Abstract
A cross-ribosome binding site (cRBS) adjusts the dynamic range of transcription factor-based biosensors (TFBs) by controlling protein expression and folding. The rational design of a cRBS with desired TFB dynamic range remains an important issue in TFB forward and reverse engineering. Here, we report a novel artificial intelligence (AI)-based forward-reverse engineering platform for TFB dynamic range prediction and de novo cRBS design with selected TFB dynamic ranges. The platform demonstrated superior in processing unbalanced minority-class datasets and was guided by sequence characteristics from trained cRBSs. The platform identified correlations between cRBSs and dynamic ranges to mimic bidirectional design between these factors based on Wasserstein generative adversarial network (GAN) with a gradient penalty (GP) (WGAN-GP) and balancing GAN with GP (BAGAN-GP). For forward and reverse engineering, the predictive accuracy was up to 98% and 82%, respectively. Collectively, we generated an AI-based method for the rational design of TFBs with desired dynamic ranges.
Keywords: Generative adversarial network, Predictive model, Cross-ribosome binding site, Glucarate biosensor, Dynamic range
Graphical Abstract
Highlights
-
•
De novo design of cRBS based on WGAN-GP model.
-
•
Building forward engineering platform to achieve precise prediction of TFB dynamic range.
-
•
Establishing a reverse engineering platform based on BAGAN-GP to design cRBS with desired dynamic range.
1. Introduction
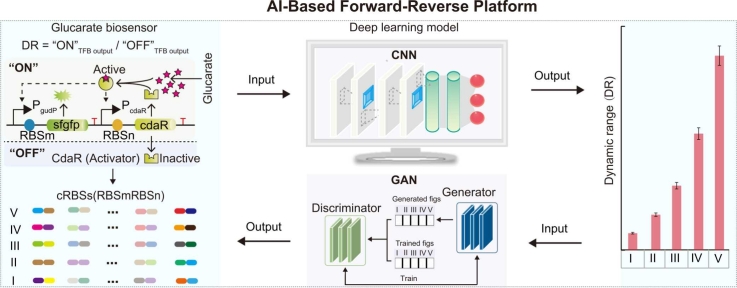
Transcription factor-based biosensors (TFBs) [1] have broad applications in metabolite detection [2], [3], high-throughput screening [4], [5], and dynamic metabolic control [6]. TFBs sense metabolite concentration signals and convert them to detectable signals, such as fluorescence and growth trends. One of the most important parameters used to characterize TFBs is dynamic range, which is defined as gene expression fold change with respect to the presence or absence of an inducer [7], [8]. An appropriate dynamic range is critical for well-designed TFBs. A previous study reported that a ribosome binding site (RBS) regulated TFB dynamic range by adjusting gene expression and protein folding [7]. The TFB included two RBSs which controlled the translation of a transcription factor and reporter. The combination of both RBSs was defined as a cross-RBS (cRBS). Thus, cRBS sequences were essential for the rational design of TFBs. Two key challenges which must be considered in rationally designing TFBs are the construction of forward and reverse engineering platforms; the former predicts TFB dynamic ranges with a given cRBS while the latter generates cRBS sequences with desired TFB dynamic ranges.
Using cRBSs with random sequences to construct TFBs often generates un-functional or low dynamic range TFBs. Hence, a large and functional cRBS library is required for forward engineering to exclude invalid constructs. Therefore, a random mutation is usually coupled with high-throughput screening to obtain cRBSs that generate functional TFBs. For example, Siedler et al. constructed and screened functional cRBS variants from a random mutation library using a droplet microfluidics technique to optimize the dynamic range of the PadR-based biosensor, and finally achieved a 130-fold increase [9]. However, screened cRBSs constitute only a relatively small library when compared with all possible functional cRBSs. Furthermore, screening functional cRBSs is both laborious and time-consuming. Therefore, a computational approach must be established to efficiently design novel functional cRBSs. Recent advances in deep learning approaches generated alternative methods for de novo DNA sequence design [10], [11]. In particular, generative adversarial networks (GANs, a generative model that learns through the game between generator and discriminator neural networks) [12], [13], [14] generating biological sequences have yielded impressive results [15]. For example, Wang et al. constructed a Wasserstein GAN (WGAN, the improved GAN model that Wassertein measures the distance between generated and real data distributions, which solves the problems of unstable training and poor diversity of generated samples) system with a gradient penalty (WGAN-GP, the improved WGAN model that proposes an interpolation method for GP to make the model meet Lipschitz constraints, which solves the problems of slow training speed and low quality of generated samples) to generate novel promoters and predict their activity. The authors showed that 70.8% of artificial intelligence (AI)-designed promoters demonstrated good activity [15]. A detailed introduction to the GAN model and its development is shown (Supplementary Note S1).
TFB forward engineering helps predict the dynamic range of given functional cRBSs. However, biosensor engineering often requires reverse engineering to design cRBSs from given dynamic ranges [16], [17]. With respect to deep learning advances, a reverse engineering strategy based on deep learning is considered an alternative TFB design approach. Conditional-GAN networks [18], [19], which are constrained distribution-based [16] generative models, are promising strategies which capture constrained sequence spaces and generate cRBSs with desired TFB dynamic ranges. In conditional-GAN development, Mariani et al. proposed the balancing GAN (BAGAN, an enhancement tool that restores the balance of unbalanced datasets and improves the accuracy of classification) network, which generates state-of-the-art minority-class images on highly unbalanced datasets [20]. However, BAGAN training is unstable when images in different classes are similar. Thus, Huang et al. optimized the loss function in the BAGAN network; the authors proposed a BAGAN network with a gradient penalty (BAGAN-GP), which overcame this issue and achieved good results in generating small sample cell images [21]. Moreover, we added a sequence autoencoder [22], [23] (Supplementary Fig. S1) to BAGAN-GP model architecture (Supplementary Fig. S2) to extract cRBS sequence information by learning the cRBS potential structure and similarity, and finally generate novel cRBS sequences through decoder. Because the BAGAN-GP model processes highly unbalanced cRBS datasets [7], this framework can be used to perform reverse engineering for designing cRBSs based on TFB dynamic ranges.
To simultaneously achieve forward and reverse TFB rational engineering, we established and trained a comprehensive platform based on our previously collected 7053 cRBSs. These cRBSs were designed and synthesized by a DNA microarray approach to tune TFB dynamic ranges [7]. Using fluorescence-activated cell sorting (FACS) and next-generation sequencing (NGS) analysis TFBs, we obtained a multi-to-one mapping dataset that included 3592, 980, 944, 596, and 941 cRBSs in sub-libraries I–V, respectively [7]. These libraries had gradient TFB dynamic ranges. In this study, using the collected datasets, we first established a forward engineering platform for de novo cRBS sequence design based on the WGAN-GP model. Subsequently, we engineered a reverse engineering platform for novel cRBS sequence generation with desired TFB dynamic ranges based on the BAGAN-GP model. Furthermore, high prediction accuracy facilitated cRBS generation with selected high TFB dynamic ranges. We then developed a filter model using the conventional neural network (CNN, a neural network designed to process data with a similar grid structure, includes convolution, pooling and fully connected layers) to reduce prediction noise in the BAGAN-GP model in cRBS generation using selected low TFB dynamic ranges. Overall, our forward-reverse engineering platform can be effectively used to design novel functional cRBSs with desired TFB dynamic ranges.
2. Results
2.1. De novo design of cRBSs using the WGAN-GP model
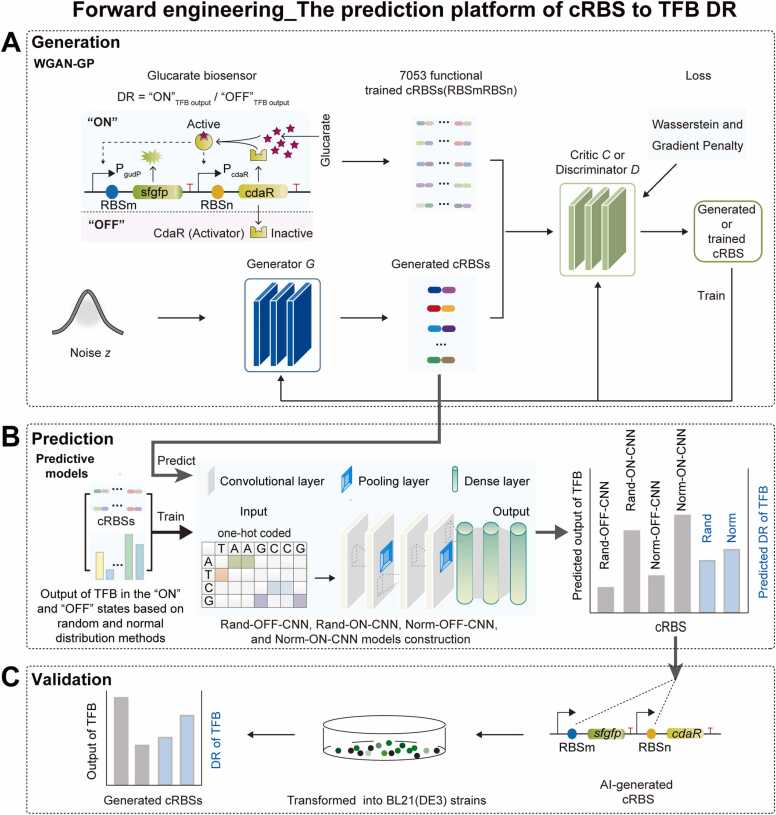
Several studies have reported forward engineering [7], [15]. In our previous glucarate biosensor study, the technical process from designing cRBSs to obtaining TFB dynamic ranges exemplified a forward engineering approach [7]. However, rapidly and precisely designing functional cRBSs to tune TFB dynamic ranges is costly for forward engineering. Thus, we introduced a forward engineering workflow to create functional cRBSs to avoid invalid constructs (Fig. 1). This included a WGAN-GP model [15], [24] for de novo functional cRBS generation (Fig. 1A) and four predictive models to predict outputs and TFB dynamic ranges controlled by WGAN-GP-generated cRBSs (Fig. 1B). Then, generated cRBSs were experimentally tested using a TFB in Escherichia coli (Fig. 1C).
Fig. 1.
Forward engineering workflow. (A) The cRBS generation stage. Inactive CdaR (“OFF” state) is activated by glucarate and simultaneously increases its own and PgudP-controlled gene (“ON” state) expression [7]. Dashed arrows represent feedback activation. First, the discriminator was trained five times using 7053 functionally trained cRBSs to generate cRBSs datasets. Then, the generator was trained once using noise z consisting of standard normal distribution datasets to capture cRBS data distribution and generate cRBSs to fool the discriminator. Subsequently, the discriminator tried to distinguish generated cRBSs from trained ones. Finally, numerous functional cRBSs were generated by a zero-sum game (Supplementary Note S1) between the discriminator and generator networks in the WGAN-GP model. (B) Predicting TFB output and dynamic ranges with WGAN-GP-generated cRBSs. First, 7053 cRBSs and TFB output datasets were built based on random and normal distribution approaches. Then, the CNN model was continuously trained to obtain four predictive models: Rand-OFF-CNN, Rand-ON-CNN, Norm-OFF-CNN, and Norm-ON-CNN. Finally, the output and dynamic range of a TFB with WGAN-GP-generated cRBSs was predicted based on the four predictive models. (C) Validation of model prediction performance. Candidate cRBSs with differential TFB outputs in “ON” and “OFF” states were selected and experimentally validated in Escherichia coli BL21 (DE3). DR: dynamic range; PcdaR: a constitutive promoter controlling cdaR; PgudP: an inducible promoter controlling sfgfp.
The GAN comprised two competing neural networks, the generator and the discriminator [13], [25]. The generator is equivalent to mapping from one functional distribution to another. Thus, the generator network generated the feature distribution of functional cRBS sequences and mapped these to generated cRBS sequences (Fig. 1A). The discriminator network evaluated differences between generated cRBSs and functionally trained cRBSs, and determined if cRBSs were generated or trained (Fig. 1A). Based on continuous game training between the generator and the discriminator (Supplementary Note S1), the GAN model generated novel cRBS sequences according to the feature distribution of functional cRBSs. However, the GAN model was often difficult to train and prone to instability, and the pattern contraction led to poor sample diversity [26]. When the GAN model identified a small number of generated cRBSs that fooled the discriminator, the generator was unable to generate other novel cRBSs. WGAN improved GAN training stability and diversity [27], [28], which was enhanced by using the Wasserstein loss function. However, the ability of WGAN to learn complex features remained insufficient [26]. Thus, WGAN-GP facilitated a more stable training process via the addition of a gradient penalty to the critic’s loss function [15], [29] (Fig. 1A), thereby capturing more cRBS sequence features.
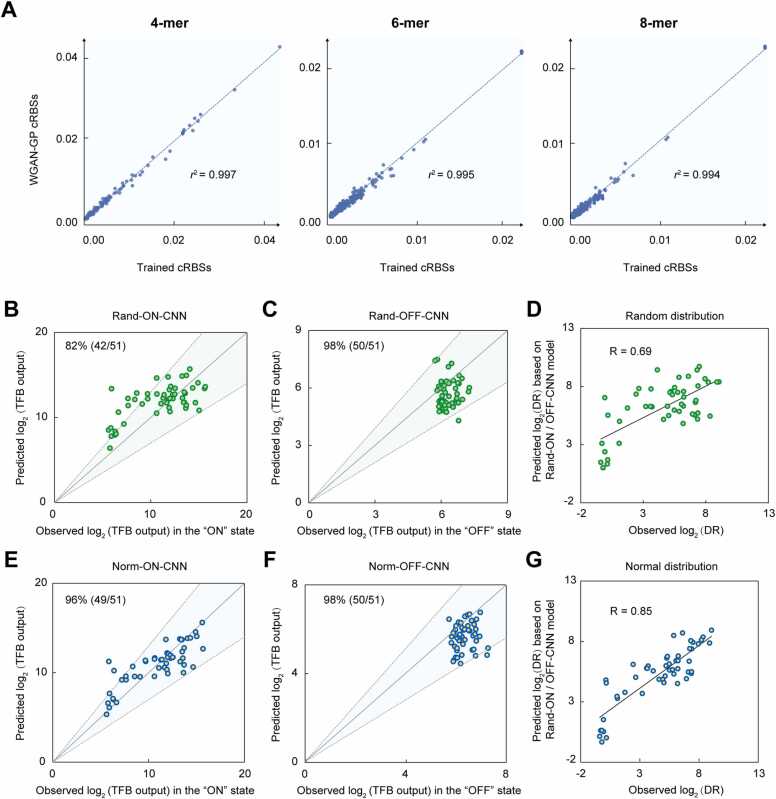
We used the WGAN-GP generative model to identify novel cRBS sequences, analyzed them using WebLogo, and observed that generated cRBS sequence distribution was the same as trained functional cRBS sequence distribution (Supplementary Fig. S3). In addition, to evaluate the similarity between the generated cRBS and the trained cRBS sequences, we used the hamming distance, which is the number of different nucleotides at the corresponding position of two equal-length cRBS sequences, to measure the distance matrix between sequences for the trained cRBS and the generated cRBS datasets, and within each cRBS (Supplementary Fig. S4A–C). Boxplots showed the distribution of the cRBS sequences distance (Supplementary Fig. S4D). Then, we performed the distance matrix summary statistics and found that the mean/standard deviation of the distance between sequences for trained cRBS and generated cRBS, trained cRBS internally, and generated cRBSs internally were 6.57/1.56, 6.33/1.55, and 6.77/1.60, respectively (Supplementary Fig. S4D). It indicated that the sequence distribution of generated and trained cRBSs was similar. And the model independently generated cRBS sequences based on learning functional cRBS sequence characteristics. To further evaluate WGAN-GP model ability toward learning cRBS sequence characteristics, we calculated the base frequencies of k-mer (4-, 6-, and 8-mer) numbers [15], [30], [31] between WGAN-GP model-generated cRBSs and trained cRBSs (Fig. 2A). We observed that the correlation of all k-mer frequencies between trained cRBSs and WGAN-GP-generated cRBSs was> 0.99, suggesting the model was highly reliable in generating cRBSs and learning the sequence characteristics of trained cRBSs.
Fig. 2.
K-mer distribution and prediction accuracy analysis of WGAN-GP-generated cRBSs. (A) Scatter plots showing 4-, 6-, and 8-mer numbers between WGAN-GP model-generated cRBSs and trained cRBSs. Each point represents a certain k-mer. The x- and y-axes represent k-mer frequencies. Verifying the output for 51 synthetic TFBs using Rand-ON-CNN (B), Rand-OFF-CNN (C), Norm-ON-CNN (E), and Norm-OFF-CNN (F) predictive models. Correlation analyses between experimentally-observed and predicted TFB dynamic ranges based on random (D) and normal (G) distribution methods. The coefficient R was used to evaluate linear correlations between predicted and experimentally observed TFB dynamic ranges. DR: dynamic range. Rand and Norm represent random and normal methods, respectively. The grey diagonal denotes y = x. Light green and blue areas represent predicted TFB outputs in “ON” and “OFF” states with a± 30% error margin. Data represent the mean and standard deviation of two replicates.
2.2. Predicting TFB outputs in “ON” and “OFF” states
We previously obtained 3592, 980, 944, 596, and 941 cRBSs and corresponding gradient TFB output ranges datasets in sub-libraries I–V, respectively [7]. Using this multi-to-one mapping dataset to accurately predict TFB outputs from generated cRBSs, we sought to construct training datasets for each cRBS mapped to a sole TFB output (one-to-one mapping dataset), rather than TFB output ranges. However, this approach was time-consuming, labor-intensive, and costly to formulate a one-to-one mapping dataset via experimentation. To overcome this, we proposed random (Supplementary Figs. S5A and S5B) and normal (Supplementary Figs. S5C and S5D) distribution methods to construct 7053 cRBSs and related TFB output datasets in “OFF” and “ON” states [7] (Fig. 1B, Supplementary Tables S1 and S2). Then, 6000 datasets were randomly selected to train the CNN model (Method 4.5). Finally, four predictive models were constructed: Rand-OFF-CNN, Rand-ON-CNN, Norm-OFF-CNN, and Norm-ON-CNN (Fig. 1B). To evaluate model performance, we analyzed correlations between predicted and observed TFB outputs in the remaining 1053 cRBSs in “ON” and “OFF” states. All four prediction models had Pearson’s correlation coefficients (PCC, a linear correlation coefficient that reflect the linear correlation degree of the predicted and observed TFB output variables) of 0.57, 0.52, 0.62, and 0.52 (Supplementary Figs. S6A–D), respectively. Thus, the four models had moderate prediction performance.
To verify the prediction accuracy of these models in predicting TFB outputs in “ON” and “OFF” states, 51 cRBSs (GSnCSn; GS, RBS of the reporter protein sfGFP; CS, RBS of the regulator CdaR; n = 1–51) (Supplementary Table S3) generated by WGAN-GP were randomly selected to replace the RBS of CdaR and sfGFP. The 51 cRBS sequences were used as inputs, and predicted TFB outputs worked as model outputs (Fig. 1B–C). When compared with experimentally-observed TFB outputs (Supplementary Table S4), the prediction accuracy of TFB outputs from Rand-OFF-CNN, Rand-ON-CNN, Norm-OFF-CNN, and Norm-ON-CNN models reached 82% (42/51) (Fig. 2B), 98% (50/51) (Fig. 2C), 96% (49/51) (Fig. 2E), and 98% (50/51) (Fig. 2F), respectively. This was significantly higher than our previous report (72.2%) [7]. In addition, Pearson’s correlation analysis was performed between predicted and experimentally-observed TFB dynamic ranges (“ON”TFB output/“OFF”TFB output) (Fig. 1A, Supplementary Fig. S7). The predicted dynamic range based on random (Fig. 2D) and normal (Fig. 2G) distributions displayed a strong correlation (PCC = 0.69 and PCC = 0.85, respectively) with experimental dynamic ranges. The higher prediction accuracy and Pearson’s correlation of normal distribution predictive models suggested better TFB outputs and dynamic range prediction performance when compared with random distribution predictive models. Our previous classification model classified the dynamic range levels of multiple cRBS inputs, often resulting in several different cRBSs belonging to the same dynamic range level. In contrast, the models established in this study achieved accurate and sole TFB outputs and dynamic range predictions with sole cRBS inputs. Overall, we efficiently designed functional cRBSs using the WGAN-GP model and accurately predicted outputs and TFB dynamic ranges.
2.3. Reverse engineering TFBs based on the BAGAN-GP model
A forward engineering system that de novo generates functional cRBSs and predicts TFB outputs and dynamic ranges can accurately evaluate genotype to phenotype [15]. However, reverse engineering from phenotype to genotype [16], [17] inverts this paradigm by starting with desired TFB dynamic ranges and seeking ideal cRBSs. Reverse engineering is also an important focus of this study to design cRBSs at an accelerated pace. Therefore, to develop a reverse engineering platform to de novo generate cRBSs with desired TFB dynamic ranges, we established a BAGAN-GP model which showed superior performance and considerable potential for processing unbalanced minority-class datasets [21].
To extract and transform cRBS sequence information, we introduced a sequence autoencoder to encode and decode cRBS sequences (Supplementary Fig. S1). cRBS sequence information was transcoded into a 64 × 64 × 1 figure using sequence encoder, and figure information was decoded into cRBS sequences using sequence decoder. To generate cRBSs with expected dynamic ranges, we introduced an AI-based reverse engineering workflow (Fig. 3) which included BAGAN-GP (Fig. 3A) and filter models (Fig. 3B) for de novo generated cRBSs with desired TFB dynamic ranges. Then, generated cRBSs were experimentally tested using a TFB in E. coli (Fig. 3C).
Fig. 3.
Reverse engineering workflow. (A) The cRBS generation stage with desired TFB dynamic ranges. First, cRBS sequences from sub-libraries I–V [7] were encoded into 64 × 64 × 1 figures based on the sequence encoder (SEn) model (Supplementary Fig. S1) (Step 1). Then, the autoencoder model was pretrained to learn the global features of Step 1-created datasets. The well-trained autoencoder model captured the global features of the sub-libraries I–V and initialized BAGAN-GP model weights (Supplementary Fig. S2). The initialized BAGAN-GP model converged faster to the desired result (Step 2). Subsequently, the initialized BAGAN-GP model was trained based on the training of the discriminator and generator. In the trained process of each BAGAN-GP step, the discriminator was trained five times using figures and corresponding TFB dynamic range label datasets. The generator was trained once to capture the figure information of the sub-libraries I–V. Novel figures were generated by the generator to fool the discriminator, and the discriminator produced validity scores to determine if the figures were generated by the generator and sub-library-matching (Step 3). Finally, the generator generated novel figures with desired sub-library features. The generated figures transformed back into cRBS sequences with desired TFB dynamic ranges based on sequence decoder (SDe) (Supplementary Fig. S1) (Step 4). (B) The filtration stage which reduces prediction noise in the BAGAN-GP model. cRBSs and five TFB dynamic range datasets [7] were used to build and evaluate a filter model consisting of CNN, which was used to learn relationships between cRBS sequence features and TFB dynamic ranges. Finally, a filter model was constructed to reduce noise and accurately predict TFB dynamic ranges. (C) Validation of model prediction performances. Candidate cRBSs with expected TFB dynamic ranges were selected and experimentally validated in Escherichia coli BL21 (DE3). DR: dynamic range; CNN: convolutional neural network.
2.4. The BAGAN-GP model captures cRBS sequence features in five sub-libraries
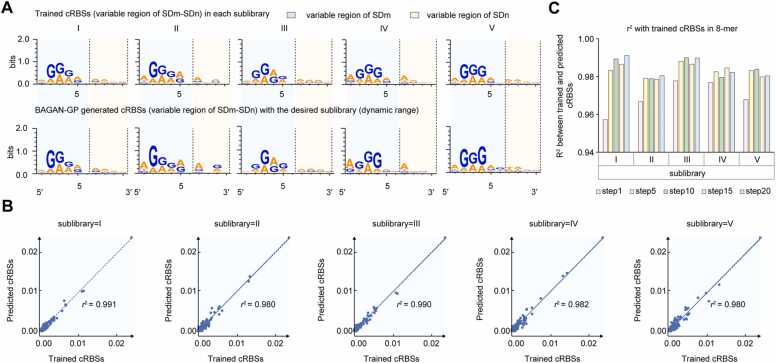
To comprehensively evaluate de novo generated cRBSs based on the BAGAN-GP model in five dynamic range classes, we first analyzed generated cRBS sequence motifs (Fig. 4A). Generated cRBS sequences from the five sub-libraries exhibited obvious sequence differences (Fig. 4A) and indicated that the BAGAN-GP model captured cRBS sequence features from these sub-libraries.
Fig. 4.
The SD motif and 8-mer distribution of BAGAN-GP-generated cRBSs. (A) The sequence logos of trained cRBSs and BAGAN-GP-generated cRBS sequences in five sub-libraries. (B) The 8-mer scatter plot showing trained cRBSs and generated cRBSs with desired TFB dynamic ranges. Each point represents a certain 8-mer. The x- and y-axes represent 8-mer frequencies in trained and generated cRBSs with selected TFB dynamic ranges. (C) R-squared analyses the correlations of 8-mer frequencies in step n (n = 1, 5, 10, 15, 20) between trained cRBSs and BAGAN-GP-generated cRBSs from sub-libraries I–V. SDm and SDn represent SD sequences controlling sfgfp and cdaR translation.
Moreover, to further evaluate how the BAGAN-GP model could learn cRBS sequence characteristics of each sub-library, we calculated the base frequency of k-mer (4-, 6-, and 8-mer) numbers [15] between BAGAN-GP-generated cRBSs and trained cRBSs in each sub-library (Fig. 4B, Supplementary Figure S8). We observed that the correlation of all k-mer frequencies between trained cRBSs and BAGAN-GP-generated cRBSs in each sub-library was> 0.98, suggesting the model was highly reliable in designing cRBSs, and learning the sequence characteristics of each sub-library. Because the Shine-Dalgarno (SD) sequence contained eight bases in our study, we then analyzed the correlation of 8-mer frequencies of each sub-library between trained cRBSs and generated cRBSs of the training steps 1, 5, 10, 15, and 20 (300 epochs/step) in the BAGAN-GP model training process (Supplementary Figure S9). We observed that the BAGAN-GP model capture of cRBS sequence characteristics of each sub-library remained the same after step 5 (Fig. 4C), and created a good performance BAGAN-GP model to generate cRBSs with desired TFB dynamic ranges. Taken together, the BAGAN-GP model captured cRBS sequence features in highly unbalanced minority-class datasets containing 3592, 980, 944, 596, and 941 cRBSs in each sub-library.
2.5. BAGAN-GP-generated-cRBSs show a high probability of improving the dynamic range
After generating thousands of cRBS sequences based on the BAGAN-GP model, we randomly selected 140 AI-based cRBSs (GSmCSm; m=52–191) (Supplementary Table S3) from sub-libraries I–V. Then, 140 glucarate biosensors were successfully constructed where outputs were detected in “ON” and “OFF” states using FACS (Supplementary Table S5), and dynamic ranges were calculated (Supplementary Figure S10). Then, to evaluate the prediction performance of the BAGAN-GP model, we analyzed desired and experimentally observed TFB dynamic ranges of generated cRBSs and found that the BAGAN-GP model had prediction accuracies of 43% (Fig. 5A), 32% (Fig. 5B), 68% (Fig. 5C), 57% (Fig. 5D), and 82% (Fig. 5E) in the five sub-libraries, respectively. Moreover, desired and experimentally observed TFB dynamic ranges of generated cRBSs had a strong correlation (R = 0.94) (Fig. 5F). These results demonstrated BAGAN-GP model effectiveness in generating cRBSs with desired TFB dynamic ranges.
Fig. 5.
Evaluating 140 AI-generated cRBSs with desired TFB dynamic ranges and improving prediction accuracy. (A–E) Prediction accuracy analyses of the BAGAN-GP model for 28 synthetic glucarate biosensors in sub-libraries I–V, respectively. We suggested that the synthetic TFB was constructed by generated cRBSs in sub-library V, whose dynamic range was more extensive than trained ones (247-fold [7]). (F) Correlation analysis between desired and experimentally-observed dynamic ranges of TFBs controlled by BAGAN-GP-generated cRBSs in each sub-library. (G) The improved prediction accuracy of TFB dynamic ranges in sub-libraries I and II using the constructed filter model. DR: dynamic range. GS and CS are RBSs of the reporter sfGFP and regulator CdaR, respectively. The dotted box represents cRBSs whose desired and observed TFB dynamic ranges had a± 30% error margin. Data represent the mean and standard deviation of two replicates.
2.6. Establishing a filter model to reduce prediction noise in the BAGAN-GP model
The BAGAN-GP model de novo generated cRBSs with desired TFB dynamic ranges by learning sequence feature distributions of trained cRBSs in five sub-libraries. Therefore, the prediction accuracy of sub-libraries I and II was relatively low due to the high similarity of sequence distribution between both sub-libraries. This made it difficult for the BAGAN-GP model to determine which TFB dynamic ranges generated cRBSs belonged to. In other words, prediction noise existed in the BAGAN-GP model. Therefore, we introduced a filter model to reduce BAGAN-GP prediction noise in sub-libraries I and II.
Here, we chose the CNN model to construct a filter that accurately identified the TFB dynamic ranges to which generated cRBS belonged. Firstly, 80% of cRBSs and their sequence features in each sub-library were randomly selected to train the filter model. Then, the remaining 20% of cRBS sequences and corresponding features were used to evaluate the performance of the filter model for predicting TFB dynamic ranges. In the five sub-libraries, the area under the curve (AUC) was 0.81, 0.93, 0.71, 0.81, and 0.80, respectively, and the average AUC was 0.87 (Supplementary Figure S11). Thus, the filter displayed good performance in predicting TFB dynamic ranges.
To reduce the prediction noise of BAGAN-GP-generated cRBSs in sub-libraries I and II, we filtered out cRBS sequences that were not generated in sub-libraries I and II based on the constructed filter model. Finally, the prediction accuracy increased to 65% and 54% in sub-libraries I and II, respectively (Fig. 5G). These results suggested that the BAGAN-GP-filter reverse engineering platform generated cRBSs with desired TFB dynamic ranges in E. coli.
3. Discussion
Currently, to rationally design TFBs, two enormous challenges must be overcome: predicting TFB dynamic ranges with given cRBSs for forward engineering, and generating cRBS sequences with selected TFB dynamic ranges for reverse engineering [1]. To achieve TFB rational design, we developed a forward-reverse engineering platform based on WGAN-GP and BAGAN-GP models to de novo design cRBSs and generate cRBSs with desired TFB dynamic ranges.
To intelligently predict a phenotype based on a given genotype and generate biological components based on an expected phenotype, a forward-reverse engineering research approach is vital. In previous study, we obtained cRBS sequences and TFB output datasets from five sub-libraries based on the DNA microarrays, FACS, and NGS techniques [7]. However, for forward engineering, to predict the output and dynamic range of TFBs that were controlled by WGAN-GP-generated cRBSs, many one-to-one mapping datasets had to be constructed to train the predictive model, which would have been time-consuming, labor-intensive, and costly. To address this, we proposed obtaining these mapping datasets by random and normal distributions. Our predictive models, developed based on random and normal distribution methods, showed good performance in predicting TFB outputs and dynamic ranges. This approach could help researchers obtain one-to-one mapping experimental datasets and avoid time-consuming, laborious, and costly research caused by repeated bench work. It also provides new perspectives for the acquisition of one-to-one datasets.
From the perspective of supervised pattern recognition [32], [33], [34], the translation mechanisms of cRBSs tuning TFB dynamic ranges could be considered as molecular classifiers, distinguishing generated cRBS sequences from trained cRBSs in each library. Therefore, generated cRBSs must have similar characteristics to trained cRBSs in each library to recruit translation mechanisms to tune TFB dynamic ranges. However, unbalanced data classification is a ubiquitous natural phenomenon [19]. For example, a significant imbalance in cRBS datasets containing 3592, 980, 944, 596, and 941 cRBSs was identified in sub-libraries I–V, respectively, from our previous study [7], among which, classes II–V belonged to minority-class cRBSs. Thus, finding a suitable supervised generative model to navigate unbalanced biological datasets is of great importance. Fortunately, BAGAN-GP is superior to other state-of-the-art GANs in generating minority-class images for unbalanced datasets [21], [35]. Moreover, BAGAN-GP automatically extracts crucial transcoded cRBS figure features from each library, mimics cRBS distribution, and generates high-quality cRBS sequences. Thus, the BAGAN-GP model could be used to augment unbalanced sequence classification datasets and restore its balance.
The BAGAN-GP model accomplishes generative tasks by learning cRBS sequence distributions in each sub-library. Therefore, when this distribution between sub-libraries were similar, it was difficult for the BAGAN-GP model to predict which sub-library generated cRBSs belonged to. Thus, to reduce prediction noise, a filter was added to improve the prediction accuracy of the BAGAN-GP model. This filter model improved the design accuracy of generated cRBSs with desired TFB dynamic ranges, laid a solid foundation for using reverse engineering systems in TFBs, and provided a window for the future design of biological elements with desired phenotypes.
In conclusion, we proposed an AI-based forward-reverse engineering platform to de novo generate cRBSs with expected TFB dynamic ranges. The platform showed a powerful functionality based on experimental verification in vivo. Our work provided new perspectives for the de novo generation of biological elements with desired biological properties. Thus, deep learning approaches have the potential to explore relationships between genotype and phenotype and the sequence features of generated elements even with limited prior knowledge.
4. Materials and methods
4.1. Strains and culture conditions
All study strains are listed in Supplementary Table S6. Escherichia coli (E. coli) JM109 and E. coli BL21 (DE3) were used for plasmid cloning and protein expression, respectively. Luria-Bertani (LB) medium supplemented with 100 µg/mL ampicillin was used for cell culture at 37 °C or 30 °C. A sterile phosphate-buffered saline (PBS) solution containing NaCl (8 g/L), KCl (0.2 g/L), Na2HPO4 (1.44 g/L), and KH2PO4 (0.24 g/L) was used to wash cells three times before fluorescence intensity measurements.
4.2. Glucarate biosensor construction
All study plasmids and primers are listed in Supplementary Tables S6 and S7, respectively. To evaluate and verify WGAN-GP, CNN, BAGAN-GP, and filter models’ performance, 191 AI-generated cRBSs were cloned into glucarate biosensor vectors. The primer pairs: F/R-GSn; F/R-GSm; F/R-CSn; F/R-CSm; n = 1–51; m= 52–191 were designed based on different cRBS sequences (Supplementary Table S3). The modified glucarate biosensor plasmid pJKR-H-RBSn1-cdaR-RBSm1 [7] was used as a template for whole-plasmid polymerase chain reaction (PCR). PCR products were then purified and digested using Dpn I at 37 °C for 30 min and transformed into E. coli JM109 cells for screening by colony PCR and Sanger sequencing. Thus, 51 pJKR-H-CSn-cdaR-GSn plasmids and 140 pJKR-H-CSm-cdaR-GSm plasmids were successfully constructed.
4.3. Fluorescence assays
E. coli BL21 (DE3) cells expressing the glucarate biosensor were cultured to saturation phase and incubated at a concentration of 2% in 24-well plates containing fresh LB medium at 300 rpm and 37 °C. After 3 h, 0 or 20 g/L glucarate was added to 24-well plates, and incubation resumed for 12 h to assess the fluorescence intensity of sfGFP (TFB output). The minimum TFB output was determined when cells were induced with 0 g/L glucarate (“OFF” state) [7] (Fig. 1A). The maximum TFB output was determined when cells were induced with 20 g/L glucarate (“ON” state) [7] (Fig. 1A). TFB dynamic ranges were defined as fold change in TFB outputs in “ON” and “OFF” states [7] (Fig. 1A). Before measurements, induced cultures were diluted in 0.01 M PBS (pH 7.4) and incubated on ice until evaluation using a BD FACS AriaII cell sorter (Becton Dickinson). At least 100,000 events were captured for each sample. BD FACS-Diva software was then used to analyze TFB outputs in “ON” and “OFF” states (Bandpass filter = 530/30 nm; blue laser = 488 nm). FlowJo software was used to calculate the dynamic range of glucarate biosensors.
4.4. Data construction using normal and random data distribution approaches
We proposed two methods to build the one-to-one mapping dataset, including normal and random data distribution. For the normal distribution method, 7053 TFB outputs in “ON” and “OFF” states were obtained based on the mean and standard deviation of TFB outputs using the truncated normal distribution function in the SciPy module in Python3. Similarly, for the random distribution approach, 7053 TFB outputs in “ON” and “OFF” states were obtained based on TFB output ranges using the random function in the NumPy module in Python3. Then, cRBSs with large counts were assigned as high TFB outputs based on published next-generation sequencing (NGS) results [7]. Subsequently, 7053 cRBSs and their corresponding TFB outputs in “ON” and “OFF” states were obtained based on normal and random distribution methods. Finally, we developed models to predict the TFB outputs of each given cRBS in “ON” and “OFF” states. Thus, the dynamic range of the glucarate biosensor was calculated based on predicted TFB outputs.
4.5. Introduction and training the predictive model
We trained CNN [36], [37] and developed predictive models for TFB “ON” and “OFF” states based on constructed one-to-one mapping data. Datasets from our previous work contained 7053 cRBSs with corresponding TFB outputs in “ON” and “OFF” states [7]. For the trained data of TFB “ON” and “OFF” states built based on the normal and random methods, the batch size was set to 64 and 128, respectively. Batch size, defined as the number of cRBSs selected during the training process, affected the optimization degree and speed of the predictive models. We used the Leaky Rectified Linear Unit (LeakyReLU, alpha = 0.1) as the activation function to efficiently learn cRBS sequence features and stochastic gradient descent (learning rate = 0.0001) as the optimization approach to improve the training rate. We trained this predictive model using 5400 cRBSs and corresponding TFB outputs as the training set, 600 cRBSs and corresponding TFB outputs as the validation set, and others as the testing set.
4.6. Introduction and training the encoder model
Rumelhart et al. first proposed the autoencoder model [23], while several recent studies described autoencoder applications in molecular biology [22], [38], [39]. The autoencoder contained encoder and decoder networks [40]. By adjusting the number of neurons in the hidden layer of a vast neural network, sequences were changed into any size or dimension vector and coding processes completed. Thus, the sequence encoder model took sequences into a 64 × 64 × 1 figure format, and the sequence decoder model decoded the figure format to the sequence. Training datasets were from the reported 7053 cRBSs [7] and the optimizer used Adam with a learning rate equal to 0.0001, beta 1 equal to 0.9, and beta 2 equal to 0.99 for training.
4.7. Training the BAGAN-GP model
Training datasets contained 7053 experimentally identified cRBS sequences for the glucarate biosensor [7]. We used all cRBS sequences in the dataset as trained samples. In the BAGAN-GP model, the batch size was 64, and we trained the sequence autoencoder (Supplementary Figure S1) network for 100 epochs. Then, cRBS sequences were encoded as figures by sequence encoder. The 5642 figures were randomly selected as the training set to train the BAGAN-GP model (Supplementary Figure S2), and others were used as the testing set.
We first trained the autoencoder network for 200 epochs in the BAGAN-GP model and then transferred the weight from autoencoder to the GAN model. In this model, we trained five times for the discriminator and once for the generator in each batch training. We used RMSprop optimizer for training. The batch size was 64, and the step was 20. Each step trained 300 epochs, and the learning rate was 0.0002. The best result was identified at step 20, so we selected this parameter for the reverse engineering design of cRBSs with desired TFB dynamic ranges. Finally, generated cRBS sequences were obtained based on sequence decoder.
4.8. Statistics
The area under the receiver operating characteristic (ROC) curve (AUC) was calculated using the metrics.auc function in the “sklearn” Python package. Two replicates were used for each glucarate biosensor strain. All statistical analyses were performed using SciPy (1.5.0), NumPy (1.18.5), and scikit-learn (0.23.1) Python packages. Deep learning was conducted using TensorFlow (2.2.0) and Keras (2.3.1) frameworks. Model plots were generated in Python 3.8 using matplotlib (3.2.2) plotting libraries. Statistical test details are described in corresponding figure legends.
CRediT authorship contribution statement
Nana Ding: Conceptualization, Methodology, Formal analysis, Data curation, Visualization, Investigation, Writing – original draft. Writing – review & editing. Guangkun Zhang: Conceptualization, Methodology, Formal analysis, Data curation, Software, Investigation, Writing – review & editing. Linpei Zhang: Supervision, Investigation, Formal analysis, Writing. Ziyun Shen: Conceptualization, Methodology, Formal analysis. Lianghong Yin: Conceptualization, Writing – review & editing. Shenghu Zhou: Conceptualization, Supervision, Project administration, Writing – review & editing. Yu Deng: Conceptualization, Project administration, Funding acquisition, Writing – review & editing.
Author contributions
N.D., G.Z., L.Z., and Z.S. performed experiments and analyzed the data. N.D., G.Z., S.Z., and Y.D. analyzed the data and wrote the manuscript. N.D., L.Y., S.Z., and Y.D. conceived the project, analyzed the data, and wrote the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2019YFA0905502), Distinguished Young Scholars of Jiangsu Province (BK20220089), The Key R&D Project of Jiangsu Province (Modern Agriculture) (BE2022322), the National Natural Science Foundation of China (21877053 and 31900066), Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-KJGG-015), and the Scientific Research Development Foundation of Zhejiang A&F University (2023LFR020).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.04.026.
Contributor Information
Shenghu Zhou, Email: zhoush@jiangnan.edu.cn.
Yu Deng, Email: dengyu@jiangnan.edu.cn.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Data Availability
The computer source codes for generating novel cRBSs with desired TFB dynamic ranges can be found at https://github.com/YuDengLAB/AI_based_cRBS_design. Flow cytometry data for this study was deposited at Flow Repository and is directly accessible at http://flowrepository.org/id/FR-FCM-Z5N4.
References
- 1.Mitchler M.M., Garcia J.M., Montero N.E., Williams G.J. Transcription factor-based biosensors: a molecular-guided approach for natural product engineering. Curr Opin Biotechnol. 2021;69:172–181. doi: 10.1016/j.copbio.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kang Z., Zhang M., Gao K., Zhang W., Meng W., Liu Y., et al. An l-2-hydroxyglutarate biosensor based on specific transcriptional regulator LhgR. Nat Commun. 2021;12(1):3619. doi: 10.1038/s41467-021-23723-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J.W., Barajas J.F., Burdu M., Ruegg T.L., Dias B., Keasling J.D. Development of a transcription factor based lactam biosensor. ACS Synth Biol. 2017;6(3):439–445. doi: 10.1021/acssynbio.6b00136. [DOI] [PubMed] [Google Scholar]
- 4.Cheng F., Tang X.L., Kardashliev T. Transcription factor-based biosensors in high-throughput screening: advances and applications. Biotechnol J. 2018;13(7) doi: 10.1002/biot.201700648. [DOI] [PubMed] [Google Scholar]
- 5.Lin J.L., Wagner J.M., Alper H.S. Enabling tools for high-throughput detection of metabolites: Metabolic engineering and directed evolution applications. Biotechnol Adv. 2017;35(8):950–970. doi: 10.1016/j.biotechadv.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Doong S.J., Gupta A., Prather K.L.J. Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli. Proc Natl Acad Sci U S Ame. 2018;115(12):2964–2969. doi: 10.1073/pnas.1716920115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding N.N., Yuan Z.Q., Zhang X.J., Chen J., Zhou S.H., Deng Y. Programmable cross-ribosome-binding sites to fine-tune the dynamic range of transcription factor-based biosensor. Nucleic Acids Res. 2020;48(18):10602–10613. doi: 10.1093/nar/gkaa786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F.Z., Carothers J.M., Keasling J.D. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol. 2012;30(4):354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- 9.Siedler S., Khatri N.K., Zsohár A., Kjærbølling I., Vogt M., Hammar P., et al. Development of a bacterial biosensor for rapid screening of yeast p-coumaric acid production. ACS Synth Biol. 2017;6(10):1860–1869. doi: 10.1021/acssynbio.7b00009. [DOI] [PubMed] [Google Scholar]
- 10.Mahmud M., Kaiser M.S., Hussain A., Vassanelli S. Applications of deep learning and reinforcement learning to biological data. Ieee T Neur Net Lear. 2018;29(6):2063–2079. doi: 10.1109/tnnls.2018.2790388. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q.C., Yang L.T., Chen Z.K., Li P. A survey on deep learning for big data. Inf Fusion. 2018;42:146–157. doi: 10.1016/j.inffus.2017.10.006. [DOI] [Google Scholar]
- 12.Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., et al. Generative adversarial networks. Commun Acm. 2020;63(11):139–144. doi: 10.1145/3422622. [DOI] [Google Scholar]
- 13.Goodfellow I.J., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., et al. Generative adversarial networks. Adv Neural Inf Process Syst. 2014;3:2672–2680. doi: 10.48550/arXiv.1406.2661. [DOI] [Google Scholar]
- 14.Yi X., Walia E., Babyn P. Generative adversarial network in medical imaging: A review. Med Image Anal. 2019;58 doi: 10.1016/j.media.2019.101552. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y., Wang H., Wei L., Li S., Liu L., Wang X. Synthetic promoter design in Escherichia coli based on a deep generative network. Nucleic Acids Res. 2020;48(12):6403–6412. doi: 10.1093/nar/gkaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong Y., Li D., Zhang C., Wu C., Wang H., Xin M., et al. Inverse design of two-dimensional graphene/h-BN hybrids by a regressional and conditional GAN. Carbon. 2020;169:9–16. doi: 10.1016/j.carbon.2020.07.013. [DOI] [Google Scholar]
- 17.Sanchez-Lengeling B., Aspuru-Guzik A. Inverse molecular design using machine learning: Generative models for matter engineering. Science. 2018;361(6400):360–365. doi: 10.1126/science.aat2663. [DOI] [PubMed] [Google Scholar]
- 18.Yuan H., Cai L., Wang Z.Y., Hu X., Zhang S.T., Ji S.W. Computational modeling of cellular structures using conditional deep generative networks. Bioinformatics. 2019;35(12):2141–2149. doi: 10.1093/bioinformatics/bty923. [DOI] [PubMed] [Google Scholar]
- 19.Zheng M., Li T., Zhu R., Tang Y.H., Tang M.J., Lin L.L., et al. Conditional Wasserstein generative adversarial network-gradient penalty-based approach to alleviating imbalanced data classification. Inf Sci. 2020;512:1009–1023. doi: 10.1016/j.ins.2019.10.014. [DOI] [Google Scholar]
- 20.Mariani G., Scheidegger F., Istrate R., Bekas C., Malossi C. 2018. BAGAN: Data augmentation with balancing GAN. [DOI] [Google Scholar]
- 21.Huang G., Jafari A.H. Enhanced balancing GAN: minority-class image generation. Neural Comput Appl. 2021:1–10. doi: 10.1007/s00521-021-06163-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eraslan G., Simon L.M., Mircea M., Mueller N.S., Theis F.J. Single-cell RNA-seq denoising using a deep count autoencoder. Nat Commun. 2019;10:390. doi: 10.1038/s41467-018-07931-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rumelhart D., Hinton G.E., Williams R.J. Learning representations by back propagating errors. Nature. 1986;323(6088):533–536. doi: 10.1038/323533a0. [DOI] [Google Scholar]
- 24.Gao X., Deng F., Yue X.H. Data augmentation in fault diagnosis based on the Wasserstein generative adversarial network with gradient penalty. Neurocomputing. 2020;396:487–494. doi: 10.1016/j.neucom.2018.10.109. [DOI] [Google Scholar]
- 25.Xu Y.G., Zhang Z.G., You L., Liu J.J., Fan Z.W., Zhou X.B. scIGANs: single-cell RNA-seq imputation using generative adversarial networks. Nucleic Acids Res. 2020;48(15) doi: 10.1093/nar/gkaa506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong Y., Hwang U., Yoo J., Yoon S. How generative adversarial networks and their variants work: An overview. Acm Comput Surv. 2019;52(1):1–43. doi: 10.1145/3301282. [DOI] [Google Scholar]
- 27.Gupta A., Zou J. Feedback GAN for DNA optimizes protein functions. Nat Mach Intell. 2019;1(2):105–111. doi: 10.1038/s42256-019-0017-4. [DOI] [Google Scholar]
- 28.Yang Q.S., Yan P.K., Zhang Y.B., Yu H.Y., Shi Y.Y., Mou X.Q., et al. Low-dose CT image denoising using a generative adversarial network with Wasserstein distance and perceptual loss. Ieee T Med Imaging. 2018;37(6):1348–1357. doi: 10.1109/tmi.2018.2827462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C.Y., Xu C., Yao X., Tao D.C. Evolutionary generative adversarial networks. Ieee T Evol Comput. 2019;23(6):921–934. doi: 10.1109/tevc.2019.2895748. [DOI] [Google Scholar]
- 30.Liu X.Y., Gupta S.T.P., Bhimsaria D., Reed J.L., Rodríguez-Martínez J.A., Ansari A.Z., et al. De novo design of programmable inducible promoters. Nucleic Acids Res. 2019;47(19):10452–10463. doi: 10.1093/nar/gkz772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen A.A., Voigt C.A. Deep learning to predict the lab-of-origin of engineered DNA. Nat Commun. 2018;9(1):1–10. doi: 10.1038/s41467-018-05378-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feldmann J., Youngblood N., Wright C.D., Bhaskaran H., Pernice W.H.P. All-optical spiking neurosynaptic networks with self-learning capabilities. Nature. 2019;569(7755):208–215. doi: 10.1038/s41586-019-1157-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain A.K., Duin R.P.W., Mao J.C. Statistical pattern recognition: A review. Ieee T Pattern Anal. 2000;22(1):4–37. doi: 10.1109/34.824819. [DOI] [Google Scholar]
- 34.Havlicek V., Corcoles A.D., Temme K., Harrow A.W., Kandala A., Chow J.M., et al. Supervised learning with quantum-enhanced feature spaces. Nature. 2019;567(7747):209–212. doi: 10.1038/s41586-019-0980-2. [DOI] [PubMed] [Google Scholar]
- 35.Jing X.Y., Zhang X.Y., Zhu X.K., Wu F., You X.G., Gao Y., et al. Multiset feature learning for highly imbalanced data classification. Ieee T Pattern Anal. 2021;43(1):139–156. doi: 10.1109/tpami.2019.2929166. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J., Troyanskaya O.G. Predicting effects of noncoding variants with deep learning–based sequence model. Nat Methods. 2015;12(10):931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen K.M., Cofer E.M., Zhou J., Troyanskaya O.G. Selene: a PyTorch-based deep learning library for sequence data. Nat Methods. 2019;16(4):315–318. doi: 10.1038/s41592-019-0360-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng J.J., Xue H.S., Wei Z.Y., Tuncali I., Hao J.Y., Shang X.Q. Integrating multi-network topology for gene function prediction using deep neural networks. Brief Bioinform. 2021;22(2):2096–2105. doi: 10.1093/bib/bbaa036. [DOI] [PubMed] [Google Scholar]
- 39.Ding J.R., Condon A., Shah S.P. Interpretable dimensionality reduction of single cell transcriptome data with deep generative models. Nat Commun. 2018;9:2002. doi: 10.1038/s41467-018-04368-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinton G.E., Salakhutdinov R.R. Reducing the dimensionality of data with neural networks. Science. 2006;313(5786):504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Data Availability Statement
The computer source codes for generating novel cRBSs with desired TFB dynamic ranges can be found at https://github.com/YuDengLAB/AI_based_cRBS_design. Flow cytometry data for this study was deposited at Flow Repository and is directly accessible at http://flowrepository.org/id/FR-FCM-Z5N4.