Abstract
In eubacteria, there are three slightly different pathways for the synthesis of m-diaminopimelate (m-DAP), which is one of the key linking units of peptidoglycan. Surprisingly, for unknown reasons, some bacteria use two of these pathways together. An example is Corynebacterium glutamicum, which uses both the succinylase and dehydrogenase pathways for m-DAP synthesis. In this study, we clone dapD and prove by enzyme experiments that this gene encodes the succinylase (Mr = 24082), initiating the succinylase pathway of m-DAP synthesis. By using gene-directed mutation, dapD, as well as dapE encoding the desuccinylase, was inactivated, thereby forcing C. glutamicum to use only the dehydrogenase pathway of m-DAP synthesis. The mutants are unable to grow on organic nitrogen sources. When supplied with low ammonium concentrations but excess carbon, their morphology is radically altered and they are less resistant to mechanical stress than the wild type. Since the succinylase has a high affinity toward its substrate and uses glutamate as the nitrogen donor, while the dehydrogenase has a low affinity and incorporates ammonium directly, the m-DAP synthesis is another example of twin activities present in bacteria for access to important metabolites such as the well-known twin activities for the synthesis of glutamate or for the uptake of potassium.
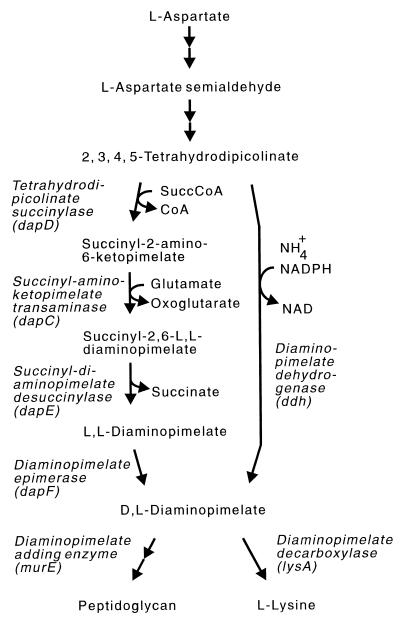
The amino acid meso-diaminopimelate (m-DAP) is one of the key intermediates of peptidoglycan synthesis. It serves to link the glycan backbones in the cell walls of many bacteria, giving them their shape and rigid structure. In addition, the synthesis of m-DAP is required for protein synthesis, since after decarboxylation, it yields l-lysine. Due to this vital role of m-DAP for bacteria and the facts that mammals neither synthesize nor require m-DAP and that many proven antibiotics act in preventing bacterial cell wall synthesis, the m-DAP biosynthesis pathway is an attractive target for the rational design of new drugs. Accordingly, m-DAP analogs were assayed for inhibition of enzyme activity (9), and the crystal structures of several enzymes of m-DAP synthesis were resolved (2, 3, 19). Interestingly, there are three slightly different pathways of m-DAP synthesis. They all share the first steps for aspartate and pyruvate conversion to l-2,3,4,5-tetrahydrodipicolinate, but they vary in the subsequent conversion to yield m-DAP (Fig. 1): pathway 1, a four-step reaction with succinylated intermediates; pathway 2, another four-step reaction involving acetylated intermediates; and pathway 3, a one-step reaction catalyzed by an ammonium-incorporating dehydrogenase. An example of the respective variant is Escherichia coli (pathway 1), Bacillus subtilis (pathway 2), and Bacillus sphaericus (pathway 3). Each variant pathway (hereafter termed the succinylase, acetylase, and dehydrogenase variant, respectively) has its specific cofactor demand. However, it is still not known whether there is any special reason for this wide range of m-DAP synthesis, whether the respective pathway, for instance, corresponds to a particular need of the bacterium in question.
FIG. 1.
The split pathway of m-DAP synthesis for the formation of m-DAP and l-lysine as present in C. glutamicum. On the left is shown the succinylase variant, on the right the dehydrogenase variant. Some bacteria have only the succinylase or dehydrogenase variant. The third acetylase variant is comparable to the succinylase variant but uses acetyl groups instead of succinyl groups. SuccCoA, succinyl coenzyme A; CoA, coenzyme A.
There is an unusual situation in the gram-positive bacterium Corynebacterium glutamicum. The succinylase and dehydrogenase variants operate in this organism side by side (Fig. 1). Due to this situation, C. glutamicum is an ideal candidate to be assayed on the relevance of the variant pathways established in bacteria. The m-DAP pathway in C. glutamicum has been intensively investigated, since mutants of this bacterium are used to produce the m-DAP-derived l-lysine on a scale of more than 3 × 105 tons/year (t/y) (10). We have studied almost all the enzymes of the m-DAP pathway of l-lysine synthesis (5, 23) and used them for flux increase (5), and we have shown by nuclear magnetic resonance studies that in vivo both the succinylase and dehydrogenase variants contribute to m-DAP formation (12, 27). The relative use of both pathways is dependent on the ammonium concentration. To study a particular function of the two variant pathways in C. glutamicum, we had already inactivated the dehydrogenase-encoding ddh gene and found reduced l-lysine yields with overproducers, whereas the growth rates were not affected (24). However, it proved difficult to clone the genes for the enzymes of the four-step reaction. Although we were able to clone the desuccinylase-encoding gene dapE (28) and we also obtained a DNA fragment which led to succinylase activity, its structure did not correspond to that in the chromosome. As we demonstrated, this can be attributed to the adjacent aroP gene which encodes an aromatic amino acid transporter (29). In this study, we describe the cloning of dapD (succinylase), as well as the structure of a related gene, and apply gene inactivation to enable physiological experiments. This allows conclusions to be drawn on the use and function of the dehydrogenase and succinylase variants of m-DAP synthesis.
MATERIALS AND METHODS
Strains, plasmids, and growth.
The bacterial strains and the most relevant plasmids are given in Table 1. C. glutamicum was grown at 30°C on minimal medium CGXII (16) or brain heart infusion (BHI; Difco), with the indicated supplements. For enzyme determinations, cells were grown on complex medium CGIII (13). E. coli strains were grown at 37°C, except for RDD32 dapD, which was grown at 30°C. When appropriate, ampicillin and kanamycin were used at concentrations of 40 and 50 μg ml−1, respectively. Growth was monitored by measuring absorbance at 600 nm of diluted cultures (PM6; Zeiss, Oberkochen, Germany). The isolation of clones complementing the dapD mutation in E. coli RDD32 was performed as described previously (28).
TABLE 1.
Strains and selected plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference |
|---|---|---|
| E. coli | ||
| DH5 | Cloning strain | |
| S17-1 | Mobilizing donor strain | 26 |
| RDD32 | dapD::MuCts | 17 |
| C. glutamicum | ||
| ATCC 13032 | Type strain | |
| ATCC 13032 dapD | dapD::pEMdapDint Kmr | This work |
| ATCC 13032 dapE | dapE::pEMdapEint Kmr | This work |
| ATCC 13032 orf2 | orf2::pEMorf2int Kmr | This work |
| ATCC 13032 Δorf3 | Deletion of the 479-bp KpnI fragment | This work |
| Plasmids | ||
| pJC1 | Shuttle vector, Kmr | 4 |
| pEM1 | Integration vector, mobilizable, nonreplicative in C. glutamicum, Kmr | 23 |
| pK18mobsacB | Replacement vector, mobilizable, nonreplicative in C. glutamicum, Kmr | 21 |
| pJCdapD1.55 | pJC1::dapD | This work |
| pJCorf3/Bam3.4 | pJC1::orf3/Bam3.4 | This work |
Genetic techniques, sequencing, and hybridization.
Standard procedures were used for the isolation and in vitro recombination of DNA. E. coli DH5 and S17-1 were transformed by the CaCl2 method, whereas electroporation was used to transform E. coli RDD32 and the C. glutamicum strains (11). To transfer nonreplicative plasmids into C. glutamicum, conjugation was done as previously described (20), and clones with integrated vectors were selected by resistance to kanamycin (15 μg ml−1).
The DNA sequence was determined by the dideoxy-chain termination method, using the AutoRead Sequencing kit and the A.L.F. sequencer from Pharmacia (Freiburg, Germany). The sequences were derived from deletion clones, but two fluorescein-labelled primers (5′-CGCGGTCGGCGTCACCG-3′ and 5′-GCCTCCTCAACAATGTCGT-3′) were made to verify the sequence of a region with an extraordinarily high G+C content. The structural integrity of cloned fragments and of the appropriate chromosomal deletions was verified by DNA hybridization, using digoxigenin-labelled DNA as a hybridizing probe.
Construction of inactivation and deletion mutants.
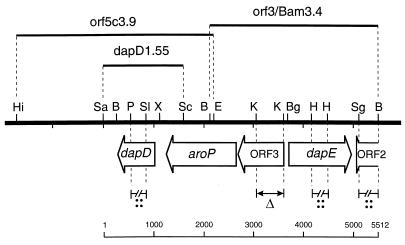
To construct the orf3 deletion mutant, the 479-bp KpnI fragment was deleted from the 2.5-kb EcoRI fragment of the cluster carrying orf3 as well as the adjacent 5′ regions of aroP and dapE (Fig. 2). This 2.0-kb Δorf3 fragment was ligated with pK18mobsacB, and the resulting plasmid was used for intergeneric conjugation to recombine with the chromosome of C. glutamicum. Using the sacB gene of the integrated vector, clones were selected for a second recombination (21). An attempt was made to construct a dapD deletion mutant. A pK18mobsacB derivative harboring a dapD gene with the 457-bp Eco47III-NsiI fragment deleted was made for this purpose. However, of 24 recombinants tested, the second recombination always yielded the reconstituted wild-type situation. This is indicative of a selective pressure for the presence of dapD. Therefore, this gene was disrupted by use of the internal 330-bp PstI-SalI dapD fragment which was ligated to pEM1 (Table 1). To construct the dapE inactivation mutant, the internal 298-bp HindII dapE fragment was ligated to pEM1 and used for conjugation. Similarly, orf2 was inactivated by use of the internal 321-bp SgrAI-BamHI orf2 fragment ligated to pEM1.
FIG. 2.
Overview of the dapDE locus of C. glutamicum. The thick black line shows the general arrangement with selected restriction sites. Selected fragments used in this study are shown over the line, and the regions used to construct specific mutants, either by gene disruption (::) or gene deletion (▵), are shown under the line. The scale at the bottom of the figure shows length (in base pairs). Abbreviations: B, BamHI; Bg, BglII; E, EcoRI; H, HindII; Hi, HindIII; K, KpnI; P, PstI; Sa, SacI; Sc, ScaI; Sg, SgrAI; Sl, SalI; X, XhoI.
Cell disruption and enzyme assays.
Cells of C. glutamicum were washed with 0.9% NaCl, resuspended in 0.1 M potassium phosphate buffer, pH 7, and the optical density was adjusted to an absorbance at 600 nm of 100. Sonication was done for 10 min with a microtip-equipped Branson sonifier at an output of 2.5 and with the duty cycle set to 20%. The homogenate was centrifuged for 30 min (12,000 × g, 4°C), and the protein concentration was determined by the biuret reaction. The tetrahydrodipicolinate succinylase (EC 2.3.1.117), N-succinyl-diaminopimelate desuccinylase (EC 3.5.1.18), and diaminopimelate epimerase (EC 5.1.1.7) activities were quantified as described previously (23).
Quantification of amino acids and ammonium.
Cells were separated from the medium and inactivated by silica oil centrifugation with the additional extraction and neutralization procedure previously described (23), yielding cellular and extracellular fractions. Amino acids were quantified as their ortho-phthaldialdehyde derivatives by automatic precolumn derivatization and separation by reversed-phase chromatography with fluorescence detection. Quantification of ammonium concentrations was done enzymatically by using glutamate dehydrogenase.
Electron microscopy.
Cells were grown on Luria-Bertani medium (LB) containing 10% sucrose and 1.6% agar. After growth for 18 h at 30°C, the cells were resuspended in 200 μl of water to give a final absorbance at 600 nm of 4 to 6. One microliter of this cell suspension was placed onto a 5-mm2 piece of a filter (Nucleopore, Sartorius, Germany), which was dried overnight. The cells were fixed with 2.5% glutaraldehyde, washed twice with water, and subsequently dehydrated in a graded series of solutions with increasing concentrations of acetone (20, 40, 60, 80, and 100%). After the cells were coated with gold, the morphology of the specimen was resolved with a scanning electron microscope (S360; Leika Cambridge, Mass.).
Nucleotide sequence accession number.
The nucleotide sequence of dapD and its flanking regions has been deposited in the EMBL database under accession number AJ004934.
RESULTS
Cloning and chromosomal localization of dapD.
In previous work we had isolated cosmid pDE07 carrying C. glutamicum DNA, which complemented the dapD mutation in E. coli RDD32 (28). Although the insert was structurally altered, the complementing function was subcloned and sequenced in an attempt to obtain access to dapD which is of particular interest, since its gene product initiates entry into the succinylase variant (Fig. 1). Surprisingly, the complementing function consisted of sequences identical to the first part (109 amino acids) of orf3 at the dapE locus of C. glutamicum (Fig. 2) but fused with unknown sequences. To explain this puzzling information, we carried out intensive Southern blot analyses, eventually resulting in the structurally intact fragment orf5c3.9. Subcloning and enzyme experiments yielded a 1.55-kb SacI-ScaI fragment, which gave rise to an 11-fold increase in succinylase activity on plasmid pJC1 in C. glutamicum (Table 2). The nucleotide sequence of this fragment revealed an open reading frame of 690 nucleotides. In accord with the genetic and functional experiments, it was designated dapD. This C. glutamicum gene has an unusually high G+C content of 61%, while the neighboring sequences have the genome’s G+C content of 54%. This analysis shows that dapD is close to orf3 and dapE. The chromosomal organization of the entire dapDE locus of 5,512 bp thus derived is shown in Fig. 2.
TABLE 2.
Tetrahydrodipicolinate succinylase (DapD) and succinyldiaminopimelate desuccinylase (DapE) activities in C. glutamicum strains derived from the wild type
| C. glutamicum strain | Sp act (μmol min−1 mg of protein−1
|
|
|---|---|---|
| Succinylase | Desuccinylase | |
| ATCC 13032(pJC1) | 0.008 | 0.021 |
| ATCC 13032(pJCdapD1.55) | 0.093 | NDa |
| ATCC 13032 dapD | <0.001 | ND |
| ATCC 13032 dapE | ND | <0.001 |
| ATCC 13032(pJCorf3/Bam3.4) | 0.009 | ND |
| ATCC 13032 Δorf3 | 0.012 | 0.018 |
ND, not done.
Similarity and function of DapD and other proteins encoded by genes of the dap locus.
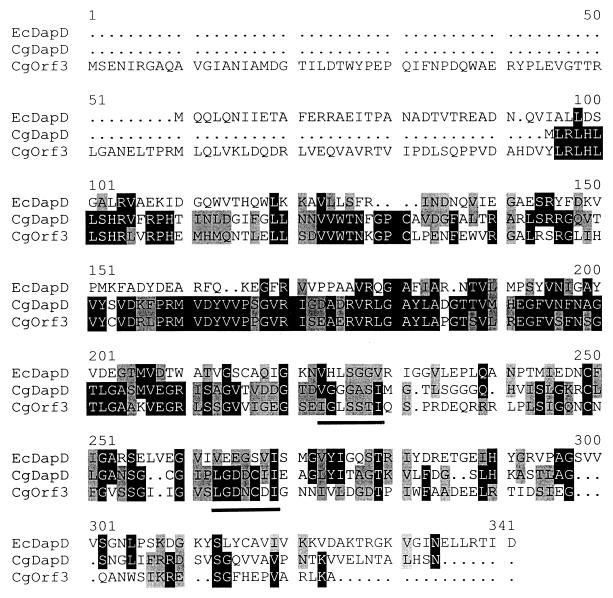
The deduced amino acid sequence of dapD has only 21% identical amino acid residues with DapD of E. coli (Fig. 3), while other structural homologs of the E. coli polypeptide have more than 60% identical amino acids (18). Astonishingly, the two gene products derived from dapD and orf3 of C. glutamicum have a high degree (46%) of identity with each other (Fig. 3). Even on the nucleotide level, a high degree of identity is present (not shown), which was apparently one prerequisite for the artificial clone pDE07 being a fusion of both genes. Both the dapD- and orf3-derived polypeptides display the imperfect tandem-repeated heptapeptide sequence (I/L/V)-G-X4-I. This encodes a left-handed parallel β helix characteristic of most acyltransferases (2). However, orf3-carrying fragments do not result in succinylase activity (Table 2). Nevertheless, a transcript initiating 74 nucleotides in front of orf3 was formed (Northern and primer extension analyses not shown). An orf3 deletion mutant was constructed as well as an orf2 insertion mutant (for details, see Materials and Methods). These mutants exhibited no phenotype. These mutants were not altered in succinylase, N-succinylaminoketopimelate transaminase, or diaminopimelate epimerase activity, thus excluding further genes of the succinylase pathway from being located at the dapDE locus analyzed.
FIG. 3.
Direct comparison of three sequences. The deduced amino acid sequences of the dapD gene product of C. glutamicum (CgDapD) and of E. coli (EcDapD) and of the orf3 gene product of C. glutamicum (CgOrf3) are compared. Amino acids that are identical (black boxes) or similar (shaded boxes) are indicated. Although the DapD polypeptides from C. glutamicum and E. coli both represent functional tetrahydrodipicolinate succinylases, their structural similarity is less than that of the DapD and Orf3 polypeptides from C. glutamicum. The bars represent the imperfect tandem-repeated heptapeptide sequence characteristic of acyltransferases (2).
An inactive succinylase pathway results in higher ammonium requirements.
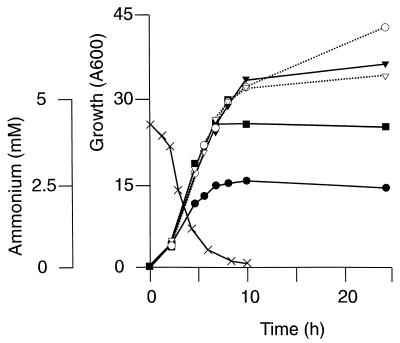
To examine the function of the succinylase variant, we used vector integration to disrupt dapD in the wild-type strain of C. glutamicum. The enzyme assay revealed the absence of succinylase activity in the C. glutamicum dapD mutant constructed (Table 2). Similarly, a C. glutamicum dapE mutant strain was constructed which was proven by enzyme activity determinations to be without desuccinylase activity. Both mutants and the wild type were grown on salt medium CGXII as well as on complex medium BHI. On both these media, neither the growth rate nor the final cell density was affected (not shown). However, when we added 4% glucose to BHI, we noted that growth of both mutants was arrested at a cell density (A600) of 15, whereas with the wild type, a much higher cell density of 50 was obtained. Since the salt medium CGXII contains 300 mM ammonium but BHI is not supplemented with ammonium, we studied growth of the dapD mutant as a function of the ammonium concentration (Fig. 4). In BHI plus 4% glucose, the addition of increasing ammonium concentrations resulted in increasing cell yields of the dapD mutant. At 100 mM ammonium sulfate, the cell yield was comparable to that of the wild type, indicating that the dehydrogenase variant can compensate for the loss of the succinylase activity only at high ammonium concentrations. A direct determination of ammonium in the culture of the dapD mutant (BHI without ammonium added) confirms that initially a low concentration of about 5 mM ammonium is present, which is reduced to about 0.45 mM when the dapD mutant ceases growth (Fig. 4). With the dapE mutant, a comparable growth dependence on the ammonium supply was detected (not shown). These data conclusively show that the succinylase variant is required for the use of organic nitrogen compounds of the complex medium. Accordingly, on a salt medium where free ammonium was replaced by 5 mM l-glutamate as a nitrogen source, the dapD and dapE mutants of C. glutamicum were unable to grow within 2 days, whereas growth of the wild type was completed after 15 h (not shown).
FIG. 4.
Growth of two strains of C. glutamicum. The wild-type strain (open symbols) and the dapD mutant of C. glutamicum (solid symbols) are shown. Cells were grown in rich BHI medium plus 4% glucose without (NH4)2SO4 (circles), with 20 mM (NH4)2SO4 (squares), and with 100 mM (NH4)2SO4 (triangles). Growth was measured by monitoring A600. In addition, the decrease in the concentration of free ammonium (×) for the dapD mutant without (NH4)2SO4 is shown.
Intracellular amino acid concentrations.
The restricted growth and its correlation with the ammonium concentration could be due to either a limiting l-lysine supply or a shortage of m-DAP (Fig. 1). Intracellular amino acid concentrations were determined in an attempt to quantify these amino acids. For this purpose cells, were grown on BHI plus glucose for 10 h when growth had already been arrested (Fig. 4). In all strains analyzed (dapE mutant, dapD mutant, and wild type), l-lysine was present at a high concentration of 69 ± 12 mM, whereas m-DAP was below the detection limit of 0.4 mM (intracellular). Therefore, l-lysine is available for the cell. It was apparently taken up via the well-known lysine import carrier of C. glutamicum. In the complex medium, l-lysine is present as a proteinogenic amino acid, whereas this is not the case for m-DAP. Therefore, m-DAP was added, but it did not result in increased growth of either of the mutants, which is not surprising due to the apparent inability of C. glutamicum to take up m-DAP (31). Interestingly, in the cytosol of the dapE mutant, a new amino acid was detected; this amino acid was not found in the wild type or the dapD mutant. Comparison with standards identified this derivative as l,l-N-succinyl-DAP, which reached a concentration as high as 115 mM after 5 h of growth on BHI plus glucose. This shows the high affinity of the succinylase toward its substrate tetrahydrodipicolinate (25), although no further flux through the succinylase variant is possible in the dapE mutant analyzed.
Cell wall integrity of the dapD mutant.
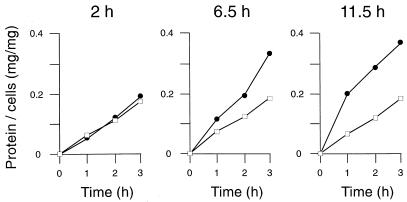
Since we noted higher protein yields with the dapD mutant than with the wild type during preparation of cell extracts for enzyme assays, we quantified the efficiency of cell disruption by sonication. For this purpose, cells of the dapD mutant grown on BHI were removed at three different times from the culture, and aliquots were disrupted for either 1, 2, or 3 min. As quantified by the protein released from the disrupted cells (Fig. 5), the efficiency of disruption of the mutant is identical with the control when the strains were grown for 2 h. At this time point, the growth rates of the strains are comparable (Fig. 4). However, an approximately twofold increase in the disruption efficiency of the dapD mutant is perceptible when the strains were grown for 6.5 h. This sensitivity of the dapD mutant to mechanical stress is increased even more when the ammonium concentration in the growth medium at the cultivation time was 0.45 mM. The dapE mutant behaved similarly (not shown).
FIG. 5.
Kinetics of cell disruption. Cell disruption was measured by the release of protein from cells of the wild type (□) and the dapD mutant (•) of C. glutamicum. Cells were harvested after 2, 6.5, and 11.5 h of growth in rich BHI medium plus glucose (4%). The cells were disrupted by sonication.
Morphology of the dapD and dapE mutants.
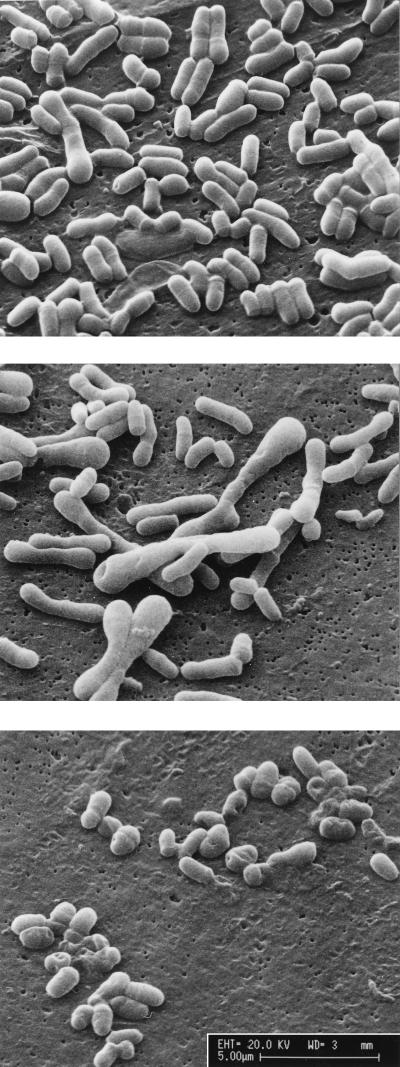
Cells grown on rich medium with excess carbon were prepared for raster electron microscopy. The wild type is rod shaped and about 1.4 to 2 μm long (Fig. 6). In contrast, cells of the dapD mutant are elongated (up to 6 μm long) and often club shaped at their ends. The dapE mutant is also altered in its morphology. In this case, the mutation results in a rather coccoidal cell form. This altered cell morphology is again indicative of a substantially altered peptidoglycan structure.
FIG. 6.
Electron micrographs of wild-type and mutant strains of C. glutamicum. Wild-type (top) and dapD (middle) and dapE (bottom) mutants grown under identical conditions are shown. Bar, 5 μm.
DISCUSSION
There are only a few cases where two synthesis pathways for a cellular building block exist in one organism. As can be concluded from individual enzyme measurements (1, 15), m-DAP can, however, be synthesized in some bacteria via two variant pathways simultaneously. A nuclear magnetic resonance analysis for C. glutamicum indicated that in vivo, both the succinylase and dehydrogenase variants of m-DAP synthesis contribute to the synthesis of m-DAP (27). In the dapD and dapE mutants with no succinylase pathway, growth is clearly affected, as well as the cell wall rigidity and the shape. These latter criteria are indicative of altered cross-linking of the peptidoglycan, which in C. glutamicum is exclusively via m-DAP (22). These consequences of the succinylase variant inactivation on the structure of C. glutamicum are comparable to that resulting from antibiotics applied to E. coli and interfering with peptidoglycan synthesis. The difference in the morphology of the two C. glutamicum mutants is not surprising, since many cytoplasmatic and membrane-located steps are involved in murein synthesis, including synthesis, hydrolysis, and turnover of murein and its precursors to enable controlled enlargement of the sacculus during growth (8). One reason for the different morphology of the dap mutants could be the extremely high accumulation of l,l-N-succinyl-DAP in the cytosol of the dapE mutant, which is, of course, not the case in the dapD mutant.
The fact that possession of the dehydrogenase variant alone has severe consequences on the growth of C. glutamicum shows that this variant is not suited for all growth conditions of the cell, such as very low ammonium concentrations. This is in agreement with the low affinity (Km of 34 mM) of the dehydrogenase toward ammonium (14). Thus, this variant is not suitable for providing the basic metabolite flux toward m-DAP formation. In fact, even at the very high ammonium concentration of 600 mM assayed, the flux via the dehydrogenase contributes only 51% to total m-DAP formation (27). In contrast to the situation with the dehydrogenase, possession of the succinylase alone has less-pronounced effects. In ddh mutants, we found decreased accumulation of final l-lysine (23) and occasionally noted a prolonged lag phase after inoculation, especially with poor carbon sources like acetate. Although there is still no experimental evidence, it is conceivable that energetic aspects determine the flux partition. Note that in several aspects, the two pathways of nitrogen incorporation into m-DAP resemble other systems where two pathways exist: the systems Trk (low affinity) and Kdp for potassium uptake or glutamate dehydrogenase (low affinity) and glutamine synthetase for ammonium assimilation. These systems share the properties that one enzyme has a low affinity but is constitutively formed, whereas the other has a high affinity and is energetically more costly. The same holds for m-DAP formation, where the dehydrogenase has the low affinity and the enzyme is constitutively formed in the two different organisms measured (6, 15), but the succinylase has the high affinity and is the one which is energetically more costly (25). Although the glutamate dehydrogenase in E. coli was originally considered to be a dispensable enzyme, it was demonstrated only recently that with high ammonium availability and limited energy, a mutant without glutamate dehydrogenase has a growth disadvantage (7). In view of this, the presence of the two pathways of m-DAP synthesis together represent another case where the flexibility of the cell increases in response to a limited supply of energy.
How is m-DAP made in bacteria other than C. glutamicum? The following situations are shown to exist: only one variant (the succinylase, acetylase, or dehydrogenase variant) is present or two variants (the succinylase and dehydrogenase variants or the acetylase and dehydrogenase variants) are present. The existence of two variants together is not restricted to C. glutamicum, but it is also the case in Bacillus macerans, for instance (1). In accordance with the present analysis, there is of course a special problem with those bacteria that make m-DAP exclusively via the dehydrogenase variant. This is the case, for example, in B. sphaericus or Bacillus pasteurii. In fact, it is noted that B. pasteurii required an elevated level of ammonium supply for growth (22) and that organic nitrogen sources could not easily replace ammonium (30).
In conclusion, the availability of m-DAP is critical to decide on the structure and finally the proliferation of the cell. Therefore, it is not surprising that there are several ways to provide this important molecule in bacteria. In particular, the possession of two pathways together ensures supply of m-DAP under a variety of conditions. This is the case in C. glutamicum and probably also in several other bacteria (1, 15). The dehydrogenase variant operates at a high concentration of free ammonium. Its exclusive presence restricts growth in environments with low concentrations of free ammonium. Instead, environments rich in organic nitrogen require the succinylase pathway to be active for optimal growth. Since the succinylase variant is energetically more expensive than the dehydrogenase variant, the use of the dehydrogenase variant could be more favorable in a situation where energy is limited. Thus, both pathways together enable the optimal adaption of the cell to different environments and the dynamic response to changes in the environmental conditions.
ACKNOWLEDGMENTS
We thank K. Krumbach for enzyme activity determinations, H. P. Bochem for electron microscopy, and M. Pátek (Prague) for RNA analyses.
The work was supported in part by the Federal Ministry of Education, Science and Technology (grant 031 062 6).
REFERENCES
- 1.Bartlett A T M, White P J. Species of Bacillus that make a vegetative peptidoglycan containing lysine lack diaminopimelate epimerase but have diaminopimelate dehydrogenase. J Gen Microbiol. 1985;131:2145–2152. [Google Scholar]
- 2.Beaman T W, Binder D A, Blanchard J S, Roderick S L. Three dimensional structure of tetrahydrodipicolinate N-succinyltransferase. Biochemistry. 1997;36:489–494. doi: 10.1021/bi962522q. [DOI] [PubMed] [Google Scholar]
- 3.Blickling S, Renner C, Laber B, Pohlenz H, Holak T, Huber R. Reaction mechanism of Escherichia coli dihydrodipicolinate synthase investigated by X-ray crystallography and NMR spectroscopy. Biochemistry. 1997;36:24–33. doi: 10.1021/bi962272d. [DOI] [PubMed] [Google Scholar]
- 4.Cremer J, Eggeling L, Sahm H. Cloning of the dapA dapB cluster of the lysine-secreting bacterium Corynebacterium glutamicum. Mol Gen Genet. 1990;220:478–480. [Google Scholar]
- 5.Cremer J, Eggeling L, Sahm H. Control of the lysine biosynthesis sequence in Corynebacterium glutamicum as analyzed by overexpression of the individual corresponding genes. Appl Environ Microbiol. 1991;57:1746–1752. doi: 10.1128/aem.57.6.1746-1752.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cremer J, Treptow C, Eggeling L, Sahm H. Regulation of enzymes of lysine biosynthesis in Corynebacterium glutamicum. J Gen Microbiol. 1988;134:3221–3229. doi: 10.1099/00221287-134-12-3221. [DOI] [PubMed] [Google Scholar]
- 7.Helling R B. Why does Escherichia coli have two primary pathways for synthesis of glutamate? J Bacteriol. 1994;176:4664–4668. doi: 10.1128/jb.176.15.4664-4668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Höltje J. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1988;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam L K P, Arnold L D, Kalantar T H, Kelland J G, Lane-Bell P M, Palcicand M M, Pickard M A, Vederas J C. Analogs of diaminopimelic acid as inhibitors of meso-diaminopimelate dehydrogenase and LL-diaminopimelate epimerase. J Biol Chem. 1988;263:11814–11819. [PubMed] [Google Scholar]
- 10.Leuchtenberger W. Amino acids—technical production and use. Products of primary metabolism. Biotechnology. 1966;6:455–502. [Google Scholar]
- 11.Liebl W, Bayerl A, Stillner U, Schleifer K H. High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol Lett. 1989;65:299–304. doi: 10.1016/0378-1097(89)90234-6. [DOI] [PubMed] [Google Scholar]
- 12.Marx A, Striegel K, de Graaf A A, Sahm H, Eggeling L. Response of the central metabolism of Corynebacterium glutamicum to different flux burdens. Biotechnol Bioeng. 1997;56:168–180. doi: 10.1002/(SICI)1097-0290(19971020)56:2<168::AID-BIT6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 13.Menkel E, Thierbach G, Eggeling L, Sahm H. Influence of increased aspartate availability on lysine formation by a recombinant strain of Corynebacterium glutamicum and utilization of fumarate. Appl Environ Microbiol. 1989;55:684–688. doi: 10.1128/aem.55.3.684-688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Misono H, Soda K. Properties of meso-diaminopimelate D-dehydrogenase from Bacillus sphaericus. J Biol Chem. 1980;255:10599–10605. [PubMed] [Google Scholar]
- 15.Misono H, Togawa H, Yamamoto T, Soda K. meso-Diaminopimelate d-dehydrogenase: distribution and the reaction product. J Bacteriol. 1979;137:22–27. doi: 10.1128/jb.137.1.22-27.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morbach S, Kelle R, Winkels S, Sahm H, Eggeling L. Engineering the homoserine dehydrogenase and threonine dehydratase control points to analyse flux towards L-isoleucine in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 1996;45:612–620. [Google Scholar]
- 17.Richaud F, Richaud C, Haziza C, Patte J-C. Isolement et purification de genes d’Escherichia coli K12 impliques dans la biosynthese de la lysine. C R Acad Sci Ser III. 1981;293:507–512. [PubMed] [Google Scholar]
- 18.Richaud F, Richaud C, Martin C, Haziza C, Patte J-C. Regulation of expression and nucleotide sequence of the Escherichia coli dapD gene. J Biol Chem. 1984;259:14824–14828. [PubMed] [Google Scholar]
- 19.Scapin G, Reddy S G, Blanchard J S. Three-dimensional structure of meso-diaminopimelic acid dehydrogenase from Corynebacterium glutamicum. Biochemistry. 1996;35:13540–13551. doi: 10.1021/bi961628i. [DOI] [PubMed] [Google Scholar]
- 20.Schäfer A, Kalinowski J, Simon R, Seep-Feldhaus A, Pühler A. High-frequency conjugal plasmid transfer from gram-negative Escherichia coli to various gram-positive coryneform bacteria. J Bacteriol. 1990;172:1663–1666. doi: 10.1128/jb.172.3.1663-1666.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 22.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schrumpf B, Schwarzer A, Kalinowski J, Pühler A, Eggeling L, Sahm H. A functionally split pathway for lysine synthesis in Corynebacterium glutamicum. J Bacteriol. 1991;173:4510–4516. doi: 10.1128/jb.173.14.4510-4516.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schrumpf B, Eggeling L, Sahm H. Isolation and prominent characteristics of an L-lysine hyperproducing strain of Corynebacterium glutamicum. Appl Microbiol Biotechnol. 1992;37:566–571. [Google Scholar]
- 25.Simms S A, Voiges W H, Gilvarg C. Purification and characterization of succinyl-CoA: tetrahydrodipicolinate N-succinyltransferase from Escherichia coli. J Biol Chem. 1984;259:2734–2741. [PubMed] [Google Scholar]
- 26.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 27.Sonntag K, Eggeling L, De Graaf A A, Sahm H. Flux partitioning in the split pathway of lysine synthesis in Corynebacterium glutamicum. Quantification by 13C- and 1H-NMR spectroscopy. Eur J Biochem. 1993;213:1325–1331. doi: 10.1111/j.1432-1033.1993.tb17884.x. [DOI] [PubMed] [Google Scholar]
- 28.Wehrmann A, Eggeling L, Sahm H. Analysis of different DNA fragments of Corynebacterium glutamicum complementing dapE of Escherichia coli. Microbiology (Reading) 1994;140:3349–3356. doi: 10.1099/13500872-140-12-3349. [DOI] [PubMed] [Google Scholar]
- 29.Wehrmann A, Morakkabati S, Krämer R, Sahm H, Eggeling L. Functional analysis of sequences adjacent to dapE of Corynebacterium glutamicum reveals the presence of aroP, which encodes the aromatic amino acid transporter. J Bacteriol. 1995;177:5991–5993. doi: 10.1128/jb.177.20.5991-5993.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White P J, Lotay H K. Minimal nutritional requirements of Bacillus sphaericus NCTC 9602 and 26 other strains of this species: the majority grow and sporulate with acetate as sole major source of carbon. J Gen Microbiol. 1980;118:13–19. [Google Scholar]
- 31.Yeh P, Sicard A, Sinskey A. General organization of the genes specifically involved in the diaminopimelate-lysine biosynthetic pathway of Corynebacterium glutamicum. Mol Gen Genet. 1988;212:105–111. doi: 10.1007/BF00322451. [DOI] [PubMed] [Google Scholar]