Abstract
Advances in transcriptomic technologies have deepened our understanding of the cellular gene expression programs of multicellular organisms and provided a theoretical basis for disease diagnosis and therapy. However, both bulk and single-cell RNA sequencing approaches lose the spatial context of cells within the tissue microenvironment, and the development of spatial transcriptomics has made overall bias-free access to both transcriptional information and spatial information possible. Here, we elaborate development of spatial transcriptomic technologies to help researchers select the best-suited technology for their goals and integrate the vast amounts of data to facilitate data accessibility and availability. Then, we marshal various computational approaches to analyze spatial transcriptomic data for various purposes and describe the spatial multimodal omics and its potential for application in tumor tissue. Finally, we provide a detailed discussion and outlook of the spatial transcriptomic technologies, data resources and analysis approaches to guide current and future research on spatial transcriptomics.
Keywords: Spatial transcriptomic technologies
1. Introduction
Multicellular organisms, including human beings, are composed of tissues and organs, which are all specialized in a single biological process and are composed of a large diversity of cells. Although hundreds of types of cells that comprise multicellular organisms all share the same genome, their gene expression patterns can be quite distinct. This difference is caused not only by internal gene regulation but also by signals from the external tissue microenvironment. The development of transcriptomic technologies from bulk RNA sequencing to single-cell RNA sequencing (scRNA-seq) [1], [2] advances knowledge of cellular gene expression to the single-cell level. These transcriptomic sequencing techniques have accumulated a wealth of information on cell type-specific gene regulation, but the original tissue structure is inevitably destroyed. Hence, the single cells in the suspension lose the spatial location information of tissue, which leads to changes in gene expression profiles of some single cells during the enzymatic hydrolysis process or after leaving the specific microenvironment. Additionally, certain cells of special morphology, such as neurons and glial cells with complex structures in the brain, are difficult to access by tissue enzymolysis [3], [4]. The connection between individual cells is still broken, which is difficult to restore the real sample state and leads to a limited understanding of the interactions between cells and the external environment. Therefore, a series of spatial transcriptomic technologies have been developed to profile the expression of genes in tissues more comprehensively and intuitively, which were chosen as the most valuable annual method of 2020 by Nature Methods [5].
Spatial transcriptomic technology is used to parse RNA-seq data at the spatial level, which can simultaneously obtain the spatial context and transcriptional pattern of cells to improve our understanding of tissue architecture. As the basic units of an organism, cells interact with the microenvironment at a specific spatial location to play their specific biological functions, so spatial location information is particularly important for studying and understanding the genesis mechanism of cell biology [6], tumor biology [7], developmental biology [8] and other disciplines [9]. The development of microscopy (including super-resolution and single-molecule imaging), multiplex fluorescence in situ hybridization and other techniques have deepened our understanding of the structure and function of cells and tissues. Sequencing technologies have enabled us to perform a quantitative and qualitative analysis of the expression landscape of genes in unknown cells or tissues. Spatial transcriptomes can combine microscopic imaging and sequencing technologies to obtain gene expression data while preserving the spatial location information of samples to the greatest extent, enabling researchers to find more valuable results. It is the basis of biological research to reveal the function and heterogeneity of spatial organization structure. This demand has driven the development of many spatial transcriptomic technologies.
Thanks to the reduction in sequencing costs, the improvement in microscopy and imaging and the increase in computing power, the spatial transcriptome is rapidly evolving and improving. Spatial transcriptomic technologies based on different approaches have been developed and improved, which have been generating and accumulating massive amounts of unstructured data containing spatial information and gene expression information. Meanwhile, a lot of computational methods for analyzing spatial transcriptomic data are also emerging. However, the variety and complexity of spatial transcriptomic technologies, data resources and analysis approaches all make researchers unfamiliar with the field dizzy and unable to get started quickly. Thus, we put effort to compile a simple atlas of spatial transcriptomics, which could serve as a guidance of spatial transcriptomic technologies, data resources and analysis approaches for researchers. Specifically, we firstly classified 22 diverse spatial transcriptomic technologies into four categories based on their technical foundations and detailed each of them to help researchers choose the best-suited technology in terms of aims. Secondly, we manually collected 791 spatial transcriptomic datasets in 479 articles and integrated these available data to facilitate data accessibility and subsequent analysis. Thirdly, we marshaled 70 computational approaches for analyzing spatial transcriptomic data together in total, and carefully arranged them for a variety of research purposes, such as dimensionality reduction, clustering and cell-cell interaction. Fourthly, we discussed the significance of spatial transcriptomics in the context of molecular biology and the trend towards spatial multimodal omics and described the value of future applications of spatial multimodal omics in tumor tissue. Finally, we summarized and provided an outlook of the spatial transcriptomic technologies, data resources and analysis approaches to guide current and future spatial transcriptomics research.
2. Spatial transcriptomic technologies
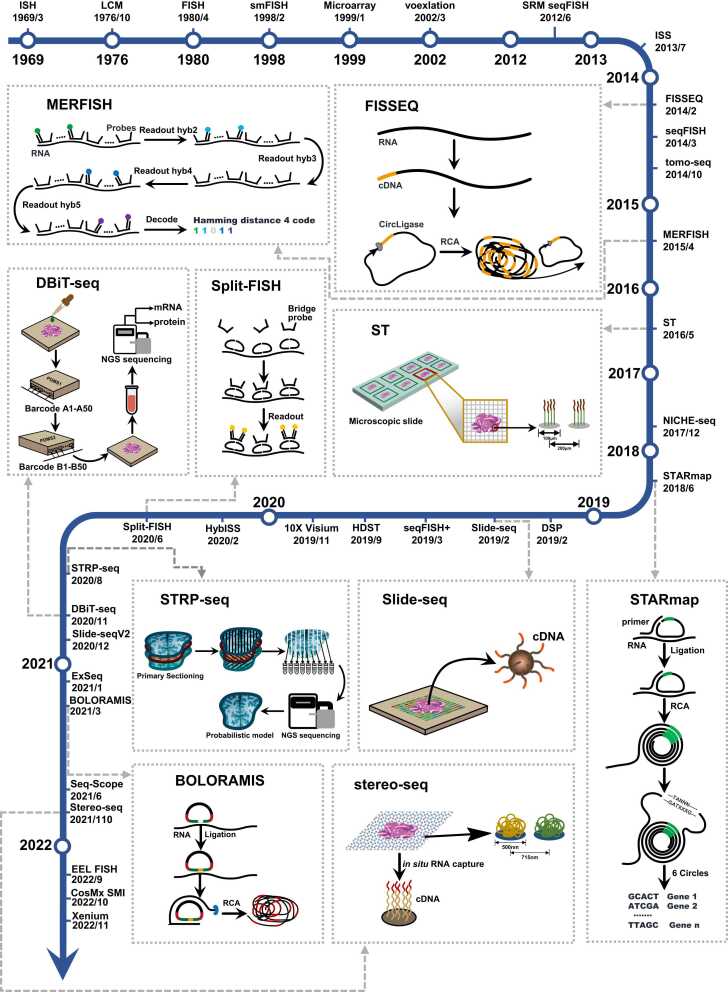
Spatial transcriptomic technologies have evolved in different technological trajectories, with each route of approach reflecting the characteristics of their respective technologies. The choice of spatial transcriptomic technology requires a trade-off between gene throughput, sequence information, sensitivity, resolution, area size and feasibility [10]. Here, we classified the spatial transcriptomic technologies into four categories based on basic types of technologies: in situ hybridization (ISH)-based technologies, in situ sequencing (ISS)-based technologies, next generation sequencing (NGS)-based technologies and the spatial information reconstruction technologies [11], [12]. A summary of these technologies for spatial transcriptomics is shown in Fig. 1 and Table 1.
Fig. 1.
Map of spatial transcriptomics and related technological developments. The dates marked are referenced to the date of acceptance of the paper, whereas preprint articles are referenced to the date of online publication. Visium is referenced to the press release on the official 10X Geonomics website. The ten technologies selected are representative of the development of spatial transcriptomic technologies, and together with the explanations in the text, they provide an accurate picture of the evolution of spatial transcriptomes.
Table 1.
Technical highlights of four technical routes in spatial transcriptomics.
| In situ hybridization (ISH)-based technologies | ||||
|---|---|---|---|---|
| Method | Application | Detection efficiency | Features | Refs |
| seqFISH | Fresh-frozen | 84 % | Targeted, expensive experiments | [9], [42] |
| MERFISH | Fresh-frozen | 80 % | Targeted, high robustness of probe design method, expensive experiments | [9], [17] |
| seqFISH+ | Fresh-frozen | 49 % | Targeted, expensive experiments | [9], [18] |
| DSP | Fresh-frozen or FFPE | NA | Targeted, commercially available | [19] |
| Split-FISH | Fresh-frozen | NA | Targeted, no tissue clearance required,expensive experiments | [9], [20] |
| EEL FISH | Fresh-frozen | 13.2 % | Targeted, transferring RNA using electrophoresis, low-cost experiments | [21] |
| SMI | Fresh-frozen or FFPE | One or two copies per cell | Targeted, high signal to noise ratio detection, commercially available | [22] |
| In situ sequencing (ISS)-based technologies | ||||
| Method | Application | Detection efficiency | Features | Refs |
| ISS | Fresh-frozen or FFPE | < 1 % | Targeted, low throughput, commercially available | [42], [103] |
| FISSEQ | Fresh-frozen or FFPE | < 0.005 % | Unbiased, whole transcriptome, low capture efficiency,commercially available | [42], [103], [187] |
| STARmap | Fresh-frozen | higher than single-cell RNA sequencing | Targeted, reverse transcription-free | [25] |
| HybISS | Fresh-frozen or FFPE | higher than ISS | Targeted, higher throughput than ISS | [26] |
| BOLORAMIS | Cell lines | 10–30 % | Targeted, reverse transcription-free | [29] |
| Next generation sequencing (NGS)-based technologies | ||||
| Method | Application | Resolution | Features | Refs |
| ST/Visium | Fresh-frozen/Fresh-frozen or FFPE | 100/55 µm | Unbiased, whole transcriptome, low capture efficiency, commercially available | [31], [188], [189] |
| slide-seq | Fresh-frozen | 10 µm | Unbiased, whole transcriptome, low capture efficiency | [33] |
| HDST | Fresh-frozen | 2 µm | Unbiased, whole transcriptome, low capture efficiency | [34] |
| DBiT-seq | Fresh-frozen or FFPE | 10 µm | Unbiased, whole transcriptome, low capture efficiency | [35] |
| slide-seqV2 | Fresh-frozen | 10 µm | Unbiased, whole transcriptome, higher capture efficiency than slide-seq | [36] |
| Seq-Scope | Fresh-frozen | 0.5–0.8 µm | Unbiased, whole transcriptome, low capture efficiency | [37] |
| stereo-seq | Fresh-frozen | 500/715 nm | Unbiased, whole transcriptome, low capture efficiency, largest detection area | [38] |
| Spatial information reconstruction technologies | ||||
| Method | Application | Algorithm | Features | Refs |
| tomo-seq | Fresh-frozen | Iterative Proportional Fitting (IPF) | Unbiased, imaging-free, whole transcriptome, low capture efficiency | [40] |
| STRP-seq | Fresh-frozen | Tomographer | Unbiased, imaging-free, whole transcriptome, low capture efficiency | [41] |
2.1. In situ hybridization (ISH)-based technologies
ISH-based spatial transcriptomic techniques are a category of techniques that can profile transcriptomes while preserving the spatial locations of transcripts by hybridizing labeled complementary probes to mRNAs of interest. The technical basis for ISH emerged when Gall et al. [13] achieved the visualization of gene expression in oocytes of the toad Xenopus using ribosomal RNA probes labeled with radioactive tritium. Non-radioactive fluorescent ISH was developed for fluorescence microscopical localization of specific DNA sequences based on the covalent binding of fluorochromes to the 3′-terminus of RNA [14]. A highly multiplexed approach termed single-molecule fluorescence in situ hybridization (smFISH) was developed to simultaneously measure transcripts with high resolution of subcellular spatial localization in healthy mouse kidney cells, and was regarded as the “gold standard” in terms of sensitivity [15]. Lubeck et al. [16] extended smFISH and adopted a sequential barcoding strategy, called sequential fluorescence in situ hybridization (seqFISH), to uniquely identify mRNAs with a drastic increase in the detection capacity for multiplex genes. Another smFISH-based technique, namely multiplexed error-robust fluorescence in situ hybridization (MERFISH) [17], was designed and developed to overcome detection errors and achieve a dramatic increase in the detection of RNA species and quantity in individual cells (Fig. 1). MERFISH combines combinatorial markers and a well-conceived encoding scheme to upgrade the FISH method, making it necessary to read at least four bits for a single code word to be read in error [17]. SeqFISH+ was proposed to be an upgraded version of seqFISH, and barcodes in seqFISH+ were encoded with up to 60 channels. Allowing for more coded samples in sequential hybridization with standard confocal microscopy imaging, seqFISH+ shorten imaging times [18]. GeoMx Digital Spatial Profiling (DSP) was proposed to address the problems encountered in proteomic and RNA transcriptomics. DSP is a commercially available platform in which detection relies in part on unique indexing oligonucleotides with UV linkers assigned to specific target regions [19]. Split-FISH was presented to use a split probe (Fig. 1). This method is another brilliant idea for probe design after MERFISH. Instead of directly pairing with the target base, the probe with a fluorescent marker can specifically bind to a designed bridge probe. A bridge probe is sensitive to the target base and can bind to it efficiently. This design option allows for the accurate detection of the target without tissue removal [20]. Enhanced Electric Fluorescence in situ Hybridization (EEL FISH) uses electrophoresis to transfer negatively charged RNA to the tissue surface and achieves a median gene detection efficiency of 13.2% [21]. This method also has the merits of reduced tissue background noise, less lateral spread and lower experimental costs. NanoString has released the CosMx spatial molecular imager (SMI), a commercially available multimodal omics technology platform [22]. The probe design for RNA detection is based on the MERFISH design approach, which results in low RNA decoding errors. For achieving high signal-to-noise ratios, CosMx SMI uses UV lysis of PC joints as applied in DSP technology.
ISH-based techniques are typically efficient in terms of detection but costly in terms of experimentation [11]. The upgrade of ISH-based technologies has been a process of improvement in probe design and hybridization read methods. The unique design of the probe for MERFISH was a peak in the uniqueness of the probe design scheme for this type of technology, followed by a similar coding scheme for seqFISH+ .
2.2. In situ sequencing (ISS)-based technologies
Sequencing RNA molecules directly in tissues is the technical task of the spatial transcriptome, and ISS-based approaches yield spatial localization of the transcriptome by sequencing. ISS was based on padlock probes and rolling circle amplification (RCA) [23], which preserves spatial information. In fluorescence in situ RNA sequencing (FISSEQ), the amplicons of cDNA are generated inside the cells [24]. Roughly, the single cycle of the experimental process is firstly reverse transcription of RNA, followed by cyclization of cDNA, rolling-circle amplification (RCA), introduction of fluorescence for labeling when appropriate, and finally reading the imaging data (Fig. 1). An RNA sequencing technique applied to intact tissues is called Spatially-resolved Transcript Amplicon Readout Mapping (STARmap) [25] (Fig. 1). This approach proposes two new ways to complete the two key steps in the entire sequencing process, namely, Specific Amplification of Nucleic Acids via Intramolecular Ligation (SNAIL) and Sequencing with Error-reduction by Dynamic Annealing and Ligation (SEDAL) [25]. The ISS method was modified to a new version called Hybridization-based In Situ Sequencing (HybISS). The upgrade of probes allows the new method to be applied to a wide range of tissues. The most intuitive manifestation of HybISS is in sequencing, which abandons the original sequence-by-ligation (SBL) [26] methods and improves a barcode system for probe readout. Both Split-FISH and the HybISS technique of the same period used bridge probes to enhance detection accuracy. In 2021, an expansion sequencing (ExSeq) method combining FISSEQ and expansion microscopy (ExM) [27] allowed for a clearer viewing field [28]. Another method named barcoded oligonucleotides ligated on RNA amplified for multiplexed and parallel in situ analyses (BOLORAMIS) [29] discards the reverse transcription step, which is present in all previous methods (Fig. 1). The probes were directly hybridized with RNA molecules without reverse transcription to form cDNA, and cDNA is looped by SplintR Ligase and further amplified to form amplicons, which are later immobilized in the cell to maintain spatial location. 10X Genomics has launched Xenium, a commercial in situ sequencing-based technology [23] that uses roll-for-amplification to amplify RNA signals within tissues and maintain tissue integrity, enabling subsequent detection of RNA and protein on the same slice [30].
ISS-based techniques are designed to detect minute amounts of transcriptomic signals by cycling and amplifying them. ISS-based experiments were upgraded to include roll-loop generation and signal reading, with STARmap minimizing background interference and BOLORAMIS reducing errors by reducing the number of reverse transcription steps.
2.3. Next generation sequencing (NGS)-based technologies
The spatial information in the tissue can also be localized by precise pre-selection, and then the tissue to be sequenced can be obtained by sectioning. Tissue sections are placed on a specially designed slide capable of capturing RNA and then sequenced for analysis. The process of Spatial Transcriptomics (ST) [31] involves placing the section on a slide containing 1007 captured sites and then permeabilize the section to capture mRNA and perform reverse transcription and fluorescent labeling (Fig. 1). This technology was later upgraded to Visium [32]. Slide-seq used a different strategy to capture RNA in tissues. The biggest change is the adoption of DNA-barcoded beads [33] that characterize the location rather than specially designed probes to capture RNA information in tissues. Each bead has a diameter of 10 µm, thus enabling a large throughput and high resolution of detection (Fig. 1). During the same period, another high-resolution method was developed under the name of High-Definition Spatial Transcriptomics (HDST) [34]. In this method, each bead is placed in a 2 µm well, and a higher definition than Slide-seq is achieved by capturing and decoding millions of beads.
A method called deterministic barcoding in tissue for spatial omics sequencing (DBiT-seq) extends the measurement of biological macromolecules to proteins, allowing simultaneous observation of RNA and proteins [35] (Fig. 1). The principle of this method is similar to creating a Cartesian coordinate system on a thin tissue section to form a 50 × 50 grid, and obtain spatial location information by matrix-like positioning of each small grid. After spatial labeling of the tissue, the tissue is disassembled to extract the base information. Slide-seqV2 is an upgraded version of Slide-seq [36]. The improvements are focused on three aspects: first, the barcoded bead synthesis, second, the array sequencing pipeline, and third, the enzymatic processing of cDNA [36]. Improvements in these areas have led to improvements in the sensitivity and capture efficiency of this method. Seq-Scope achieves an ultra-high resolution comparable to the resolution of optical microscopy [37]. It does not directly measure mRNA in tissues, but does so by indirect means. Specifically, the first sequencing achieves capture and spatial localization of the target transcripts, and the second achieves decoding and reverse transcription of the first sequencing information to read the information from the cDNA. A method for handling large-scale tissue (13.2 cm × 13.2 cm), called spatial enhanced resolution omics-sequencing (Stereo-seq) [38] was developed based on DNA nanoball (DNB) and in situ RNA capture in the same period (Fig. 1). The diameter of the capture point has reached 500 nm, and this method has a trend of commercialization.
The NGS-based methods have transcriptome-wide capture capability, but are not as efficient as the FISH-based methods [11]. The methods differ in the size and resolution of the sections they can hold. From ST in 2016 to Stereo-seq, the resolution has been increased from 100 µm to 500 nm [31], [38]. The implementation paths of Slide-seq and DBiT-seq represent different resolution enhancement methods, while Beijing Genomics Institute has used lithography to build capture arrays on silicon chips to achieve the highest resolution.
2.4. Spatial information reconstruction technologies
The idea of "whole-part-whole" is reflected in revealing the spatial characteristics of gene expression. A method called voxelation [39] was published to reconstruct the entire gene expression of the mouse brain using a method similar to medical imaging. The tomo-seq [40], a new technique combining RNA sequencing and digital image reconstruction, was applied to zebrafish embryos during development and established a spatial 3D map of gene expression. An iterative proportional fitting (IPF) algorithm was used to construct the relationship between zebrafish gene expression and the spatial location of embryo slices [40]. In the STRP-seq, cellular gene expression in the brain was regarded as a regionally continuous non-mutational pattern so that adjacent slices can be considered to have the same spatial gene expression pattern when the tissue is finely sectioned (14 µm per slice) (Fig. 1). The authors' team named this method Spatial Transcriptomics by Reoriented Projections and sequencing (STRP-seq) [41]. Inspired by computed tomography algorithms, STRP-seq reconstructs sequencing results from different angle slices in three dimensions to derive spatial information. Its algorithm is superior to tomo-seq in establishing the spatial pattern of a given gene [41]. STRP-seq is a further development of the tomo-seq method where the spatial information of the transcriptome in the tissue is lost after tissue dissociation using a computational approach to reconstruct the transcriptome.
In summary, spatial transcriptomic technologies have different advantages depending on their principles. ISH-based technologies offer subcellular resolution [43] and are more efficient than others, for example the detection efficiency of seqFISH [42] and MERFISH [17] is 84 % and 80 %, respectively. NanoString has released two commercial platforms that support both RNA and protein detection, but both technologies do not support simultaneous detection of RNA and protein on the same slice currently. The probe design of the SMI is inspired by the MERFISH idea, so the robustness of the detection is higher than that of the DSP [22]. These techniques are generally characterized by being expensive, time-consuming and difficult to perform, but this has changed with the advent of EEL FISH. Not only does EEL FISH cost much less for a single experiment than sequencing-based methods, but it is also able to detect 10 cm2 in 61 h. Electrophoresis reduces lateral diffusion and allows for more accurate RNA localization [21]. ISS-based techniques are generally less efficient in terms of detection due to the uncertainty in the reverse transcription step [29]. The commercial ISS platform Xenium embodies cellular resolution coupled with a non-destructive approach to tissue, enabling the detection of RNA and protein on the same slice [30]. NGS-based technologies are less efficient in detection but are transcriptome-wide. Visium is currently the most successful technology in commercial use, driving third-party packages for downstream data analysis into dominant adaptation [44], [45], [46]. Stereo-seq has the largest detection area of 13.2 cm2 [38], thus enabling the analysis of animal embryos to be completed [8], [38], [47]. The spatial information reconstruction technologies offer another way of thinking about obtaining spatial positions. Although the techniques in question are not used on a wide scale, probably because of the low resolution, this still provides a practical solution to a real problem by providing a useful idea of an indirect way to restore information. There is no perfect technique for solving all spatial transcriptomic problems yet, and a good strategy when exploring tissue space is to combine, for example, single-cell techniques with spatial transcriptomic techniques and a combination of multiple spatial transcriptomic techniques [30].
3. Spatial transcriptomic data resources
With the development and application of spatial transcriptomic technologies in diverse studies, a large amount of spatial transcriptomic data have been generated. However, the generated data resources are usually scattered worldwide, and are not efficiently organized and utilized in real time.
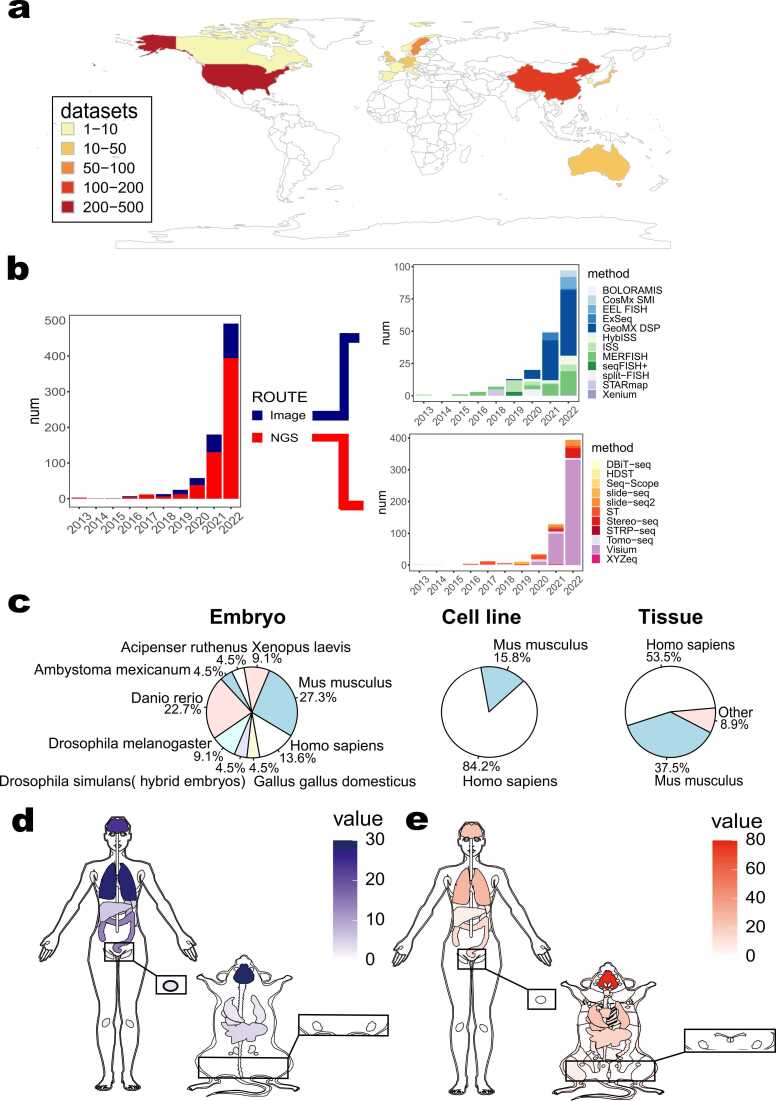
We manually collected 791 datasets in 479 articles spanning 22 countries from 2013 to the present, and carefully identified and classified the attributes of the collected data to demonstrate the development of spatial transcriptomics (Supplementary Table 1). We collected data from PubMed and bioRxiv as well as other databases [11], [48], [49] and used Publish or Perish [50] in the selection process. The spatial transcriptomic data comprehensively compiled from multiple sources will facilitate the accessibility, availability and integration of spatial data. According to the mechanics of these technologies, we further classified current spatial transcriptomic technologies into two broad categories: image-based methods including ISH-based and ISS-based technologies, and sequencing-based methods involving NGS-based technologies and spatially informative reconstruction techniques [32], [51], [52]. A total of 192 image-based datasets and 599 sequencing-based datasets were included in the data collection. Various degrees of spatial transcriptomic research have been conducted worldwide with large datasets being generated in the United States, China and Europe (Fig. 2a). In the last decade, research into sequencing-based and image-based technologies has been on the rise, with sequencing-based datasets growing faster (Fig. 2b). Especially, sequencing-based ST and Visium and image-based DSP methods have been developing rapidly, and these methods will be the focus of attention of spatial transcriptomic researchers in the future. Furthermore, we counted the kind of sources of experimental material, including embryos, cell lines and tissue slices from 34 species. As a result, spatial transcriptomic research is widely applied to various species and organs, with most current studies focusing mainly on Homo sapiens and Mus musculus (Fig. 2c, Supplementary Fig). Additionally, we tailed the human and mouse tissue sections that were used in the spatial transcriptomic studies by the researchers and found that the brain tissue is the most extensively studied in both human and mouse (Fig. 2d, e). Due to the Corona Virus Disease 2019 (COVID-19) pandemic, image-based techniques have been more widely used in the human lung [11] during the last three years (Fig. 2d). Overall, the statistics of the data provide an overall picture of the applications of spatial transcriptomics, so that researchers can make use of some of these experiences. Although our data resource is a relatively comprehensive collection of spatial transcriptomic data generated before December 31, 2022, it is able to reflect the overall evolution and trend of spatial transcriptomic technologies over time.
Fig. 2.
Statistics of spatial transcriptomic datasets. a, Differences in the distribution of papers in the database across countries. b, Trend graphs of the number of articles issued in both technical lines. Data are available until 31 December, 2022. c, Categories of experimental material used in spatial transcriptomic experiments. d, Percentage of human and mouse organs or tissues used in imaging-based technologies. e, Percentage of human and mouse organs or tissues used in sequencing-based technologies.
4. Spatial transcriptomic data analysis
Despite technological differences, the key goal of the spatial transcriptomic analysis is to integrate information of gene expression and spatial locations to allow the mining of useful biological information from experimentally obtained data. The whole process of data analysis can be usually divided into pre-processing and downstream data analysis [11], [53]. A series of available third-party software packages have been developed to facilitate the data analysis process, which is usually goal-oriented [11], [32], [45], [51]. Several reviews have recently demonstrated the process of spatial transcriptomics data analysis and the capabilities of related software packages in data analysis [10], [32], [45], [51], [54], [55], [56]. Here, we present the workflow of spatial transcriptomic data analysis including a specific data analysis process (Supplementary File), and collected the corresponding computational tools for a variety of research purposes (Supplementary Table 2) to facilitate the subsequent optimization of the data analysis process and algorithm development.
4.1. Pre-processing
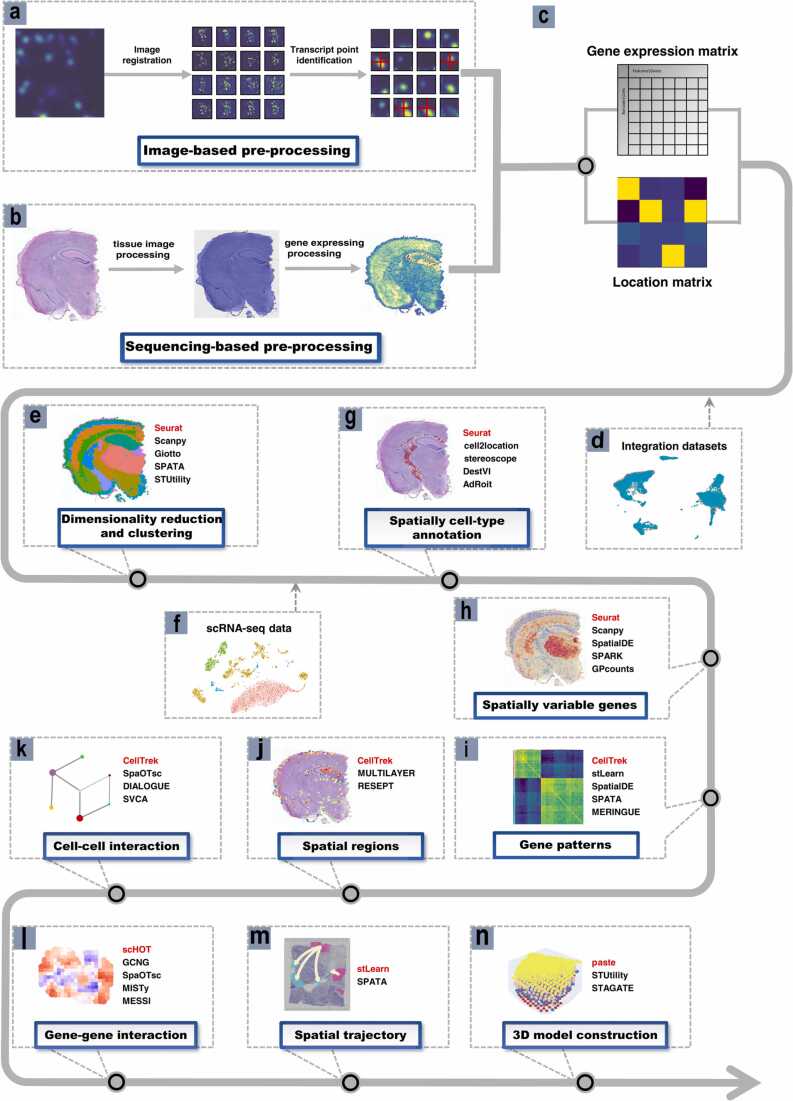
Pre-processing of the image-based data typically involves image registration, transcript spot identification and cell segmentation [11] (Fig. 3a). The detection, counting and localization of fluorescence signals constitute fundamental steps in the pre-processing of data derived from image-based spatial transcriptomic techniques. Because of variations in background brightness and spot quality of images, accurate, high-throughput signal detection and localization remain the main objectives of current image data analysis methods. Several software packages, such as DeepBlink [57], BarDensr [58], graph-ISS [184], and ISTDECO [59], have been developed to address these challenges. They identify the location, type and expression of RNA in the image, and generate the related location index matrix and gene expression matrix. To generate spatial single-cell data, the image needs to be segmented to group the detected RNA into individual cells, and the following packages can be used to achieve cell segmentation: Sparcle [60], Spage2vec [61], JSTA [62], Baysor [151], SSAM [152], etc.
Fig. 3.
Workflow of data analysis. a, Pre-processing processes of imaging-based technologies. b, Pre-processing processes based on NGS based data, using Visium data as an example. c, The gene location matrix and the gene expression matrix are gotten after pre-processing. d, Combination of different data sources and batch effect correction. e, The data is dimensioned down to reduce the number of metrics to be analyzed while retaining as much information as possible. Spots with similar expression patterns are then clustered, and the clustering results are visualized by non-linear dimensionality reduction (t-SNE or UMAP). f, Prediction of cell types by combining spatial transcriptomic data with single-cell sequencing data from the same sample. g, Prediction of cell types from spatial transcriptomic data by deconvolution or integration in combination with annotated homologous single-cell data. h, Calculation of spatially variable genes using spatial dimensional information. i, Co-expression analysis of gene expression information is performed to mine the correlation patterns of different gene expression. j, Defining spatial regions and recognizing possible tissue sub-regions by combining gene expression patterns with a cell type annotation. k, Analysis of intercellular interactions through spatial dimensional information of spatial transcriptomic data. l, Analysis of gene interactions using spatial information from gene expression. m, Reshaping the trajectory of cells in space over time and inferring the evolution and differentiation processes between cells using spatial dimensional information. n, Aligning and integrating spatial transcriptomic data and visualizing 3D models using gene expression similarities and spatial distances between loci in sequential or paired slices.
The pre-processing of sequencing-based spatial transcriptomic data revolves around the processing and integration of sequencing results and tissue images [23], [24], [25], [26], [27], [28], [29], [30], [31]. Pre-processing can be largely divided into three steps: tissue image processing, gene expression processing and relating gene expression to tissue images. Tissue image processing requires image registration, and then divides the entire tissue area into different spots to generate a location index matrix corresponding to each spot region. For gene expression processing, sequenced reads are aligned to the reference genome to generate a gene expression matrix. The coding information of the two matrices is matched to combine gene expression information with the corresponding spatial locations. For 10X Visium technology, SpaceRanger [63] is designed to accomplish these missions (Fig. 3b).
Finally, the pre-processing yields a well-prepared gene expression matrix and location index matrix, which served as the starting point for the downstream analysis (Fig. 3c).
4.2. Downstream analysis
The downstream analysis can be divided into the following parts: batch-effect correction (Fig. 3d), dimensionality reduction and (spatial) clustering (Fig. 3e), spatial cell-type annotation (Fig. 3g) with scRNA-seq data (Fig. 3f), spatially variable genes (Fig. 3h), gene pattern (Fig. 3i), spatial region (Fig. 3j), cell-cell interaction (Fig. 3k), gene-gene interaction (Fig. 3l), spatial trajectory (Fig. 3m) and three-dimensionality model (Fig. 3n). New methods and tools for the downstream analysis of spatial transcriptomic data have exploded recently. Here, we summarize and give an introduction of the tools for each analysis, and Table 2 shows some of the easy-to-use tools.
Table 2.
Recommended packages for each analysis step.
| Package | Language | Analysis step | URL | Refs |
|---|---|---|---|---|
| BarDensr | Python | Preprocess | https://github.com/jacksonloper/bardensr | [58] |
| Space Ranger | Shell | Preprocess | https://support.10xgenomics.com/spatial-gene-expression/software/pipelines/latest/what-is-space-ranger | [63] |
| Seurat | R | Batch-effect correction; Dimensionality reduction and clustering; Spatial cell-type annotation; Spatially variable genes | https://github.com/satijalab/seurat | [44] |
| Scanpy | Python | Dimensionality reduction and clustering; Spatial cell-type annotation; Spatially variable genes | https://scanpy.readthedocs.io/en/stable/ | [153] |
| Harmony | R | Batch-effect correction | https://github.com/immunogenomics/harmony | [65] |
| Scanorama | Python | Batch-effect correction | https://github.com/brianhie/scanorama | [66] |
| cell2location | Python | Spatial cell-type annotation; Spatial regions | https://github.com/BayraktarLab/cell2location/ | [85] |
| stLearn | Python | Dimensionality reduction and clustering; Spatially variable genes; Spatial trajectory | https://github.com/BiomedicalMachineLearning/stLearn | [93] |
| Giotto | R | Dimensionality reduction and clustering; Spatial cell-type annotation; Spatially variable genes | https://github.com/RubD/Giotto | [89] |
| CellTrek | R | Gene patterns; Spatial regions; Cell-cell interaction | https://github.com/navinlabcode/CellTrek | [84] |
| scHOT | R | Gene-gene interaction | https://bioconductor.org/packages/release/bioc/html/scHOT.html | [97] |
| STUtility | R | Dimensionality reduction and clustering; Spatially variable genes; 3D model construction | https://github.com/jbergenstrahle/STUtility | [98] |
| paste | Python | 3D model construction | https://github.com/raphael-group/paste | [99] |
4.2.1. Batch-effect correction
Because we may obtain more than one slice by sequencing or want to combine our slices with data from the database, we need to merge the expression data of multiple slices before the formal analysis (Fig. 3d). The advantage of multi-slice data integration analysis is that the experimental effect can be found by analyzing the differences between slices. However, the data of different slices may have batch effect, and simple merging will cause the experimental effect to be masked by batch effect. We need to use specialized integration tools to correct the batch effect and make the data fusion better so that the experimental effects can be displayed more clearly. Seurat [64] provides the downstream analysis process as well as the batch correct method, which is very convenient for researchers using the seurat process for analysis. The harmony [65] is developed using the R language, which is also easy to integrate into seurat's analysis process. For researchers using python to analyze data, batch effect correction tools such as scanorama [66], LIGER [185], scGen [186], and scVI [67] are also available. Dozens of batch effect correction tools have been developed. Some researchers have tested these tools and made recommendations for different situations [68], [69].
4.2.2. Dimensionality reduction and clustering
Dimensionality reduction and (spatial) clustering are two key analytical steps that underpin all downstream analyses (Fig. 3e). Dimensionality reduction usually precedes clustering and is designed to eliminate data noise to allow for more accurate clustering. There are already many mature methods for dimensionality reduction such as weighted PCA [70], PCA, t-SNE [71], and UMAP [72], which are widely used in various software packages. Recently, some advanced methods of dimensionality reduction have also been proposed, such as Scanpy [153], STUtility [98], SpatialPCA [73], Spaniel [155], SpatialCPie [156], lisaClust [157] and Squidpy [183]. These methods can extract a low-dimensional low dimensional representation of the spatial transcriptomic data with enriched biological signals and preserved spatial correlation structure. Spatial clustering in spatial transcriptomics is designed to use spatial transcriptomic information to cluster tissue locations into multiple spatial clusters, effectively splitting the entire tissue into multiple tissue domains. In addition to traditional machine learning clustering methods such as Louvain clustering [74], Leiden clustering [75], k-means clustering [76], hierarchical clustering, spectral clustering [77] and Gaussian mixture models [78], more sophisticated algorithms, and neural networks have been suggested recently. These advanced methods include SC-MEB [79] using the hidden Markov random field based on Empirical Bayes, BayesSpace [80] based on a fully Bayesian statistical method, the graph convolutional network (GCN) based approach SpaGCN [81], DR-SC [82] leveraging low dimensional embeddings with spatial information to perform spatial clustering using an HMRF, etc.
4.2.3. Spatially cell-type annotation
Spatial transcriptomic methods alone cannot generate deep single-cell-level resolution transcriptomics. Successful integration of scRNA-seq data (Fig. 3 f) and spatial transcriptomic data is essential to understand the architecture of the cell-type distribution and to analyse the putative mechanisms of intercellular communication that underlie this architecture (Fig. 3g). There are two primary approaches for integrating scRNA-seq and spatial data: mapping and deconvolution [83]. Mapping is a method of assigning scRNA-seq-based cell types to each cell by migrating the annotations already established in scRNA-seq to the spatial transcriptomic data. The Mapping method have the potential to assign cells from scRNA-seq data to spatial locations in histological sections [46]. This approach is more commonly used for data generated via image-based technologies. There are some approaches to executing mapping strategies, some of which integrate single-cell data and spatial transcriptomic data by anchoring the integration workflow, such as Seurat [44]. For spatial transcriptomic data that do not reach a single-cell resolution, there are also frameworks such as CellTrek [84]. These approaches map individual cells directly back to spatial coordinates in tissue slices using machine learning methods such as co-embedding and random forest to combine single-cell data and spatial barcoding data. The deconvolution strategy is most often used by sequencing-based techniques that produce spatial barcoding data and do not reach a single-cell resolution. This strategy disentangles discrete cellular subtypes from a single capture spot by using traditional machine learning, deep learning and statistical models to predict the proportion of each cell type in the slice data points, the proportion of each cell type and the gene expression level of each cell. Many advanced approaches of this strategy have also been proposed recently. Cell2location [85] established a trained Bayesian model to accurately map cell types, and DSTG [86] used semi-supervised graphical convolutional neural networks to train the exact composition of predictive spatial transcriptomic data.Many other packages can also be applied to spatially cell-type annotation, such as stereoscope [158], DestVI [159], AdRoit [160], RCTD [161], SpatialDecon [162], SpaceFlow [163], SPOTlight [164], FICT [165], SpatialDWLS [166], STRIDE [167], STdeconvolve [168], Tangram [169] and smfishHmrf [170].
4.2.4. Spatially variable genes
In spatial transcriptomics, the traditional method for identifying SVGs (spatially variable genes) is to perform a differential expression analysis on different presentations directly (Fig. 3h). Recently, a series of new computational approaches using statistics and machine learning including deep learning has been proposed to solve this problem, such as SpatialIDE [87], SPARK [88], SpaGCN [81], Giotto [89], GPcounts [171], BOOST-GP [172], SOMDE [173], RayleighSelection [174], trendsceek [175], singleCellHaystack [176], scGCO [177], SPARK-X [178], sepal [179], etc. In these advanced approaches, SpatialIDE [87] is based on Gaussian process regression and SPARK [88] is based on a spatial generalized linear mixed model with multiple spatial kernels. Besides, the more representative methods using deep learning are SpaGCN [81] based on the graph convolutional network approach and Giotto [89] based on spatial dependence modeling using HMRF (hidden Markov random field model).
4.2.5. Gene patterns
After identifying SVGs, researchers need to use spatial visualization and existing single-cell processes and data mining methods to analyze gene expression patterns to infer gene functions (Fig. 3i). These methods can not only identify gene modules by calculating spatially weighted gene co-expression matrix or weighted network analysis, which are relatively mature methods in the single-cell analysis process, such as CellTrek [84], but also build mathematical models and other data mining methods to calculate gene expression patterns, such as MERINGUE [90].
4.2.6. Spatial regions
The location of cells in the tissue microenvironment influences gene expression, and the spatial transcriptome provides excellent conditions for exploring tissue regions (Fig. 3j). MULTILAYER [91] compares spatial localization to infer the degree of co-expression after all patterns of overexpressed genes have been detected. The degree of co-expression reveals functionally relevant spatial gene co-expression and divides the initial spatial transcriptome slice into functionally relevant spatial community regions. The spatial transcriptomic data contain a variety of information about space. Critical spatial location matrices, corresponding to the staining of HE regions of sections, and relative distances between individual cells are implicit in the gene expression data. All of this information can be used to delineate spatial regions. To make full use of the spatial dimensional information, RESEPT [92] accurately infers and visualizes the organizational structure by mapping transcriptomic data to RGB images and using a supervised convolutional neural network model for spatial region segmentation. Alternatively, HE-stained images can be combined in stLearn [93] to assist in optimizing the clustering process. On the basis of traditional clustering, stLearn divides clusters into subclusters based on the location of each cluster. SpaGCN [81] merges the HE staining information provided by ST data into a spatial neighborhood map, which can be combined with gene expression from Spot via a graph convolutional network. CellTrek [84] reclassifies cell type regions by comparing gene expression differences between co-expressed gene modules calculated according to gene pattern.
4.2.7. Cell-cell interaction
Cells in tissue interact with each other all the time, and even more so when they are spatially close to each other. Gene relationship databases and methods for studying cellular communication in scRNA-seq have been developed in sufficient abundance (Fig. 3k). The spatial information provided by the spatial transcriptome makes it possible to analyze cell-cell interactions based on the location of the cells. SpaOTsc [94] combines the two by developing a map between the spatial transcriptomic and single-cell transcriptomic datasets, thus using the spatial data to reconstruct spatial metrics of single-cell data in scRNA-seq data. Multiple cells interact with each other to maintain the homeostasis of the tissue microenvironment. DIALOGUE [95] treats coordinated cells within a region as higher-order functional units at the tissue level. From the perspective of functional multicellular populations, DIALOGUE [95] identifies multicellular programs associated with different phenotypes through co-regulatory gene sets across cell types. SVCA [96] based on the Gaussian process assesses the impact of cellular interactions on the expression of individual molecules by disentangling the effects that affect gene expression.
4.2.8. Gene-gene interaction
Gene interactions are divided into intracellular gene interactions and intercellular gene interactions (Fig. 3l). Cells communicate with each other through a variety of signal transduction mechanisms, which are essential interactions between genes. The advent of spatial transcriptomic technologies has provided information not only on intracellular gene expression but also on the spatial relationships between cells, providing a basis for the analysis of large-scale extracellular interactions. By integrating and reconstructing information from gene expression and positional matrices, greater insight can be gained into the molecular mechanisms behind cellular interactions. There are many advanced methods proposed to solve this problem, such as GCNG [180], which represents data from spatial transcriptomics as graphs of relationships between cells and infers the gene interactions involved in cellular communication from spatial single-cell data. The regulatory mechanisms behind gene expression are complex. ScHOT [97] seeks to examine higher-order structural changes in gene expression, such as correlations between genes in differentiation pseudotimes, between discrete populations and across spatial regions. MISTy [181] uses views from different spatial contexts to model expressed intercellular interactions. MESSI [182] utilises signaling to predict gene expression in cells.
4.2.9. Spatial trajectory
Cell trajectory analysis can help infer the evolution and differentiation process between cells by analysing the dynamic changes in gene expression between cells (Fig. 3m). Based on the spatial location information provided by the spatial transcriptome, cell trajectories can be visualized in the tissue slice. StLearn [93] offers an algorithm called pseudo-space-time (PST) trajectory analysis that infers biological process from gradients in the transcriptional state across tissues. SPATA [154] analyzes dynamic changes by using monocle3, a package that implies pseudo-time inference.
4.2.10. 3D model construction
All the current technological approaches are focused on capturing two-dimensional spatial information. With the development of spatial transcriptomics, 3D structures are constructed based on the alignment and integration of serial sections of tissue from a single sample, providing opportunities to explore tissue structure (Fig. 3n). The growth of traditional machine learning methods and deep learning frameworks provides many advanced computational approaches to address this problem. The most common is the utilization of machine learning and statistical methods. For example, STUtility [98] aligns slices with ICP (iterative nearest point algorithm) to visualize them on a 3D model; PASTE [99] uses Wasserstein Optimal Transport (FGW-OT) [100] based on Optimal Transport theory to construct 3D alignment. Certainly, there are other approaches, such as STAGATE [101] using neural networks. By using graph attention autoencoders, STAGATE [101] integrates spatial information and gene expression profiles to learn low-dimensional latent embeddings, accurately identify spatial domains and extract 3D expression domains.
The practicability in data analysis depends on the researcher's biological literacy (especially in cell biology and molecular biology), familiarity with programming languages (the widely used Python and R), choice of software package algorithms and the hardware facilities in the laboratory. The actual analysis process may not involve practising all of the data analysis covered in this article, but rather a selection of items depending on the experiment purpose. We have collated 70 software packages (Supplementary Table 2) according to the purpose of the downstream analysis and have selected those that we consider easy to use or learn. We have chosen 13 easy-to-use packages in Table 2, which cover the whole range of data analysis steps in this article. It is relatively easy to complete the data analysis tasks successfully, but understanding and grasping the context and meaning implied by them precisely requires a long period of practice. The widespread use of the same technology will contribute to the choice of developers to join the technology-led spatial transcriptomics ecosystem. Competition between technologies and software packages for downstream analysis will foster the development of more universal and easy-to-use spatial transcriptomic data analysis, which will benefit the future participation of spatial transcriptomics in clinical applications.
5. Coming of spatial multimodal omics
Spatial multimodal omics will be the next hot spot in the academic community [102], [103], [104], [105]. This is already well known, but a longer-term perspective is more insightful in understanding such developments. The double helix structure of DNA and the establishment of the genetic central dogma initiated the era of molecular biology. The Polymerase Chain Reaction (PCR) enables the rapid amplification of DNA, which deeply influences the progress of the life sciences. Under the genetic central dogma, RNA and protein detection techniques are now becoming more sophisticated, contributing to the development and flourishing of spatial multimodal omics [103], [104], [106]. As they become commercially available, these technologies will become increasingly common in basic research, allowing researchers to explore the mechanisms of change in biological tissues from a more microscopic and holistic perspective.
Spatial transcriptomics is the basis of multimodal omics [10]. Spatial transcriptomic techniques, which now have unique advantages in revealing cell fate regulation and tissue microenvironment interactions, may become as fundamental as PCR in the future to be able to perform in every laboratory. On the other hand, on the grounds of spatial transcriptomics, spatial multimodal omics is crying out to emerge. Spatial multimodal omics is an instantaneous demonstration of the genetic central dogma in the regulation of life, as it enables the simultaneous detection of RNA and proteins in tissues as well as the analysis of linkage processes in different processes in the regulation of life processes. The genetic central dogma stops at proteins, but proteins need to be modified to assume their functions [107], and the development of spatial multimodal omics will extend the content of the genetic central dogma.
Biochemical changes within tissues can be detected by single-cell techniques. Spatial transcriptomic techniques can accurately locate the regions where such changes occur and infer the cell-cell interactions within the tissue [52]. Spatial proteomics assays can provide further insights into the regulatory transitions revealed by the spatial transcriptome. DISCO-MS, an automated spatial proteomics technology, has made a mark in the detection of cellular heterogeneity in tissues [108]. Multimodal omics technologies can combine the strengths of each technique to provide a snapshot of the valid biological information within a tissue, thus interpreting the information at the data processing stage. In fact, protein post-translational modifications are very complex and the linkage of their regulatory processes with genes is yet to be further discovered. Among the commercially available technologies to detect both RNAs and proteins, protein detection is inferior to that of RNA, but there is no doubt that this situation will improve significantly in the coming years.
In the detection of tumor tissue, multimodal omics technologies can better characterize tumor heterogeneity [109] and the tumor microenvironment [110] than previous single-cell sequencing and spatial transcriptomic technologies, so that we can understand the function mechanism of tumors at a deeper level. The understanding of tumors is constantly evolving, and the current understanding of tumors is focused on the interpretation of tumor heterogeneity and the tumor microenvironment [111], [112], [113], [114], [115]. A tumor can be regarded as a special, vital organ [116]. The production of a tumor implies a severe failure of the immune system. Aggressive malignant tumors change the microenvironment in which they exist during their developmental phase, thus making it more favorable for them to proliferate [115], [117]. The determination of the border between tumor cells and normal cells is important in the diagnosis of tumors, and metastasis is one of the main reasons why tumors are difficult to kill. The proliferation of tumors causes changes in the entire body's internal environment, for example, changes in the enzyme activity in blood tests. Exploring tumor interactions with its microenvironment is not possible with conventional stained pathology, but this is possible with the support of spatial multimodal omics techniques. The combination of single-cell sequencing technology and Visium can reveal the molecular mechanisms of immune concordance in breast cancer, the cellular basis of intratumoral heterogeneity and the influence of the microenvironment on cell subtypes [118]. New hormone receptors and signaling regulators in breast cancer development were identified in the work of A. Janesick et al. [30].
6. Summary and outlook
Spatial transcriptomic technologies can help understand tissue architecture and biological mechanisms not available with traditional techniques, such as bulk RNA-seq and scRNA-seq. We summarized spatial transcriptomic technologies to help researchers understand the technical characteristics and choose the best-suited technology in terms of research tasks. Spatial transcriptomic technology is undoubtedly a powerful tool in the study of the true gene expression of cells in situ in tissues and provides important research means for many fields, such as oncology [119], immunology [120], developmental biology [121], [122], reproductive biology [123] and neurobiology [3], [124]. Nonetheless, spatial transcriptomics is a thriving discipline, where there is still a lot of room for the development of technologies. Firstly, there is no single best solution available but often a trade-off in terms of transcriptome-wide profiling, high resolution, high throughput and high detection efficiency [1], [38], [40] until now. Therefore, researchers are still seeking to develop new ideal technologies to meet all these objectives, and future improvements will help explore the detailed cellular and molecular interactions in tissue. Furthermore, it is important to standardize the way the technology is evaluated, so that key metrics such as the RNA capture efficiency and resolution can be assessed in a unified way [125]. Secondly, future spatial transcriptomic technologies should add the dynamic processes of the organism in the temporal dimension to the previous two or three-dimensional spatial transcriptomics. Spatiotemporal transcriptomics will enable the dynamic investigation of development and disease progression, and ZipSeq [43] and SPACECAT [126] are good starting points for developing methods in the temporal dimension. Thirdly, spatial transcriptomics will benefit from the advances in spatial technologies, and spatial technologies will be increasingly applied to other multimodal omics, such as spatial genomics [127], spatial epigenomics [128] and spatial proteomics [129], which can complement spatial transcriptomics. Spatial multimodal omics will be the research hot spot in life science and be of paramount importance to give researchers a more systematic and comprehensive understanding of the biological process, cellular component and molecular function [102], [104], [105]. Fourthly, the plethora of new spatial technologies and the rapidity of technological updates require that researchers undergo constant knowledge updates at a high cost of study time. Like the popularization of Visium currently, several outstanding spatial technologies may outperform other technologies and would become the mainstream technologies in the future. Finally, the current use of spatial transcriptomic technologies as a useful and effective tool for exploring life processes is mainly for academic purposes, and there is still a long way to go from large-scale clinical applications. Spatial transcriptomics has expanded our knowledge of cellular and molecular changes in diseases and cancerous heterogeneity. Looking forward, we anticipate that spatial multimodal omics combining genomics, epigenomics, transcriptomics, proteomics and metabonomics will be an up-and-coming solution to provide more detailed information on diseases to help improve clinical outcomes efficiently, such as disease diagnosis, drug development and medical treatment.
Spatial transcriptomic technologies have generated and accumulated massive amounts of data with various data structures and formats, but these data have not yet been integrated into comprehensive databases. STOmicsDB proposed a platform standard for archiving spatial transcriptomic data, and the data already collected can be easily used [49]. However, the data collection on this platform is incomplete, and data sharing needs to be made easier. Therefore, we manually collected and comprehensively compiled 791 spatial transcriptomic datasets from multiple sources to facilitate the accessibility, availability and integration of spatial data. The current data mainly focus on Homo sapiens and Mus musculus, and thus there is great potential for more investigations of other species in the future. The integration of data generated by different spatial transcriptomic technologies can leverage their respective advantages to better achieve transcriptome-wide coverage, high resolution and high detection efficiency. Besides, single-cell transcriptomic data can play a complementary role, and we can get more accurate spatial transcriptomic information of the target tissue by integrating data of single-cell and spatial transcriptomic. Furthermore, the transcriptome is only one aspect of cell function, so spatial transcriptome alone can only provide a partial picture of gene regulatory networks. Future data integration of single-cell or spatial multimodal omics, such as genome, epigenome, proteome, metabolome and 3D chromatin conformation, which can complement spatial transcriptome, will facilitate the subsequent analysis to allow a fuller understanding of cell identity and state. In particular, the Human Cell Atlas (HCA) [130] and the BRAIN Initiative Cell Census Consortium (BICCC) [131] are integrating data across diverse modalities to construct a multimodal atlas for humans or mice, including spatial transcriptomic data. The Human Reference Atlas (HRA) [132] contributed by 16 consortia, including the Human Biomolecular Atlas Program (HuBMAP) [133], the Kidney Precision Medicine Project (KPMP) [134], [135], the Human Tumor Atlas Network (HTAN) [136] and the Chan Zuckerberg Initiative (CZI) [130], [137], [138], the Genotype-Tissue Expression project (GTEx) [139] and the GenitoUrinary Developmental Molecular Anatomy Project (GUDMAP) [140], aims to create a benchmark reference supporting spatial queries to advance biomedical and clinical research. In the future, the establishment of a unified database for integrating, querying, and visualizing multi-source data may enable the comprehensive analysis of data resources.
It is particularly important and necessary to analyze and interpret spatial transcriptomic data and transform it into useful information and knowledge by using computational tools efficiently, but the multitude of computational tools for data analysis can bewilder beginners. Therefore, we detailed the workflow of spatial transcriptomic data analysis and marshaled 70 computational tools to analyse spatial transcriptomic data for a variety of purposes, facilitating researchers to select appropriate data analysis tools or develop new packages. There is much room for improvement in the existing computational tools of data analysis. The development of algorithms should avoid the limitations inherent in the original technology and integrate data from different sources across technologies to complement each other. The massive amounts of existing bulk and single-cell RNA sequencing data may also help predict or complement spatial transcriptomic data, and there are already algorithms for generating a spatially resolved single-cell expression profile from bulk RNA-seq data [141] and integration analysis of data from 10X Visium with scRNA-seq [83], [142], [143]. Besides, given tissue sections in most spatial transcriptomic technologies, it is of great importance to develop powerful and efficient computational tools that will integrate and align the spatial transcriptomic data of multiple sections from multiple individuals or developmental stages to enable a three-dimensional or spatiotemporal analysis of life activities. The recent proposal of probabilistic alignment of ST experiments (PASTE) [99] was a successful attempt at the alignment and integration of spatial transcriptomics data. Particularly, there is great potential for computational approaches that conduct analyses in both spatial and temporal dimensions simultaneously in the future, and spatiotemporal analysis will be important for spatial developmental trajectory and disease progression and can provide novel and dynamic insights into spatial heterogeneity within the tissue microenvironment. Several computational tools have recently emerged for scRNA-seq-based analysis, such as developmental trajectory [144], [145], [146] and RNA velocity [147], [148], [149], and some spatial transcriptomic data have been successfully analyzed using these scRNA-seq-based approaches [150]. Further development of algorithms for spatiotemporal analysis will allow measurements of cell states in four dimensions. Moreover, novel computational tools for integrating spatial transcriptomics data with data of single-cell or spatial multimodal omics will be a major future direction and have an urgent need to be developed. Through spatial multimodal omics analysis, tissue structures, gene expression patterns, cellular interactions and molecular interactions will be better understood, which will be fundamental to giving a fuller picture of cellular function. With the rapid development of artificial intelligence (AI) techniques, such as deep learning, transfer leaning, reinforcement learning, ensemble learning and meta learning, AI has been applied to more and more biomedical researches including spatial transcriptomics. Although it is still difficult to systematically apply AI to specific problems in the biomedical field, we believe AI, especially multimodal learning [190], [191], will play an important role in spatial multimodal omics and provide a good solution for computational analysis in the future. Finally, an integrated, scalable, multi-compatible and unified computational platform for data analysis that supports both easy-to-use command line and graphical interfaces will greatly simplify spatial transcriptomics data analysis, facilitating researchers to focus more on deciphering the code of life for biological discovery.
Funding
This work was supported in part by the National Key Research and Development Project of China (2021YFC2302400 and 2021YFC2302403), National Natural Science Foundation of China (81830101 and 31901064), General Project of Chongqing Natural Science Foundation of China (CSTB2022NSCQ-MSX1059), and Intelligent Medicine Research Project of Chongqing Medical University (ZHYXQNRC202103).
CRediT authorship contribution statement
Liangchen Yue: Formal analysis, Investigation, Resources and Writing - Original Draft. Feng Liu: Formal analysis, Investigation, Resources and Writing - Original Draft. Jiongsong Hu: Formal analysis, Investigation, Resources and Writing - Original Draft. Pin Yang: Investigation and Resources. Yuxiang Wang: Visualization. Junguo Dong: Visualization. Wenjie Shu: Conceptualization, Supervision and Project administration. Xingxu Huang: Supervision and Project administration. Shengqi Wang: Supervision and Project administration.
Author contributions
W. S. designed, conceived, and supervised the work. S. W. and X. H. co-supervised the work. L. Y., F. L., J. H., and P. Y. summarized all the literature and analysis the data. Y. W., J. D. helped with the figures. W. S., L. Y., F. L., J. H., and P. Y. wrote the manuscript. All authors discussed and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.01.016.
Contributor Information
Wenjie Shu, Email: shuwj@bmi.ac.cn.
Xingxu Huang, Email: huangxx@shanghaitech.edu.cn.
Shengqi Wang, Email: sqwang@bmi.ac.cn.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Tang F., et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 2.Haque A., et al. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9(1):75. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maynard K.R., et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat Neurosci. 2021;24(3):425–436. doi: 10.1038/s41593-020-00787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lacar B., et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016;7:11022. doi: 10.1038/ncomms11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marx V. Method of the year: spatially resolved transcriptomics. Nat Methods. 2021;18(1):9–14. doi: 10.1038/s41592-020-01033-y. [DOI] [PubMed] [Google Scholar]
- 6.Foreman R., Wollman R. Mammalian gene expression variability is explained by underlying cell state. Mol Syst Biol. 2020;16(2) doi: 10.15252/msb.20199146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji A.L., et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell. 2020;182(2):497–514. doi: 10.1016/j.cell.2020.05.039. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu C., et al. Spatiotemporal mapping of gene expression landscapes and developmental trajectories during zebrafish embryogenesis. Dev Cell. 2022;57(10):1284–1298. doi: 10.1016/j.devcel.2022.04.009. e5. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L., et al. Clinical and translational values of spatial transcriptomics. Signal Transduct Target Ther. 2022;7(1):111. doi: 10.1038/s41392-022-00960-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao A., et al. Exploring tissue architecture using spatial transcriptomics. Nature. 2021;596(7871):211–220. doi: 10.1038/s41586-021-03634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moses L., Pachter L. Museum of spatial transcriptomics. Nat Methods. 2022;19(5):534–546. doi: 10.1038/s41592-022-01409-2. [DOI] [PubMed] [Google Scholar]
- 12.Asp M., Bergenstråhle J., Lundeberg J. Spatially resolved transcriptomes-next generation tools for tissue exploration. Bioessays. 2020;42(10) doi: 10.1002/bies.201900221. [DOI] [PubMed] [Google Scholar]
- 13.Gall J.G., Pardue M.L. Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc Natl Acad Sci USA. 1969;63(2):378–383. doi: 10.1073/pnas.63.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauman J.G., et al. A new method for fluorescence microscopical localization of specific DNA sequences by in situ hybridization of fluorochromelabelled RNA. Exp Cell Res. 1980;128(2):485–490. doi: 10.1016/0014-4827(80)90087-7. [DOI] [PubMed] [Google Scholar]
- 15.Femino A.M., et al. Visualization of single RNA transcripts in situ. Science. 1998;280(5363):585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 16.Lubeck E., et al. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11(4):360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen K.H., et al. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348(6233) doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eng C.L., et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature. 2019;568(7751):235–239. doi: 10.1038/s41586-019-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merritt C.R., et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol. 2020;38(5):586–599. doi: 10.1038/s41587-020-0472-9. [DOI] [PubMed] [Google Scholar]
- 20.Goh J.J.L., et al. Highly specific multiplexed RNA imaging in tissues with split-FISH. Nat Methods. 2020;17(7):689–693. doi: 10.1038/s41592-020-0858-0. [DOI] [PubMed] [Google Scholar]
- 21.Borm L.E., et al. Scalable in situ single-cell profiling by electrophoretic capture of mRNA using EEL FISH. Nat Biotechnol. 2022 doi: 10.1038/s41587-022-01455-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He S., et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol. 2022 doi: 10.1038/s41587-022-01483-z. [DOI] [PubMed] [Google Scholar]
- 23.Ke R., et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 2013;10(9):857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- 24.Lee J.H., et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343(6177):1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 2018;361:6400. doi: 10.1126/science.aat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gyllborg D., et al. Hybridization-based in situ sequencing (HybISS) for spatially resolved transcriptomics in human and mouse brain tissue. Nucleic Acids Res. 2020;48(19) doi: 10.1093/nar/gkaa792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen F., Tillberg P.W., Boyden E.S. Optical imaging. Expansion microscopy. Science. 2015;347(6221):543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alon S., et al. Expansion sequencing: Spatially precise in situ transcriptomics in intact biological systems. Science. 2021;371:6528. doi: 10.1126/science.aax2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S., et al. Barcoded oligonucleotides ligated on RNA amplified for multiplexed and parallel in situ analyses. Nucleic Acids Res. 2021;49(10) doi: 10.1093/nar/gkab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janesick A., et al. High resolution mapping of the breast cancer tumor microenvironment using integrated single cell, spatial and in situ analysis of FFPE tissue. bioRxiv. 2022 doi: 10.1038/s41467-023-43458-x. 2022.10.06.510405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ståhl P.L., et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353(6294):78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 32.Charitakis N., Ramialison M., Nim H.T. In: Transcriptomics in Health and Disease. Passos G.A., editor. Springer International Publishing; Cham: 2022. Comparative analysis of packages and algorithms for the analysis of spatially resolved transcriptomics data; pp. 165–186. [Google Scholar]
- 33.Rodriques S.G., et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363(6434):1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vickovic S., et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat Methods. 2019;16(10):987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y., et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell. 2020;183(6):1665–1681. doi: 10.1016/j.cell.2020.10.026. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stickels R.R., et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat Biotechnol. 2021;39(3):313–319. doi: 10.1038/s41587-020-0739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho C.S., et al. Microscopic examination of spatial transcriptome using Seq-Scope. Cell. 2021;184(13):3559–3572. doi: 10.1016/j.cell.2021.05.010. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen A., et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell. 2022;185(10):1777–1792. doi: 10.1016/j.cell.2022.04.003. e21. [DOI] [PubMed] [Google Scholar]
- 39.Brown V.M., et al. Multiplex three-dimensional brain gene expression mapping in a mouse model of Parkinson's disease. Genome Res. 2002;12(6):868–884. doi: 10.1101/gr.229002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Junker J.P., et al. Genome-wide RNA Tomography in the zebrafish embryo. Cell. 2014;159(3):662–675. doi: 10.1016/j.cell.2014.09.038. [DOI] [PubMed] [Google Scholar]
- 41.Schede H.H., et al. Spatial tissue profiling by imaging-free molecular tomography. Nat Biotechnol. 2021;39(8):968–977. doi: 10.1038/s41587-021-00879-7. [DOI] [PubMed] [Google Scholar]
- 42.Shah S., et al. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron. 2016;92(2):342–357. doi: 10.1016/j.neuron.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu K.H., et al. ZipSeq: barcoding for real-time mapping of single cell transcriptomes. Nat Methods. 2020;17(8):833–843. doi: 10.1038/s41592-020-0880-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hao Y., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587. doi: 10.1016/j.cell.2021.04.048. e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng Z., et al. Statistical and machine learning methods for spatially resolved transcriptomics data analysis. Genome Biol. 2022;23(1):83. doi: 10.1186/s13059-022-02653-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B., et al. Benchmarking spatial and single-cell transcriptomics integration methods for transcript distribution prediction and cell type deconvolution. Nat Methods. 2022;19(6):662–670. doi: 10.1038/s41592-022-01480-9. [DOI] [PubMed] [Google Scholar]
- 47.Wang M., et al. High-resolution 3D spatiotemporal transcriptomic maps of developing Drosophila embryos and larvae. Dev Cell. 2022;57(10):1271–1283. doi: 10.1016/j.devcel.2022.04.006. e4. [DOI] [PubMed] [Google Scholar]
- 48.Fan Z., Chen R., Chen X. SpatialDB: a database for spatially resolved transcriptomes. Nucleic Acids Res. 2020;48(D1) doi: 10.1093/nar/gkz934. D233-d237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Z., et al. STOmicsDB: a database of spatial transcriptomic data. bioRxiv. 2022 2022.03.11.481421. [Google Scholar]
- 50.Harzing, A.W.,2007. Publish or Perish, available from 〈https://harzing.com/resources/publish-or-perish〉.
- 51.Dries R., et al. Advances in spatial transcriptomic data analysis. Genome Res. 2021;31(10):1706–1718. doi: 10.1101/gr.275224.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams C.G., et al. An introduction to spatial transcriptomics for biomedical research. Genome Med. 2022;14(1):68. doi: 10.1186/s13073-022-01075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luecken M.D., Theis F.J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019;15(6) doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heydari A.A., Sindi S.S. Deep learning in spatial transcriptomics: learning from the next next-generation sequencing. bioRxiv. 2022 doi: 10.1063/5.0091135. 2022.02.28.482392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kleino I., et al. Computational solutions for spatial transcriptomics. Comput Struct Biotechnol J. 2022;20:4870–4884. doi: 10.1016/j.csbj.2022.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palla G., et al. Spatial components of molecular tissue biology. Nat Biotechnol. 2022;40(3):308–318. doi: 10.1038/s41587-021-01182-1. [DOI] [PubMed] [Google Scholar]
- 57.Eichenberger B.T., et al. deepBlink: threshold-independent detection and localization of diffraction-limited spots. Nucleic Acids Res. 2021;49(13):7292–7297. doi: 10.1093/nar/gkab546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen S., et al. BARcode demixing through non-negative spatial regression (BarDensr) PLoS Comput Biol. 2021;17(3) doi: 10.1371/journal.pcbi.1008256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andersson A., et al. ISTDECO: in situ transcriptomics decoding by deconvolution. bioRxiv. 2021 2021.03.01.433040. [Google Scholar]
- 60.Prabhakaran S., Nawy T., Pe’er D. Sparcle: assigning transcripts to cells in multiplexed images. bioRxiv. 2021 doi: 10.1093/bioadv/vbac048. 2021.02.13.431099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Partel G., Wählby C. Spage2vec: unsupervised representation of localized spatial gene expression signatures. FEBS J. 2021;288(6):1859–1870. doi: 10.1111/febs.15572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Littman R., et al. Joint cell segmentation and cell type annotation for spatial transcriptomics. Mol Syst Biol. 2021;17(6) doi: 10.15252/msb.202010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.10xGenomics. Space Ranger and Loupe Browser. 2020; Available from: 〈https://support.10xgenomics.com/spatial-gene-expression/software/overview/welcome〉.
- 64.Stuart T., et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902. doi: 10.1016/j.cell.2019.05.031. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Korsunsky I., et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12):1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hie B., Bryson B., Berger B. Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat Biotechnol. 2019;37(6):685–691. doi: 10.1038/s41587-019-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lopez R., et al. Deep generative modeling for single-cell transcriptomics. Nat Methods. 2018;15(12):1053–1058. doi: 10.1038/s41592-018-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tran H.T.N., et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020;21(1):12. doi: 10.1186/s13059-019-1850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luecken M.D., et al. Benchmarking atlas-level data integration in single-cell genomics. Nat Methods. 2022;19(1):41–50. doi: 10.1038/s41592-021-01336-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hong D.K., et al. Optimally weighted PCA for high-dimensional heteroscedastic data. arXiv: Stat Theory. 2018 doi: 10.1016/j.jmva.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maaten Lvd, Hinton G.E. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 72.McInnes L., Healy J. UMAP: uniform manifold approximation and projection for dimension reduction. ArXiv. 2018 abs/1802.03426. [Google Scholar]
- 73.Shang L., Zhou X. Spatially aware dimension reduction for spatial transcriptomics. bioRxiv. 2022 doi: 10.1038/s41467-022-34879-1. 2022.01.19.476966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blondel V.D., et al. Fast unfolding of communities in large networks. J Stat Mech: Theory Exp. 2008;2008:P10008. [Google Scholar]
- 75.Traag V.A., Waltman L., Eck N.Jv. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep. 2019;9 doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hartigan, J.A. and M.A. Wong. A k-means clustering algorithm. 1979.
- 77.Ng A., Jordan M.I., Weiss Y. On spectral clustering: analysis and an algorithm. NIPS. 2001 [Google Scholar]
- 78.Fraley C., Raftery A.E. Model-based clustering, discriminant analysis, and density estimation. J Am Stat Assoc. 2002;97:611–631. [Google Scholar]
- 79.Yang Y., et al. SC-MEB: spatial clustering with hidden Markov random field using empirical Bayes. Brief Bioinform. 2022;23:1. doi: 10.1093/bib/bbab466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao E., et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat Biotechnol. 2021;39(11):1375–1384. doi: 10.1038/s41587-021-00935-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu J., et al. SpaGCN: integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network. Nat Methods. 2021;18(11):1342–1351. doi: 10.1038/s41592-021-01255-8. [DOI] [PubMed] [Google Scholar]
- 82.Liu W., et al. Joint dimension reduction and clustering analysis of single-cell RNA-seq and spatial transcriptomics data. Nucleic Acids Res. 2022;50(12) doi: 10.1093/nar/gkac219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Longo S.K., et al. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat Rev Genet. 2021;22(10):627–644. doi: 10.1038/s41576-021-00370-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wei R., et al. Spatial charting of single-cell transcriptomes in tissues. Nat Biotechnol. 2022 doi: 10.1038/s41587-022-01233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kleshchevnikov V., et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat Biotechnol. 2022;40(5):661–671. doi: 10.1038/s41587-021-01139-4. [DOI] [PubMed] [Google Scholar]
- 86.Song Q., Su J. DSTG: deconvoluting spatial transcriptomics data through graph-based artificial intelligence. Brief Bioinform. 2021;22:5. doi: 10.1093/bib/bbaa414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Svensson V., Teichmann S.A., Stegle O. SpatialDE: identification of spatially variable genes. Nat Methods. 2018;15(5):343–346. doi: 10.1038/nmeth.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sun S., Zhu J., Zhou X. Statistical analysis of spatial expression patterns for spatially resolved transcriptomic studies. Nat Methods. 2020;17(2):193–200. doi: 10.1038/s41592-019-0701-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dries R., et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 2021;22(1):78. doi: 10.1186/s13059-021-02286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miller B.F., et al. Characterizing spatial gene expression heterogeneity in spatially resolved single-cell transcriptomic data with nonuniform cellular densities. Genome Res. 2021;31(10):1843–1855. doi: 10.1101/gr.271288.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moehlin J., et al. Inferring biologically relevant molecular tissue substructures by agglomerative clustering of digitized spatial transcriptomes with multilayer. Cell Syst. 2021;12(7):694–705. doi: 10.1016/j.cels.2021.04.008. e3. [DOI] [PubMed] [Google Scholar]
- 92.Chang Y., et al. Define and visualize pathological architectures of human tissues from spatially resolved transcriptomics using deep learning. bioRxiv. 2021 doi: 10.1016/j.csbj.2022.08.029. 2021.07.08.451210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pham, D., et al., stLearn: integrating spatial location, tissue morphology and gene expression to find cell types, cell-cell interactions and spatial trajectories within undissociated tissues. 2020.
- 94.Cang Z., Nie Q. Inferring spatial and signaling relationships between cells from single cell transcriptomic data. Nat Commun. 2020;11(1):2084. doi: 10.1038/s41467-020-15968-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jerby-Arnon, L. and A. Regev, Mapping multicellular programs from single-cell profiles. bioRxiv, 2020: p. 2020.08.11.245472.
- 96.Arnol D., et al. Modeling cell-cell interactions from spatial molecular data with spatial variance component analysis. Cell Rep. 2019;29(1):202–211. doi: 10.1016/j.celrep.2019.08.077. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ghazanfar S., et al. Investigating higher-order interactions in single-cell data with scHOT. Nat Methods. 2020;17(8):799–806. doi: 10.1038/s41592-020-0885-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bergenstråhle J., Larsson L., Lundeberg J. Seamless integration of image and molecular analysis for spatial transcriptomics workflows. BMC Genom. 2020;21(1):482. doi: 10.1186/s12864-020-06832-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zeira R., et al. Alignment and integration of spatial transcriptomics data. Nat Methods. 2022;19(5):567–575. doi: 10.1038/s41592-022-01459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lin Y., Yang J.Y.H. 3D reconstruction of spatial expression. Nat Methods. 2022;19(5):526–527. doi: 10.1038/s41592-022-01476-5. [DOI] [PubMed] [Google Scholar]
- 101.Dong K., Zhang S. Deciphering spatial domains from spatially resolved transcriptomics with an adaptive graph attention auto-encoder. Nat Commun. 2022;13(1):1739. doi: 10.1038/s41467-022-29439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eisenstein M. Seven technologies to watch in 2022. Nature. 2022;601(7894):658–661. doi: 10.1038/d41586-022-00163-x. [DOI] [PubMed] [Google Scholar]
- 103.Lewis S.M., et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods. 2021;18(9):997–1012. doi: 10.1038/s41592-021-01203-6. [DOI] [PubMed] [Google Scholar]
- 104.Wu Y., et al. Spatial omics: navigating to the golden era of cancer research. Clin Transl Med. 2022;12(1) doi: 10.1002/ctm2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang J., et al. Spatiotemporal Omics-refining the landscape of precision medicine. Life Med. 2022 [Google Scholar]
- 106.Raufaste-Cazavieille V., Santiago R., Droit A. Multi-omics analysis: paving the path toward achieving precision medicine in cancer treatment and immuno-oncology. Front Mol Biosci. 2022;9 doi: 10.3389/fmolb.2022.962743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie P., et al. Neddylation of PTEN regulates its nuclear import and promotes tumor development. Cell Res. 2021;31(3):291–311. doi: 10.1038/s41422-020-00443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bhatia H.S., et al. Spatial proteomics in three-dimensional intact specimens. Cell. 2022;185(26):5040–5058. doi: 10.1016/j.cell.2022.11.021. e19. [DOI] [PubMed] [Google Scholar]
- 109.Turajlic S., et al. Resolving genetic heterogeneity in cancer. Nat Rev Genet. 2019;20(7):404–416. doi: 10.1038/s41576-019-0114-6. [DOI] [PubMed] [Google Scholar]
- 110.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 111.Lyubetskaya A., et al. Assessment of spatial transcriptomics for oncology discovery. Cell Rep Methods. 2022;2(11) doi: 10.1016/j.crmeth.2022.100340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Melzer C., et al. Cancer stem cell niche models and contribution by mesenchymal stroma/stem cells. Mol Cancer. 2017;16(1):28. doi: 10.1186/s12943-017-0595-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Galeano Niño J.L., et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. 2022;611(7937):810–817. doi: 10.1038/s41586-022-05435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fang W., et al. Intratumoral heterogeneity as a predictive biomarker in anti-PD-(L)1 therapies for non-small cell lung cancer. Mol Cancer. 2021;20(1):37. doi: 10.1186/s12943-021-01331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hinshaw D.C., Shevde L.A. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79(18):4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hu B., et al. A review of spatial profiling technologies for characterizing the tumor microenvironment in immuno-oncology. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.996721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Perco P., Mayer G. The kidney-more than the sum of its cellular parts. Kidney Int. 2022;102(6):1217–1219. doi: 10.1016/j.kint.2022.08.032. [DOI] [PubMed] [Google Scholar]
- 118.Wu S.Z., et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021;53(9):1334–1347. doi: 10.1038/s41588-021-00911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Andersson A., et al. Spatial deconvolution of HER2-positive breast cancer delineates tumor-associated cell type interactions. Nat Commun. 2021;12(1):6012. doi: 10.1038/s41467-021-26271-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Thrane K., et al. Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III cutaneous malignant melanoma. Cancer Res. 2018;78(20):5970–5979. doi: 10.1158/0008-5472.CAN-18-0747. [DOI] [PubMed] [Google Scholar]
- 121.Asp M., et al. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell. 2019;179(7):1647–1660. doi: 10.1016/j.cell.2019.11.025. e19. [DOI] [PubMed] [Google Scholar]
- 122.Misra A., et al. Characterizing neonatal heart maturation, regeneration, and scar resolution using spatial transcriptomics. J Cardiovasc Dev Dis. 2021;9:1. doi: 10.3390/jcdd9010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Garcia-Alonso L., et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat Genet. 2021;53(12):1698–1711. doi: 10.1038/s41588-021-00972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hasel P., et al. Neuroinflammatory astrocyte subtypes in the mouse brain. Nat Neurosci. 2021;24(10):1475–1487. doi: 10.1038/s41593-021-00905-6. [DOI] [PubMed] [Google Scholar]
- 125.Tian L., Chen F., Macosko E.Z. The expanding vistas of spatial transcriptomics. Nat Biotechnol. 2022 doi: 10.1038/s41587-022-01448-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Genshaft A.S., et al. Live cell tagging tracking and isolation for spatial transcriptomics using photoactivatable cell dyes. Nat Commun. 2021;12(1):4995. doi: 10.1038/s41467-021-25279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Takei Y., et al. Integrated spatial genomics reveals global architecture of single nuclei. Nature. 2021;590(7845):344–350. doi: 10.1038/s41586-020-03126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Deng Y., et al. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature. 2022 doi: 10.1038/s41586-022-05094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lundberg E., Borner G.H.H. Spatial proteomics: a powerful discovery tool for cell biology. Nat Rev Mol Cell Biol. 2019;20(5):285–302. doi: 10.1038/s41580-018-0094-y. [DOI] [PubMed] [Google Scholar]
- 130.Regev A., et al. The human cell atlas. eLife. 2017;6 doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ecker J.R., et al. The BRAIN initiative cell census consortium: lessons learned toward generating a comprehensive brain cell atlas. Neuron. 2017;96(3):542–557. doi: 10.1016/j.neuron.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Börner K., et al. Anatomical structures, cell types and biomarkers of the human reference atlas. Nat Cell Biol. 2021;23(11):1117–1128. doi: 10.1038/s41556-021-00788-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.HuBMAP Consortium. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature, 2019. 574(7777): p. 187–192. [DOI] [PMC free article] [PubMed]
- 134.El-Achkar T.M., et al. A multimodal and integrated approach to interrogate human kidney biopsies with rigor and reproducibility: guidelines from the kidney precision medicine project. Physiol Genom. 2021;53(1):1–11. doi: 10.1152/physiolgenomics.00104.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Himmelstein D.S., et al. Systematic integration of biomedical knowledge prioritizes drugs for repurposing. eLife. 2017;6 doi: 10.7554/eLife.26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Srivastava S., et al. The making of a precancer atlas: promises, challenges, and opportunities. Trends Cancer. 2018;4(8):523–536. doi: 10.1016/j.trecan.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 137.Moghe I., Loupy A., Solez K. The human cell atlas project by the numbers: relationship to the banff classification. Am J Transpl. 2018;18(7):1830. doi: 10.1111/ajt.14757. [DOI] [PubMed] [Google Scholar]
- 138.Rozenblatt-Rosen O., et al. The human cell atlas: from vision to reality. Nature. 2017;550(7677):451–453. doi: 10.1038/550451a. [DOI] [PubMed] [Google Scholar]
- 139.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet, 2013. 45(6): p. 580–585. [DOI] [PMC free article] [PubMed]
- 140.McMahon A.P., et al. GUDMAP: the genitourinary developmental molecular anatomy project. J Am Soc Nephrol. 2008;19(4):667–671. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- 141.Liao J., et al. De novo analysis of bulk RNA-seq data at spatially resolved single-cell resolution. Nat Commun. 2022;13(1):6498. doi: 10.1038/s41467-022-34271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Qi J., et al. Single-cell and spatial analysis reveal interaction of FAP(+) fibroblasts and SPP1(+) macrophages in colorectal cancer. Nat Commun. 2022;13(1):1742. doi: 10.1038/s41467-022-29366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.La Manno G., et al. Molecular architecture of the developing mouse brain. Nature. 2021;596(7870):92–96. doi: 10.1038/s41586-021-03775-x. [DOI] [PubMed] [Google Scholar]
- 144.Trapnell C., et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wolf F.A., et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019;20(1):59. doi: 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bendall S.C., et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157(3):714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bergen V., et al. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020;38(12):1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 148.Qiu X., et al. Mapping transcriptomic vector fields of single cells. Cell. 2022;185(4):690–711. doi: 10.1016/j.cell.2021.12.045. e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.La Manno G., et al. RNA velocity of single cells. Nature. 2018;560(7719):494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Abdelaal, T., et al., SIRV: Spatial inference of RNA velocity at the single-cell resolution. 2021. [DOI] [PMC free article] [PubMed]
- 151.Petukhov V., et al. Cell segmentation in imaging-based spatial transcriptomics. Nat Biotechnol. 2022;40(3):345–354. doi: 10.1038/s41587-021-01044-w. [DOI] [PubMed] [Google Scholar]
- 152.Park J., et al. Cell segmentation-free inference of cell types from in situ transcriptomics data. Nat Commun. 2021;12(1):3545. doi: 10.1038/s41467-021-23807-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kueckelhaus J., et al. Inferring spatially transient gene expression pattern from spatial transcriptomic studies. bioRxiv. 2020 2020.10.20.346544. [Google Scholar]
- 155.Queen R., et al. Spaniel: analysis and interactive sharing of Spatial Transcriptomics data. bioRxiv. 2019 [Google Scholar]
- 156.Bergenstråhle J., Bergenstråhle L., Lundeberg J. SpatialCPie: an R/Bioconductor package for spatial transcriptomics cluster evaluation. BMC Bioinform. 2020;21(1):161. doi: 10.1186/s12859-020-3489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Patrick E., et al. Spatial analysis for highly multiplexed imaging data to identify tissue microenvironments. bioRxiv. 2021 doi: 10.1002/cyto.a.24729. 2021.08.16.456469. [DOI] [PubMed] [Google Scholar]
- 158.Andersson A., et al. Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography. Commun Biol. 2020;3(1):565. doi: 10.1038/s42003-020-01247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lopez R., et al. Multi-resolution deconvolution of spatial transcriptomics data reveals continuous patterns of inflammation. bioRxiv. 2021 2021.05.10.443517. [Google Scholar]
- 160.Yang T., et al. AdRoit is an accurate and robust method to infer complex transcriptome composition. Commun Biol. 2021;4(1):1218. doi: 10.1038/s42003-021-02739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cable D.M., et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat Biotechnol. 2022;40(4):517–526. doi: 10.1038/s41587-021-00830-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Danaher P., et al. Advances in mixed cell deconvolution enable quantification of cell types in spatial transcriptomic data. Nat Commun. 2022;13(1):385. doi: 10.1038/s41467-022-28020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ren H., et al. Identifying multicellular spatiotemporal organization of cells with SpaceFlow. Nat Commun. 2022;13(1):4076. doi: 10.1038/s41467-022-31739-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Elosua-Bayes M., et al. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res. 2021;49(9) doi: 10.1093/nar/gkab043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Teng H., Yuan Y., Bar-Joseph Z. Clustering spatial transcriptomics data. Bioinformatics. 2021;38(4):997–1004. doi: 10.1093/bioinformatics/btab704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dong R., Yuan G.C. SpatialDWLS: accurate deconvolution of spatial transcriptomic data. Genome Biol. 2021;22(1):145. doi: 10.1186/s13059-021-02362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sun D., et al. STRIDE: accurately decomposing and integrating spatial transcriptomics using single-cell RNA sequencing. Nucleic Acids Res. 2022;50(7) doi: 10.1093/nar/gkac150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Miller B.F., et al. Reference-free cell type deconvolution of multi-cellular pixel-resolution spatially resolved transcriptomics data. Nat Commun. 2022;13(1):2339. doi: 10.1038/s41467-022-30033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Biancalani T., et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat Methods. 2021;18(11):1352–1362. doi: 10.1038/s41592-021-01264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhu Q., et al. Identification of spatially associated subpopulations by combining scRNAseq and sequential fluorescence in situ hybridization data. Nat Biotechnol. 2018 doi: 10.1038/nbt.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.BinTayyash N., et al. Non-parametric modelling of temporal and spatial counts data from RNA-seq experiments. Bioinformatics. 2021 doi: 10.1093/bioinformatics/btab486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li Q., et al. Bayesian modeling of spatial molecular profiling data via Gaussian process. Bioinformatics. 2021 doi: 10.1093/bioinformatics/btab455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hao M., Hua K., Zhang X. SOMDE: a scalable method for identifying spatially variable genes with self-organizing map. Bioinformatics. 2021 doi: 10.1093/bioinformatics/btab471. [DOI] [PubMed] [Google Scholar]
- 174.Govek K.W., Yamajala V.S., Camara P.G. Clustering-independent analysis of genomic data using spectral simplicial theory. PLOS Comput Biol. 2019;15(11) doi: 10.1371/journal.pcbi.1007509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Edsgärd D., Johnsson P., Sandberg R. Identification of spatial expression trends in single-cell gene expression data. Nat Methods. 2018;15(5):339–342. doi: 10.1038/nmeth.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vandenbon A., Diez D. A clustering-independent method for finding differentially expressed genes in single-cell transcriptome data. Nat Commun. 2020;11(1):4318. doi: 10.1038/s41467-020-17900-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang K., Feng W., Wang P. Identification of spatially variable genes with graph cuts. bioRxiv. 2018 doi: 10.1038/s41467-022-33182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhu J., Sun S., Zhou X. SPARK-X: non-parametric modeling enables scalable and robust detection of spatial expression patterns for large spatial transcriptomic studies. Genome Biol. 2021;22(1):184. doi: 10.1186/s13059-021-02404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Anderson A., Lundeberg J. sepal: identifying transcript profiles with spatial patterns by diffusion-based modeling. Bioinformatics. 2021;37(17):2644–2650. doi: 10.1093/bioinformatics/btab164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yuan Y., Bar-Joseph Z. GCNG: graph convolutional networks for inferring gene interaction from spatial transcriptomics data. Genome Biol. 2020;21(1):300. doi: 10.1186/s13059-020-02214-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tanevski J., et al. Explainable multiview framework for dissecting spatial relationships from highly multiplexed data. Genome Biol. 2022;23(1):97. doi: 10.1186/s13059-022-02663-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Li D., Ding J., Bar-Joseph Z. Identifying signaling genes in spatial single-cell expression data. Bioinformatics. 2021;37(7):968–975. doi: 10.1093/bioinformatics/btaa769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Palla G., et al. Squidpy: a scalable framework for spatial omics analysis. Nat Methods. 2022;19(2):171–178. doi: 10.1038/s41592-021-01358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Partel G., et al. Automated identification of the mouse brain’s spatial compartments from in situ sequencing data. BMC Biol. 2020;18(1):144. doi: 10.1186/s12915-020-00874-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Welch J.D., et al. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell. 2019;177(7):1873–1887. doi: 10.1016/j.cell.2019.05.006. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lotfollahi M., Wolf F.A., Theis F.J. scGen predicts single-cell perturbation responses. Nat Methods. 2019;16(8):715–721. doi: 10.1038/s41592-019-0494-8. [DOI] [PubMed] [Google Scholar]
- 187.Lee J.H. Quantitative approaches for investigating the spatial context of gene expression. Wiley Inter Rev Syst Biol Med. 2017;9:2. doi: 10.1002/wsbm.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Meylan M., et al. Tertiary lymphoid structures generate and propagate anti-tumor antibody-producing plasma cells in renal cell cancer. Immunity. 2022;55(3):527–541. doi: 10.1016/j.immuni.2022.02.001. e5. [DOI] [PubMed] [Google Scholar]
- 189.Maynard K.R., et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. bioRxiv. 2020 doi: 10.1038/s41593-020-00787-0. 2020.02.28.969931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tan H.H., Bansal M. LXMERT: learning cross-modality encoder representations from transformers. ArXiv. 2019 abs/1908.07490. [Google Scholar]
- 191.Baltrušaitis T., Ahuja C., Morency L.P. Multimodal machine learning: a survey and taxonomy. IEEE Trans Pattern Anal Mach Intell. 2019;41(2):423–443. doi: 10.1109/TPAMI.2018.2798607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material