Abstract
CCDC88B is a risk factor for several chronic inflammatory diseases in humans and its inactivation causes a migratory defect in DCs in mice. CCDC88B belongs to a family of cytoskeleton-associated scaffold proteins that feature protein:protein interaction domains. Here, we identified the Rho/Rac Guanine Nucleotide Exchange Factor 2 (ARHGEF2) and the RAS Protein Activator Like 3 (RASAL3) as CCDC88B physical and functional interactors. Mice defective in Arhgef2 or Rasal3 show dampened neuroinflammation, and display altered cellular response and susceptibility to colitis; ARHGEF2 maps to a human Chromosome 1 locus associated with susceptibility to IBD. Arhgef2 and Rasal3 mutant DCs show altered migration and motility in vitro, causing either reduced (Arhgef2) or enhanced (Rasal3) migratory properties. The CCDC88B/RASAL3/ARHGEF2 complex appears to regulate DCs migration by modulating activation of RHOA, with ARHGEF2 and RASAL3 acting in opposite regulatory fashions, providing a molecular mechanism for the involvement of these proteins in DCs immune functions.
Subject terms: Cell migration, Inflammation, Protein-protein interaction networks, Inflammatory bowel disease
CCDC88B physically interacts with ARHGEF2 and RASAL3; defective mice show dampened neuroinflammation, altered susceptibility to colitis and altered DCs motility by modulating RHOA, with ARHGEF2 and RASAL3 acting in opposite regulatory fashions.
Introduction
Neuroinflammation refers to inflammation of the brain and of the spinal cord. Acute neuroinflammation is a severe and rapidly fatal condition encountered in microbial or auto-immune encephalitis1,2. Neuroinflammation is a common feature and contributor of chronic inflammatory, autoimmune and neurodegenerative diseases affecting the brain and the spinal cord such as in multiple sclerosis (MS), Alzheimer’s disease and Parkinson’s disease3. Chronic neuroinflammation is a major driver of pathogenesis in these conditions, and a better understanding of the underlying cellular and molecular mechanism is required to develop new treatment strategies. The mouse model of experimental cerebral malaria (ECM) induced by infection with Plasmodium berghei ANKA (PbA) has been used extensively to demonstrate the critical role of myeloid cells (neutrophils, dendritic cells, macrophages), CD4+ and CD8+ T cells and NK cells in the pathogenesis of acute neuroinflammation, and to establish the role of pro-inflammatory mediators (e.g., TNF, type I and type II IFN, IL1b, MIP1a, MIP1b/CCL4, CXCL1, CXCL9, CXCL10)4. On the other hand, the mouse model of experimental autoimmune encephalomyelitis (EAE) has been used to study cell types and molecules involved in chronic neuroinflammation with associated autoimmunity5.
We have used genome wide mutagenesis in mice to identify novel genes that modulate neuroinflammation, and for which mutational inactivation protects against lethal encephalitis during infection with Plasmodium berghei ANKA6–9. One of the genes uncovered was Ccdc88b, a gene expressed exclusively in hematopoietic organs (spleen, bone marrow, lymph nodes and thymus), and in lymphoid and myeloid cells derived from them6. Immunophenotyping studies indicated that abrogation of Ccdc88b function causes loss of T lymphocyte function, with decreased maturation, impaired activation and reduced cytokine production in response to pro-inflammatory signals or T-cell receptor engagement6. In myeloid cells, loss of Ccdc88b impairs several aspects of dendritic cell (DC) function, suggesting that the CCDC88B protein is essential for cellular and molecular pathways common to these two cell types. Finally, we have shown that CCDC88B is required for in vitro mobility and in vivo migration of DCs, including their capacity to activate T lymphocytes10.
In humans, the CCDC88B gene maps on Chromosome 11q13 within a locus associated with vulnerability to several common inflammatory diseases including psoriasis, primary biliary cirrhosis, sarcoidosis, MS, and inflammatory bowel disease (IBD)11–13. In mouse models, we have shown that Ccdc88b-deficient mice are protected against DSS-induced intestinal colitis. Likewise, in a T cell transfer model, Ccdc88b-deficent T cells do not induce colitis in immunocompromised Rag1−/− mice13. Parallel studies have shown that the CCDC88B protein and mRNA are elevated in the colon of IBD patients. Furthermore, expression of CCDC88B mRNA is regulated by cis-acting variants in CD14+ myeloid cells, with CCDC88B mRNA expression correlated positively with disease risk in a cohort of Crohn’s disease patients13. Together, these findings have established a key role for CCDC88B in the pathogenesis of inflammatory conditions of the brain and of the gut.
CCDC88B belongs to the Hook-related protein family that includes, CCDC88A (GIV, Girdin, HkRP1), CCDC88B (Gipie, HkRP3) and CCDC88C (DAPLE, HkRP2). These Hook proteins are defined by shared secondary structure motifs that include an N-terminal Hook-related microtubule-binding domain (MBD), a central coiled coil domain (CCD), and a C-terminal domain involved in binding different subcellular organelles14. Whereas the MBD and CCD domains show sequence conservation, the divergent C-terminal domain is thought to confer protein-specific functions14,15. The study of these proteins has shown that they are cytoplasmic scaffold proteins that function in different physiological and biochemical pathways, some of which have been revealed by proteins they recruit to specific molecular scaffolds. CCDC88A binds to the Akt kinase and to the actin cytoskeleton and is involved in migration of cancer cells, and in angiogenesis16. CCDC88C is a cytoplasmic scaffold protein essential for transducing Wnt signaling pathways17, and mutations in CCDC88C have been linked to transformation and metabolism of cancer cells18,19; familial autosomal recessive truncations of CCDC88C was also found in severe congenital hydrocephalus20. CCDC88B was shown to interact with the CDC42 guanine nucleotide exchange factor DOCK 821.
Here, we have used a systematic proteomics approach to identify CCDC88B protein interactors in primary thymocytes and in the T cell line BI-141. These studies identified the Rho/Rac Guanine Nucleotide Exchange Factor 2 (ARHGEF2) and the RAS Protein Activator Like 3 (RASAL3) as specific CCDC88B interactors, and this was further validated by co-immunoprecipitation and by double immunofluorescence with confocal microscopy. In addition, mice defective in Rasal3 or Arhgef2 share a protective phenotype in mouse models of inflammation similar to that exhibited by Ccdc88b mutants: Arhgef2 and Rasal3 deficient mice are protected against neuroinflammation in the EAE model and, to a lesser extent, the ECM model. Interestingly, loss of Rasal3 or Arhgef2 also exacerbates the effect of DSS induced colitis. Finally, mutations in Rasal3 and in Arhgef2 affect the motility and migration of DCs, a function also affected in Ccdc88b mutant primary DCs10. These studies identify the CCDC88B/RASAL3/ARHGEF2 molecular scaffold as playing a critical role in the migratory properties of DCs.
Results
RASAL3 and ARHGEF2 physically interact with CCDC88B
We previously established the cellular basis for the ECM and IBD protective effect of Ccdc88b inactivation13. Yet, the underlying mechanistic basis of these effects remains unknown. Primary sequence analysis of CCDC88B points to several family-specific sequence motifs associated with protein-protein interaction and organelles binding or localization. To identify CCDC88B-dependent biochemical pathways important for the function of myeloid and lymphoid cells, we sought to identify proteins that physically and functionally interact with CCDC88B in these cells6,10,13.
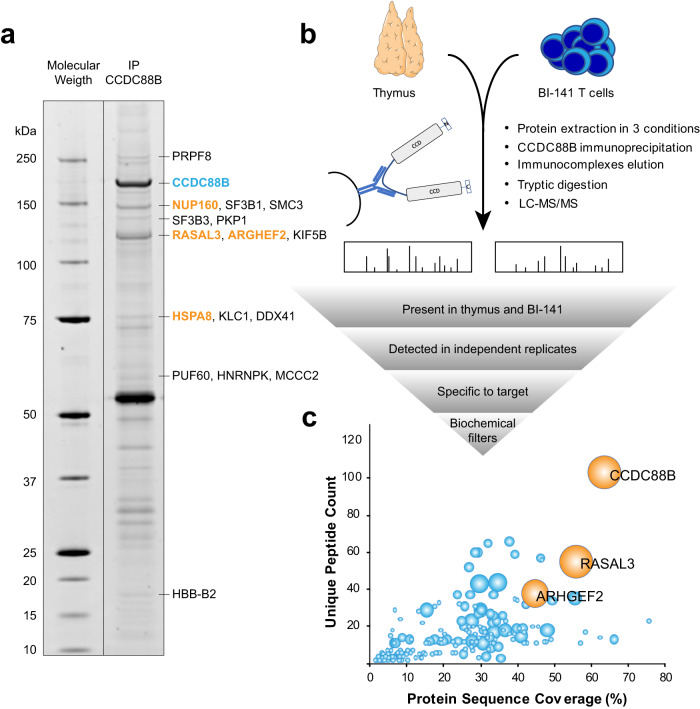
We first performed an experiment where CCDC88B was immunoprecipitated from murine thymic extracts, followed by separation of captured proteins by SDS-PAGE. Nine Sypro stained bands were excised and analyzed by LC-MS/MS (Fig. 1a and Supplementary Data 1), and four different putative CCDC88B interactors were detected with a high spectral count: RAS Protein Activator Like 3 (RASAL3), Rho/Rac Guanine Nucleotide Exchange Factor 2 (ARHGEF2), Heat Shock Protein Family A Member 8 (HSPA8) and Nucleoporin 160 (NUP160). Of note, these proteins were excised at their corresponding molecular mass and were easily visible on SDS-PAGE. CCDC88B was also detected as the most abundant protein at its expected molecular mass (Fig. 1a).
Fig. 1. Identification of CCDC88B-interacting proteins by LC-MS/MS.
a Co-immunoprecipitation of CCDC88B from whole thymus extract with anti-CCDC88B anti-serum analyzed by gel electrophoresis. The gel was stained with Sypro, nine bands were excised and submitted to LC-MS/MS (see “Results”). The four most abundant proteins (spectral count >20) are identified in orange. b Schematic representation of the experimental procedure used for proteomics analysis. Proteins were extracted from whole thymus or BI-141 T cell line using three different lysis buffers, then submitted to immunoprecipitation using an anti-CCDC88b polyclonal anti-serum. Eluates were subjected to LC-MS/MS to identify and quantify enriched proteins. The results were analyzed using multiple filtering parameters to identify the most probable CCDC88B interactors. An independent experiment was performed in duplicate on whole thymic extracts. c Graph presenting LC-MS/MS quantitative results. The percentage of protein length that was covered by MS/MS identified peptides is plotted against the number of unique peptides identified, whereas the bubble size is proportional to a score (arbitrary units) based on filtering parameters (see Supplementary Data 1); the two best candidates (along with CCDC88B) are drawn in orange. Parts of (b) were drawn by using pictures from Servier Medical Art. Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License (https://creativecommons.org/licenses/by/3.0/).
We next carried out co-immunoprecipitations of CCDC88B from cryo-milled BI-141 T cells and mouse thymi, followed by identification of pulled down proteins by LC-MS/MS (Fig. 1b); 253 proteins were detected (Supplementary Data 1) and filters taking into account abundance (number of peptides and protein coverage), relative intensity, retention of interaction under different co-immunoprecipitation conditions, and presence in both BI-141 cells and in primary thymic co-immunoprecipitates in independent tests were applied. Because CCDC88B is a cytoplasmic protein, nuclear proteins, DNA-binding proteins and species commonly found in protein:protein interaction studies22 were discarded. A priority score was assigned to the different species detected (Fig. 1b; Supplementary Data 1 and Supplementary Fig. 1), resulting in the identification of RASAL3 and ARHGEF2 as the two top candidate CCDC88B interactors (Fig. 1c). RASAL3 is a 114.7 kDa cytosolic protein, member of the Ras GTPase-activating proteins (RasGAP) family with characteristic pleckstrin homology (PH), C2, and Ras GTPase-activation protein (RasGAP) domains. This protein family may act as a negative regulator of Ras signaling23. ARHGEF2 (also known as GEF-H1) is a 133.7 kDa cytosolic protein member of the Rho/Rac Guanine Nucleotide Exchange Factor family known to activate Rho-GTPases by promoting the exchange of GDP for GTP24.
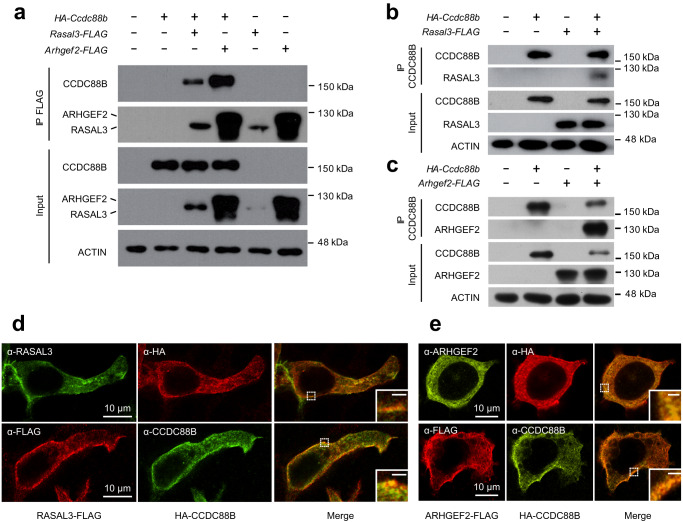
Physical interaction between CCDC88B, RASAL3 and ARHGEF2 was further investigated following pairwise transient expression of the 3 proteins in HEK293T cells (Fig. 2). Cell extracts were subjected to immunoprecipitation and immunoblotting with pairs of antibodies; CCDC88B (detected by immunoblotting with anti-HA) could be pulled down by either RASAL3 or ARHGEF2 when immunoprecipitated with anti-FLAG antibody (Fig. 2a). Conversely, CCDC88B immunoprecipitated both RASAL3 (Fig. 2b) and ARHGEF2 (Fig. 2c) detected by anti-FLAG. In addition, interaction between CCDC88B, RASAL3, and ARHGEF2 was analyzed by double immunofluorescence following expression of the proteins in HEK293T cells transfected as above. These experiments identified a strong overlapping staining between CCDC88B and RASAL3 (Fig. 2d) or ARGHEF2 (Fig. 2e). Staining for the 3 proteins expressed in HEK293T cells was very similar, appearing primarily associated with both the cell membrane and with intracellular punctate/filamentous compartment.
Fig. 2. Validation of physical interaction between CCDC88B, ARHGEF2 and RASAL3.
a Co-immunoprecipitation demonstrating interactions between CCDC88B, RASAL3 and ARHGEF2. HEK293T cells were transiently transfected with the indicated expression vectors and cells extracts were subjected to immunoprecipitation with an anti-FLAG monoclonal antibody. Immunoprecipitated proteins (IP FLAG) and total lysates (Input) were analyzed by gel electrophoresis (SDS-PAGE) and subjected to immunoblotting using an anti-HA monoclonal antibody to reveal HA-tagged CCDC88B. b, c HEK293T cells transfected with the indicated expression vectors were subjected to immunoprecipitation with an anti-CCDC88B polyclonal anti-serum. Immunoprecipitated proteins (IP CCDC88B) and total lysates (Input) were analyzed by gel electrophoresis and subjected to immunoblotting using an anti-FLAG monoclonal antibody to reveal FLAG tagged RASAL3 and ARHGEF2 (b, c respectively). d, e Double immunofluorescence of HA-CCDC88B and RASAL3-FLAG or ARHGEF2-FLAG co-expressed in transiently transfected HEK293T cells, respectively. Tagged proteins were revealed by combinations of anti-FLAG, anti-CCDC88B, anti-HA, and anti-ARHGEF2 (with corresponding fluorophores) as indicated in the boxes. Images were acquired by confocal microscopy, insert scale bar = 1 μm. Closeups in inserts show the extent of colocalization near the cell membrane.
Taken together, these studies strongly suggest that CCDC88B forms a cellular complex with RASAL3 and ARHGEF2 in T cells. This complex appears associated with the cell membrane and with an intracellular reticulated compartment. These findings are in agreement with our recent observation that CCDC88B co-localizes with intracellular actin filaments10 and suggest that the CCDC88B/ARHGEF2/RASAL3 complex may be associated with the cytoskeleton.
Absence of RASAL3 and ARHGEF2 protects against neuroinflammation
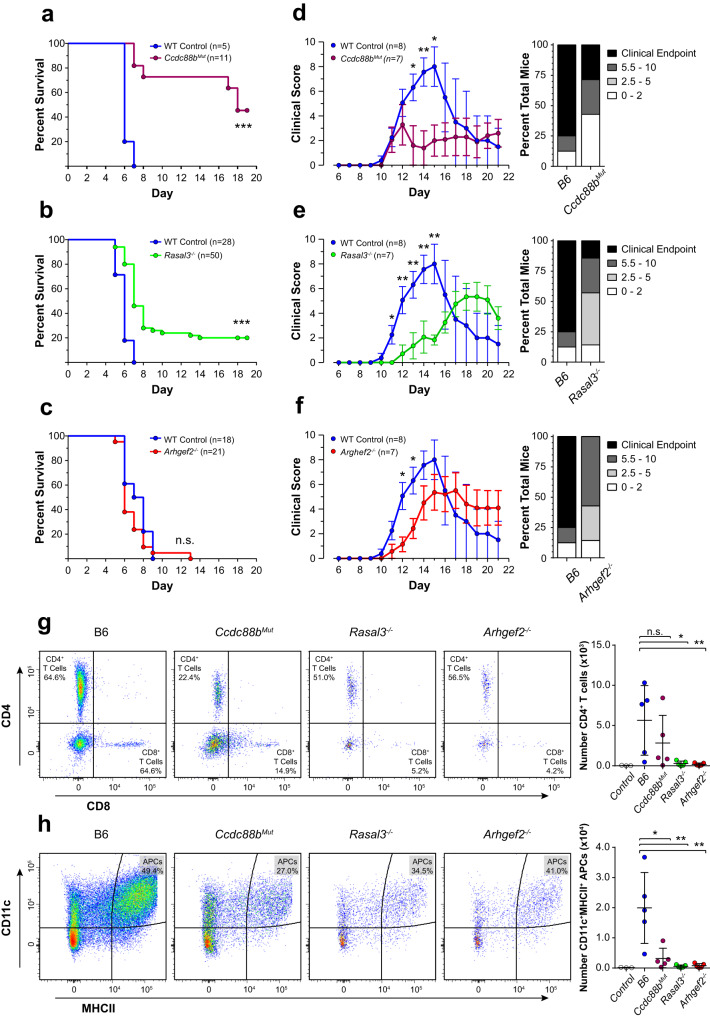
We next tested whether the proximity and physical interaction detected between CCDC88B, ARHGEF2 and RASAL3 translates into a functional relationship. Having previously shown that Ccdc88b inactivation protects against neuroinflammation (ECM model)6, we evaluated a possible role for ARHGEF2 and RASAL3 in neuroinflammation using mouse mutants inactivated at each locus. For this, we created an Arhgef2 loss-of-function mutant by CRISPR-Cas9 gene editing in a C57BL/6J (B6) genetic background (Supplementary Fig. 2). Rasal3−/− mutant mice have been previously described25. Control B6, along with Ccdc88bMut, Rasal3−/− and Arhgef2−/− mutant mice were infected with PbA, monitored for appearance of neurological symptoms and survival was recorded (Fig. 3a–c). Ccdc88bMut mice were highly resistant to the cerebral phase of the disease (d5–d9), with greater than ~70% survival at d16 (Fig. 3a). On the other hand, Rasal3−/− mice showed a moderate but significant level of resistance over controls, with ~20% survival beyond d16 (Fig. 3b). Arhgef2−/− mutant mice were as susceptible to ECM as controls (Fig. 3c).
Fig. 3. Loss of RASAL3 and ARHGEF2 reduces susceptibility to neuroinflammation.
Survival of B6, Ccdc88bMut (a), Rasal3−/− (b), and Arhgef2−/− (c) mutant mice following infection with Plasmodium berghei ANKA (PbA). The total number of mice in each group is indicated; animals surviving beyond day 8 are considered resistant to PbA-induced lethal neuroinflammation (n.s. non-significant, ***p < 0.001, Gehan-Breslow-Wilcoxon test). d–f Experimental autoimmune encephalomyelitis (EAE) was induced by combined treatment with pertussis toxin and myelin oligodendrocyte protein and the extent of pathogenesis (clinical score) was assessed daily as described in “Materials and methods”. Clinical scores (left) and EAE incidence, representing the highest score reached by each individual mouse over the course of the experiment (right), of B6, Ccdc88bMut, Rasal3−/− and Arhgef2−/− mice are shown, along with the number of mice used in each group (clinical scores shown as means ± SEM; *p < 0.05, **p < 0.01; Mann–Whitney test). Data from (d) to (f) are representative of at least three independent experiments where each group were compared to the same controls within the same experiment. g, h EAE was induced in B6, Ccdc88bMut, Rasal3−/− and Arhgef2−/− mice and the spinal cords harvested at day 11. Total CD45+CD3+CD4+ T cells (g) and total CD45+CD11cHighMHCIIHigh antigen presenting cells (h) were analyzed by flow cytometry. Representative flow cytometry data (normalized for 10 million viable cells; left) and quantification of individual mice (right) for each genotype are shown; data from (g) are gated first on total viable CD45+CD3+ cells and data from (h) are gated first on total viable CD45+ cells (see Supplementary Fig. 3). Control: B6 mice without EAE induction.
We also investigated Ccdc88bMut, Arhgef2−/− and Rasal3−/− mutants in a non-microbial model of neuroinflammation (Fig. 3d–f). In EAE, neuroinflammation and axonal damage are induced by autoimmune response to myelin oligodendrocyte glycoprotein which is co-administered with pertussis toxin. Response to EAE is evaluated by appearance and severity of symptoms assessed by a clinical score and by overall survival. In the EAE model, Ccdc88bMut (Fig. 3d), Rasal3−/− (Fig. 3e), and Arhgef2−/− (Fig. 3f) mutants showed significant protection when compared to B6 controls, including lower clinical scores and increased survival, as identified by the fraction of animals reaching clinical endpoint (Fig. 3d–f). Clinical scores for individual mice revealed that all mutants displayed a much milder phenotype than controls.
To better characterize the basis of EAE resistance seen in Ccdc88bMut, Rasal3−/− and Arhgef2−/− mice, we investigated the infiltration of pro-inflammatory cells in the spinal cord of mutant and controls at day 11, a time point that corresponds to appearance of neurological symptoms (Fig. 3d–f). Flow cytometry analysis (see Supplementary Fig. 3) of the cellular content of Rasal3−/− and Arhgef2−/− spinal cord identified a significant reduction in the numbers of infiltrating CD45+CD3+CD4+ T cells (Fig. 3g) and CD45+CD11c+MHCII+ antigen presenting cells (Fig. 3g) compared to B6 controls. The decreased infiltration of antigen presenting cells was also noted in the spinal cord of Ccdc88bMut mice (Fig. 3h). These results strongly suggest that EAE resistance in Rasal3−/−, Arhgef2−/− and Ccdc88bMut is associated with reduced/delayed infiltration of lymphoid and myeloid cells in the spinal cord, with possible concomitant dampening of pathological neuroinflammation.
These results show that the Ccdc88bMut, Arhgef2−/− and Rasal3−/− mutants display significant resistance in one or both models of neuroinflammation models tested. Hence, the physical interaction detected between the CCDC88B, ARHGEF2 and RASAL3 proteins appears to underlie functional relationship when tested in vivo during neuroinflammation.
Rasal3−/− and Arhgef2−/− mice are highly susceptible to DSS-induced colitis
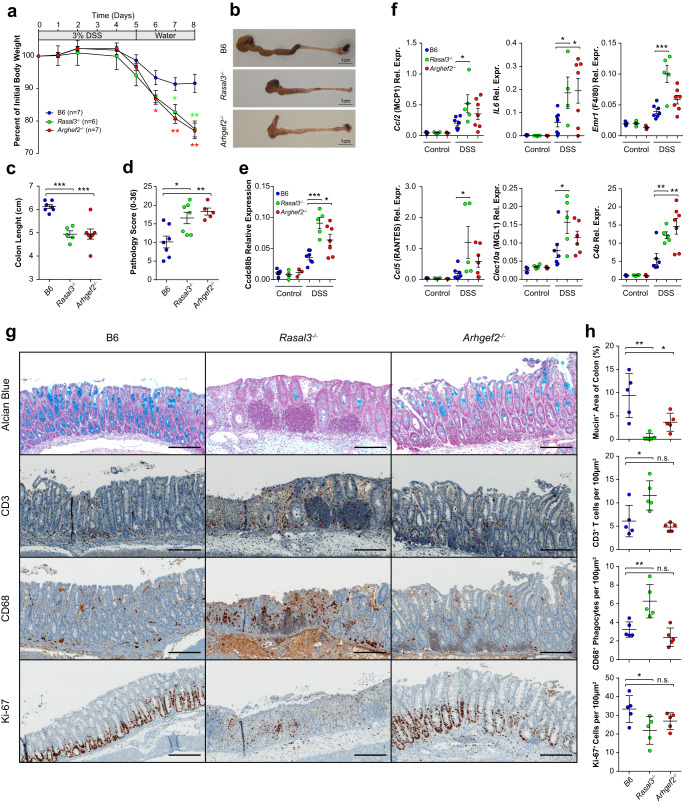
We have previously shown that the protection of Ccdc88bMut mice against intestinal colitis is due to reduced intestinal inflammation and decreased leukocyte infiltration13. We therefore tested the response of Arhgef2−/− and Rasal3−/− mutant to DSS-induced colitis. We noticed that both mutant mice lost significantly more weight than the B6, an indicator of the pathology progression (Fig. 4a). Moreover, the colons from both Arhgef2−/− and Rasal3−/− mice were significantly shorter than B6 controls (Fig. 4b, c). Histological examination of colon sections also showed a quantitatively higher pathology score in both Arhgef2−/− and Rasal3−/− mice compared to B6, based on criteria that included inflammatory cell infiltration, submucosal edema and surface epithelial degeneration (Fig. 4d). Increased cellular infiltration of cells in inflamed colons of Arhgef2−/− and Rasal3−/− mice (seen in the histological pathology scoring) was concomitant to increased expression of Ccdc88b mRNA (Fig. 4e), a marker of myeloid and lymphoid cells13. Furthermore, expression of mRNAs for pro-inflammatory cytokines (IL6), chemoattractant (MCP1, RANTES), markers of cellular infiltration (MGL1, F4/80), and C4b, were significantly elevated in the colon of DSS-treated Rasal3−/− and, to a lesser degree in Arhgef2−/− mice compared to B6 (Fig. 4f). Lastly, immunohistological studies of colon sections revealed that both Rasal3−/− and Arhgef2−/− DSS-treated mice show reduced level of mucin (Fig. 4g, h). Quantification of cellular infiltrates (2 distalmost fields of view of each colon) revealed a significantly increased infiltration of CD3+ T cells and CD68+ mononuclear phagocytes, as well as a decreased number of KI-67+ proliferating cells in DSS-treated Rasal3−/− mice compared to B6, all in agreement with greater susceptibility in Rasal3−/− mice (Fig. 4g, h). Interestingly, cellular infiltrates in the colons of DSS-treated Arhgef2−/− mice were quantitatively similar to those seen in B6 controls (Fig. 4g, h), and this despite a stronger inflammatory environment (Fig. 4f), suggesting possible differences in the mechanistic basis of DSS-susceptibility in Rasal3−/− and Arhgef2−/− mutants.
Fig. 4. Loss of RASAL3 or ARHGEF2 enhances pathogenesis of DSS-induced colitis.
Control B6, as well as Rasal3−/− and Arhgef2−/− mutant mice were given 3% DSS for 5 days followed by 3 days of water and were then sacrificed. a Body weight loss, expressed as percent of initial weight, for each genotype (means ± SEM, *p < 0.05, **p < 0.01; two-tailed Student’s t test). b Representative images of colons from B6, Rasal3−/− and Arhgef2−/− mice at day 8. b Quantification of colons length (means ± SEM. ***p < 0.001; two-tailed Student’s t test). Data from (a) to (c) are representative of three independent experiments. d Pathology scores evaluating inflammatory cell infiltration, submucosal edema, gland loss and surface epithelial erosion/ulceration (mean ± SEM; *p < 0.05, **p < 0.01; two-tailed Student’s t test). qPCR data for the relative expression for Ccdc88b (e) as well as indicated genes coding for cytokines, chemokines, and myeloid cell markers (f) in distal colons of non-treated (Control) or at day 8 following DSS treatment (DSS). Data represent expression relative to Hprt which was used as an internal control (means ± SEM, *p < 0.05, **p < 0.01; ***p < 0.001; two-tailed Student’s t test). g Immunohistochemistry staining of colons of B6, Rasal3−/− and Arhgef2−/− mice at day 8 following DSS treatment for the indicated markers. Insert bar: 100 µm. Data are representative of at least 5 colons per group. h Quantification of (g), for the average of the two most distal field of view of each colon (at 10X, using automated counting, see “Materials and methods”). Each data point represents an individual mouse (means ± SEM, n.s. non-significant; *p < 0.05, **p < 0.01; two-tailed Student’s t test).
To further investigate the contribution of lymphoid cells to the exacerbated colitis phenotype seen in Rasal3−/− and Arhgef2−/− mice, we tested the ability of B6, Rasal3−/− and Arhgef2−/− naïve CD4+ CD25−CD45RBHi T cells to induce colitis upon adaptive transfer into lymphopenic Rag1−/− mice. Seven weeks post-transfer, histology of colon sections revealed no difference between B6 and Arhgef2−/−, including a similar CD3+ T cells infiltration (Supplementary Fig. 4a) and pathology score (Supplementary Fig. 4b), suggesting that the exacerbated colitis response to DSS seen in Arhgef2−/− mice (Fig. 4) was not lymphoid in nature. Conversely, transfer of Rasal3−/− naive T cells failed to induce colitis when compared to B6, exhibiting both a decrease in the amount of infiltrating CD3+ T cells (Supplementary Fig. 4a) and a lower pathology score (Supplementary Fig. 4b). This suggests that RASAL3 is require for the proper function of T lymphocytes during colitis, in agreement with recently published studies25,26.
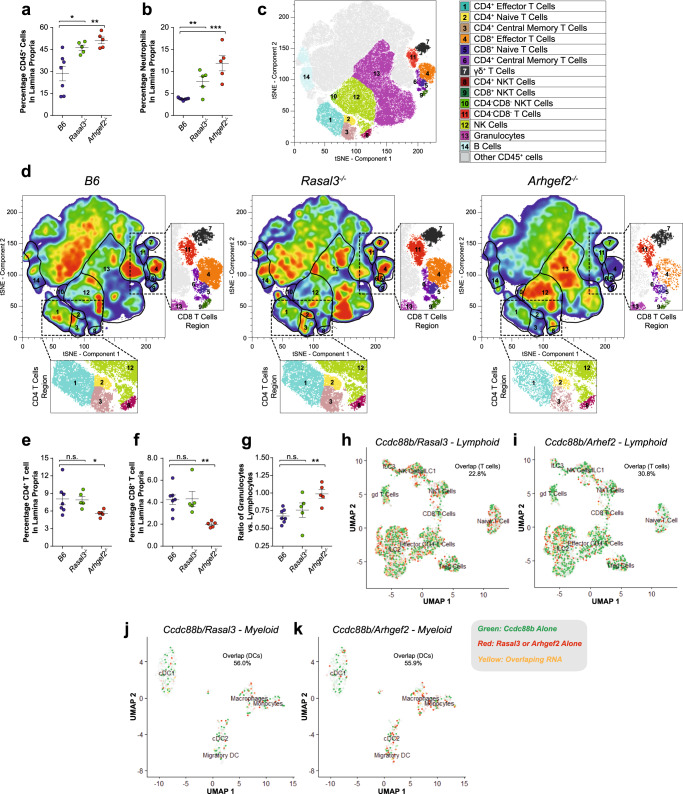
To better understand the susceptibility of Arhgef2−/− and Rasal3−/− mice to colitis, we conducted cellular immunophenotyping of colon cell populations by flow cytometry following DSS treatment. These showed a greater infiltration of CD45+ cells in lamina propria of Arhgef2−/− and Rasal3−/− mice (Fig. 5a), with an important contribution from CD11b+/Ly6G+ neutrophils in both mutants compared to controls (Fig. 5b). Moreover, unsupervised tSNE visualization in Arhgef2−/− mice revealed a significantly reduced proportion and number of both CD4+ and CD8+ T cells (populations 1 to 6, Fig. 5c, d) in this infiltrate compared to B6 and Rasal3−/− mice (Fig. 5e, f) This abnormal cellular infiltration leads to a higher granulocytes to lymphocytes ratio in Arhgef2−/− mice (Fig. 5g), suggesting further gene-specific effects of the mutation on the type of cellular infiltrate at the site of inflammation/tissue damage. Taken together, these results indicate an altered response of Rasal3−/− and Arhgef2−/− mice to acute DSS-induced colitis, with higher susceptibility and increased pathology and enhanced inflammatory response. Furthermore, we note unique differences in the composition of the cellular infiltrates in the gut of Arhgef2−/− mutant mice. These results point to both a requirement for RASAL3 and ARHGEF2 in neuroinflammation (in the EAE model), while their absence is associated with increased inflammation in the gut following DSS-induced colitis.
Fig. 5. Cellular infiltrate in the lamina propria of RASAL3−/− and ARHGEF2−/− mice following DSS-induced colitis.
Control B6, as well as Rasal3−/− and Arhgef2−/− mutant mice were given 3% DSS for 5 days followed by 3 days of water and were then sacrificed. a, b Percentage of total CD45+ cells and neutrophils in the lamina propria from B6, Rasal3−/− and Arhgef2−/− mice colon at day 8 following DSS treatment, respectively (mean ± SEM; n.s. non-significant, *p < 0.05, **p < 0.01, ***p < 0.001; two-tailed Student’s t test). c tSNE-based visualization of the different CD45+ cell populations found in the lamina propria, colored and numbered as per the legend (pooled analysis from all genotypes). d Same visualization as in (c), but for B6 (left), Rasal3−/− (middle) and Arhgef2−/− (right) mice colon at day 8 following DSS treatment, with the highest cellular density in red. Inserts depict regions of the tSNE plots corresponding to the CD8 and CD4 T cells clusters. e, f Percentage of CD4+ and CD8+ T cells from (d), respectively (mean ± SEM; *p < 0.05, **p < 0.01; two-tailed Student’s t test). g Ratio of total number of granulocytes vs. total number of lymphocytes from (d) (mean ± SEM; n.s. non-significant, **p < 0.01; two-tailed Student’s t test). tSNE-based visualization of single-cell RNAseq data from B6 lamina propria cells obtained at steady state for indicated lymphoid (h, i) and myeloid (j, k) populations. Expression of Ccdc88b is indicated in green and level of expression of either Rasal3 (h, j) or Arhgef2 (i, k) in red, as indicated in the legend. Indicated percentages represent the fraction of cells co-expressing RNA for Rasal3 or Arhgef2 with Ccdc88b, for the indicated populations.
We have previously observed that in other single gene models of immunodeficiency, dysregulated inflammatory response causes not only protection against acute neuroinflammation but also causes increased susceptibility to colitis-associated colorectal cancer (CA-CRC)27. Hence, we tested the impact of loss of function for Rasal3 and Arhgef2 on response to CA-CRC induced by combined treatment with DSS and the carcinogen azoxymethane (AOM). We note that as opposed to Arhgef2−/− mice that behaved like controls, Rasal3−/− mice showed a significantly greater susceptibility to CA-CRC; this susceptibility was expressed both by an increase in the total number of tumors per colon (Supplementary Fig. 5a, b), and an increase total tumor surface area (Supplementary Fig. 5c). Immunohistological studies of colon sections also revealed an exacerbated response in Rasal3−/− mice (as shown by H&E and pSTAT3 staining), with a higher cellular proliferation (identified by KI67+ cells) and greater infiltration of CD3+ T cells (Supplementary Fig. 5d). Overall, Rasal3−/− mice display an enhanced susceptibility to AOM DSS-induced CA-CRC model when compared to B6, with an increase cellular proliferation and T cell infiltration in the gut which is not seen in Arhgef2−/− mice.
Finally, to explore the co-expression of Ccdc88b with Rasal3 or Arhgef2 mRNAs in colon immune cells we performed single-cell RNA sequencing at steady state. In lymphoid cells (Fig. 5h, i), a large fraction of T cells expressing Ccdc88b also express Rasal3 (22.8% of Ccdc88b+ cells, Fig. 5h) and Arhgef2 (30.8% of Ccdc88b+ cells, Fig. 5i). In myeloid cells, an even larger proportion of cells that express Ccdc88b also express Rasal3 (56.0% of Ccdc88b+ cells, Fig. 5j) and Arhgef2 (55.9% of Ccdc88b+ cells, Fig. 5k). Co-expression of both Ccdc88b with Rasal3 and Arhgef2 in DCs is of particular interest, given that CCDC88B expression was shown to be essential for the mobility and inflammatory function of these cells, both in vitro and in vivo10, raising the possibility that RASAL3 and ARHGEF2 may also be involved in migratory properties of these cells.
Rasal3−/− and Arhgef2−/− dendritic cells exhibit altered mobility in vivo
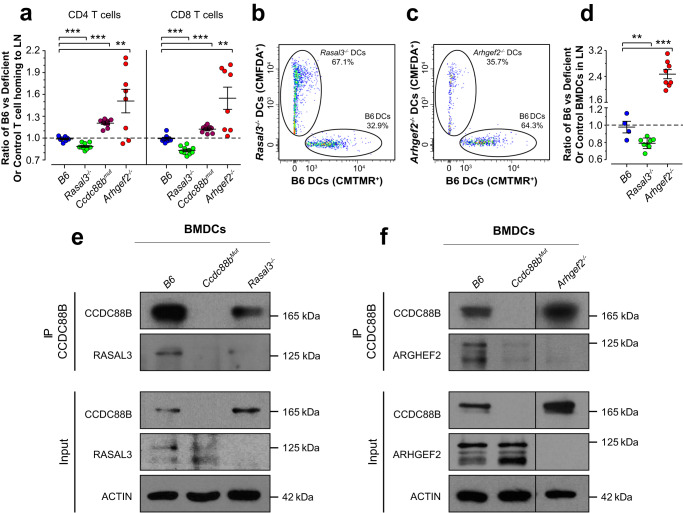
We have shown that CCDC88B is required for mobility of myeloid (DCs) and lymphoid cells in vivo10. We thus assessed the loss of Rasal3−/− and Arhgef2−/− function on mobility of T cells (Fig. 6a). These studies revealed that CD4+ and CD8+ T cells from Ccdc88bmut and Arhgef2−/− mutants showed a reduced capacity to home to the inguinal LN. Surprisingly, CD4+ and CD8+ T cells from Rasal3−/− showed a greater homing capacity than B6 cells in this model (Fig. 6a). Overall, these results suggest that a functional CCDC88B/ARHGEF2/RASAL3 complex is required for mobility of T lymphocytes in vivo.
Fig. 6. RASAL3 and ARHGEF2 are required for migration of dendritic cells in vivo.
a Total spleen cells from control B6 and either Ccdc88bmut, Rasal3−/− or Arhgef2−/− mutants were stained with CMFDA or CMTMR dye, mixed at a 1:1 ratio and injected intravenously into B6 mice. Inguinal lymph nodes (LNs) were harvested 6 h later and analyzed by flow cytometry for number of CD4+ or CD8+ T cells. Numbers are expressed as a fold enrichment of dyed B6 T cells over dyed cells from Ccdc88bmut, Rasal3−/−, Arhgef2−/− mutants or control B6 (means ± SEM, **p < 0.01, ***p < 0.001; two-tailed Student’s t test). b, c Control B6 and either Rasal3−/− or Arhgef2−/− LPS pulsed BMDCs were stained with CMFDA or CMTMR dye, mixed together at a 1:1 ratio and injected sub-cutaneously in the footpad of B6 mice. Draining popliteal LNs were harvested 48 h later and analyzed by flow cytometry (plot gated on CMFDA+ and CMTMR+ dyed cells). d Quantification from (b) and (c), expressed as a fold enrichment of dyed B6 BMDCs over dyed Rasal3−/−, Arhgef2−/− or control B6 BMDCs (means ± SEM, **p < 0.01, ***p < 0.001; two-tailed Student t test). Data from (a) to (d) are representative of three independent experiments. e, f Lysates from B6, Ccdc88bMut, Rasal3−/− and Arhgef2−/− BMDCs were immunoprecipitated with an anti−CCDC88B polyclonal anti-serum, and RASAL3 or ARHGEF2 proteins were detected by immunoblotting. Data from (e) and (f) are representative of two independent experiments.
We also assessed the role of ARHGEF2 and RASAL3 in migration of DCs in vivo. For this, primary BMDCs from B6 controls and from Rasal3−/− or Arhgef2−/− mutant mice were pulsed with LPS in vitro, followed by labeling with different fluorescent dyes (Fig. 6b, c). The ratio of labeled B6 to Rasal3−/− DCs was found to be significantly reduced, suggesting enhanced migration of Rasal3−/− DCs compared to B6 (Fig. 6b, d). Conversely, the ratio of B6 to Arhgef2−/− DCs was found to be significantly higher, in agreement with decreased migration of Arhgef2−/− DCs (Fig. 6c, d), similar to what we previously reported for Ccdc88bmut DCs10. Taken together, these results indicate that similarly to CCDC88B, both ARHGEF2 and RASAL3 are required for proper migration of DCs in vivo.
Next, given that RASAL3, ARHGEF2 and CCDC88B all play a role in DCs migration and that they do physically interact in T lymphocytes, we evaluated if these proteins also interact together in DCs. Immunoprecipitation of CCDC88B in BMDCs extracts from the B6, Ccdc88bMut, Rasal3−/− and Arhgef2−/−showed that both RASAL3 (Fig. 6e) and ARHGEF2 (Fig. 6f) were pulled down, indicating that both proteins interact with CCDC88B in DCs. A similar immunoprecipitation using Ccdc88bMut DCs did not pull down either protein (Fig. 6e, f and Supplementary Fig. 6a, b), demonstrating the specificity of the interaction. These results show that the CCDC88B/ARHGEF2/RASAL3 complexes, initially identified in thymocytes and T cells, are required for proper migration of DCs and T cells.
Rasal3 and Arhgef2 modulate dendritic cells motility in vitro through RHOA activation
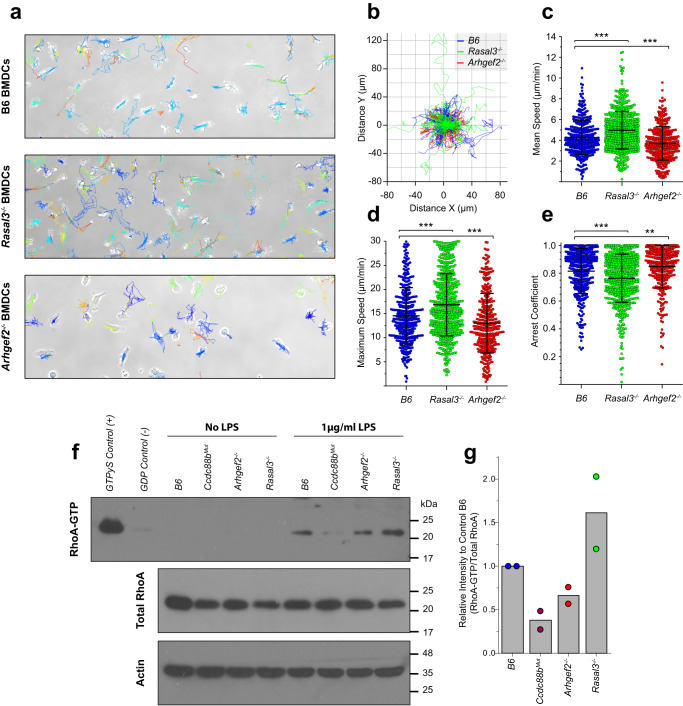
We next investigated whether the physical interaction detected between CCDC88B, RASAL3 and ARHGEF2 in DCs translates into functional interaction at the cellular level, including a possible role in DC cells mobility in vitro. For this, we used an in vitro patrolling assay at steady state (Fig. 7a); similar to the reduced mobility phenotype previously observed in Ccdc88bMut DCs10, Arhgef2−/− DCs were found to travel a significantly shorter distance than control DCs (Fig. 7a, b). Conversely, Rasal3−/− DCs displayed the opposite phenotype with an increased traveled distance compared to controls (Fig. 7a, b). This was further reflected by the higher mean speed and maximum speed of Rasal3−/− DCs (5.0 ± 1.8 µm/min and 16.9 ± 6.5 µm/min, respectively) when compared to B6 DCs (4.3 ± 1.7 µm/min and 14.6 ± 5.8 µm/min, respectively); contrariwise, the mean and maximum speeds of Arhgef2−/− DCs were significantly slower (3.7 ± 1.6 µm/min and 13.0 ± 6.2 µm/min, respectively) (Fig. 7c, d). Likewise, the arrest coefficient, calculated as the fraction of time that the cell does not move using a threshold of 7 µm/min, was respectively reduced for Rasal3−/− DCs (0.76 ± 0.17 and increased for Arhgef2−/− DCs (0.85 ± 0.15), when compared to B6 control cells (0.82 ± 0.16) (Fig. 7e).
Fig. 7. Altered motility parameters of Rasal3−/− and Arhgef2−/− dendritic cells.
BMDCs generated from B6, Rasal3−/− and Arhgef2−/− mutants were imaged by bright-field time-lapse microscopy for 90 min and tracked using the TrackMate plugin from the ImageJ software, with manual correction. a Representative field of view of B6, Rasal3−/− and Arhgef2−/− BMDC cultures with the tracking results over the course of the experiment. b Normalized Rose plot depicting 25 randomly chosen track of individual B6 controls (blue), Rasal3−/− (green) or Arhgef2−/− (red) DCs. c Average velocity of individual BMDCs from control B6, Rasal3−/− and Arhgef2−/− DCs mutants (means ± SD, ***p < 0.001; two-tailed Student’s t test). d Maximum speed reached by individual DCs (means ± SD, ***p < 0.001; two-tailed Student’s t test). e Arrest coefficient using a threshold of 7 µm/min for actively patrolling DCs (means ± SD, **p < 0.01, ***p < 0.001; two-tailed Student’s t test). Data from (a) to (e) were processed using a homemade MatLab script and are representative of three independent experiments, taking at least two separated field of view (0.314 mm2 per field of view) per DCs culture. f BMDCs from B6, Ccdc88bMut, Rasal3−/− and Arhgef2−/− mutants were stimulated with LPS and protein lysate analyzed for the level of active RhoA-GTP using Rhotekin-RBD (Rho Binding Domain) beads. As control, some BMDCs were loaded with high concentration of non-hydolyzable GTP (GTPγS, positive control) or GDP (negative control) prior to pull-down. g Quantification of (f), normalized to the total amount of RhoA expression, for two independent experiments.
ARHGEF2 is known to be required for the regulation of the small GTPase RHOA, which is itself a master regulator of cytoskeleton rearrangement and proper cellular movement28. Thus, we next tested the activation status of RHOA-GTP in B6, Ccdc88bMut, Rasal3−/− and Arhgef2−/− DCs following stimulation with LPS, using Rhotekin-coupled beads (Fig. 7f and Supplementary Fig. 6c). Stimulation of B6 DCs with LPS leads to a rapid and strong activation of RHOA, indicated by the high level of RHOA-GTP (Fig. 7f). On the other hand, absence of ARHGEF2 leads to decrease level of activation of RHOA in DCs (Fig. 7f, g). Furthermore, Ccdc88bMut DCs completely failed to activate RHOA (Fig. 7f, g). Lastly, and in opposition to Arhgef2−/− DCs, Rasal3−/− DCs exhibited an over-activation of RHOA, with a higher level of RHOA-GTP than B6 control (Fig. 7f, g). These results indicate that while CCDC88B is required for the activation of RHOA, proper level of activation is also regulated by ARHGEF2 and, inversely, RASAL3.
Considering our previous demonstration of the requirement of CCDC88B for DC cells mobility10, our results strongly suggest that CCDC88B, RASAL3 and ARHGEF2 form part of a multi-protein complex that plays an important role in DCs movement, where CCDC88B acts as a protein scaffold for the recruitment of ARHGEF2 and RASAL3 to the proper localization where they are required to modulate the motility of DCs and T-cells. In agreement with our in vivo data, the opposite effect of inactivation of Rasal3 and Arhgef2 on cell mobility raises the possibility that the 2 proteins play an opposite functional or regulatory role in the biochemical activity of this complex. This model is further supported by biochemical data showing that absence of ARHGEF2 (or CCDC88B) leads to reduced RHOA activation (possibly through loss of GTPase activity of ARHGEF2), while absence of RASAL3 leads to over-activation of RHOA (possibly through absence of the RasGAP activity of RASAL3).
Discussion
CCDC88B was discovered as a gene which inactivation protects against acute neuroinflammation in models of cerebral malaria6. Subsequently, we showed that (1) Ccdc88b inactivation also protects against inflammation in experimental colitis13, (2) CCDC88B is the human gene underlying the 11q13 locus associated with susceptibility to several inflammatory conditions, including IBD6,13, and (3) CCDC88B mRNA is regulated by cis-acting elements in human myeloid cells, and CCDC88B mRNA expression is positively correlated with disease risk in Crohn’s disease patients6,13. Subsequently, we showed that CCDC88B contributes to normal inflammatory response by regulating mobility and migration of myeloid cells (DCs) to the site of inflammation in vivo, and in vitro10. At the molecular level, the mechanism by which CCDC88B regulates leukocyte migration remains unknown.
Using a proteomics approach with CCDC88B-expressing primary thymocytes and a T-lymphocyte cell line, we identified ARHGEF2 and RASAL3 as CCDC88B interactors (Fig. 1). The physical interaction between CCDC88B and ARHGEF2/RASAL3 was validated by reciprocal co-immunoprecipitation and by double immunofluorescence (Fig. 2). Their functional interaction was established in vivo (Figs. 3 and 4): Arhgef2−/− and Rasal3−/− mutants display resistance to neuroinflammation in EAE (alike Ccdc88bmut mice), and show altered inflammatory response during DSS induced colitis. Furthermore, Ccdc88b, Rasal3,and Arhgef2 are co-expressed in T cells and DCs (Fig. 5). Finally, mutations in Arhgef2 and Rasal3 have a dramatic but opposite effect on the migratory properties of myeloid and lymphoid cells in vivo (Fig. 6) and in vitro (Fig. 7). These studies establish that CCDC88B, ARHGEF2 and RASAL3 are critical for leukocytes mobility and activation of immune and inflammatory responses.
Recent studies have established a close partnership between ARHGEF2 and the Rho family of small GTPases (RHOA) known to regulate contractibility and cell movements29–32. ARHGEF2 is a microtubule-associated RHOA-specific nucleotide exchange factor (stimulates conversion of RHOA-GDP to RHOA-GTP)33 that couples microtubule depolarization with Rho-mediated actin stress fiber formation and cell contraction34,35. The activation of ARHGEF2 in response to inflammatory, mitogenic, and morphogenic signals promotes activation of RHOA and stress fiber formation34. ARHGEF2 has been implicated in migratory function of fibroblasts, neutrophils, epithelial cells, and T lymphocytes29–32,36. In T cells, activation of ARHGEF2 by PPP2R2A is required for Th1/Th17 differentiation, and PPP2R2A deficiency in T cells dampens the ARHGEF2/RHOA/ROCK pathway activation, decreasing EAE pathogenesis37. In neutrophils, loss of ARHGEF2 function leads to reduced spreading, crawling, and migration in response to sheer stress29. More broadly, ARHGEF2-dependent regulation of cytoskeletal rearrangements have secondary effects on phagocytosis, intracellular pathogens recognition, interaction with bacterial effectors, response to viral RNAs, activation of IRF3, and production of type I interferon38–40. Finally, ARHGEF2 overexpression has been associated with tumor progression including hepatocellular carcinoma, high-grade melanomas and malignant megakaryocytes34,41,42. Disruption of ARHGEF2/RHOA signaling (removal of BNIP-2 scaffold) affects breast carcinoma cells migration43.
In agreement with this published work, we observed that ARHGEF2 forms part of a protein complex with CCDC88B that is required for migration and inflammatory functions of DCs. Indeed, Arhgef2 mutant DCs show a migratory defect in vivo (homing assay) and show reduced mobility in vitro. Arhgef2 mutants also show resistance to neuroinflammation in EAE6,10. Conversely, Arhgef2 deficiency causes susceptibility to colitis, expressed as increased pathology score, increased production of inflammatory cytokines (IL6, MCP1, RANTES, C4b), increased infiltration of CD45+ cells in the lamina propria, with a marked abundance of CD11b+/Ly6G+ neutrophils. Colitis susceptibility in Arhgef2 mutants is opposite to colitis resistance seen in Ccdc88b mutants13. The reason for this seemingly contradictory result can only be speculated on at the moment, but could involve different mutation-specific effects on cellular responses at the site of inflammation, with cellular infiltrates of different leukocytes composition including strong granulocytes content in Arhgef2 vs. Ccdc88b. The compensatory granulocytic response during colitis in Arhgef2−/− mice is similar to other mouse mutants with DC-deficiency, including Irf1 mutants27.
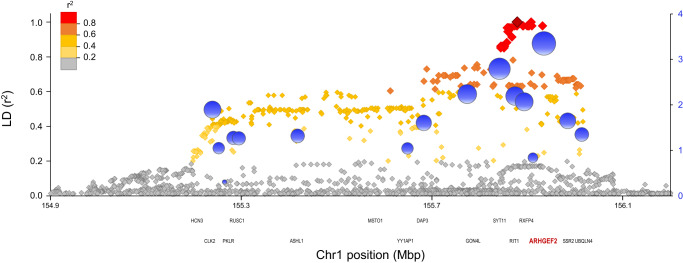
Finally, a locus on 1q22 containing 16 genes in linkage disequilibrium is associated with susceptibility to IBD44. The reported top-ranking marker (SNP rs670523) in this interval maps close to and is in almost complete LD with SNPs within or proximal to ARHGEF2 (r2 score >0.96). Annotation of this locus using a Myeloid Inflammation Score (MIS)6,45 shows that Arhgef2 has the highest MIS (3.4) in the locus interval (Fig. 8). These analyses together strongly suggest that ARHGEF2 is the morbid gene at 1q22 locus (position 155 Mb). Hence, CCDC88B (11q13) and AHRGEF2 (1q22) physically and functionally interact, and are genetically linked to IBD susceptibility in humans.
Fig. 8. ARHGEF2 containing IBD risk locus on chromosome 1 (1q22).
The single nucleotide polymorphism (SNP) rs670523 was previously associated to IBD by GWAS44. The graph shows the linkage disequilibrium structure of the proxy SNPs to rs670523 (dark gray) calculated using LDlink64 as r2 values based on the 1000 Genomes phase 3v5 CEU + GBR European reference haplotypes (left y axis). The proxy SNPs are color-coded according to their r2 values and positioned at their chromosomal location (x-axis) over the genes found at this locus. The right y-axis shows the results of the MIS (Myeloid Inflammatory Score) epigenetic scoring method that we previously described6 and designed to prioritize candidate genes involved in inflammatory processes in myeloid cells based on functional genomics information. The MIS for each gene is shown by a blue bubble over its transcriptional start site with a diameter proportional to the MIS. ARHGEF2 is found in strong LD with rs670523 and has the highest MIS within this IBD risk locus.
RASAL3, is a member of the family of RAS GTPase-activating proteins (GAP), which negatively regulates RAS signaling by stimulating hydrolysis of RAS-GTP to RAS-GDP23. It has been shown to negatively regulate Ras/Erk signaling by stimulating p21RAS and Rac2 GTPase activity23. Rasal3−/− mutant show decreased T cells and NKT cells numbers, display increased Erk phosphorylation and reduced production of cytokines by NKT cells46. In T cells, RASAL3 may repress TCR-signaling (ERK phosphorylation) which is required for T cells survival in vivo25, and Rasal3−/− mutants display dampened inflammatory allergic dermatitis26.
Here, we show that RASAL3 forms a complex with CCDC88B and ARHGEF2 that plays an important role in the migration of DCs. Rasal3−/− mutant DCs show both increased migration in vivo and enhanced mobility in vitro. This is opposite of Ccdc88b and Arhgef2 mutant DCs measured in a similar way, suggesting that RASAL3 may act to negatively regulate cell migration in the context of the CCDC88B/RASAL3/ARHGEF2 complex (Fig. 9). These findings are similar to studies of FAM49B/CYRI47,48; CYRI binds to RAC1 in the GTP bound form and negatively regulates RAC1 GTPase activity. Inactivating CYRI mutations cause (1) increased mobility/migration of myeloid cells in vitro, (2) increased resistance to Salmonella typhimurium, (3) increased susceptibility to Mycobacterium tuberculosis and Listeria monocytogenes. It is tempting to speculate that RASAL3 may play a role functionally similar to CYRI in Rho/Rac signaling in regulating cell mobility.
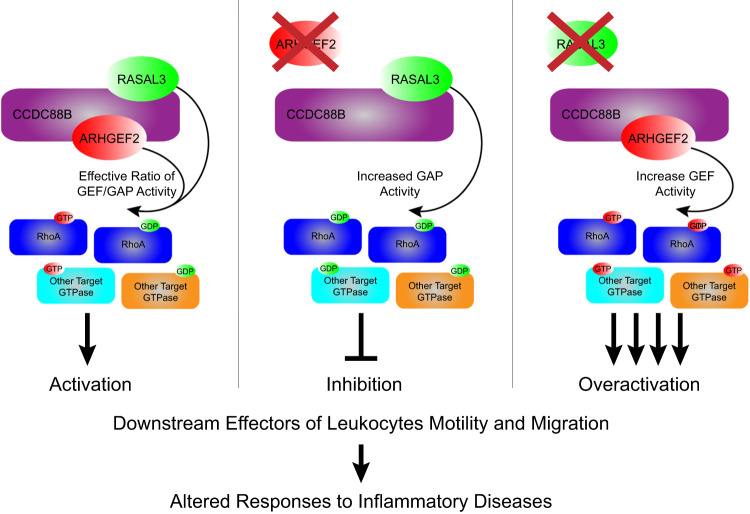
Fig. 9. CCDC88B, ARHGEF2 and RASAL3 complex.
Proposed interaction of CCDC88B with ARHGEF2 and RASAL3 in the context of cellular mobility (left), or in the absence of ARGHEF2 (middle) or RASAL3 (right), and their downstream target, RhoA. GEF guanine nucleotide exchange factor, GAP GTPase-activating protein.
Rasal3 inactivation causes resistance to neuroinflammation in the ECM and EAE models (alike Ccdc88b and Arhgef2 mutants) (Fig. 3). In addition, mutant Rasal3−/− mice show enhanced susceptibility to DSS-colitis, which is similar to Arhgef2 mutants (Fig. 4), including enhanced pathological scores, expression of inflammatory cytokine markers, and presence of CD45+ cellular infiltrate in the lamina propria and dominated by CD11b+/Ly6G+ granulocytes. Hence, although mutations in Arhgef2 and Rasal3 have opposite effects on DC migration, they cause similar pathological effects in DSS-colitis, albeit with different leukocytes infiltrates. Also, the Rasal3 mutation causes a unique susceptibility to colitis-associated colorectal cancer phenotype not seen in the Ccdc88b or Arhgef2 mutants (Supplementary Fig. 5). Such gene-specific effects could reflect unique function of the protein in T cells, DCs or other cells, or unique aspects of secondary response to inflammatory stimulus.
We propose a model where CCDC88B, ARHGEF2 and RASAL3 form a protein complex functionally critical for migration of lymphoid and myeloid cells. Although RASAL3 and ARHGEF2 are expressed in many cell types, CCDC88B is expressed exclusively in leukocytes possibly providing cell-specific function. Based in part on (1) the known but opposite regulatory role of ARHGEF2 and RASAL3 in Ras/Rho/Rac GTPase and associated signaling, (2) the known role of Rho/Rac GTPase in dynamic cytoskeletal structure, and function in cell migration, and (3) the phenotypic expression and inverse impact of loss of function at either Arhgef2, and Rasal3 on DC cell mobility (Figs. 6 and 7), our findings suggest that ARHGEF2 and RASAL3 play an agonist and antagonist role in regulating the activity of this protein complex in cell movement by modulating the activation status of RHOA, and potentially other GTPases (Fig. 9). Finally, our findings in mouse models together with results from genetic association studies in humans for at least 2 of its components (CCDC88B, ARHGEF2) strongly suggest that this complex and genetic variants within its constituents may impact migration of inflammatory cells, and possibly impact genetic vulnerability to chronic inflammatory diseases in humans.
Materials and methods
Mice
Wild type C57BL/6J (B6) mice, 8–12 weeks of age, were obtained from the Jackson Laboratory (Bar Harbor, ME). Except were indicated, both male and female animals were used indiscriminately. The Ccdc88bm1PGrs mutant mouse strain (referred to as Ccdc88bMut) was generated by genome-wide chemical mutagenesis6. Rasal3−/− mice were provided by Dr. H. Suzuki, from the National Center for Global Health and Medicine, Chiba (Japan), and is described elsewhere25. Arhgef2−/− mice were generated by CRISPR/CAS9 by the Transgenic Core of the Life Sciences Complex at McGill University, using the experimental protocol and characterization strategy summarized in Supplementary Fig. 2. All mice were housed under specific pathogen-free conditions at the animal care facility of the Goodman Cancer Research Centre, McGill University, and the animal studies were conducted using protocols approved by the McGill Institutional Review Board (protocol number 5287) and following guidelines and regulations of the Canadian Council of Animal Care. We have complied with all relevant ethical regulations for animal use.
Co-immunoprecipitation for SDS-PAGE and mass spectrometry analyses
Initial anti-CCDC88B co-immunoprecipitations from mouse primary thymocytes (Fig. 1a and Supplementary Data 1: MS/MS Expt #1) were carried out essentially as previously described49, using: (1) 200 mg of cryo-milled thymocytes; (2) 20 mM HEPES, pH 7.4, 0.5% (v/v) Triton X-100, 200 mM NaCl; and (3) an affinity-purified rabbit anti-CCDC88B polyclonal anti-serum6 coupled to Dynabeads M270 Epoxy in-house (adapted from ref. 50). This strategy was maintained for subsequent co-immunoprecipitations with changes as described herein. To prepare thymocytes, ~3 g of mice thymi were harvested, passed through a 70 µm cell strainer using a syringe plunger and washed with PBS. Cell suspensions were then centrifuged inside a syringe, the supernatant was removed, and the pellet was injected directly into liquid nitrogen. Two additional duplicate anti-CCDC88B co-immunoprecipitations from thymi were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, prior to co-immunoprecipitation optimization screening. To further explore potential missing information in our IPs, an anti-CCDC88B co-immunoprecipitation based interaction screen51 was carried out on BI-141 T cells at ~50 mg per reaction, and the results are curated on www.copurification.org/. From this screen, three conditions were selected for further anti-CCDC88B co-immunoprecipitations, in triplicate, and label-free quantitative (LFQ) MS analysis from mouse primary thymocytes and BI-141 T cells: (1) 20 mM HEPES, pH 7.4, 200 mM NaCl, 0.5% (v/v) Triton X-100; (2) 20 mM HEPES, pH 7.4, 150 mM NaCl, 10 mM deoxy-Big CHAP; (3) 200 mM ammonium acetate, pH 7.0, 1% (v/v) Triton X-100. The criteria for selection of these conditions was based on visual inspection of the stained gel as follows: prominent staining of the band associated with CCDC88B and a relatively discrete pattern of other protein bands with some staining intensity differences between them.
Sample workup for LC-MS/MS
Co-immunoprecipitated fractions were eluted in 1.1x NuPAGE sample loading buffer (Thermo Fisher Scientific #NP007) at 70 °C, reduced with DTT, alkylated with iodoacetamide, and run ~ 1 h at 200 V on a 1 mm 4–12% Bis-Tris NuPAGE gel followed by Sypro Ruby staining and protein band excision (Fig. 1a and Supplementary Data 1: MS/MS Expt #1) or as 4–6 mm gel-plugs, prior to Coomassie G-250 staining, peptide workup and LC-MS-MS (essentially as described in refs. 52,53). Gel bands or plugs were excised, cut into 1 mm cubes, de-stained, and digested overnight with 3.1 ng/μl trypsin (Promega, Madison, WI, #V5280) in 25 mM ammonium bicarbonate. Peptides were extracted from the gel in two incubations of 1 h each with 1.7% v/v formic acid, 67% v/v acetonitrile at room temperature with agitation. After partial evaporation by vacuum centrifugation to remove acetonitrile, digests were desalted on Stage Tips54. Stage Tip eluates were concentrated by vacuum centrifugation, loaded onto an Easy-Spray column (ES800, Thermo Fisher Scientific) and gradient-eluted (Solvent A = 0.1% v/v formic acid in water, Solvent B = 0.1% v/v formic acid in acetonitrile, flow rate 300 nl/min) into an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific) acquiring data-dependent CID fragmentation spectra.
MS data analysis
Files were submitted to MaxQuant55 for protein identification and LFQ (label-free quantification). Searches were performed against mouse protein sequences, exogenous contaminants, and a decoy database of reversed protein sequences. LFQ was performed separately for each of three buffer conditions. Non-mouse proteins and proteins that scored worse than any hit in the decoy database were removed from the MaxQuant output file “proteingroups.txt”. LFQ intensities were log2 transformed and samples were grouped by both cell type (thymocytes or BI-141 cells) and buffer condition. For each of the three immunoprecipitation conditions, respectively, proteins showing a LFQ intensity below three in at least one cell type were not considered. Missing values were imputed from a normal distribution with a down-shifted median relative to the measured data distribution, in order to simulate intensity near the detection limit. Triplicate immunoprecipitates from thymocytes and BI-141 cells in each immunoprecipitation condition were compared using two-sided Student’s t test as follow. Statistical significance was determined by a modified t-test that controls the relative importance of the Student’s t test p value and the fold change56,57 with a threshold for FDR (permutations) of 0.01. The RAW and MaxQuant processed files are available for download via ProteomeXchange with identifier PXD023779. Furthermore, a series of filters were used in a decision tree to prioritize possible CCDC88B interactors for further validation. First, candidate CCDC88B interactors were given priority if they were detected in at least 2 independent experiments, in different elution conditions, and if they were detected with immunoprecipitates from both primary thymus and BI-141 cells. Peptide coverage and abundance (peptide counts) in the extracts and relative intensity were also considered to prioritize interactors (Supplementary Fig. 1). Abundant proteins often found in co-immunoprecipitates (CRAPome v1.1) were excluded22. Since CCDC88B is known to be a cytoplasmic protein with microtubule interacting domains6,13, all nuclear proteins and DNA-binding proteins were also excluded from the analysis. A ponderation score was established (Supplementary Data 1 and Supplementary Fig. 1) based on these criteria and identified RASAL3 and ARHGEF2 as two prioritized CCDC88B interactors.
Immunoprecipitation and immunoblotting
HEK293T cells were transiently transfected with a combination of expression plasmids encoding HA-CCDC88B6, RASAL3-FLAG (EX-Mm25984-M14, GeneCopoeia) or ARHGEF2-FLAG (EX-Mm19287-M14, GeneCopoeia) epitope tagged proteins, using the lipofectamine 2000 system (Thermo Fisher). Twenty-four hours later, transfected cell pellets were harvested and resuspended in RIPA lysis buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA 1 mM EGTA 1% NP-40, 2.5 mM sodium pyrophosphate), supplemented with protease inhibitors (complete ultra protease inhibitor, Roche). Samples were clarified by centrifugation and incubated with monoclonal primary antibodies anti-FLAG (Sigma), or anti-HA (Santa Cruz Biotechnology), or IgG used as a negative control; immune complexes were recovered using protein G beads (Dynabeads). Beads were washed extensively, and protein eluted using boiling Laemmli buffer (2% SDS, 10% glycerol, 5% 2-mercaptoethanol) and analyzed by SDS-PAGE. In some experiments, bone marrow-derived dendritic cells (BMDCs) were generated from B6, Ccdc88bmut, Rasal3−/−, Arhgef2−/− bone marrow as described previously10. These cells were lysed in 20 mM HEPES, pH7.4, 300 mM NaCl, 1% NP-40 (supplemented with protease inhibitors). Samples were clarified by centrifugation, pre-cleared and incubated overnight with affinity-purified rabbit anti-CCDC88B hyperimmune serum6; immune complexes were recovered using Sera-Mag Speedbeads Protein A/G Magnetic Particles (Sigma) according to the manufacturer protocol. For immunoblotting, proteins were separated by SDS-PAGE and transferred onto nitrocellulose membrane (GE-Healthcare), followed by incubation with rabbit anti-CCDC88B hyperimmune serum, followed by detection with HRP anti-rabbit antibodies (GE-Healthcare) and ECL, as previously described6. In some experiments, an affinity-purified rabbit anti-RASAL3 hyperimmune serum25 or anti-ARHGEF2 antibody (Cell Signaling Technology) were used (both at 1:300 dilution). Unprocessed original Western Blot analyses can be found in Supplementary Fig. 6.
Immunofluorescence
Transfected HEK293T cells expressing HA-CCDC88B, in combination with either RASAL3-FLAG or ARHGEF2-FLAG, were fixed for 5 min with methanol at −20 °C, followed by permeabilization for 4 min at −20 °C with 30% methanol/70% acetone. Fixed cells were blocked with 3% BSA in PBS for 20 min and incubated overnight with the primary antibody anti-CCDC88B (rabbit polyclonal6, 1:200) or anti-HA (Santa Cruz Biotechnology, 1:1000) with either anti-RASAL3 (rabbit polyclonal, provided by Dr. Suzuki25, 1:200], anti-ARHGEF2 (Cell Signaling Technology, 1:200) or anti-FLAG (Sigma, 1:1000). Bound antibodies were detected using either Alexa488-coupled anti-rabbit IgG (Invitrogen) and Alexa594-coupled anti-mouse IgG (Invitrogen) secondary antibodies at a 1:000 dilution. In some experiments, cells were also counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen) to visualize the nuclei. Images were acquired using a Zeiss LSM710 Meta Laser Scanning Confocal microscope (100x lens) and processed with ImageJ (Fiji) with a maximum intensity projection.
Plasmodium berghei ANKA Infection
Infections with P. berghei ANKA (PbA) were performed as we previously described9. Parasites were originally provided by the Malaria Reference and Research Reagent Resource Center (MR4) and were kept frozen at −80 °C in RPMI 1640 with 15% glycerol. Freshly thawed parasites were passaged once into B6 mice (7 days) and parasitemia in freshly harvested blood was monitored on stained thin blood smears (Diff-Quick reagents staining; Fisher Scientific) to prepare the infectious inoculum. PbA infection was performed using 106 parasitized red blood cells (pRBCs) injected intravenously. Mice were then monitored for the appearance of neurological symptoms associated with cerebral malaria three times daily and euthanized when reaching clinical endpoints. All remaining animals were euthanized on day 19 post-infection (experimental endpoint).
Experimental autoimmune encephalomyelitis
Experimental autoimmune encephalomyelitis (EAE) was induced as described elsewhere9. Briefly, female mice were injected subcutaneously with a short peptide of myelin oligodendrocyte glycoprotein (MOG, amino acids 35–55, 150 µg) emulsified in complete Freund’s adjuvant (50 μg/mouse). The same day and 2 days later, mice received 300 ng of pertussis toxin injected intraperitoneally. Mice were monitored daily for progressive paresis of tail and limbs; severely impaired animals were euthanized. Score was evaluated individually for the tail, each hind limb and each front limb on a scale from 0 to 3 (0, no symptoms; 1, weak; 2, full paresis; 3, no movement), for a possible maximum score of 10. The EAE incidence score was recorded as the highest score reached by each individual mouse over the course of the experiment. In some experiment, the mice were sacrificed at day 11, perfused, and the spinal cord harvested by flushing the spinal column. The spinal cord was then cut into small pieces and digested for 45 min in DNAseI (50 µg/ml, New England Biolabs) and collagenase V (1 mg/ml, Sigma). Single cells suspension was then prepared by passing through a 70 µm cell strainer using a syringe plunger and washed with PBS. Finally, the cells were stained with vital dye Zombie Aqua dye (1:400 dilution, Biolegend) then surface stained with the following fluorescently-labeled antibodies: 1:200 FITC anti-NK1.1 (clone PK136, eBioscience), 1:300 PerCP-Cy5.5 anti-Ly6G (clone 1A8, Biolegend), 1:300 APC anti-CD8α (clone 53-6.7, eBioscience), 1:400 APC-Fire 750 anti-CD45 (clone 30-F11, Biolegend), 1:1000 BV421 anti-Ly6C (clone HK1.4, Biolegend), 1:300 BV605 anti-CD11b (clone M1/70, Biolegend), 1:400 BV711 I-A/I-E anti-MHCII (clone M5/114.15.2, Biolegend), 1:300 BV785 anti-CD11c (clone N418, Biolegend), 1:400 PE anti-CD4 (clone RM4-5, Biolegend), 1:200 PE-Dazzle 594 anti-CD3 (clone 17A2, Biolegend) and 1:400 PE-Cy7 anti-F4/80 (clone BM8, Biolegend). Stained samples were processed using a Fortessa flow cytometer (BD Biosciences) and the results were analyzed using FlowJo software (Tree Star Inc). Gating strategy can be found in Supplementary Fig. 3, and each sample were normalized for 10 million living cells.
Mouse model of intestinal colitis
The mouse model of DSS induced intestinal colitis was performed as we previously described13. Briefly, male mice were treated with 3% DSS (w/v, dextran sodium sulfate; MP Biomedicals) in their drinking water for 5 days, followed by 3 days of water. Mice were weighed daily throughout the experiments and euthanized at day 8. The entire colon was removed, measured and fixed in 10% neutral-buffered formalin overnight. Samples were embedded in paraffin, cut into 4 µm sections followed by staining with eosin and hematoxylin. To avoid bias, a trained pathologist scored each colon blindly on a scale of 36, with up to 4 points given in each of those categories: inflammatory cell infiltration, inflammatory cell depth, submucosal edema, increased mucosal thickening, surface epithelial degeneration, gland epithelial apoptosis, gland epithelial degeneration/abscesses, gland goblet/enterocyte ratio decrease and gland loss. In some experiments, colons from control and treated mice were instead dissociated with Lamina Propria Dissociation Kit (Miltenyi Biotec), according to the manufacturer instructions. Single-cell suspensions were prepared from colon, and stained with vital dye Zombie Aqua dye (1:400 dilution, Biolegend) then surface stained with the following fluorescently-labeled antibodies: 1:200 FITC anti-NK1.1 (clone PK136, eBioscience), 1:300 PerCP-Cy5.5 anti-CD4 (clone RM4-5, Biolegend), 1:200 PE anti-TCRγδ (clone GL3, eBioscience), 1:200 PE-Dazzle 594 anti-CD3 (clone 17A2, Biolegend), 1:200 PE-Cy7 anti-CD44 (clone IM7, eBioscience), 1:300 APC anti-CD62L (clone MEL-14, eBioscience), 1:200 AlexaFluor 700 anti-CD8α (clone 53-6.7, eBioscience), 1:400 APC-Fire 750 anti-CD45 (clone 30-F11, Biolegend) and 1:300 eFluor 450 anti-CD19 (clone eBio1D3, eBioscience). In some case, a myeloid panel was used instead: 1:300 FITC anti-CD64 (clone X54-5/7.1, Biolegend), 1:300 PerCP-Cy5.5 anti-Ly6G (clone 1A8, Biolegend), 1:200 PE anti-SiglecF (clone E50-2440, BDBioscience), 1:200 PE-Dazzle 594 anti-CD103 (clone 2E7, Biolegend), 1:400 PE-Cy7 anti-CD45 (30-F11, Biolegend), 1:200 APC anti-CD317 (clone 927, Biolegend), 1:800 APC-Cy7 I-A/I-E anti-MHCII (clone M5/114.15.2, Biolegend), 1:1000 BV421 anti-Ly6C (clone HK1.4, Biolegend), 1:300 BV605 anti-CD11b (clone M1/70, Biolegend), 1:300 BV785 anti-CD11c (clone N418, Biolegend) and 1:400 Zombie Aqua Fixable Viability (Biolegend). Stained samples were processed using a Fortessa flow cytometer (BD Biosciences) and the results were analyzed using FlowJo software (Tree Star Inc).
Colon histology and immunochemistry
The fixed colon tissues were embedded in paraffin after dehydration, and 4 μm paraffin sections were prepared. After dewaxing and hydration, H&E staining was performed. The paraffin sections were scored as previously described13. To visualize goblet cells and mucin, an Alcian blue-PAS staining was performed. Sections were stained with Alcian blue at pH 2.5 (AB) followed by a staining step with the periodic acid-Schiff (PAS). For the IHC, paraffin-embedded tissue sections were de-waxed and rehydrated, incubated in Diva Decloaker antigen retrieval solution (Biocare) and boiled for 20 min in a pressure cooker; Enzyme Block (DAKO) was also used for 15 min to block peroxidase. Slides were stained with either 1:100 anti-Ki-67 (D3B5; Cell signaling), anti-CD68 (KP1, abcam), 1:250 anti-pSTAT3(Tyr705/D3A7, cell signaling), 1:100 anti-CD3 (SP7, Abcam) for 1 h. All antibodies were used in automated immunohistochemistry assays in accordance with the protocols recommended by the manufacturer (VENTANA BenchMark ULTRA instrument). Bound antibodies were detected using peroxidase-goat anti-rabbit secondary antibodies (AB_2307391, Jackson laboratories) used at a 1:500 dilution (VENTANA DISCOVERY series instruments). Slides were counter-stained with hematoxylin, then mounted. In some experiment, slides were then scanned and process using Aperio ImageScope (Version 12.4.3, Leica Biosystems Imaging Inc). The 2 most distal field of view (at 10x) were then systematically processed to quantify infiltration of immune cell populations using Fiji (Version 1.53f5158) using the following main operators: Thresholding (Otsu Binary), Automatic Smoothing/Sharpening, Watersheding and Analyze particle.
Mouse model of T cell specific colitis
Colitis was induced as we previously described13. Briefly, spleen from B6, Arhgef2−/− and Rasal3−/− were enriched for naive CD4+ CD45RBhi T cells (CD4+ T Cell Isolation Kit; Miltenyi Biotec) and single-cell suspensions was prepared. Cells were then then surface stained with the following fluorescently-labeled antibodies: 1:300 FITC anti-CD4 (GK1.5, eBioscience), 1:250 PE anti-CD25 (PC61.5,eBioscience) and 1:1500 APC anti-CD45RB (C363.16A, eBioscience). Naive cells where then purified by cell sorting (FACSAriaII, BD Biosciences) and 5 × 105 CD4+ CD45RBhi T cells were then injected intravenously into Rag1−/− mice. Mice were sacrificed 7 weeks after injection, their colon harvested and processed as above.
Gene expression and cytokine quantitation in colon tissues
RNA from distal colons flash-frozen in liquid nitrogen was prepared and analyzed as described elsewhere13. Briefly, RNA was purified using a FastPrep 24 homogenizer (MP Biomedicals) with lysing matrix D beads (MP Biomedicals) and RNeasy kits (QIAGEN). MMLV reverse transcriptase was used with a cocktail of Oligo dT and random hexamers (Invitrogen) for cDNA synthesis. The primer set (5′-CCGGGAGCTTCGAGGCCAAC-3′ and 5′-CCTATCTGGCAAGCGGGGC-3′) was used to quantify the relative expression of Ccdc88b mRNA. Cytokines and chemokines RNA transcripts were quantified by RT-qPCR using Perfecta SYBR Green PCR kit and the following primers sets: Mcp-1 (5′-AGGTGTCCCAAAGAAGCTGTA-3′ and 5′-TCTGGACCCATTCCTTCTTG-3′), Il-6 (5′-GAAGTAGGGAAGGCCGTGG-3′ and 5′-GAAGTAGGGAAGGCCGTGG-3′), Emr1 (F4/80) (5’-CTTTGGCTATGGGCTTCCAGTC-3’and 5’-GCAAGGAGGACAGAGTTTATCGTG-3), Rantes (5′-GCAAGTGCTCCAATCTTGCA-3′ and 5′-CTTCTCTGGGTTGGCACACA-3′). Mgl1 (5’-GAGAAAGGCTTTAAGAACTGGG-3’ and 5’-GACCACCTGTAGTGATGTGGG-3’) and C4b (5’-GATGAGGTTCGCCTGCTATT-3’ and 5’-GACTTGGGTGATCTTGGACTC-3’) Gene expression was normalized to Hprt (5′-TCAGTCAACGGGGGACATAAA-3′ and 5′-GGGGCTGTACTGCTTAACCAG-3′) and relative expressions was calculated using the ΔΔCT method59.
Mouse model of colitis-associated colorectal cancer
For the colitis-associated colorectal cancer model (CA-CRC), mice were injected intraperitoneally with azoxymethane (MP Biomedical; 7 mg/kg) followed by three 4-day cycles of 2% DSS in the drinking water, with each cycle 17 days apart. Mice were monitored at regular intervals and weighed weekly. Colons were collected after 14 weeks, fixed in formalin and analyzed for the number of tumors per colon, as well as their total tumor surface area (calculated from diameter of individual tumors). In some experiments, colon sections were also analyzed by immunohistochemistry as above.
Single-cell RNA sequencing
Colons from B6 mice were collected, cut into small pieces, and washed in gut buffer (1X Hank’s Balanced Salt Solution (HBSS) containing 2% heat-inactivated fetal bovine serum (FBS) and 15 mM of HEPES. Epithelial cells were removed by shaking in gut buffer with 5 mM EDTA for 30 min at 37 °C, with occasional vortexing. Finally, tissues were treated with collagenase IV (20 mg/ml) and DNase I (10 mg/ml) in RPMI-1640 supplemented with 5% FBS and 15 mM of HEPES for 20 min at 37 °C and passed through a 70 μM cell strainer to collect lamina propria cells. Cells were stained for viability using Zombie Viability Dye V500 (1:400), and surface stained with anti-CD45 APC (1:400, clone 30F11, Biolegend). Stained cell suspensions were sorted on a BD FACSAriaIII Cell Sorter (BD Biosciences) to obtain viable CD45+ cells. Freshly sorted cells were washed and resuspended in 0.04% BSA in PBS for loading on the 10x Chromium chip. Single-cell capture and cDNA preparation was done according to the 10x Single-Cell 3’ (version 3.1) protocol, with 8000 cells targeted for capture per sample. Libraries were sequenced on the NovaSeq 6000 Sequencing System (Illumina). Raw sequencing data can be found on the Gene Expression Omnibus Repository (GEO)60 with the accession number GSE249342. Raw gene expression matrices were generated for each sample by the Cell Ranger, and the output was analyzed using the Seurat package. Low-quality reads were filtered based on three criteria: number of detected genes per cell, number of UMIs expressed per cell and mitochondrial content, using the following threshold parameters: nGene (between 200 and 7500), nUMI (between 500 and 75,000), and percentage of mitochondrial genes expressed (<7.5%). Doublets were then identified by finding cells expressing markers of two cell lineages simultaneously, as well as using the DoubletFinder package. After removal of low-quality cells and doublets, we captured 12,475 CD45+ cells from B6 colonic lamina propria (total from 2 samples). Gene expression matrices were normalized using the NormalizeData function and scaled using the ScaleData function, regressing out effects of cell cycle and percentage of expressed mitochondrial genes. Identification of highly variable features, linear dimension reduction by PCA transformation, UMAP dimensionality reduction, and cell clustering were performed using standard Seurat package workflows. Myeloid cells were integrated for further sub-clustering. Normalization, scaling, PCA, UMAP and clustering were performed as described above.
T cell homing assay
Spleen were harvested, processed into single-cell suspensions and red blood cells lysed using ammonium chloride containing buffer (ACK; Fisher scientific). Splenocytes were stained with either 2 µM of CellTracker green (5-chloromethylfluorescein diacetate or CMFDA, ThermoFisher) or 20 µM of CellTracker orange (5-(and-6)-(((4-chloromethyl)benzoyl)amino)tetramethylrhodamine or CMTMR, Thermo Fisher) for 15 min at 37 °C. Stained cells were extensively washed with PBS, counted twice, and a mixture (1:1 ratio) of independently stained splenocytes (total of 1 × 107 cells) were injected intravenously into the tail vein of B6 mice. Inguinal lymph nodes (LNs) were harvested 6 h later and processed for analysis of cell composition by flow cytometry using the following fluorescently-labeled antibodies in addition to CMFDA and CMTMR: 1:300 PerCP-Cy5.5 anti-CD4 (clone RM4-5, Biolegend), 1:400 APC anti-CD45 (clone 30-F11, eBioscience), 1:300 APC-Fire anti-CD8a (clone 53-6.7, eBioscience), 1:200 PE-Dazzle 594 anti-CD3 (clone 17A2, Biolegend) and Zombie Aqua Fixable Viability (Biolegend). The relative amount of CMFDA+ T cells compared to CMTMR+ T cells was calculated as follows: (% of CMFDA+ T cells in sample/% of CMFDA+ T cells in injection mix)/(% of CMTMR+ T cells in sample/% of CMTMR+ T cells in injection mix), as we described10.
In vivo migration assay
Migration assays were performed as previously described10. LPS-treated bone marrow-derived dendritic cells (BMDC) were stained with either 2 µM of CellTracker green (CMFDA, Thermo Fisher) or 20 µM of CellTracker orange (CMTMR, Thermo Fisher) for 15 min at 37 °C, extensively washed with PBS, counted twice, and a mixture (1:1 ratio) of independently stained cells (total of 1 × 106 cells) were injected in the footpad of B6 mice. Popliteal LNs were harvested 48 h later, and the ratio of CMFDA and CMTMR labeled CD11c+ cells was analyzed by flow cytometry. The relative amount of CMFDA+ DCs compared to CMTMR+ DCs was calculated as above.
In vitro patrolling assay
Patrolling assays were performed as previously described10. BMDCs were cultured in Nunc Lab-Tek 8-wells cover-glass chambers (Thermo Fisher) for 3 h at 37 °C and 5% CO2 in RPMI supplemented with 10% fetal bovine serum (HyClone) before imaging. Two representative fields of view per sample were acquired every minute for 90 min by brightfield microscopy using a Zeiss LSM700. Images were processed with ImageJ (Fiji58) and analyzed using the built-in TrackMate plugin61 in addition to a homemade MatLab script (MathWorks). Single tracks produced by the TrackMate plugin were manually removed when tracking cellular debris or non-adherent cells.
RHOA activation assay
Bone marrow-derived dendritic cells (BMDCs) were produced as described previously10, serum starved for 24 h and stimulated for 5 min at 37 °C in RPMI supplemented with 10% fetal bovine serum with 1 µg/ml LPS. BMDCs were then wash extensively with ice cold PBS and processed rapidly to a Rho Activation Assay Biochem Kit (Cytoskeleton Inc) accordingly to the manufacturer instructions. Briefly, BMDCs were lysed in 50 mM Tris pH 7.5, 10 mM MgCl2, 0.5 M NaCl, and 2% Igepal (Cytoskeleton Inc), and 800 µg of lysates submitted to immunoprecipitation with Rhotekin RBD beads IP for 1h15 at 4 °C with 50 µg beads. Lysate was then washed, eluted in 2x Laemmli sample buffer, boiled for 10 min and analyzed by SDS-PAGE and Western blot analysis using Anti-RHOA monoclonal IgM antibody (1:500 Cytoskeleton Inc), as described above. For this assay, positive control was also produced by saturating BMDCs lysate with 200 μM GTPγS and negative control was produced by saturating BMDCs with 1 mM GDP, before immunoprecipitation. Western blot images were scanned, processed with ImageJ (Fiji58) and analyzed for the relative intensity of RHOA-GTP when compared to total RHOA in the input.
Statistical analysis and reproducibility
Results are presented as mean ± SEM, unless otherwise indicated. Prism5 (GraphPad) software was used for all statistical tests, using parametric tests when criteria for normality is satisfied and non-parametric otherwise, as indicated. Differences were considered statistically significant when p ≤ 0.05. p values are indicated by *p ≤ 0.05, **p < 0.01, ***p < 0.001. Data are independent biological replicate, where each individual data point in each plots represent a single mouse or primary cell line derived from an individual mouse, except when otherwise indicated; number of independent experiments carried out with similar results are identified in each corresponding Figure legend and were all reproducible.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Geneviève Perreault, Suzan Gauthier, the Cell Vision Core Facility and the Advanced BioImaging Facility (ABIF) at McGill University. This work was supported in part by research grants to P.G. from the Canadian Institutes for Health Research (CIHR, Foundation grant), and the Canadian Cancer Research Society. P.G. is supported by a Distinguished James McGill Professorship award from McGill University. D.L. is supported by a research grant from the CIHR and a Junior Investigator award from the Fonds de Recherche du Québec Santé (FRQS). This work benefited from collaboration with the National Center for Dynamic Interactome Research (www.ncdir.org) leveraging funds from the National Institute of General Medical Sciences of the National Institutes of Health (NIH), grant P41GM109824; J.L. is funded in-part by NIH grant R01GM126170.
Author contributions
J.-F.O., D.L., J.L., P.G. and N.F. designed the study; J.-F.O., D.L., T.J., M.J.P., L.G., H.J., K.R.M., G.X., J.L. and N.F. performed the experiments; J.-F.O., D.L., T.J., R.C., H.J., J.L. and N.F. analyzed the data; J.-F.O., D.L., J.L., P.G. and N.F. wrote the initial draft and J.-F.O., D.L., R.C., H.S., J.L., P.G. and N.F. contributed to the final draft of the manuscript and provided expertise.
Peer review
Peer review information
This manuscript has been previously reviewed at another Nature Portfolio journal. The manuscript was considered suitable for publication without further review at Communications Biology. Handling Editor: Christina Karlsson Rosenthal.
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE62 partner repository with the dataset PXD023779. The protein interactions from this publication have been submitted to the IMEx (http://www.imexconsortium.org) consortium through IntAct63 and assigned the identifier IM-28785. The raw single-cell RNA sequencing data can be found on the Gene Expression Omnibus Repository (GEO)60 with the accession code GSE249342. Any other data that support the findings of this study, including additional MS/MS data (see Supplementary Data 1), are available within the article, its Supplementary Information and from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jean-Frederic Olivier, David Langlais.
These authors jointly supervised this work: John LaCava, Philippe Gros, Nassima Fodil.
Change history
1/24/2024
In this article the url and its hyperlink provided for www.copurifcation.org/ in the sentence beginning ‘To further explore potential missing information in our IPs,...’ was incorrect. The original article has been corrected.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-023-05751-9.
References
- 1.Lamb TJ, Brown DE, Potocnik AJ, Langhorne J. Insights into the immunopathogenesis of malaria using mouse models. Expert Rev. Mol. Med. 2006;8:1–22. doi: 10.1017/S1462399406010581. [DOI] [PubMed] [Google Scholar]
- 2.Mishra SK, Newton CR. Diagnosis and management of the neurological complications of falciparum malaria. Nat. Rev. Neurol. 2009;5:189–198. doi: 10.1038/nrneurol.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogers J, Mastroeni D, Leonard B, Joyce J, Grover A. Neuroinflammation in Alzheimer’s disease and Parkinson’s disease: are microglia pathogenic in either disorder? Int. Rev. Neurobiol. 2007;82:235–246. doi: 10.1016/S0074-7742(07)82012-5. [DOI] [PubMed] [Google Scholar]
- 4.Torre S, Langlais D, Gros P. Genetic analysis of cerebral malaria in the mouse model infected with Plasmodium berghei. Mamm. Genome. 2018;29:488–506. doi: 10.1007/s00335-018-9752-9. [DOI] [PubMed] [Google Scholar]
- 5.Karpus WJ. Cytokines and chemokines in the pathogenesis of experimental autoimmune encephalomyelitis. J. Immunol. 2020;204:316–326. doi: 10.4049/jimmunol.1900914. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy JM, et al. CCDC88B is a novel regulator of maturation and effector functions of T cells during pathological inflammation. J. Exp. Med. 2014;211:2519–2535. doi: 10.1084/jem.20140455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bongfen SE, et al. An N-ethyl-N-nitrosourea (ENU)-induced dominant negative mutation in the JAK3 kinase protects against cerebral malaria. PLoS ONE. 2012;7:e31012. doi: 10.1371/journal.pone.0031012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torre S, et al. THEMIS is required for pathogenesis of cerebral malaria and protection against pulmonary tuberculosis. Infect. Immun. 2015;83:759–768. doi: 10.1128/IAI.02586-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torre S, et al. USP15 regulates type I interferon response and is required for pathogenesis of neuroinflammation. Nat. Immunol. 2017;18:54–63. doi: 10.1038/ni.3581. [DOI] [PubMed] [Google Scholar]
- 10.Olivier JF, et al. CCDC88B is required for mobility and inflammatory functions of dendritic cells. J. Leukoc. Biol. 2020;108:1787–1802. doi: 10.1002/JLB.3A0420-386R. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, et al. Discovery of six new susceptibility loci and analysis of pleiotropic effects in leprosy. Nat. Genet. 2015;47:267–271. doi: 10.1038/ng.3212. [DOI] [PubMed] [Google Scholar]
- 12.Fischer A, et al. A novel sarcoidosis risk locus for Europeans on chromosome 11q13.1. Am. J. Respir. Crit. Care Med. 2012;186:877–885. doi: 10.1164/rccm.201204-0708OC. [DOI] [PubMed] [Google Scholar]
- 13.Fodil N, et al. CCDC88B is required for pathogenesis of inflammatory bowel disease. Nat. Commun. 2017;8:932. doi: 10.1038/s41467-017-01381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le-Niculescu H, Niesman I, Fischer T, DeVries L, Farquhar MG. Identification and characterization of GIV, a novel Galpha i/s-interacting protein found on COPI, endoplasmic reticulum-Golgi transport vesicles. J. Biol. Chem. 2005;280:22012–22020. doi: 10.1074/jbc.M501833200. [DOI] [PubMed] [Google Scholar]
- 15.Simpson F, et al. A novel hook-related protein family and the characterization of hook-related protein 1. Traffic. 2005;6:442–458. doi: 10.1111/j.1600-0854.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- 16.Weng L, Enomoto A, Ishida-Takagishi M, Asai N, Takahashi M. Girding for migratory cues: roles of the Akt substrate Girdin in cancer progression and angiogenesis. Cancer Sci. 2010;101:836–842. doi: 10.1111/j.1349-7006.2009.01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oshita A, et al. Identification and characterization of a novel Dvl-binding protein that suppresses Wnt signalling pathway. Genes Cells. 2003;8:1005–1017. doi: 10.1111/j.1365-2443.2003.00692.x. [DOI] [PubMed] [Google Scholar]
- 18.Ear J, et al. Two isoforms of the guanine nucleotide exchange factor, Daple/CCDC88C cooperate as tumor suppressors. Sci. Rep. 2019;9:12124. doi: 10.1038/s41598-019-48420-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ara H, et al. Role for Daple in non-canonical Wnt signaling during gastric cancer invasion and metastasis. Cancer Sci. 2016;107:133–139. doi: 10.1111/cas.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drielsma A, et al. Two novel CCDC88C mutations confirm the role of DAPLE in autosomal recessive congenital hydrocephalus. J. Med. Genet. 2012;49:708–712. doi: 10.1136/jmedgenet-2012-101190. [DOI] [PubMed] [Google Scholar]
- 21.Ham H, Huynh W, Schoon RA, Vale RD, Billadeau DD. HkRP3 is a microtubule-binding protein regulating lytic granule clustering and NK cell killing. J. Immunol. 2015;194:3984–3996. doi: 10.4049/jimmunol.1402897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellacheruvu D, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King PD, Lubeck BA, Lapinski PE. Nonredundant functions for Ras GTPase-activating proteins in tissue homeostasis. Sci. Signal. 2013;6:re1. doi: 10.1126/scisignal.2003669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodge RG, Ridley AJ. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 2016;17:496–510. doi: 10.1038/nrm.2016.67. [DOI] [PubMed] [Google Scholar]
- 25.Muro R, et al. The Ras GTPase-activating protein Rasal3 supports survival of naive T cells. PLoS ONE. 2015;10:e0119898. doi: 10.1371/journal.pone.0119898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muro R, Nitta T, Kitajima M, Okada T, Suzuki H. Rasal3-mediated T cell survival is essential for inflammatory responses. Biochem. Biophys. Res. Commun. 2018;496:25–30. doi: 10.1016/j.bbrc.2017.12.159. [DOI] [PubMed] [Google Scholar]
- 27.Jeyakumar T, et al. Inactivation of interferon regulatory factor 1 causes susceptibility to colitis-associated colorectal cancer. Sci. Rep. 2019;9:18897. doi: 10.1038/s41598-019-55378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joo E, Olson MF. Regulation and functions of the RhoA regulatory guanine nucleotide exchange factor GEF-H1. Small GTPases. 2021;12:358–371. doi: 10.1080/21541248.2020.1840889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fine N, et al. GEF-H1 is necessary for neutrophil shear stress-induced migration during inflammation. J. Cell Biol. 2016;215:107–119. doi: 10.1083/jcb.201603109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nalbant P, Chang YC, Birkenfeld J, Chang ZF, Bokoch GM. Guanine nucleotide exchange factor-H1 regulates cell migration via localized activation of RhoA at the leading edge. Mol. Biol. Cell. 2009;20:4070–4082. doi: 10.1091/mbc.e09-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsapara A, et al. The RhoA activator GEF-H1/Lfc is a transforming growth factor-beta target gene and effector that regulates alpha-smooth muscle actin expression and cell migration. Mol. Biol. Cell. 2010;21:860–870. doi: 10.1091/mbc.e09-07-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heasman SJ, Carlin LM, Cox S, Ng T, Ridley AJ. Coordinated RhoA signaling at the leading edge and uropod is required for T cell transendothelial migration. J. Cell Biol. 2010;190:553–563. doi: 10.1083/jcb.201002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren Y, Li R, Zheng Y, Busch H. Cloning and characterization of GEF-H1, a microtubule-associated guanine nucleotide exchange factor for Rac and Rho GTPases. J. Biol. Chem. 1998;273:34954–34960. doi: 10.1074/jbc.273.52.34954. [DOI] [PubMed] [Google Scholar]
- 34.Krendel M, Zenke FT, Bokoch GM. Nucleotide exchange factor GEF-H1 mediates cross-talk between microtubules and the actin cytoskeleton. Nat. Cell Biol. 2002;4:294–301. doi: 10.1038/ncb773. [DOI] [PubMed] [Google Scholar]
- 35.Chang YC, Nalbant P, Birkenfeld J, Chang ZF, Bokoch GM. GEF-H1 couples nocodazole-induced microtubule disassembly to cell contractility via RhoA. Mol. Biol. Cell. 2008;19:2147–2153. doi: 10.1091/mbc.e07-12-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birkenfeld J, Nalbant P, Yoon SH, Bokoch GM. Cellular functions of GEF-H1, a microtubule-regulated Rho-GEF: is altered GEF-H1 activity a crucial determinant of disease pathogenesis? Trends Cell Biol. 2008;18:210–219. doi: 10.1016/j.tcb.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Pan W, et al. The regulatory subunit PPP2R2A of PP2A enhances Th1 and Th17 differentiation through activation of the GEF-H1/RhoA/ROCK signaling pathway. J. Immunol. 2021;206:1719–1728. doi: 10.4049/jimmunol.2001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiang HS, et al. GEF-H1 controls microtubule-dependent sensing of nucleic acids for antiviral host defenses. Nat. Immunol. 2014;15:63–71. doi: 10.1038/ni.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meiri D, et al. Mechanistic insight into GPCR-mediated activation of the microtubule-associated RhoA exchange factor GEF-H1. Nat. Commun. 2014;5:4857. doi: 10.1038/ncomms5857. [DOI] [PubMed] [Google Scholar]
- 40.Wang H, et al. Guanine nucleotide exchange factor -H1 promotes inflammatory cytokine production and intracellular mycobacterial elimination in macrophages. Cell Cycle. 2017;16:1695–1704. doi: 10.1080/15384101.2017.1347739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng IK, et al. GEF-H1 over-expression in hepatocellular carcinoma promotes cell motility via activation of RhoA signalling. J. Pathol. 2012;228:575–585. doi: 10.1002/path.4084. [DOI] [PubMed] [Google Scholar]
- 42.Gao Y, et al. Role of RhoA-specific guanine exchange factors in regulation of endomitosis in megakaryocytes. Dev. Cell. 2012;22:573–584. doi: 10.1016/j.devcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan M, et al. BNIP-2 retards breast cancer cell migration by coupling microtubule-mediated GEF-H1 and RhoA activation. Sci. Adv. 2020;6:eaaz1534. doi: 10.1126/sciadv.aaz1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radovanovic I, et al. Genetic control of susceptibility to Candida albicans in SM/J mice. J. Immunol. 2014;193:1290–1300. doi: 10.4049/jimmunol.1400783. [DOI] [PubMed] [Google Scholar]
- 46.Saito S, et al. RASAL3, a novel hematopoietic RasGAP protein, regulates the number and functions of NKT cells. Eur. J. Immunol. 2015;45:1512–1523. doi: 10.1002/eji.201444977. [DOI] [PubMed] [Google Scholar]
- 47.Fort L, et al. Fam49/CYRI interacts with Rac1 and locally suppresses protrusions. Nat. Cell Biol. 2018;20:1159–1171. doi: 10.1038/s41556-018-0198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuki KE, et al. CYRI/FAM49B negatively regulates RAC1-driven cytoskeletal remodelling and protects against bacterial infection. Nat. Microbiol. 2019;4:1516–1531. doi: 10.1038/s41564-019-0484-8. [DOI] [PubMed] [Google Scholar]
- 49.LaCava, J., Jiang, H. & Rout, M. P. Protein complex affinity capture from cryomilled mammalian cells. J. Vis. Exp.118, 54518 (2016). [DOI] [PMC free article] [PubMed]
- 50.Cristea IM, Chait BT. Conjugation of magnetic beads for immunopurification of protein complexes. Cold Spring Harb. Protoc. 2011;2011:pdb prot5610. doi: 10.1101/pdb.prot5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hakhverdyan Z, et al. Rapid, optimized interactomic screening. Nat. Methods. 2015;12:553–560. doi: 10.1038/nmeth.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang H, Taylor MS, Molloy KR, Altukhov I, LaCava J. Identification of RNase-sensitive LINE-1 ribonucleoprotein interactions by differential affinity immobilization. Bio Protoc. 2019;9:e3200. doi: 10.21769/BioProtoc.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 54.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 55.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 56.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tyanova S, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 58.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 60.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tinevez JY, et al. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 62.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Orchard S, et al. The MIntAct project-IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE62 partner repository with the dataset PXD023779. The protein interactions from this publication have been submitted to the IMEx (http://www.imexconsortium.org) consortium through IntAct63 and assigned the identifier IM-28785. The raw single-cell RNA sequencing data can be found on the Gene Expression Omnibus Repository (GEO)60 with the accession code GSE249342. Any other data that support the findings of this study, including additional MS/MS data (see Supplementary Data 1), are available within the article, its Supplementary Information and from the corresponding author upon reasonable request.