elevated. Our results reinforce the notion that the survival of E. coli cells during carbon starvation is proportional to the capacity of the strains to degrade rRNA (15, 20). However, the observed degradation of rRNA cannot explain the concomitant changes in peptidyltransferase activity. Evidently, the catalytic activity is independent of the initial concentration of functional ribosomes or puromycin, because it is defined by the kmax/Ks ratio. Moreover, the coexistence of intact 23S and 16S rRNA with fragmented rRNA suggests that complex C is probably formed by intact rRNA. Otherwise, more than one kinetic species of complex C should participate in peptide bond formation, giving nonlinear double-reciprocal plots (30). Nevertheless, the concentration of functional ribosomes is an essential factor which contributes to the rate value of peptide bond formation. The extent of rRNA fragmentation seems to regulate the number of functional ribosomes. Although this number is reduced after prolonged starvation in sea salts medium, the remaining functional ribosomes may be essential not only for the expression of required genes in the nongrowing state but also for efficient recovery from starvation when nutrients become available.
Another point of interest is the correlation of changes in peptidyltransferase activity with stoichiometric aberrations in ribosomal proteins appearing during starvation. The method applied cannot detect small changes in the protein content of ribosomes. Nevertheless, these experiments preclude gross changes occurring to ribosomal proteins during the 7 days of starvation. The only apparent alteration is a decrease in the value of the L12/L7 molar ratio, occurring during the initial phase of starvation (up to 24 h). However, an interesting finding during this phase of starvation is that the replacement of starved by exponential-phase L7/L12 protein restores the peptidyltransferase activity to a substantial level, comparable to that shown in exponential-phase ribosomes (Table 1). In addition, this replacement reverses the ribosome deficiency in binding AcPhe-tRNA (Fig. 1), evidently not only at the initial phase but throughout the starvation period. For an unknown reason(s), similar treatment of ribosomes isolated from 72-h- or 168-h-starved cells does not improve the peptidyltransferase activity of reconstituted ribosomes. Since the ribosomal proteins L7 and L12 differ solely in the presence of an amino-terminal acetyl group on L7 (33), it is reasonable to assume that the acetylation level of L12 protein may act as a modulator of ribosome efficiency during starvation. We cannot exclude, however, the existence of additional changes in the structure or in the conformation of the L7/L12 dimer, occurring upon starvation and affecting the peptidyltransferase activity. The significance of L7/L12 protein in ribosomal functions has been well demonstrated in the past (14, 21, 36).
Undoubtedly, the gap in our understanding of the exponential phase as opposed to the starvation phase in natural bacterial isolates is evident, and the unraveling of the corresponding relative regulatory mechanisms needs further investigation.
ACKNOWLEDGMENTS
We thank C. Dimakopoulos for expert technical assistance with densitometry and D. Spathas and D. Synetos for reviewing the manuscript.
This work was supported in part by MED POL grant TRNS 030203/GRE-147(IV) from the World Health Organization.
REFERENCES
- 1.Albertson N H, Nystrom T. Effects of starvation for exogenous carbon on functional mRNA stability and rate of peptide chain elongation in Escherichia coli. FEMS Microbiol Lett. 1994;117:181–188. doi: 10.1111/j.1574-6968.1994.tb06762.x. [DOI] [PubMed] [Google Scholar]
- 2.Andrieux E, Cozzone A J. Conformational changes in bacterial polysomes induced by amino acid starvation. Int J Biochem. 1984;16:113–116. doi: 10.1016/0020-711x(84)90060-0. [DOI] [PubMed] [Google Scholar]
- 3.Barritault D, Expert-Bezancon A, Guerin M F, Hayes D. The use of acetone precipitation in the isolation of ribosomal proteins. Eur J Biochem. 1976;63:131–135. doi: 10.1111/j.1432-1033.1976.tb10215.x. [DOI] [PubMed] [Google Scholar]
- 4.Boothroyd J C, Wang A, Campbell D A, Wang C C. An unusually compact ribosomal DNA repeat in the protozoan Giardia lamblia. Nucleic Acids Res. 1987;15:4065–4085. doi: 10.1093/nar/15.10.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daniel W W. Biostatistics: foundation for analysis in the health sciences. New York, N.Y: Wiley; 1978. pp. 284–303. [Google Scholar]
- 6.Davis B D, Luger S M, Tai P C. Role of ribosome degradation in the death of starved Escherichia coli cells. J Bacteriol. 1986;166:439–445. doi: 10.1128/jb.166.2.439-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drainas D, Kalpaxis D L. Bimodal action of spermine on ribosomal peptidyltransferase at low concentration of magnesium ions. Biochim Biophys Acta. 1994;1208:55–64. doi: 10.1016/0167-4838(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 8.Dresden M H, Hoagland M B. Polyribosomes of Escherichia coli. Breakdown during glucose starvation. J Biol Chem. 1967;242:1065–1068. [PubMed] [Google Scholar]
- 9.Fersht A. Enzyme structure and mechanism. New York, N.Y: W. H. Freeman & Co.; 1985. p. 103. [Google Scholar]
- 10.Flärdh K, Cohen P S, Kjelleberg S. Ribosomes exist in large excess over the apparent demand for protein synthesis during carbon starvation in marine Vibrio sp. strain CCUG 15956. J Bacteriol. 1992;174:6780–6788. doi: 10.1128/jb.174.21.6780-6788.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groat R G, Schultz J E, Zychlinsky E, Bockman A, Matin A. Starvation proteins in Escherichia coli: kinetics of synthesis and role in starvation survival. J Bacteriol. 1986;168:486–493. doi: 10.1128/jb.168.2.486-493.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamel E, Koka M, Nakamoto T. Requirement of an Escherichia coli 50S ribosomal protein component for effective interaction of the ribosome with T and G factors and with guanosine triphosphate. J Biol Chem. 1972;247:805–814. [PubMed] [Google Scholar]
- 13.Josaitis C A, Gaal T, Gourse R L. Stringent control and growth-rate-dependent control have nonidentical promoter sequence requirements. Proc Natl Acad Sci USA. 1995;92:1117–1121. doi: 10.1073/pnas.92.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalpaxis D L, Karahalios P, Papapetropoulou M. Growth phase and growth rate dependence of ribosomal peptidyltransferase activity status in E. coli. Biochimie. 1995;77:963–971. doi: 10.1016/0300-9084(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan R, Apirion D. The fate of ribosomes in Escherichia coli cells starved for a carbon source. J Biol Chem. 1974;250:1854–1863. [PubMed] [Google Scholar]
- 16.Legault L, Jeantet G, Gros F. Inhibition of in vitro protein synthesis by ppGpp. FEBS Lett. 1972;27:71–75. doi: 10.1016/0014-5793(72)80412-5. [DOI] [PubMed] [Google Scholar]
- 17.Li K, Subramanian A R. Selective separation procedure for determination of ribosomal proteins L7 and L12. Anal Biochem. 1975;64:121–129. doi: 10.1016/0003-2697(75)90413-3. [DOI] [PubMed] [Google Scholar]
- 18.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 68–202. [Google Scholar]
- 19.Maruyama H, Ono M, Mizuno D. Ribosome degradation and the degradation products in starved Escherichia coli. III. Ribosomal RNA degradation during the complete deprivation of nutrients. Biochim Biophys Acta. 1970;199:176–183. doi: 10.1016/0005-2787(70)90706-9. [DOI] [PubMed] [Google Scholar]
- 20.Matin A, Auger E A, Blum P H, Schultz J E. Genetic basis of starvation survival in nondifferentiating bacteria. Annu Rev Microbiol. 1989;43:293–316. doi: 10.1146/annurev.mi.43.100189.001453. [DOI] [PubMed] [Google Scholar]
- 21.Nag B, Tewari D S, Traut R R. Monoclonal antibodies to epitopes in both C-terminal and N-terminal domains of Escherichia coli ribosomal protein L7/L12 inhibit elongation factor binding but not peptidyl transferase activity. Biochemistry. 1987;26:461–465. doi: 10.1021/bi00376a018. [DOI] [PubMed] [Google Scholar]
- 22.Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 23.Nyström T. Glucose starvation stimulon of Escherichia coli: role of integration host factor in starvation survival and growth phase-dependent protein synthesis. J Bacteriol. 1995;177:5707–5710. doi: 10.1128/jb.177.19.5707-5710.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ofverstedt L G, Zhang K, Tapio S, Skoglund U, Isaksson L A. Starvation in vivo for aminoacyl-tRNA increases the spatial separation between the two ribosomal subunits. Cell. 1994;79:629–638. doi: 10.1016/0092-8674(94)90548-7. [DOI] [PubMed] [Google Scholar]
- 25.Pain V M, Lewis J A, Huvos P, Henshaw E C, Clemens M J. The effects of amino acid starvation on regulation of polypeptide chain initiation in Ehrlich ascites tumor cells. J Biol Chem. 1980;255:1486–1491. [PubMed] [Google Scholar]
- 26.Ramagopal S. Metabolic changes in ribosomes of Escherichia coli during prolonged culture in different media. Eur J Biochem. 1984;140:353–361. doi: 10.1111/j.1432-1033.1984.tb08108.x. [DOI] [PubMed] [Google Scholar]
- 27.Ron E Z, Kohler R E, Davis B D. Polysomes extracted from Escherichia coli by freeze-thaw-lysozyme lysis. Science. 1966;153:1119–1120. doi: 10.1126/science.153.3740.1119. [DOI] [PubMed] [Google Scholar]
- 28.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 29.Schweder T, Lee K H, Lomovskaya O, Matin A. Regulation of Escherichia coli starvation sigma factor by ClpXP protease. J Bacteriol. 1996;178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segel I H. Enzyme kinetics. New York, N.Y: Wiley Interscience; 1975. pp. 64–71. [Google Scholar]
- 31.Siegele D A, Kolter R. Life after log. J Bacteriol. 1992;174:345–348. doi: 10.1128/jb.174.2.345-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorensen M A, Jensen K F, Pedersen S. High concentrations of ppGpp decrease the RNA chain growth rate: implications for protein synthesis and translational fidelity during amino acid starvation in Escherichia coli. J Mol Biol. 1994;236:441–454. doi: 10.1006/jmbi.1994.1156. [DOI] [PubMed] [Google Scholar]
- 33.Spirin A. Ribosome structure and protein biosynthesis. Menlo Park, Calif: The Benjamin/Cummings Publishing Co. Inc.; 1986. p. 126. [Google Scholar]
- 34.Svitil A L, Cashel M, Zyskind J W. Guanosine tetraphosphate inhibits protein synthesis in vivo. J Biol Chem. 1993;268:2307–2311. [PubMed] [Google Scholar]
- 35.Synetos D, Coutsogeorgopoulos C. Studies on the catalytic rate constant of ribosomal peptidyltransferase. Biochim Biophys Acta. 1987;923:275–285. doi: 10.1016/0304-4165(87)90014-6. [DOI] [PubMed] [Google Scholar]
- 36.Traut R R, Oleinikov A V, Makarov E, Jokhadze G, Perroud B, Wang B. Structure and function of Escherichia coli ribosomal protein L7/L12: effect of cross-links and deletions. In: Nierhaus K H, Franceschi F, Subramanian A R, Erdmann V A, Wittman-Liebold B, editors. The translational apparatus: structure, function, regulation, evolution. New York, N.Y: Plenum Press; 1993. pp. 521–532. [Google Scholar]
- 37.Vogel U, Jensen K F. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J Bacteriol. 1994;176:2807–2813. doi: 10.1128/jb.176.10.2807-2813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wada A, Igarashi K, Yoshimura S, Aimoto S, Ishihama A. Ribosome modulation factor: stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem Biophys Res Commun. 1995;214:410–417. doi: 10.1006/bbrc.1995.2302. [DOI] [PubMed] [Google Scholar]
- 39.Wada A, Yamazaki Y, Fujita N, Ishihama A. Structure and probable genetic location of a “ribosome modulation factor” associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc Natl Acad Sci USA. 1990;87:2657–2661. doi: 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner R, Theissen G, Zacharias M. Regulation of ribosomal RNA synthesis and control of ribosome formation in E. coli. In: Nierhaus K H, Franceschi F, Subramanian A R, Erdmann V A, Wittmann-Liebold B, editors. The translational apparatus: structure, function, regulation, evolution. New York, N.Y: Plenum Press; 1993. pp. 119–130. [Google Scholar]
- 41.Yamagishi M, Matsushima H, Wada A, Sakagami M, Fujita N, Ishihama A. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase- and growth rate-dependent control. EMBO J. 1993;12:625–630. doi: 10.1002/j.1460-2075.1993.tb05695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida M, Travers A, Clark B F C. Inhibition of translation initiation complex formation by MS1. FEBS Lett. 1972;23:163–166. doi: 10.1016/0014-5793(72)80331-4. [DOI] [PubMed] [Google Scholar]