Abstract
This study aimed to investigate the performance of 18F-DCFPyL positron emission tomography/computerized tomography (PET/CT) models for predicting benign-vs-malignancy, high pathological grade (Gleason score > 7), and clinical D'Amico classification with machine learning. The study included 138 patients with treatment-naïve prostate cancer presenting positive 18F-DCFPyL scans. The primary lesions were delineated on PET images, followed by the extraction of tumor-to-background-based general and higher-order textural features by applying five different binning approaches. Three layer-machine learning approaches were used to identify relevant in vivo features and patient characteristics and their relative weights for predicting high-risk malignant disease. The weighted features were integrated and implemented to establish individual predictive models for malignancy (Mm), high path-risk lesions (by Gleason score) (Mgs), and high clinical risk disease (by amico) (Mamico). The established models were validated in a Monte Carlo cross-validation scheme. In patients with all primary prostate cancer, the highest areas under the curve for our models were calculated. The performance of established models as revealed by the Monte Carlo cross-validation presenting as the area under the receiver operator characteristic curve (AUC): 0.97 for Mm, AUC: 0.73 for Mgs, AUC: 0.82 for Mamico. Our study demonstrated the clinical potential of 18F-DCFPyL PET/CT radiomics in distinguishing malignant from benign prostate tumors, and high-risk tumors, without biopsy sampling. And in vivo 18F-DCFPyL PET/CT can be considered a noninvasive tool for virtual biopsy for personalized treatment management.
Supplementary Information
The online version contains supplementary material available at 10.1007/s43657-023-00108-y.
Keywords: Prostate cancer, 18F-DCFPyL positron emission tomography/computerized tomography, Radiomics, Three layer-machine learning, High-risk tumor
Introduction
Prostate cancer (PCa) is the second leading lethal cancer in men (McGuire 2016). In clinical routine, identification of malignant prostate tumors and path-risk stratification of PCa patients is of great significance for subsequent management, especially in the context of personalized diagnosis and treatment (Bratan et al. 2013; Preisser et al. 2020).
The determination of prostate and clinical aggressive phenotypes in PCa remains a clinical challenge. Currently, clinical PCa screening relies on the serum level of prostate-specific antigen (PSA) (D'Amico et al. 1998). The biopsy Gleason score (GS) is the most important tissue-based marker for prostate cancer patients. However, the major challenges in biopsy-based pathological stratification of prostate cancer includes sampling error (e.g., failing to biopsy the area with the highest GS) and variations in biopsy interpretation. Clinical risk stratification in patients with primary prostate cancer is mainly built on clinical stage, PSA, and GS on transrectal biopsy sampling, which might inadequately demonstrate the heterogeneity of tumors (Hsieh et al. 2022). Biopsy-derived GS drives risk stratification and treatment management for PCa patients and the Gleason grading system remains the most powerful prognostic predictor for patients with prostate cancer. For PCa patients stratification, D'Amico classification was developed according to the clinical tumor node metastasis (TNM) stage, biopsy GS, and preoperative prostate-specific antigen level in clinical practice (Litwin and Tan 2017; Valdez-Vargas et al. 2021). This comprehensive analysis could help clinicians in the treatment management of patients with PCa (Ghezzo et al. 2022).
Despite some advantages in the diagnosis and staging of PCa (Tan et al. 2022), conventional imaging techniques, such as computed tomography (CT), magnetic resonance imaging (MRI), are limited due to their low specificity, which usually leads to false-positive results and unnecessary biopsies (Borofsky et al. 2018; Le et al. 2015). In this scenario, positron emission tomography (PET) has played an increasingly vital role via multiple disease-targeting tracers. Of note, radionuclide-labeled prostate-specific membrane antigen (PSMA) has been widely applied to the localization of primary PCa and metastasis. This is because PSMA is overexpressed on most PCa epithelial cells and its expression level strikingly elevates with higher carcinoma grading and staging or restaging (Kyriakopoulos et al. 2018; Obek et al. 2017; Perera et al. 2016).
Radiomics based on PET images has been well established as a promising quantitative approach to indicate the heterogeneity within tumors and help classify risk-based patients in diverse clinical settings (Chen et al. 2017a, 2017b; Lambin et al. 2017; Perandini et al. 2016). Furthermore, radiomics features, which include intensity, shape, texture and wavelet, can provide comprehensive information such as cancer phenotypes, the tumor microenvironment, and so on (Gatenby et al. 2013). Strikingly, with the combination of the patient-derived data, the imaging-based radiomics' capacity will be augmented in the robust improvement of the clinical decision support system (CDSS) (Aerts 2016). Advances in machine learning modeling as applied to clinical challenging may enable the discovery and prediction of clinical cancer pathological and biological characteristics.
In this present study, we aim to explore the predictive performance of radiomics in the risk stratification of PCa patients based on 18F-DCFPyL PET/CT imaging. Through machine learning analysis, we seek to screen out the radiomics features to differentiate the malignancy of PCa and stratify the primary lesions by GS, and D'Amico risk into low/intermediate, vs high-risk of biochemical recurrence after surgery based on clinical TNM stage, GS, and PSA level.
Materials and Methods
Patients Data
Inclusion criteria for this study were: (1) Abnormally elevated serum PSA value. (2) Prostate lesions were found or not found by other examinations (digital rectal examination, ultrasound, CT, MRI, etc.). (3) Before PET examinations, the patients had not received chemotherapy, radiotherapy, endocrine therapy or other therapies. Subjects who met the above three criteria simultaneously are included in our study. The patients without exhibiting relevant radiotracer uptake above background in the prostate on 18F-DCFPyL PET/CT imaging were excluded. This was a monocentric pilot study approved by the Ethical Committee of Qilu Hospital Shandong University. Written informed consents were obtained from all the patients.
18F-DCFPyL PET/CT Imaging
All the patients underwent 18F-DCFPyL PET/CT imaging without fasting. All PET/CT examinations were performed at a whole-body PET/CT scanner (General Electric, Discovery STE16). 18F-DCFPyL, the PSMA-targeting tracer produced by the same PET/CT center, was intravenously injected into patients at a standardized dose of 3.7 MBq/kg. After 18F-DCFPyL administration, the patients rested in a quiet room for about 60 min before scanning.
The image acquisition ranged from parietal to femoral. Sixteen row helical CT scan row helical CT scan was used with the following conditions: tube voltage (120 kv), tube current (300 mA), 3.25 mm thick. PET collection time of every bed was three minutes, the whole body scanning needed six to seven beds. Using the viewpoint method, the scan data was reconstructed into CT, PET and PET/CT fusion images. PET matrix was 128×128, and CT matrix was 512×512. The voxel size was 3.75×3.75×3.75 mm3.
Image Analysis
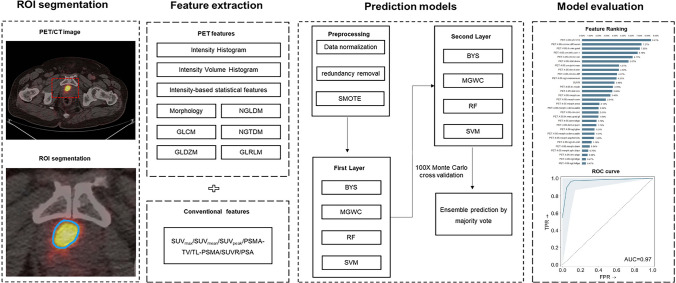
The image analysis was done by two board-certified physicians with four and six years of experience in reading PET/CT. After that, they used LIFEx software Version 6.3.0. to delineate prostate lesions as volumes of interest (VOIs) on PET images, who are blind to each other (Nioche et al. 2018). The initial lesion delineations were cross-examined and corrected manually, the final VOIs were confirmed by a senior nuclear medicine specialist together with a board-certified urologist. To normalize the standardized uptake max value (SUVmax), an extra reference location in the gluteus muscle was contoured, and ratio of standardized uptake max value (SUVR, lesion SUVmax/Gluteus SUVmax) was calculated.
The PET/CT images were primarily evaluated by visual inspection, we evaluated the images presenting metal artifacts and respiratory mobility. Then we analyzed the characteristics of the PET/CT images (such as location, morphology, number, density, and PSMA uptake level) and then selected typical images. Subsequently, through quantitative analysis, regions of interest (ROIs) of lesions were manually placed. Conventional standardized uptake values, including standardized uptake max value (SUVmax), standardized uptake mean value (SUVmean), total volume of PSMA (PSMA-TV), total lesion of PSMA (TL-PSMA), standardized uptake peak value (SUVpeak), SUVR, were merged with the extracted 137 radiomics features to compose a 143 long feature vector for each patient.
Radiomics Analysis
Patients
For the malignancy differentiation, the input dataset was composed of 138 patients, including 109 prostate cancer patients and 29 patients with benign prostate disease. All patients were treated according to guideline recommendations. According to the institution's standard pathologic procedures, all the histological samples were processed according to the institution's standard pathologic procedures. Grading and staging were performed, respectively, according to the International Society of Urological Pathology of World Health Organization (WHO/ISUP) 2016 system and Union for International Cancer Control (UICC) TNM classification.
For the risk stratification, 109 prostate cancer patients were divided into two subgroups according to the GS. The subgroup with GS > 7 included 59 patients, while the subgroup with GS ≤ 7 included 50 patients, and these prostate cancer patients were also divided into another two subgroups according to D'Amico Score, including 34 patients with low-medium and 75 patients with high D'Amico grades.
For the malignancy differentiation analysis and D'Amico grading analysis, to avoid class imbalance, the Synthetic Minority Over-sampling Technique (SMOTE) was employed to synthesize new instances of the minority class between preexisting class members (Wang et al. 2021).
Feature Extraction
Following the Imaging Biomarker Standardization Initiatives (IBSI) guidelines, radiomics features with “very strong” and “strong” consensus values were extracted from each image using an in-house pipeline. Radiomics features extracted from primary lesions were used for machine learning (ML) modeling. A total of 137 radiomics features were extracted from each image (Supplementary figures). Finally, the 137 radiomics features combined with patient PSA serum level and conventional SUV PET (SUVmax, SUVmean, PSMA-TV, TL-PSMA, SUVpeak, and SUVR) were merged to form feature vector.
The image was resampled to 2.0 × 2.0 × 2.0 uniform voxel resolution. Following the Imaging Biomarker Standardization Initiatives (IBSI) guidelines, radiomics features with “very strong” and “strong” consensus values were extracted from each image to support biomarker standardization and study repeatability. The 137 radiomics features were extracted from each image including Gray-level cooccurrence matrix (GLCM), Gray-level distance zone matrix (GLDZM), Neighboring grey level dependence matrix (NGLDM), Neighborhood grey tone difference matrix (NGTDM), Gray-level run-length Matrix (GLRLM), intensity and morphology features.
Feature Redundancy Reduction
Features collected from patients were normalized and redundant features were removed using correlation matrix analysis. The features with an absolute value of Pearson correlation coefficient greater than 0.85 are considered redundant, and the data after removing redundant features are used for the next machine learning modeling (Supplementary files). Preprocessing resulted in average 26 features across all Monte Carlo (MC) folds in the Mm, and average 19 features and 21 features across all MC folds in Mgs and Mamico, respectively.
Machine Learning and Cross-Validation
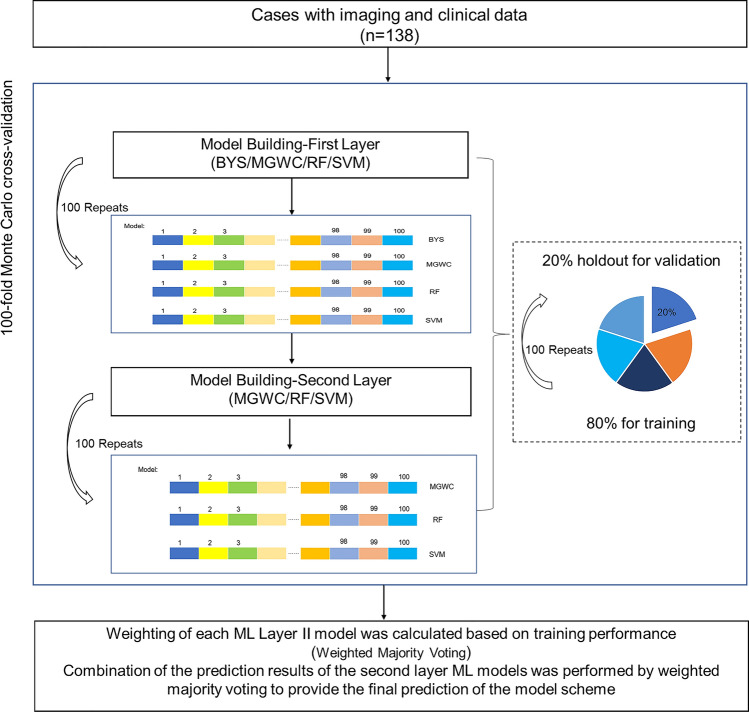
Preprocessing step algorithms as well as their parameter values performed in all Monte Carlo folds before ML. To minimize the effect of method bias and improve the prediction performance, Ensemble Learning was used to build the model, including the following four algorithms: Bayesian classification (BYS), multi-Gaussian weighting (MGWC), random forest (RF) and support vector machine (SVM). The integrated learning model was divided into three layers. Meta-training dataset is generated by evaluating the training results of the first layer for the second layer training, and the final model decision was obtained by weighted tickets of the predicted results of the three models. The importance of each feature in the prediction model was estimated by calculating an R-squared ranking on each cross-validation.
Each model was cross-validated using 100-fold Monte Carlo, using a sample ratio of 80% for the training set and 20% for the validation set. Each cross-validation was re-sampled to ensure that the sample labels in the validation set were balanced. Confusion Matrix analysis was used to estimate model performance, including accuracy (ACC), sensitivity (SENS), specificity (SPEC), positive predictive value (PPV), negative predictive value (NPV), and area under the receiver operating characteristic curve (AUC).
Various machine learning algorithms were established in each fold to minimize the effect of algorithm bias. Each model was trained by randomly selecting 80% of the preprocessed training data per MC fold.
Meta-training sets were created by evaluating the samples of the preprocessed training set in each MC fold by the trained models in the first ML layer. To create the meta-training set, the prediction results of each trained model in the first ML layer were handled as feature values of the given training sample. The meta-training set was the input for training the second ML layer prediction models. These models were trained to identify patterns in the prediction of the first ML layer models to result in mixed super learners.
A combination of the prediction results of the second ML layer models was performed by weighted majority voting to provide the final prediction of the model scheme. The weighting of each second ML Layer model was calculated based on training performance. In addition, ML Layer second models having less training performance than the median of all the second ML layer model training performances had a weight zero in the final vote (Figs. 1 and 2). All detailed ML-methods were described in supplementary files.
Fig. 1.
Radiomics pipeline depicts the data processing and operations to build a radiomics ML model with validation. Firstly, manual segmentation of ROI on PET/CT. Then feature extraction was performed after preprocessing. Data analysis was performed using machine learning. A predictive, prognostic and diagnostic model was built and validated
Fig. 2.
100-fold Monte Carlo cross-validation. Each model was cross-validated using 100-fold Monte Carlo, using a sample ratio of 80% to 20% for the training set and validation set. BYS, Bayesian Classifier; MGWC, Multi-Gaussian Weighted Classifier; RF, Random Forest Classifier; SVM, Support Vector Machine Classifier
Results
Patients
One hundred and thirty-eight patients were enrolled in the study; in these patients, PET/CT, PSA values, histopathology, GS and D'Amico scores were documented (Table 1).
Table 1.
Characteristics of the 138 patients with primary prostate cancer involved in this study
| Patient characteristics (n = 138) | Value |
|---|---|
| Age (years), median (IQR) | 67.4 (64–73) |
| PSA (ng/mL), median (IQR) | 15.8 (8.0–25.2) |
| Pathologic results, n (ratio%) | 138 |
| Adenocarcinoma | 109 (79) |
| Hyperplasia | 26 (19) |
| Inflammation | 3 (2) |
| Pathologic T staging, n (ratio%) | 109 |
| T1c–T2a | 25 (23) |
| T2b | 25 (23) |
| ≥ T2c | 59 (54) |
| Primary Gleason pattern, n (ratio%) | 109 |
| 3 | 23 (21) |
| 4 | 61 (56) |
| 5 | 25 (23) |
| Secondary Gleason pattern, n (ratio%) | 109 |
| 3 | 35 (32) |
| 4 | 53 (50) |
| 5 | 21 (18) |
| Total Gleason Score, n (ratio%) | 109 |
| 6 | 6 (5) |
| 7 | 44 (40) |
| ≥ 8 | 59 (55) |
| Amico Score, n (ratio%) | 109 |
| 0 | 1 (1) |
| 1–3 | 33 (29) |
| 4–12 | 75 (70) |
Cross-Validation Performance
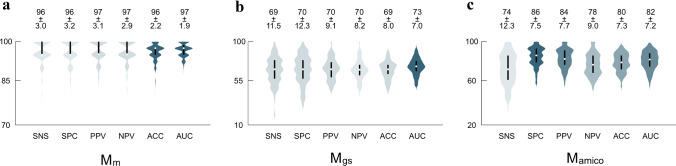
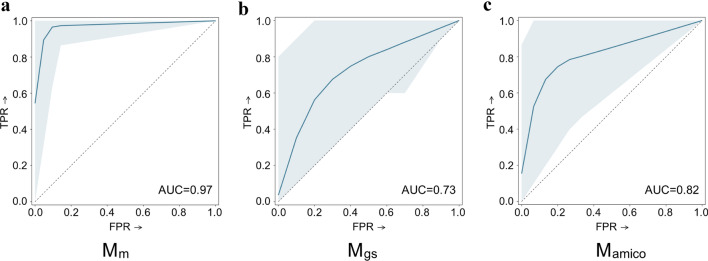
The cross-validated malignancy differentiation, GS and D'Amico risk stratification prediction model performances across all MC-folds yielded in average of 96%, 69%, 74% sensitivity, 96%, 70%, 86% specificity, 97%, 70%, 84% positive predictive value, 97%, 70%, 78% negative predictive value and 96%, 69%, 80% accuracy, respectively (Fig. 3). The area under the receiver operating curve (AUC) of the models was 0.97, 0.73, and 0.82, respectively (Fig. 4).
Fig. 3.
The cross-validated performance of a Mm, b Mgs, c Mamico. SNS, Sensitivity; SPC, Specificity; PPV, Positive Predictive Value; NPV, Negative Predictive Value; ACC, Accuracy; AUC, Area Under the Receiver Operating Characteristic Curve. Performance values as percentages (%). Detailed values were listed in supplementary files
Fig. 4.
The cross-validated Receiver Operator Characteristics (ROC) curves of a Mm, b Mgs, c Mamico. The thick blue line corresponds to the mean ROC curve, while the light blue shaded area represents the spread of all 100 ROC curves generated across the validation folds. AUC, Area Under the Receiver Operating Characteristics Curve (mean of all 100 AUCs); FPR, False Positive Rate; TPR, True Positive Rate
Feature Importance Ranking
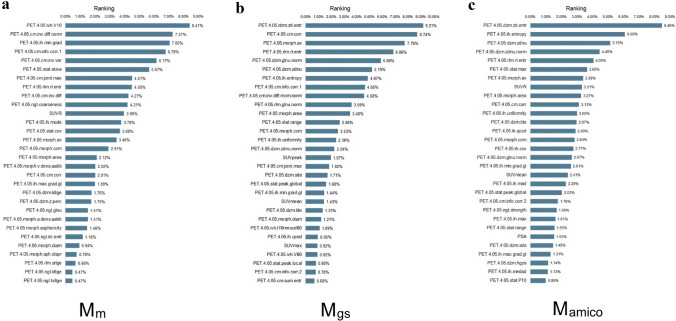
Feature ranking and selection were performed as part of the data preprocessing steps of each fold. The final feature importance was calculated as the mean of all feature rankings across the MC folds. The three most predictive features of Mm were PET.4.05.ivh.V10 (8.41%), PET.4.05. cm.inv.diff.norm (7.27%) and PET.4.05.ih.min.grad (7.05%) (Fig. 5a). In the Mgs prediction model, three most predictive features were PET.4.05.dzm.zd.entr (9.21%), PET.4.05.cm.corr (8.74%) and PET.4.05.morph.av (7.76%) (Fig. 5b). While in the prediction model of Mamico, the most predictive feature was PET.4.05.dzm.zd.entr (8.48%), followed by PET.4.05.ih.entropy (6.09%) and PET.4.05.dzm.zdnu (5.15%) (Fig. 5c). Note that the details of the highest-ranking 30 features are shown in the supplemental files. Representative three patients with benign prostate hyperplasia and low-grade and high-grade prostate cancer were illustrated, the corresponding values of the highest important features were recorded (Fig. 6).
Fig. 5.
The 30 most predictive features of a Mm, b Mgs, c Mamico. Detailed feature values are listed in supplemental files
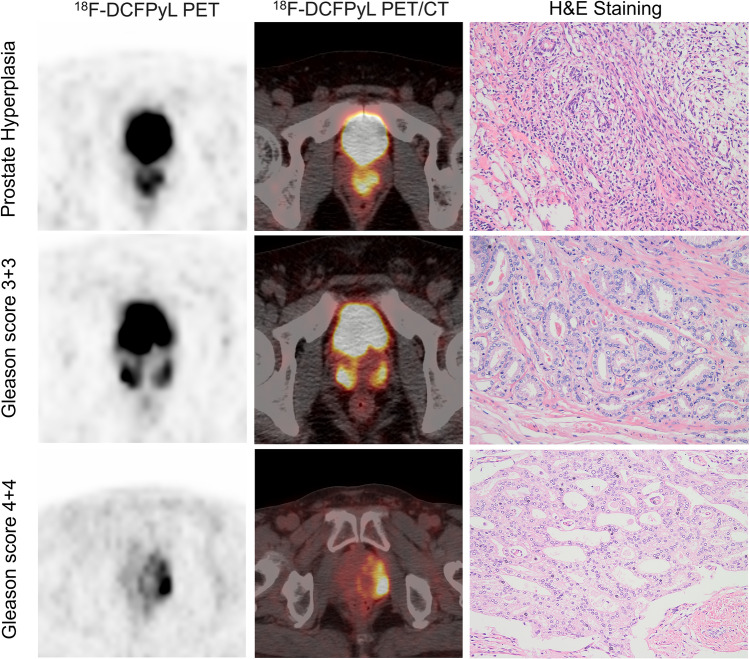
Fig. 6.
Representative 18F-DCFPyL PET/CT scans of three patients who presented prostate hyperplasia, low-medium and high Gleason grade tumors
Discussion
PSMA PET imaging shown promise for patient screening and management in metastatic castration-resistant prostate cancer prior to radioligand therapy. Although PSMA PET imaging is appreciated for playing a vital role in diagnosing and grading prostate cancer, but it has been restricted to baseline characterization and grading in the primary tumor. Furthermore, conventional biopsy-derived Gleason grade frequently underestimates the pathological risk. The radiomics ML-analysis of PSMA PET imaging is potentially feasible for tailoring PCa patients' management strategies quantitatively and effectively in our previous findings. In this study, we recruited 79% of patients with a diagnosis of malignancy and 54.1% patients with GS > 7 disease based on conventional TRUS-biopsies we constructed and validated the feasibility of machine-learned radiomics models built on 18F-PSMA PET/CT for predicting prostate lesion-specific benign vs malignancy and low vs high risk.
Application of radiomics ML models to patients with primary prostate cancer demonstrated model performance that may enable the prediction of clinically aggressive disease, which showed excellent cross-validation performances for malignancy differentiation (AUC 0.97), GS (AUC 0.73) and D'Amico risk stratification (AUC 0.82). It resolved that radiomics features are of prominent significance in relation to the tumor risk in PCa patients, which could be employed well as a non-invasive way to predict the aggressiveness for the distinguishment of different PCa subgroups with improving the accuracy of prediction. Our current model potentially provides a non-invasive approach for PCa screening that may reduce unnecessary prostate biopsies and subsequent patients' management.
Conventional diagnostic methods, e.g., serum PSA test, digital rectal examinations (DREs), and prostate biopsy, inevitably have inherent major deficiencies. The main deficiencies are that they may lead to overdiagnosis of prostate cancer and misleading diagnosis of clinically significant cancer. 18F-DCFPyL PET, as a novel noninvasive method, has been recommended for an examination of prostate cancer and proved to be beneficial in the detection of malignancy. PSMA PET was helpful for disease staging and risk stratification of prostate cancer, and its combination with several biomarkers might reduce unnecessary biopsies. Moreover, PSMA PET contains molecular information, which has not attracted enough attention in clinical practice. Recently, artificial intelligence, such as radiomics, has shown great potential for evaluating the aggressiveness of multiple tumors.
For lesional risk assessment, the radiomics features based on PET come up significant with superiority in the importance ranking list, however, conventional uptake values are not a significant indicator to differentiate the malignant and high-risk tumor (both defined by GS or D'Amico grading) from a variety of prostate diseases, even conventional imaging parameters have been widely acknowledged to have the potential to evaluate aggressiveness.
Our results indicate that the in-vivo Gleason scoring can accurately stratify the PCa patients with discerned risks as the alternative to the biopsy-based score. Effective as it is on treatment planning and pathology scoring is limited in assessing the whole prostate and the characterization of the tumor heterogeneity as it is usually sampled from a single biopsy site (Hatt et al. 2017). Besides, our published risk classification systems have been demonstrated to potentially grade prostate cancer patients into the wrong groups (Papp et al. 2021). So, the high-throughput non-invasive approach could enhance the risk stratification of primary prostate cancer based on PET/CT radiomics and machine learning.
Non-prominent value of PSA on GS grading might be related to its nonspecific nature and the relatively low PSA values in recruited patients. Moreover, the patients with high- differentiated prostates and PSMA expression may have present PSA levels (Ladjevardi et al. 2013; Sarikaya et al. 2014).
There are some limitations to our study. Although this automated ML model has shown promise in accurately grading prostate tumor, which might achieve pathologist-level performance. The sample size is relatively small from a mono-medical center, which explains our case distribution of 80% for training the model and 20% for the validation set. Further study with a large sample size will be conducted for model validation with the cooperation of other medical centers. Secondly, our cohort includes patients with prostate adenocarcinoma, benign prostate hyperplasia, and prostatitis, which is necessary for malignancy differentiation but comparably heterogeneous for modeling. Furthermore, an alternative strategy for discriminant analysis using the k-fold strategy to split data into training and validation sets was warranted in future evaluation (Laudicella et al. 2022).
Conclusion
We developed the ML models based on 18F-DCFPyL PET/CT radiomics as a noninvasive approach for identifying malignant and relative high-risk primary tumors. We also demonstrated an independent prognostic model for hormonal therapy, which can be considered a noninvasive tool for individual treatment management. Our findings should be further validated by future larger multicenter studies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Qilu Hospital of China, Vienna General Hospital of Austria, and Evomics Medical Technology Co., Ltd. for the support.
Abbreviations
- PET
Positron emission tomography
- PCa
Prostate cancer
- GS
Gleason score
- PSA
Prostate-specific antigen
- CT
Computed tomography
- PSMA
Prostate-specific membrane antigen
- TNM
Tumor node metastasis
- MRI
Magnetic resonance imaging
- CDSS
Clinical decision support system
- SUV
Standardized uptake value
- SUVmean
Standardized uptake mean value
- SUVmax
Standardized uptake max value
- SUVpeak
Standardized uptake peak value
- SUVR
Ratio of standardized uptake max value
- VOIs
Volumes of interest
- PSMA-TV
Total volume of PSMA
- TL-PSMA
Total lesion of PSMA
- SMOTE
Synthetic minority over-sampling technique
- IBSI
Imaging biomarker standardization initiatives
- MC
Monte Carlo
- ML
Machine learning
- GLCM
Gray-level cooccurrence matrix
- GLDZM
Gray-level distance zone matrix
- NGLDM
Neighboring grey level dependence matrix
- NGTDM
Neighborhood grey tone difference matrix
- GLRLM
Gray-level run-length matrix
- BYS
Bayesian classification
- MGWC
Multi-Gaussian weighting
- RF
Random forest
- SVM
Support vector machine
- WHO
World Health Organization
- ISUP
International Society of Urological Pathology
- UICC
Union for International Cancer Control
- ACC
Accuracy
- SENS
Sensitivity
- SPEC
Specificity
- PPV
Positive predictive value
- NPV
Negative predictive value
- AUC
Area under the receiver operating characteristic curve
- DREs
Digital rectal examinations
- Mm
Predictive models for malignancy
- Mgs
Predictive models for high path-risk lesions (by Gleason score)
- Mamico
Predictive models for high clinical risk disease (by amico)
- ROI
Region of interest
Authors’ contributions
YKL performed data curation, image analysis, investigation, methodology, original and revised draft writing and uploading; SLH and SWW performed machine learning analysis and original draft writing; FL and JN performed original draft writing; PS, JFL and SH performed radioactive tracer synthesis and quality control; LLQ performed image analysis; XL and XL performed methodology, writing, reviewing and editing.
Declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Ethics approval
This study was approved by the Ethical Committee of Shandong University Qilu Hospital, Jinan, Shandong, China (KYLL-2017–573). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Data availability
The datasets generated during analysis in the current study are available from the corresponding author.
Consent to participate
Written informed consent was obtained from each participant before enrollment.
Consent for publication
The participants have consented to the submission of the data and figures to the journal.
Contributor Information
Xin Li, Email: lixin16@sdu.edu.cn.
Xiang Li, Email: xiang.li@meduniwien.ac.at.
References
- Aerts HJ. The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol. 2016;2(12):1636–1642. doi: 10.1001/jamaoncol.2016.2631. [DOI] [PubMed] [Google Scholar]
- Borofsky S, George AK, Gaur S, Bernardo M, Greer MD, Mertan FV, Taffel M, Moreno V, Merino MJ, Wood BJ, Pinto PA, Choyke PL, Turkbey B. What are we missing? False-negative cancers at multiparametric MR imaging of the prostate. Radiology. 2018;286(1):186–195. doi: 10.1148/radiol.2017152877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratan F, Niaf E, Melodelima C, Chesnais AL, Souchon R, Mege-Lechevallier F, Colombel M, Rouviere O. Influence of imaging and histological factors on prostate cancer detection and localisation on multiparametric MRI: a prospective study. Eur Radiol. 2013;23(7):2019–2029. doi: 10.1007/s00330-013-2795-0. [DOI] [PubMed] [Google Scholar]
- Chen B, Zhang R, Gan Y, Yang L, Li W. Development and clinical application of radiomics in lung cancer. Radiat Oncol. 2017;12(1):154. doi: 10.1186/s13014-017-0885-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Harmon S, Perk T, Li X, Chen M, Li Y, Jeraj R. Diagnostic classification of solitary pulmonary nodules using dual time (18)F-FDG PET/CT image texture features in granuloma-endemic regions. Sci Rep. 2017;7(1):9370. doi: 10.1038/s41598-017-08764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amico AV, Whittington R, Malkowicz SB, Schultz D, Blank K, Broderick GA, Tomaszewski JE, Renshaw AA, Kaplan I, Beard CJ, Wein A. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280(11):969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- Gatenby RA, Grove O, Gillies RJ. Quantitative imaging in cancer evolution and ecology. Radiology. 2013;269(1):8–15. doi: 10.1148/radiol.13122697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzo S, Bezzi C, Presotto L, Mapelli P, Bettinardi V, Savi A, Neri I, Preza E, Samanes Gajate AM, De Cobelli F, Scifo P, Picchio M (2022) State of the art of radiomic analysis in the clinical management of prostate cancer: a systematic review. Crit Rev Oncol Hematol 169:103544. 10.1016/j.critrevonc.2021.103544 [DOI] [PubMed]
- Hatt M, Tixier F, Pierce L, Kinahan PE, Le Rest CC, Visvikis D (2017) Characterization of PET/CT images using texture analysis: the past, the present... any future? Eur J Nucl Med Mol Imaging 44(1):151–165. 10.1007/s00259-016-3427-0 [DOI] [PMC free article] [PubMed]
- Hsieh PF, Chang TY, Lin WC, Chang H, Chang CH, Huang CP, Yang CR, Chen WC, Chang YH, Wang YD, Huang WC, Wu HC. A comparative study of transperineal software-assisted magnetic resonance/ultrasound fusion biopsy and transrectal cognitive fusion biopsy of the prostate. BMC Urol. 2022;22(1):72. doi: 10.1186/s12894-022-01011-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakopoulos CE, Chen YH, Carducci MA, Liu G, Jarrard DF, Hahn NM, Shevrin DH, Dreicer R, Hussain M, Eisenberger M, Kohli M, Plimack ER, Vogelzang NJ, Picus J, Cooney MM, Garcia JA, DiPaola RS, Sweeney CJ. Chemohormonal therapy in metastatic hormone-sensitive prostate cancer: long-term survival analysis of the randomized phase III E3805 CHAARTED Trial. J Clin Oncol. 2018;36(11):1080–1087. doi: 10.1200/JCO.2017.75.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladjevardi S, Berglund A, Varenhorst E, Bratt O, Widmark A, Sandblom G (2013) Treatment with curative intent and survival in men with high-risk prostate cancer. A population-based study of 11,380 men with serum PSA level 20–100 ng/mL. BJU Int 111(3):381–388. 10.1111/j.1464-410X.2012.11320.x [DOI] [PubMed]
- Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue R, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- Laudicella R, Comelli A, Liberini V, Vento A, Stefano A, Spataro A, Croce L, Baldari S, Bambaci M, Deandreis D, Arico D, Ippolito M, Gaeta M, Alongi P, Minutoli F, Burger IA, Baldari S. [(68)Ga]DOTATOC PET/CT radiomics to predict the response in GEP-NETs undergoing [(177)Lu]DOTATOC PRRT: the "Theragnomics" concept. Cancers (Basel) 2022;14(4):984. doi: 10.3390/cancers14040984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le JD, Tan N, Shkolyar E, Lu DY, Kwan L, Marks LS, Huang J, Margolis DJ, Raman SS, Reiter RE. Multifocality and prostate cancer detection by multiparametric magnetic resonance imaging: correlation with whole-mount histopathology. Eur Urol. 2015;67(3):569–576. doi: 10.1016/j.eururo.2014.08.079. [DOI] [PubMed] [Google Scholar]
- Litwin MS, Tan HJ. The diagnosis and treatment of prostate cancer: a review. JAMA. 2017;317(24):2532–2542. doi: 10.1001/jama.2017.7248. [DOI] [PubMed] [Google Scholar]
- McGuire S (2016) World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr 7(2):418–419. 10.3945/an.116.012211 [DOI] [PMC free article] [PubMed]
- Nioche C, Orlhac F, Boughdad S, Reuze S, Goya-Outi J, Robert C, Pellot-Barakat C, Soussan M, Frouin F, Buvat I. LIFEx: A freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 2018;78(16):4786–4789. doi: 10.1158/0008-5472.CAN-18-0125. [DOI] [PubMed] [Google Scholar]
- Obek C, Doganca T, Demirci E, Ocak M, Kural AR, Yildirim A, Yucetas U, Demirdag C, Erdogan SM, Kabasakal L, Members of Urooncology Association T (2017) The accuracy of (68)Ga-PSMA PET/CT in primary lymph node staging in high-risk prostate cancer. Eur J Nucl Med Mol Imaging 44(11):1806-1812. 10.1007/s00259-017-3752-y [DOI] [PubMed]
- Papp L, Spielvogel CP, Grubmuller B, Grahovac M, Krajnc D, Ecsedi B, Sareshgi RAM, Mohamad D, Hamboeck M, Rausch I, Mitterhauser M, Wadsak W, Haug AR, Kenner L, Mazal P, Susani M, Hartenbach S, Baltzer P, Helbich TH, Kramer G, Shariat SF, Beyer T, Hartenbach M, Hacker M. Supervised machine learning enables non-invasive lesion characterization in primary prostate cancer with [(68)Ga]Ga-PSMA-11 PET/MRI. Eur J Nucl Med Mol Imaging. 2021;48(6):1795–1805. doi: 10.1007/s00259-020-05140-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perandini S, Soardi GA, Motton M, Augelli R, Dallaserra C, Puntel G, Rossi A, Sala G, Signorini M, Spezia L, Zamboni F, Montemezzi S. Enhanced characterization of solid solitary pulmonary nodules with Bayesian analysis-based computer-aided diagnosis. World J Radiol. 2016;8(8):729–734. doi: 10.4329/wjr.v8.i8.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera M, Papa N, Christidis D, Wetherell D, Hofman MS, Murphy DG, Bolton D, Lawrentschuk N. Sensitivity, specificity, and predictors of positive (68)Ga-prostate-specific Membrane antigen positron emission tomography in advanced prostate cancer: a systematic review and meta-analysis. Eur Urol. 2016;70(6):926–937. doi: 10.1016/j.eururo.2016.06.021. [DOI] [PubMed] [Google Scholar]
- Preisser F, Cooperberg MR, Crook J, Feng F, Graefen M, Karakiewicz PI, Klotz L, Montironi R, Nguyen PL, D'Amico AV. Intermediate-risk prostate cancer: stratification and management. Eur Urol Oncol. 2020;3(3):270–280. doi: 10.1016/j.euo.2020.03.002. [DOI] [PubMed] [Google Scholar]
- Sarikaya S, Resorlu M, Oguz U, Yordam M, Bozkurt OF, Unsal A. Evaluation of the pathologic results of prostate biopsies in terms of age, Gleason score and PSA level: our experience and review of the literature. Arch Ital Urol Androl. 2014;86(4):288–290. doi: 10.4081/aiua.2014.4.288. [DOI] [PubMed] [Google Scholar]
- Tan WP, Kotamarti S, Chen E, Mahle R, Arcot R, Chang A, Ayala A, Michael Z, Seguier D, Polascik TJ. Oncological and functional outcomes of men undergoing primary whole gland cryoablation of the prostate: a 20-year experience. Cancer. 2022;128(21):3824–3830. doi: 10.1002/cncr.34458. [DOI] [PubMed] [Google Scholar]
- Valdez-Vargas AD, Sanchez-Lopez HM, Badillo-Santoyo MA, Maldonado-Valadez RE, Manzo-Perez BO, Perez-Abarca VM, Manzo-Perez G, Vanzzini-Guerrero MA, Alvarez-Canales JA (2021) Recurrence rate of localized prostate cancer after radical prostatectomy according to D'amico risk classification in a tertiary referral hospital: association study. Cir Cir 89(4):520–527. 10.24875/CIRU.200007601 [DOI] [PubMed]
- Wang S, Dai Y, Shen J, Xuan J. Research on expansion and classification of imbalanced data based on SMOTE algorithm. Sci Rep. 2021;11(1):24039. doi: 10.1038/s41598-021-03430-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during analysis in the current study are available from the corresponding author.