Abstract
Therapeutic protein, represented by antibodies, is of increasing interest in human medicine. However, clinical translation of therapeutic protein is still largely hindered by different aspects of developability, including affinity and selectivity, stability and aggregation prevention, solubility and viscosity reduction, and deimmunization. Conventional optimization of the developability with widely used methods, like display technologies and library screening approaches, is a time and cost-intensive endeavor, and the efficiency in finding suitable solutions is still not enough to meet clinical needs. In recent years, the accelerated advancement of computational methodologies has ushered in a transformative era in the field of therapeutic protein design. Owing to their remarkable capabilities in feature extraction and modeling, the integration of cutting-edge computational strategies with conventional techniques presents a promising avenue to accelerate the progression of therapeutic protein design and optimization toward clinical implementation. Here, we compared the differences between therapeutic protein and small molecules in developability and provided an overview of the computational approaches applicable to the design or optimization of therapeutic protein in several developability issues.
Keywords: Therapeutic protein, Computational approaches, Protein design, Artificial intelligence, Molecular dynamics
Graphical Abstract
1. Introduction
The advancement of molecular biology and bioengineering in the past three decades has led to the success of protein-based drugs, resulting in their widespread popularity and significant clinical effectiveness in the expanding global market [1], [2]. The shift in drug discovery has led to an increased prevalence of protein-based therapeutics, with their proportion rising significantly since the 1980 s [3]. FDA-approved protein-based drugs for cancer, autoimmune diseases, and wet macular degeneration now exceed 200 [4], [5]. Four of the top 10 global drugs sold in 2021 are protein-based, including Humira®, Keytruda®, Eylea®, Stelara®, and Opdivo®, all of which are antibodies [6]. Protein-based therapeutics offer a myriad of advantages over conventional small molecules drug, such as high affinity, specificity, and potency, as well as low toxicity and minimal adverse effects, and these advantages allow the protein-based drugs against undruggable targets for previously unresponsive small molecules [7].
Considerable efforts are being directed toward the development of protein-based therapeutics, however, the rate of new protein-based therapeutic approvals has reached a plateau, and there is a possibility of deceleration [4]. The primary cause of protein-based therapeutic development failure can be attributed to the inadequate developability of these therapeutics [8], [9], [10], [11], which encompasses factors such as affinity and selectivity [12], inherent physical and/or chemical stability [9], aggregation tendency [13], solubility and concentration, viscosity [14], manufacturability, and immunogenicity [15]. Inadequate developability will be the major cause of failures in preclinical models and clinical trials. Therefore, the development of protein-based therapeutics is essentially a multi-objective optimization process, involving the optimization of the properties mentioned above [16].
To increase the success rate and reduce costs, it is crucial to evaluate and improve the developability profile in the early stages of the drug discovery [3], [17], [18], [19], it is imperative to gain a comprehensive understanding of the physicochemical and biological attributes that govern their developability. Nevertheless, the complex nature of protein-based therapeutics, which encompasses structural and formulation intricacies, presents obstacles in elucidating their physicochemical and biological properties pertinent to drug development. The elaborate architecture of proteins renders them susceptible to numerous factors throughout the drug development process, subsequently leading to an array of downstream complications in the formulation of protein-based therapeutics for clinical application [20]. Slight modifications, such as single amino acid substitutions, can lead to unpredictable changes in the characteristics of protein drugs and formulations [21]. It is difficult to assess the developability of protein drug formulations in the early stages using existing knowledge, such as stability and aggregation kinetics [19], [22], [23]. Therefore, there is still a lack of widely recognized guidelines based on the understanding of drug entities in protein-based therapeutics to accelerate the development of protein-based therapeutics, like Lipinski's rules and the biopharmaceutics classification system exist for the small molecule formulations [24], [25].
Given the considerably larger search space of protein drugs and the complexity of protein entities, advanced methods are necessary to facilitate the development of protein-based therapeutics and optimization of the developability [26]. Experiencing rapid development, computational approaches possess strong feature extraction and modeling abilities that enable them to comprehend the complex rules governing protein properties, provide a multi-scale view for pharmaceutical scientists [27], [28], [29], [30], accelerate the development of protein-based therapeutics, and reduce costs compared to traditional experimental methods [28], [30]. Therefore, utilizing computational approaches offers great potential for changing the current paradigm of protein-based therapeutic development, and the low costs and high speed of computational approaches hold promise to accelerate the development process of protein drugs toward clinical trials.
Briefly, the computational methods in drug discovery fall into two categories: physics-based multi-scale modeling methods [31] (e.g. Rosetta [32], molecular dynamics simulation [33], [34], molecular docking [35]) and data-driven methods based on big data and data-mining (e.g. artificial intelligence, deep learning, machine learning) [36], [37], [38], [39]. These two methods have different characteristics and applicable scenarios, and a comparison of them is shown in Table 1. Aggrescan3D developed by Aleksander Kuriata et al. is an example of physics-based multi-scale modeling techniques [40]. Aggrescan3D is a structure-based computational approach to predict aggregation properties that can precisely detect protein aggregation-prone areas, surpassing conventional sequence-based method, and offer the ability to mutate predictive aggregation-prone regions to diminish aggregation tendency and improve solubility, which is an effective strategy for rational protein-based therapeutic design towards clinical applications. ProteinMPNN from David Baker’s lab is a representative of data-driven methods used in the protein drug optimization [41]. ProteinMPNN demonstrated the ability to salvage previously unsuccessful protein designs, highlighting the potential of computational methods in protein-based therapeutic development and can improve protein design efficiency with outstanding performance [41]. Apart from optimizing protein design and development, computational techniques also hold tremendous potential for biopharmaceutical process optimization and bioprocess scale-up control [42], [43].
Table 1.
Comparison of physics-based multi-scale modeling methods and data-driven methods based on big data and data-mining.
| Physics-based methods | Data-driven methods | |
|---|---|---|
| Basis | Laws of physics and chemistry | Big data |
| Accuracy | High | Highly data dependent |
| Efficiency | Low, requiring high computational resources | High, requiring relatively low computational resources in most cases |
| Scale | Low | High |
| Applicability | Target structure is necessary | Target structure is unnecessary |
| Interpretability | High, termed as “white-box” tool | Low, termed as “black-box” tool |
| Transferability | Low, requiring customization and calibration in a new system | High |
In the present review, we consider protein-based therapeutics, which include protein drugs like monoclonal antibodies, fusion proteins, interferons, interleukins, enzymes, and hormones [44], as well as protein-based drug delivery systems like albumin-bound paclitaxel [45], [46], [47], [48], antibody-drug conjugates [49], [50], and protein-based nanotherapeutics [51]. Additionally, novel protein formulations such as nanodiscs [52], nanoparticles (NPs) preincubated with serum [53], [54], and those decorated or modified with protein will also be discussed as protein-based therapeutics [55], [56], [57]. This article primarily examines the utilization of computational techniques in the development of protein-based therapeutics, starting with a comparison of small molecules and protein-based therapeutics with differing attributes and considerations for clinical viability, followed by an exploration of the applications of computational approaches in various aspects of protein-based therapeutic development, including affinity and selectivity, stability and aggregation prevention, solubility and viscosity reduction, and deimmunization, ultimately concluding with a discussion of the future direction of computational methods in the clinical application of protein-based therapeutics.
2. Comparison of developability between small molecules and protein-based therapeutics
Small molecule-based therapies have played a significant role in advancing medicine and enhancing patients' quality of life for many centuries [6], [58]. Considerable resources and endeavors have been dedicated to the advancement of small-molecule drugs, culminating in the establishment of guidelines for optimizing development and facilitating translation to clinical applications. Notable examples of such guidance include Lipinski's rules and the Biopharmaceutics Classification System (BCS) [24], [25]. These principles have linked the physicochemical and biopharmacological properties of small molecules to their developability, including solubility, hydrophobicity, penetration, stability, and crystallization properties [59]. Due to their ease of use and conceptual simplicity, these principles have become the primary indicators of developability in small molecule discovery, leading to a reduction in drug discovery and development attrition [59]. Small molecule drugs have properties such as high stability, the ability to diffuse through biological barriers, and the capability to interact with various biological targets [60], [61], [62], [63], making oral delivery possible. Therefore, the primary challenge for small molecules is to improve oral bioavailability since oral administration is the most common and preferred route due to its safety, cost-effectiveness, and ease of use [64].
The emergence of protein-based therapeutics has fundamentally changed administration routes and developability properties due to their complex structure [65]. Although they offer high therapeutic efficacy and minimal side effects, their stability in biological environments is low, resulting in poor oral bioavailability [60]. Consequently, protein drugs require invasive injection or infusion routes in the clinical practice [66]. Owing to the disparities in physicochemical and biopharmaceutical properties, as well as administration routes, substantial variations exist in the developability of protein-based therapeutics compared to small molecules. Consequently, the developability principles governing small molecule drugs cannot be directly extrapolated to protein-based therapeutics [9].
Different administration routes and formulation types have specific requirements for the developability of the protein-based therapeutics [67]. Liquid preparations are the most common formulation type, including redissolution formulations prepared through advanced industrial processes such as freeze-drying and spray-drying [68], [69]. The primary objective in developing protein drugs and formulations is to identify suitable solution conditions that maintain their activity and injectability while undergoing various processes before clinical use. The main consideration in formulation design is that proteins are sensitive to heat, pH, shear stress, organic solvents, and agitation [44]. Intravenous injection is the predominant administration route for protein drugs due to its high bioavailability. However, it has significant drawbacks, including inconvenience, medical burdens, and the risk of adverse reactions. Subcutaneous injection is a promising alternative with benefits such as improved patient convenience and acceptance, self-administration at home, and reduced healthcare costs [66], [70]. Nonetheless, it has specific developability requirements, such as low injection volume and high concentration, which may lead to aggregation, poor solubility, opalescence, and high viscosity [44], [71]. Similar requirements are needed for intramuscular injection and other specialized administration routes [14], [72], such as intravitreal injection, which also requires high-concentration formulations due to limited volume [73]. Small molecules and protein drugs require different formulation approaches due to their distinct physicochemical and biopharmaceutical properties [74]. Unlike small molecules, developing high-concentration formulations for proteins is challenging due to their vulnerable structures, diverse surface properties, and propensity for unpredictable solution behavior, resulting in several developability issues, such as high viscosity, poor stability, and aggregation.
As protein-based therapeutics become more widely investigated in preclinical and clinical studies, the optimization of their developability comes at a significant cost in terms of time and resources. Consequently, there is a growing need for strategies that can predict and enhance the developability of these therapeutics at an early stage. In the upcoming section, we will explore the diverse developability of protein-based therapeutics and the utilization of computational methods for this purpose.
3. Computational methods for the rational design of protein-based therapeutics toward the clinical stage
A myriad of computational methods has been applied to design and optimize protein-based therapeutics toward the clinical stage, and recent methods have been summarized in Table 2. In this section, we will introduce these methods with different aspects of developability in protein-based therapeutics, including affinity and selectivity, stability and aggregation prevention, solubility, viscosity reduction, and deimmunization.
Table 2.
Summary of recent computational methods to optimize the protein-based therapeutics toward the clinical stage. (DevAsp: Developability Aspects).
| DevAsp. | Name | Objectives | Methods | Ref. |
|---|---|---|---|---|
| Affinity | DeepAAI | Predicting antibody neutralizability with antigen | Graph convolutional network (GCN) and Convolutional neural network (CNN) | [75] |
| RESP | Identification of high affinity antibodies | Autoencoder | [76] | |
| Machine learning-assisted directed evolution | Machine learning is used to quickly screen a full recombination library in silico by using sequence–fitness relationships randomly sampled from the library | K-nearest neighbors, linear regression, decision trees, random forests, and multilayer perceptrons | [77] | |
| GeoPPI | Predicting the change of binding affinity upon mutations | Self-supervised learning and gradient boosting tree (GBT) | [78] | |
| CeVICA | In vitro VHH domain antibody engineering and nanobody binder selection | CDR-directed clustering analysis | [79] | |
| Building a high-quality model of a protein’s fitness landscape and screening ten million sequences via in silico directed evolution | UniRep (an unsupervised deep learning model) and Lasso-LARS/Ridge/Ridge SR/Ensembled Ridge SR | [80] | ||
| CLADE | Guiding protein engineering and directed evolution | K-means, Louvain, and the ensemble of 17 regression models | [81] | |
| Ens-Grad | Designing CDR of human Immunoglobulin G antibodies with high affinities | Machine learning | [82] | |
| DLAB | Predicting antibody–antigen binding for antigens with no known antibody binders | Structure-based deep learning | [83] | |
| ProAffiMuSeq | Predicting the binding free energy change of protein–protein complexes upon mutation | Regression model | [84] | |
| DeepRank | Classification, e.g., predicting an input PPI as biological or a crystal artifact, and regression, e.g., predicting binding affinities | Convolutional Neural Networks (CNNs) | [85] | |
| mCSM-PPI2 | Predicting effects of missense mutations in protein–protein affinity | Machine learning | [86] | |
| mCSM-AB2 | Predicting the effects of missense mutations on Ab-binding affinity | Machine learning | [87] | |
| mmCSM-AB | Predicting multi-point mutations on antigen binding affinity | Machine learning | [88] | |
| mmCSM-PPI | Predicting changes in PPI binding affinity caused by multiple point mutations | Machine learning | [89] | |
| NetTree | Predicting PPI ΔΔG | Convolutional Neural Networks and gradient-boosting trees | [90] | |
| Ymir | Calculating in silico antibody-antigen affinities | 3D-lattice-based framework | [91] | |
| MutaBind2 | Predict binding affinity changes upon single and multiple mutations | Random forest | [92] | |
| SSIPe | Quantitative estimation of the binding affinity changes (ΔΔGbind) | Protein interface profiles and a physics-based energy function | [93] | |
| EasyE and JayZ | Binding affinity estimation | Guaranteed Cost Function Network algorithms, Rosetta energy functions and Dunbrack’s rotamer library | [94] | |
| PPI-Affinity | Predicting binding affinity | Support Vector Machine | [95] | |
| TopologyNet | Predicting the protein-ligand binding affinities and protein stability changes upon mutation | Element-specific persistent homology (ESPH) method and Convolutional Neural Networks | [96] | |
| \ | Generation of high-affinity antibody sequences from low-N training data | Machine learning-based methods | [97] | |
| Predicting binder and non-binder antibodies | Convolutional Neural Networks | [98] | ||
| In silico affinity maturation | Homology Modelling and Protein Docking | [99] | ||
| Selectivity | Computational counterselection | Identifying nonspecific antibody candidates | Multi-task neural network | [100] |
| \ | Determining the polyreactivity status of a given sequence | Support vector machine (SVM) | [101] | |
| \ | Predicting antigen specificity | Deep neural networks | [102] | |
| \ | Assessing polyreactivity from protein sequence | Convolutional Neural Network and Recurrent Neural Network | [103] | |
| \ | Co-optimization of therapeutic antibody affinity and specificity | Deep Learning | [12] | |
| Stability | UniRep | Predicting the stability of natural and de novo designed proteins | Recurrent Neural Network | [104] |
| FoldArchitect | Predicting the stability of proteins | Random forest | [105] | |
| BayeStab | Predicting effects of mutations on protein stability | Graph Neural Network and Bayesian Neural Network | [106] | |
| DynaMut | Predicting the effects of missense mutations on protein stability | Random Forest with Graph-based signatures | [107] | |
| DynaMut2 | Predicting the effects of missense mutations on protein stability | Random Forest with Graph-based signatures | [108] | |
| DeepDDG | Prediction of changes in the stability of proteins due to point mutations | Neural Network | [109] | |
| Clustered tree regression | Predicting the mutation-induced protein folding free energy change | K-means and XGBoost | [110] | |
| PROST | Predicting the ΔΔG upon a single-point missense mutation | XGBoost and Extra-Trees | [111] | |
| PremPS | Evaluating the effects of missense mutations on protein stability | Random forest | [112] | |
| iStable 2.0 | Predicting protein thermal stability changes | Sequence- and structure-based tools | [113] | |
| PON-tstab | Predicting stability of protein variants | Random forests | [114] | |
| ProTstab2 | Predicting Protein Thermal Stabilities | Gradient boosting algorithm | [115] | |
| SimBa | Predicting protein stability changes upon mutations | Multilinear regression | [116] | |
| Scone | Predicting protein stability changes upon mutations | Neural network | [117] | |
| pStab | Predicting protein stability changes upon mutations | Debye–Hückel (DH) formalism | [118] | |
| \ | Predicting the spectra at the unfolding transition and denatured state | Artificial neural network | [119] | |
| \ | Predicting stability of protein variants | MM/GBSA and FEP+ | [120] | |
| \ | Understanding the Stabilizing Effect of Histidine on mAb Aggregation | All-atom molecular dynamics simulations and contact-based free energy calculations | [121] | |
| \ | Predicting protein stability change upon mutations | Variational Auto-Encoders | [122] | |
| \ | Designing mutations located at non-conserved residues with enhanced thermostability | Molecular Dynamics (MD) simulation and energy optimization methods | [123] | |
| \ | Design new sequences translating in functional proteins with enhanced thermal stability | Monte Carlo simulations and Molecular Dynamics (MD) simulation | [52] | |
| Aggregation prevention | Aggregation Time Machine | Assessing the long-term aggregation stability | Monte Carlo Analysis | [124] |
| MAPT | Prediction of antibody aggregation | \ | [125] | |
| ANuPP | Predicting Aggregation Nucleating Regions in peptides and proteins | Logistic regression and Bayesian approach | [126] | |
| MAGRE | Predicting regions prone to protein aggregation | Support Vector Machine | [127] | |
| CORDAX | Detecting APRs and predicts the structural topology and architecture of the fibril core | Logistic regression | [128] | |
| AgMata | Identification of regions involved in aggregation | Machine learning | [129] | |
| \ | Identifying aggregation rate enhancer and mitigator mutations in proteins | Support Vector Machine | [21] | |
| \ | Understanding the relationship between protein aggregation and molecular conformation | Molecular Dynamics (MD) simulation | [130] | |
| Solubility | Aggrescan3D 2.0 | Prediction and engineering of protein solubility | AGGRESCAN | [40] |
| solPredict | Antibody solubility prediction using sequence alone | Machine Learning | [131] | |
| CamSol | Predicting protein solubility | Phenomenological combination of several properties | [132] | |
| ProGAN | Predicting protein solubility | Deep Neural Network and Generative Adversarial Nets | [133] | |
| SKADE | Predicting protein solubility | Neural Network | [134] | |
| PaRSnIP | Predicting protein solubility | Gradient boosting machine | [135] | |
| DeepSol | Predicting protein solubility | Convolutional Neural Network | [136] | |
| SOLart | Predicting protein solubility | Random Forest | [137] | |
| PON-Sol2 | Identifying amino acid substitutions that increase, decrease, or have no effect on the protein solubility | Gradient Boosting algorithm | [138] | |
| SoluProt | Predicting protein solubility | Gradient Boosting algorithm | [139] | |
| SODA | Predicting protein solubility changes upon mutations | Kyte-Doolittle hydrophobicity profile and FESS | [140] | |
| \ | Predicting protein solubility | Machine Learning | [141] | |
| \ | Predicting protein solubility | Machine Learning | [142] | |
| \ | Increasing protein solubility | Variational AutoEncoders | [143] | |
| Viscosity | DeepSCM | Predicting protein viscosity | Convolutional Neural Network | [144] |
| \ | Optimization of highly viscous antibodies while maintaining binding affinity and favorable developability profile | Structure-based design | [145] | |
| \ | Predicting protein viscosity | Molecular Modeling and Machine Learning | [146] | |
| \ | Predicting protein viscosity | Multivariate Regression | [147] | |
| \ | Predicting protein viscosity | Molecular Dynamics (MD) simulation and Logistic regression | [13] | |
| \ | Predicting protein viscosity | Coarse-grained models | [148] | |
| \ | Predicting concentration-dependent viscosity curves | Stepwise linear regression | [149] | |
| \ | Selecting antibody candidates with desirable viscosity properties | Coarse-grained simulation | [150] | |
| Deimmunization and humanization | BioPhi | Humanization and humanness evaluation | Sapiens and OASis | [151] |
| Llamanade | Humanization of nanobodies | Large-scale analysis | [152] | |
| BepiPred-2.0 | Predicting B-cell epitopes | Random forest | [153] | |
| Hu-mAb | Suggesting mutations to an input sequence to reduce its immunogenicity | Random forest and LSTM | [154] | |
| iBCE-EL | Identification of B-cell epitopes (BCEs) | Extremely randomized tree (ERT) and Gradient boosting (GB) | [155] | |
| LBCEPred | Predicting B-cell epitopes | Random forest | [156] | |
| EpiSweep | Deimmunizing therapeutic protein | EpiSweep | [157] | |
| MHCEpitopeEnergy | Deimmunization | Monte Carlo-based rotamer packing and sequence design algorithm | [158] | |
| NetMHCpan-4.1 and NetMHCIIpan-4.0 | Predict binding between peptides and MHC-I and MHC-II | Machine learning | [159] | |
| \ | Deimmunization | Multi-objective combinatorial optimization | [160] | |
| \ | Predictions of antibody-specific epitope | Monte Carlo algorithm and machine learning | [161] | |
| \ | Reducing the immunogenicity | Emini surface accessibility, Parker hydrophilicity, and Karplus & Schulz flexibility methods and molecular dynamics simulation | [162] |
3.1. Affinity and selectivity
In protein-based therapeutics, the high affinity between biological targets and protein drugs is the fundamental requirement. Most of the development processes in protein-based therapeutics mainly focus on the improvement and optimization of affinity. For instance, many processes of antibody development are centered on affinity optimization, including animal immunization, hybridoma development, affinity maturation, directed evolution [163], and a variety of display methods or selection platforms [164], [165]. These powerful and effective methods for affinity optimization in protein drug development are able to produce protein drugs with nanomolar and even picomolar affinity [166], [167] or other desirable biological effects [168]. However, these technologies usually require multiple mutations of the starting protein for screening all possibilities as much as possible, which is time and cost-consuming [166], [169].
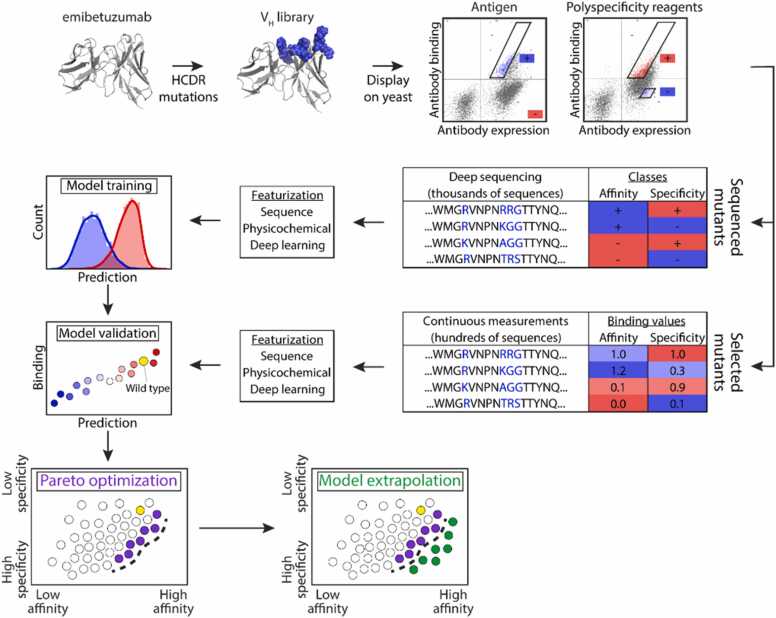
There are several studies combining the computational methods with these widely used processes for efficiency improvement and cost-saving in protein drug development or optimization. In the research of Emily K. Makowski and co-workers, they combine yeast display and machine learning for the improvement of antibody affinity without undesirable specificity (Fig. 1) [12]. The machine learning method used in this research is a powerful complementary to yeast display methods for antibody optimization. They found that the machine learning model trained by the data from yeast display can generalize to novel mutational space, meaning that researchers can firstly screen the ultra-large library of antibodies variants by time-saving machine learning method with high efficiency, and then select the candidates for subsequent processes like display screening and evaluation of other developability properties. The computational method, machine learning in this research, can explore a large range of possibilities of antibody variants, because of its time and cost-effectiveness. Similar research was finished by Derek M. Mason et al. [102]. They leveraged the deep learning model to screen a computational library of approximately 1 × 108 trastuzumab variants for high affinity against human epidermal growth factor receptor 2 and conducted multi-parameter optimization of other developability, which will be discussed later. The purpose of introducing computational methods in this research is also for time-saving and screening a large range of possibilities. Similarly, Xun Chen et al. combined the computational methods with the display method [79]. They utilized computational methods to aid the selection of the candidates of 1011 randomized sequences from ribosome display. Through a clustering algorithm based on sequence similarity, they grouped the result from ribosome display into several clusters. They assumed that each cluster represents a unique binding family, which means they can take one representative sequence in each cluster to characterize their specific downstream applications as a proof of concept and this screening mode will be efficient and provide a more comprehensive view of the landscape of binder potential. Joseph M.Taft et al. also utilized the data from display, the yeast surface display in their research, to construct a deep learning model for protein drug evaluation and optimization [170]. Unlike the previous three examples, they conduct combinatorial mutations on the receptor-binding domain (RBD, the target against SARS-CoV-2), but not on protein drug, to interrogate the impact of RBD mutations on ACE2 binding and escape from a panel of antibodies. This deep learning model can accurately predict antibody robustness to prospective SARS-CoV-2 variants and will be suitable for the evaluation of the antibody therapeutics for clinical translation. In addition to leveraging the data from display technology, computational methods can also aid the construction of the library for screening through a display or other methods [165]. GuyNimrod et al. utilized computational methods to guide the library design and conduct in vitro selection of these libraries, to design a functional antibody against the cytokine interleukin-17A (IL-17A) [171]. Through the structure of epitopes and paratopes, molecular docking, molecular dynamics simulation, and analysis of structural and energy, they rationally designed an antibody variants library, attempting to improve the complementarity with the desired epitope and enhance efficiency. There are also works using computational methods to conduct directed evolution [172], to avoid or accelerate the processes of expensive and time-consuming screening or selection of large mutational sequence space in traditional directed evolution [81]. In the study of Surojit Biswas and co-workers, they combine several computational methods, including unsupervised learning for pre-training and transfer learning with data from a few dozen mutants, to conduct the in silico directed evolution with a metropolis–hastings Markov Chain Monte Carlo algorithm to screen protein variants with improved affinity and function relative to wide type [80]. Zachary Wu et al. also introduced machine learning into the directed evolution workflow, to increase throughput with in silico modeling, reduce the expense of experimentally screening numerous candidates and improve the screening quality [173].
Fig. 1.
Overview of antibody library sorting, deep sequencing, and machine learning methods used to co-optimize the affinity and specificity of a therapeutic antibody [12]. Copyright 2022, Springer Nature.
Apart from improving the efficiency of traditional protein development processes, computational methods also revolutionize de novo protein design with high affinity, which can create novel protein drugs with the desired developability. The natural evolutionary process only sampled an infinitesimal subset of the hypothetical space of protein sequence and structure. More than 95% of protein optimization and engineering is still taking natural proteins, like an antibody from animal immunization, as starting points to mutate, modify and optimize, which can only explore the naturally occurring protein fold space and cannot fully explore the whole protein space for drug discovery [174], [175]. De novo protein design tries to explore the whole protein space and generate the proteins not found in nature, adopt desired structures, and perform novel and intriguing functions, like binding to the target with high affinity or acting as an enzyme [175]. The main goal of de novo protein design is finding a sequence folding to the desired structure satisfying the structural geometry, performing intended biological effects, and having suitable developability properties. High affinity is the primary consideration in the processes of de novo protein design and the main goal and criterion of design methods and protocols [176]. The vast search space of protein largely hinders the design of functional protein with empirical approaches or intuition, which should rely on other methods, like computational approaches [177]. In general, de novo protein design can be divided into four steps, including topology construction, backbone generation, sequence design or side chain optimization, and sequence-structure compatibility, and all of them are now performed mainly by advanced computational methods and the laws of physics and chemistry [178], [179]. Many excellent reviews summarized the computational methods used in different aspects of the de novo design [174], [178], [179], [180], [181], [182], [183], [184].
There are also some unconventional protein-based therapeutics having the requirement for high affinity, and the formulation of nanoparticles (NPs) with protein corona on the surface is one of them. A current assumption is that the formulation of NPs will influence the protein composition on NPs’ surface under the biological environment, which has a different affinity toward different receptors on cells, and therefore, the NPs with different formulations will be untaken by distinct cells and have disparate biodistribution [185]. Predicting the protein composition on the NPs’ surface or the NPs’ biological effect and biodistribution with computational methods is of significance to NP design. Leveraging a machine learning model, Zhan Ban et al. completed the prediction with a random forest model from various physicochemical parameters of NPs and environmental factors to protein composition on the NPs’ surface [186]. This model can further predict the cellular recognition of protein composition on NPs’ surfaces. James Lazarovits and co-workers developed a machine learning model to predict the biological fates of NPs in vivo (half-life, spleen accumulation, and liver accumulation) [187]. The input is the mass spectrometry protein library from the NPs isolated in multiple time points from circulation, which can predict the in vivo behaviors of NPs and dictate NPs’ interactions with cells and tissues in the body. Protein-based artificial nanosystem is another novel protein-based therapeutic with the requirement of high affinity. Yazan Haddad and co-workers developed an ellipticine-loaded ferritin with surface modification of the norepinephrine transporter (hNET) targeting peptides for tumor-targeting ability [188]. With computational methods, homology modeling, molecular docking, and molecular dynamics in this research, they found that these peptides showed an intriguing binding affinity with hNET. The hNET targeting ability of these peptides enabled rapid endocytosis of this protein-based nanosystem into neuroblastoma cells in a selective fashion, leading to apoptosis, cytotoxicity, and oncotherapy. Meiru Song et al. leveraged computational methods to study the drug vehicle of Doxorubicin (DOX), and human serum albumin (HSA) [189]. Molecular dynamics simulation elucidated the structural basis of DOX bound to HAS at different pH, providing new structural insights into pH-dependent interactions of HAS and DOX, which is useful in designing new microenvironment-responsive drug delivery systems.
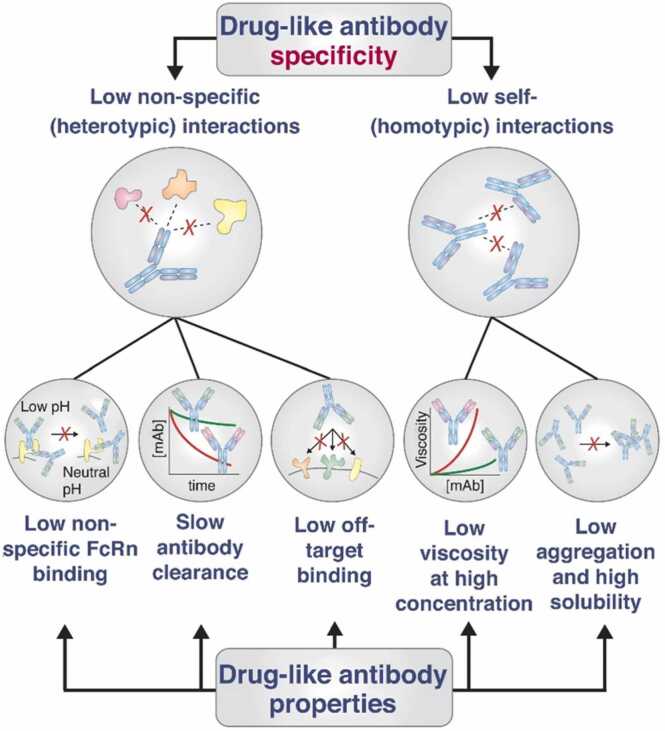
In addition to affinity, the developability of protein-based therapeutics is also significantly influenced by selectivity or specificity, which serve as crucial attributes. These factors are vital indicators for achieving success in clinical trials [8], [10]. The progression of protein-based therapeutics toward the clinical phase necessitates a delicate balance between elevated target selectivity and target binding affinity [12]. However, selectivity tends to be underemphasized in comparison to the affinity [190]. Screening out nonspecific protein drugs typically occurs late in the drug discovery process, which will lead to a waste of time and cost [100]. A prior investigation has demonstrated that selectivity serves as the most effective biophysical descriptor for predicting the successful translation of antibodies into clinical applications [10]. Selectivity refers to the relative propensity of protein drugs to interact with molecules other than their antigens or receptors, and the ideal selectivity mainly contains two aspects, low levels of antibody non-specific and self-interactions (Fig. 2) [190]. Both of them will largely influence the efficiency, safety, and other developability aspects (like viscosity and aggregation) of protein-based therapeutics toward the clinical stage [190]. For instance, the non-specific protein drug will cause off-target binding and result in limited therapeutic efficacy and safety problems or poor physicochemical properties like fast antibody clearance; the self-interactions of protein drug will lead to high viscosity, aggregation tendency, and low solubility, which will largely hinder the clinical translation and will be discussed later.
Fig. 2.
Drug-like protein drugs with high specificity have low levels of non-specific and self-interactions, which endow protein drugs with several properties including low non-specific FcRn binding, slow antibody clearance, low off-target binding, low viscosity and low aggregation, and high solubility [190]. Copyright 2019, Elsevier.
In the study conducted by Sachit Dinesh Saksena and colleagues, they employed machine learning models to facilitate computational counterselection, thereby pinpointing non-specificity sequences [100]. The computational counterselection in this research refers to a method taking sequencing data from affinity-selection experiments as data for machine learning training of nonspecific binding. Through training to jointly predict affinity to on and off-targets, the model can then be used to identify sequences that bind to the off-target molecule and remove these sequences. The efficacy of computational counterselection still needs to be validated in clinical trials in the future. In one of the previously mentioned examples in the part of affinity, Emily K. Makowski et al. not only applied machine learning to predict antibody affinity but also predict specificity (Fig. 1) [12]. To be specific, they treated the affinity between the antibody and soluble membrane proteins or ovalbumin as the index of non-specificity, because both of them are not the targets of these antibodies. Like predicting the affinity, they also acquired the data through yeast display and predicted the specificity through machine learning. Predicting both affinity and specificity enable authors to select the novel antibody with both low non-specificity and high affinity. Through the database of over 1000 polyreactive and non-polyreactive antibody sequences and bioinformatics-based analysis, Christopher T Boughter et al. successfully isolated key biophysical properties of polyreactivity, which is useful to guide the development and optimization of protein drugs [101]. They subsequently utilized these properties to design a generalizable machine learning-based classification software, which can take sequences as input and output the non-specificity property.
3.2. Stability and aggregation prevention
A prevailing tendency in the advancement of protein-based therapeutics involves low-volume and high-concentration formulations, which hold considerable significance in the biopharmaceutical sector [74]. There are numerous advantages of developing high-concentration formulation, including but not limited to the availability of subcutaneous delivery with syringes or autoinjectors, increased patient comfort, improved patient compliance, at-home delivery, and reduced healthcare costs. Even with intriguing properties, there are many severe problems in developing high-concentration formulations, such as poor stability, forming reversible or irreversible aggregates, limited solubility, and high viscosity, both of them will pose challenges in the manufacturing and delivery of protein-based therapeutics.
Proteins are complicated molecules and have a fine three-dimensional structure. This structure endows protein with many biological functions and pharmaceutical effects but makes the protein inherently unstable and sensitive to environmental conditions [9], [191]. Therefore, stability is one of the major and considerable factors in protein-based therapeutics development [192]. The basic requirement of protein drugs is highly stable so that they can tolerate the extreme conditions of manufacturing processes, like filtration and filling, and remain stable and active during transportation, storage, and administration [193]. In addition, the stability of protein will be the prerequisite for high yield in protein production and manufacture. Stability is the fundamental factor for other developability properties, and the instability will cause several issues, not only the ineffective efficacy of therapeutics but also some safety problem like immunogenic responses. Instability will also cause increased health charges, like the demand for cold chain maintenance, and therefore increase the cost per dose. Prediction, detection, mechanistic understanding of protein instabilities, and optimization of protein drug stability with relative cost and time-friendly methods in the early stage will make a difference in the drug discovery [194], [195].
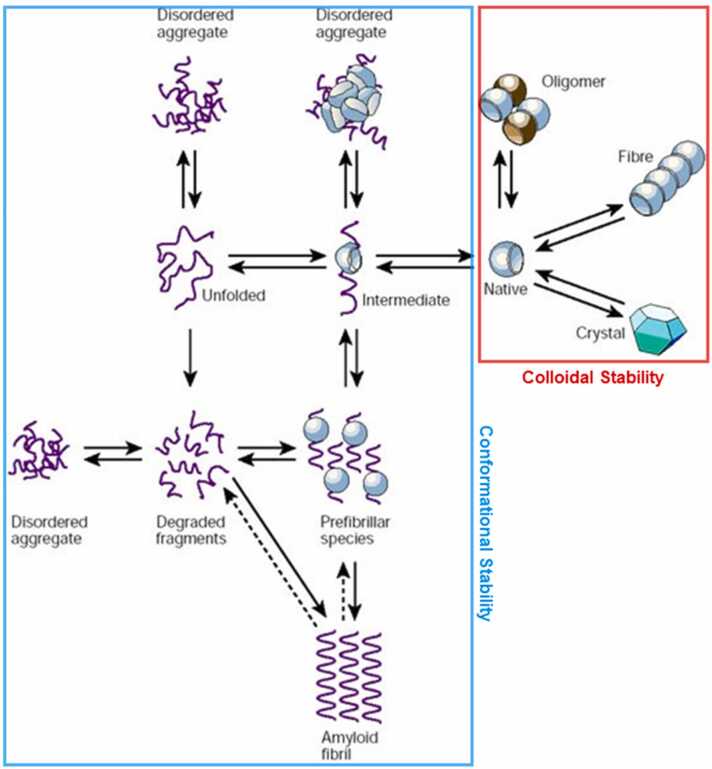
Since most protein-based therapeutics are liquid pharmaceutical preparations, the stability of the protein drugs involves colloidal stability and conformational stability (Fig. 3) [196]. Fig. 3 summarizes the equilibrium process of protein, including colloidal stability (red) and conformational stability (blue) [196]. In general, the stability of protein refers to conformational stability, in other words, conformational integrity. The protein conformation is complicated, multi-level, and inhomogeneous. Maintained by covalent bonds ionic bonding, hydrogen bonding, and Van der Waals forces, the protein conformation is sensitive and can be affected by several factors, including protein molecule, formulation gradients, composition, temperature, pH, and ionic strength [197]. Colloidal stability is the other aspect of stability in protein-based therapeutics, especially liquid pharmaceutical preparations. The colloidal stability is related to the colloidal property of proteins as simple particles, which have attractive and repulsive interactions. Even though there are some theories to describe colloidal stability, such as DLVO (Derjaguin-Landau-Verwey-Overbeek) theory [198] and electric double layer theory [199], and there are some parameters to reflect the colloidal stability, like the second virial coefficient (B22) or protein interaction parameter (kD) [200], the colloidal stability of protein drug formulation is as complicated as conformational stability and still a thorny issue in protein-based therapeutics development. Both conformational stability and colloidal stability will have considerable influences on the stability of protein-based therapeutics during manufacturing processes and long-term storage. However, it is still difficult to rational design or modifies protein-based formulation for better stability by intuition and experience, since the high complexity of protein stability. For instance, making a very subtle adjustment to the sequence or structure of a protein might have a significant effect on protein stability. There is an array of interactions controlling protein stability, making understanding the molecular mechanisms and improvement of protein stability challenging. Therefore, advanced methods are needed to predict and increase the stability in protein-based therapeutics development, and the computational method is one of the most promising approaches.
Fig. 3.
Schematic representation of the states accessible to a polypeptide chain, involving colloidal stability (red part) and conformational stability (blue part).
Adapted from ref [196].
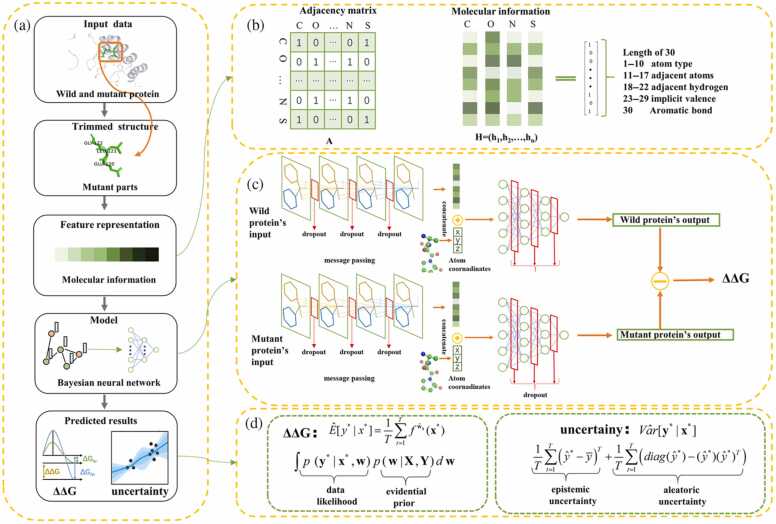
Shuyu Wang et al. developed a computational method called BayeStab, which can be utilized to predict protein thermostability change upon mutation (Fig. 4) [201]. This method combines graph neural networks and Bayesian neural networks and takes the protein data as input to predict protein mutations' ΔΔG with considerably high performance. The high generalization and symmetry performance was certified in four datasets. Huali Cao and co-workers developed a computational method called DeepDDG, a machine learning-based tool to predict the stability change of protein point mutations [109]. They trained the machine learning model with 5700 manually curated experimental data, and the performance is better than 11 other methods. In the research of Ethan C. Alley and co-workers, they carry out a pattern of unsupervised learning, next token prediction, with a recurrent neural network trained by the UniRef50 dataset with 24 million amino acid sequences [104]. This trained model is called UniRep, which is independent of structural or evolutionary data and can summarize the information in every protein sequence and convert them into fixed-length vectors. These vectors are endowed with rich information and excellent generalization ability, which can be the suitable representation of input protein sequences for downstream tasks, such as protein stability prediction. Alex Nisthal et al. combined the automated method they developed and three stability-prediction algorithms, PoPMuSiC, FoldX, and Rosetta, to obtain the most stable variants in the single-mutant landscape [202]. Hongwei Tu et al. developed a structure-independent protocol to predict the protein mutations' ΔΔG [203]. This protocol leveraged the information in sequence, physicochemical and evolutionary features, and integrated supervised (boosted tree regression) and unsupervised learning (K-means algorithm), successfully achieving high accuracy with an average PCC of 0.83. GuidoScarabelli and co-workers described a physical-based computational method to predict the thermodynamic stability of protein and compared the prediction with the actual result from diverse experiments at different pH conditions [120]. They compared the performance of several physical-based techniques, including Free Energy Perturbation (FEP), Molecular Mechanics-Generalized Born Surface Area (MM/GBSA), and the combination of MM/GBSA and molecular dynamics. The FEP got the best result, and there was a good correlation between predicted results by the FEP method and experimental results (R2 = 0.65).
Fig. 4.
The process of the BayeStab model to predict protein thermostability change upon mutation [201]. Copyright 2022 Wiley-VCH.
The issue of instability is present in some innovative and atypical protein-based therapeutics, with computational methods proving valuable for their analysis and optimization. Nanodiscs (NDs), as an example of unconventional protein-based therapeutics, consists of phospholipids and apolipoprotein A1 (ApoA1)-mimetic peptides [204], [205]. NDs offer numerous benefits, including cost-effectiveness, ease of large-scale production, an extended half-life, and passive targeting, rendering them an appealing drug delivery system. [206], [207]. Despite the potential of nanodiscs (NDs) as a drug delivery system, stability continues to pose a challenge to their clinical translation, due to their self-assembling nature. In one of our investigations, molecular dynamics simulations were employed to examine the correlation between nanodisc formulation and the conformational stability [52]. The findings indicated that both the molecular flexibility and the form of prodrug play crucial roles in determining conformational stability. In addition to exploring the mechanism of stability, molecular dynamics can also be used to design protein therapeutics with improved stability [208]. In our latest work, we developed a consensus-based normalization approach for designing reconfigurable apoA-I peptide analogs (APAs) to create tunable ND assemblies, with potential implications for structural biology and therapeutics [209]. We generated 15 divergent APAs and demonstrated that APA design influences ND diameter and stability through factors such as tandem repeats, sequence composition, and lipid-to-APA ratio. The findings reveal a strong correlation between DMPC-to-APA ratios and ND diameters, with longer APAs yielding more homogeneous particle sizes. Proline-rich substitutions contribute to both smaller and larger ND formation, while proline-tryptophan residues play a key role in forming larger NDs. Molecular simulations also indicate that basic and acidic residues in APAs enhance structural stability through hydrogen-bond and salt bridge networks. These insights advance our understanding of APA-ND assembly and offer a foundation for the computational design of APAs with desired functional and structural properties. Another novel protein-based therapeutic is Antibody-drug conjugates (ADCs). ADCs combine the selectivity and affinity of antibodies with the cytotoxicity of highly toxic small molecules and have now become intriguing protein-based therapeutics in clinical applications [49]. Nevertheless, the unstable or heterogeneous products of current production strategies of ADCs seriously hinder their clinical application. Nimish Gupta and co-workers leveraged computational methods, molecular docking, and molecular dynamics simulations to design a new ADC [210]. This new ADC will self-assemble through the non-covalent binding with active payloads without the need for modifications to the antibody structure. This method will yield homogenous ADCs with excellent stability within a short time.
Among the different stability problems affecting protein drugs, aggregation is undoubtedly the most prevalent and long-lasting one during the development of protein-based therapeutics. Aggregation may jeopardize the viability of the complete biotechnological process and is one of the major bottlenecks for the clinical translation [211]. Protein aggregation happens under a variety of physicochemical diverse supramolecular assemblies and occurs through a series of stages shown in Fig. 3, which is often initiated by the interaction between unfolded, non-native, or the native state of protein [212], [213]. Aggregation is a generic property of protein, which has strong interaction between themselves or between protein and other molecules [214]. The formation of aggregation of protein drugs may result in reduced production yields, loss of protein activities, poor solubility, and high viscosity, and cause safety issues like immune response during the production, storage, and administration of protein-based therapeutics [69], [215]. Therefore, it is necessary to understand the mechanism of protein aggregation and control it. However, since the complexity of aggregation formation, current empirical methods used in aggregation prediction and prevention are usually time and cost-consuming and ineffective. There are lots of factors determining the aggregation with diverse mechanisms, and under the circumstances, the computational methods will make a difference in stability improvement and aggregation prevention [27], [212].
There are growing numbers of computational methods to predict protein aggregation and some of them take sequences as input, and the other take 3D structure or both of them. Some reviews have summarized a wide diversity of computational methods for the protein aggregation prediction [30], [132], [212], [213], [215], [216]. Apart from methods proposed in academic papers, there are many routine methods used in the pharmaceutical industry, including SAP (Spatial aggregation propensity) [217], AggScore in Schrodinger [218], and some modules in MOE software (eg. Protein property patches). In the study of R Prabakaran and co-workers, they systematically compared nine different computational aggregation prediction methods using six diverse datasets, a variety of assessments, and providing rigorous performance analysis [219]. This analysis revealed that different methods have advantages for different prediction tasks, like solubility prediction, amyloid fibril formation prediction, etc., and the performance of computational aggregation prediction methods relies on the model architecture and training and validation datasets. This analysis will provide crucial guidance to develop aggregation prediction methods in the future.
To improve the stability or prevent the aggregation of protein-based therapeutics, the formulation method is also powerful and widely used in protein-based therapeutics development toward clinical. In the formulation of protein drugs, additives such as salts (e.g. citrates, sulfates), sugars (e.g. sorbitol, sucrose, trehalose), polymers [220], surfactants, and amino acids (e.g. glycine, arginine) are typical stabilizers in clinical [72], [221]. There are different mechanisms of these stabilizers, including directly binding with protein drugs and some complicated mechanisms involving the indirect interactions between protein drugs and stabilizers or stabilizers and stabilizers [222]. The mechanisms of the stabilization effect are complicated and still not fully understood, and the selection of stabilizers still relies on intuition, experience, and trial-and-error methods. For high stability of protein formulation, there is currently no efficient way but trying lots of pharmaceutical excipients like sugars, salt, and amino acids, until obtaining the formulation with suitable and acceptable properties. Advanced computational methods should be explored for formulation design in protein-based therapeutics.
Matthew J. Tamasi et al. reported a computational method based on active machine learning and automated polymer chemistry to design protein-stabilizing copolymers [223]. Polymer–protein hybrids are novel materials to bolster protein stability, which may be a powerful strategy in medicine. The rational design of polymer is still a big challenge because of the vast chemical and composition space. The computational method invented by the authors successfully identified the copolymers preserving or enhancing the activity of three different enzymes under the thermal denaturing conditions, which certificated the considerable generalization performance and robustness. Sunhwan Jo and co-workers applied the computational methods called SILCS (site-identification by ligand competitive saturation) to assist the rational excipient selection [224]. This method takes protein 3D structure as input and performs excipient docking and protein docking. Some results of SILCS are good indicators of experimental results, for example, the low number of the predicted binding site of excipients will be the indicator of high viscosity and poor stability. Sowmya Indrakumar et al. designed an excipients screening method integrating the microscale thermophoresis titration assays and molecular dynamics simulations, which can rank the excipients with respect to binding affinity and analyze the hotspots in proteins or peptides. The result was consistent with 1H–13C HSQC NMR titration experiments, showing its ability as a fast-screening method to rank and optimize excipients in protein or peptide formulation for stability improvement. Instead of screening excipients, Suman Saurabh et al. leveraged all-atom molecular dynamics simulations and contact-based free energy calculations to study the molecular interactions between histidine and antibodies [121]. Understanding the molecular interactions and stabilizing mechanisms of histidine or other excipients will help us screen and design protein formulation for improved protein stability and reduced protein aggregation propensity.
3.3. Solubility and viscosity reduction
Protein solubility is one of the most considerable prerequisites for the clinical translation of protein-based therapeutics, especially the highly concentrated formulations with increased demand. Some administration methods need higher requirements in protein solubility, for instance, subcutaneous injection and intramuscular injection with limited injection volume (<2/5 mL) and high dose requirement (∼500 mg) [71]. In addition, similar to stability, solubility is a critical factor of manufacturability, and poor solubility will also cause low product yield and capacity [225], [226]. Poor solubility will also cause other developability problems, especially the aggregation discussed above, and it will pose a challenge to the transportation, storage, and in vivo pharmacokinetics properties of protein drugs [214]. Measuring protein solubility is very challenging, often using surrogate parameters [167]. Traditional solubility improvement methods are also in a trial-and-error manner [227], taking lots of processes and spending a lot of time and money [228], [229]. It is still challenging to obtain a protein with high solubility efficiently in experimental methods, mainly because of the conflict between the limited preparation quality available and a large number of protein variants and formulations [230]. Like other developability, having a full understanding of protein solubility mechanism is still inaccessible, because the protein has a complicated structure and physicochemical properties, and environmental factors like pH and temperature will also influence the solubility.
JiangyanFeng et al. designed a solubility prediction method called solPredict, which can predict the proteins' apparent solubility in a histidine (pH 6.0) buffer [131]. This method is characterized by rapid, high-throughput, and cost-saving, and it only needs protein sequences as input. To overcome the limited data of solubility, they combined the pre-training and transfer learning strategy. The pre-trained model was trained on 250 million unlabeled protein sequences in an unsupervised manner, and the learned representations from this pre-trained model will contain rich and important information on protein sequences, which will be useful for predicting protein solubility. To avoid decreasing the catalytic activity when trying to increase the solubility and address the trade-off between enzyme solubility and activity, Justin R. Klesmith and co-workers developed a hybrid classification model, which can recognize the solubility-enhancing mutations that will not disrupt the wild-type function with high accuracy and without the need for high-resolution protein structure [231]. Lisanna Paladin and co-workers presented a method called SODA to predict the sequence influence on the protein solubility [140]. SODA is based on lots of physicochemical properties of proteins, including the disorder and aggregation propensities, predicted secondary structure components, and hydrophobic profile. SODA can acquire results quickly, which is an intriguing tool in protein drug development. Xuan Hana et al. proposed a strategy to estimate antibody solubility through machine learning [141]. They first constructed the data by their previously developed experimental high-throughput mAb solubility screening assay, obtaining 111 antibodies solubilities in a histidine buffer, pH 6.0. Then they acquired 3D homology models of the antibodies and calculated numerous available molecular descriptors. These descriptors were treated as the input of the machine learning model, and the solubility was the output and acquired a high accuracy for solubility prediction.
Viscosity is also a key aspect of developability, especially in the high-concentration formulation of protein drugs [232]. As mentioned earlier, most protein-based therapeutics should be administered with injection, and low viscosity is the fundamental requirement of the syringeability [233]. For injectable solutions, the upper limit of viscosity is about 50 mPa/S-1 for most situations [234]. The high viscosity will increase the force needed to deliver a solution with needles, extend the required time of injection, and sometimes make the fine needles unavailable, all of which will cause more injection pain and medical risk [74], [235]. The high viscosity will also present big challenges in production processes, including filtration, purification, mixing, and vial filling, which will not only cause the loss of raw material and high cost but also bring the risk of non-uniform products because of insufficient mixing or inaccurate filling [74], [236]. It is still difficult to understand the mechanism of the complicated viscosity behavior of protein solution, and sometimes, even a single mutation will cause different viscosity behavior [237], [238]. Numerous molecular interaction types, protein-protein interactions, and protein-excipient interactions will affect the viscosity of protein solutions [239]. Furthermore, the measurement of the viscosity of protein solution is labor-intensive and cost-consuming, requiring a large amount of protein solution (∼150 mg/mL) [12], [237]. The experimental method for viscosity measurement will hinder the high-thought and fast screening of a large amount of candidate protein drugs. Therefore, exploring computational methods with low costs and high efficacy will assist viscosity optimization in the early stage of drug discovery.
Neeraj J Agrawal and co-workers developed a novel computational method called spatial charge map (SCM), which can accurately identify highly viscous antibodies from their sequence alone [240]. The 3D structure of the protein was constructed with homology modeling, and this structure was the input of SCM. The SCM performs molecular dynamics simulation and calculated the SCM score to evaluate the viscosity performance of antibodies according to the understanding of viscosity and 3D structure. The SCM score is a good index to perform high throughput screening for protein drugs with low viscosity in the early stage. One of the disadvantages of the SCM method is time-consuming and requires computational resources since each evaluation requires molecular dynamics simulation. Based on this, Pin-Kuang Lai presented a convolutional neural network surrogate model, DeepSCM, to substitute the SCM with high efficacy without the loss of accuracy [144]. The data to train the DeepSCM model is a high-throughput MD simulation result from the original SCM, and DeepSCM can screen hundreds of protein drug candidates with only sequences as input within a few seconds. The addition of pharmaceutical excipients is a widely used method to control the viscosity of protein formulation in industry, and there are also lots of studies using computational methods to investigate excipients for viscosity reduction. In Niels Banik et al.’ study, excipient parameters calculated by in-silico methods can be treated as a screening tool for protein formulation development and viscosity reduction together with dynamic light scattering [241]. MaticProj and co-workers leveraged two computational chemistry methods to screen new viscosity-reducing agents: fingerprint similarity searching, and physicochemical property filtering. With these methods, they successfully selected 33 new agents from 94 compounds that can reduce the viscosity of two model mAbs [242].
3.4. Deimmunization and humanization
As exogenous large molecules, the potential immunogenicity is a significant problem in protein-based therapeutics development. Immunogenicity refers to the degree of host immune system can recognize and react to external agents, which include the therapeutical protein drug [243]. Immunogenicity is a considerable concern during protein drug development, which should be avoided for most therapeutic proteins apart from vaccines. The uncontrolled immune response stimulated by protein drugs will neutralize therapeutic agents, cause the loss of therapeutic efficacy, and even cause serious medical consequences like severe anaphylactic reactions or hypersensitivity reactions which can be life-threatening [243]. Therefore, there is an urgent need to develop deimmunization methods for protein drugs toward the clinical stage. Lots of deimmunization methods have been proposed in protein-based therapeutics development, aiming at mitigating immunogenicity, reducing immune-related side effects, and improving therapeutical efficacy. Among deimmunization methods, the widely used methods include shielding approaches such as PEGylation [244], XTENylation [245] or PASylation [246], humanization [247], and prediction/deletion of T cell and B cell epitopes [248]. The latter two methods are the fundamental solutions for deimmunization because the antigenic motifs in protein molecules will be modified or removed by protein engineering methods [249]. The strategies to deimmunize protein drugs, both humanization and prediction/deletion of T cell and B cell epitopes rely on the detailed understanding of the molecular and cellular mechanisms of the immune response toward protein drugs [250]. Similar to the developability problem discussed above, the current experimental methods for deimmunization are time and cost-consuming, and labor-intensive, and the computational methods will facilitate humanization and T/B cell epitope identification and deletion.
For antibody development, one of the most common generation methods of protein currently relies on the immunization of mice or another model animal (for example, Camelidae produce nanobodies). There are lots of advantages to producing protein by the model animal, including good availability, low cost, and high-efficiency [164]. For example, the antibodies developed by the model animal will experience evolution with in vivo mechanisms, like hypermutation in germinal centers, and these mechanisms will guarantee the high affinity of produced antibodies [247]. However, the products derived from non-human sources may result in severe immune response, safety problems, and reduced efficiency, which came from the anti-drug antibodies (ADAs). To avoid the formation of ADAs and subsequent immune response, the antibodies derived from non-human sources should undergo a subsequent process called humanization. The main purpose of humanization is to reduce immunogenicity and increase safety without the partial or complete loss of affinity, and during humanization, the complementarity determining regions (CDRs) of antibodies from non-human sources will be grafted onto human frameworks or the antibodies from non-human sources will be engineered to resemble human antibodies [28]. The humanized antibodies have improved clinical tolerance, accelerating the development of protein-based therapeutics. However, current traditional humanization methods are mainly based on germline sequences or natural sequence libraries of limited size, which lack enough diversity and systematicness, and may fall into a locally optimal solution for the clinical translation [151], and these methods highly rely on expensive and time-consuming experiments. There is an urgent need for advanced methods like computational methods for humanization in protein-based therapeutics development.
Claire Marks and co-workers developed a computational method called Hu-mAb, which can guide researchers to mutate the input sequences for immunogenicity reduction and humanization [154]. The construction of Hu-mAb is based on the Observed Antibody Space (OAS) database and the random forest model, and it can humanize the sequence in an optimal manner with the lowest possible number of mutations to avoid the influence of protein activity. Hu-mAb is a competitive substitute for humanization for traditional experimental methods, getting similar mutation results with experimental therapeutic humanization without the time and cost-consuming processes. David Prihoda et al. designed a computational platform called BioPhi, containing novel humanization methods (Sapiens) and humanness evaluation methods (OASis) [151]. Sapiens is an attention-based deep learning model trained on human variable region antibody sequences from the OAS database, which can produce similar results with expert methods. OASis is an accurate humanness score based on the OAS database. The high efficiency and automated humanization workflow of BioPhi make it possible to bulk process numerous sequences. In the research of Pier Paolo Olimpieri et al., they showed a web server for antibody server, which can guide the researcher to perform all the critical steps of the humanization experiment protocol, including human template selection, grafting, back-mutation evaluation, antibody modeling, and structural analysis.
The identification, prediction, and deletion of T cell and B cell epitopes are also practical methods for the deimmunization of protein-based therapeutics because they can avoid the reorganization and devitalization of immune cells or antidrug antibodies [248], [251]. In addition, epitope reorganization and removal will not only reduce the immunogenicity of protein drug but also increase the half-life, and efficacy and improve the pharmacodynamics and -kinetics properties [249]. For antibody drugs, it is also significant to recognize the epitopes, which is useful for understanding and predicting the possible cross-reactivity [252]. Lots of strategies have been applied to recognize and delete epitopes in protein drugs, including random mutagenesis and high throughput screening [253], more precise mapping using panels of antidrug antibodies [254], and structure-based design with co-crystals of antibody-antigen [255]. Both of these methods still rely on a trial-and-error process, requiring much time and cost, and computational methods are desirable and time-saving alternatives for high efficiency. There have been lots of computational methods developed to perform the identification, prediction, and deletion of T cell and B cell epitopes with high efficiency, which have been introduced in detail in other reviews [248], [249], [252], [256].
4. Future perspective
The use of computational methods to predict and optimize the developability of protein-based therapeutics has gained significant attention. In this review, we highlight recent advances in this area with regard to various aspects of developability. However, there is still much work to be done to fully harness the potential of these methods and to translate them into tangible improvements in protein-based therapeutic developability. Several obstacles must be overcome in this field.
Data-driven methods, such as ML and DL, are dependent on the quality, diversity, and quantity of datasets, especially for complex learning tasks [257]. In the discovery of protein-based therapeutics, obtaining large amounts of high-quality data is challenging [258]. For example, measuring viscosity is a time-consuming and labor-intensive process, making it difficult to gather enough data for an artificial intelligence model. Additionally, publicly available data and databases are often heterogeneous in format and structure [30], [36], resulting in non-comparable data that can impact the accuracy of predictive models and hinder data sharing [259]. High-throughput methods can also sacrifice data accuracy and relevance for increased throughput, leading to errors that can distort results [260]. Therefore, detecting anomalies in datasets is also a critical concern [36]. With the growth of AI models, the number of internal parameters increases, posing challenges to the size of datasets required for effective learning.
Multi-scale modeling techniques, such as molecular dynamics simulation, require a balance between accuracy and speed, often sacrificing one for the other. The all-atom molecular dynamics simulation offers high accuracy and detailed knowledge for mechanistic insights in protein engineering and drug discovery, but it is limited due to its high computational cost and limited speed. Coarse-Grained molecular dynamics simulation is a preferred method in high-throughput screening due to its acceptable computing resources, but it results in less accuracy and loss of detail [261]. However, it enables the simulation of multiple molecules, which is necessary for studying some in silico developability factors, such as viscosity and self-aggregation [148]. Another challenge in molecular dynamics simulation is the validity of the results, as there are assumptions and approximations made that can cause deviation from experimental results. To achieve the same thermodynamic and kinetic observables as experiments, optimization of simulation methods, including more precise force fields and topology [262], enhanced sampling methods [263], a deeper understanding of physical rules and modeling parameters, more efficient simulation methods [264], and better computational technology and speed, is necessary [265].
In developing protein-based therapeutics, the integration of multiple computational and experimental methods is necessary for optimal results [30]. Utilizing only one method can result in limited information, and no single technique can fully address all challenges in development. Integration of various computing methods and data sources is a growing trend, such as combining molecular dynamics simulation and artificial intelligence to construct prediction models in several aspects of the developability [30]. Additionally, high-efficiency computational methods can be used to accelerate or optimize high-accuracy methods [266], [267], like utilizing machine learning to accelerate the multi-scale simulation processes [263], [268] and molecular docking [269]. Besides, the integration of computational and experimental methods is an effective pattern in protein drug discovery, like the examples mentioned above using AI and experimental phase display methods to explore large protein space and optimize the protein [12], [102]. Furthermore, the incorporation of computational modeling and laboratory experiments will help researchers to get a further understanding of the drug discovery process, like drug formulation mechanisms and protein interactions with other substances [67].
Effective collaboration between computational scientists and protein drug developers is essential for significant progress in protein drug therapy with computational methods [30]. The gap between these two fields should be bridged, with computational scientists developing methods suited for protein-based therapeutics and user-friendly algorithms, while protein drug developers recognize the potential benefits of computational methods [30], [36]. Overcoming computational and skillset limitations requires cooperation between pharmaceutical and computational experts or the cultivation of interdisciplinary talent [27], [270]. Overall, computational techniques in protein-based therapeutics hold great promise for accelerating the discovery process, achieving optimal therapeutic outcomes, and reducing costs.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 82001887), Shenzhen Science and Technology Program (Grant No. JCYJ20210324115003009, JCYI202206193000001; JCYJ20220530144401004), Futian Healthcare Research Project (Grant No. FTWS2022018), and the Program of “Transverse” Research Project at Sun Yat-sen University (Grant No. K21- 75110–007).
CRediT authorship contribution statement
Conceptualization, Junqing Wang, Zhe Wang; Writing − original draft preparation, Zhidong Chen, Junqing Wang; Writing − review & editing, Zhidong Chen, Xinpei Wang, Xu Chen, Juyang Huang; Visualization, Zhidong Chen, Junqing Wang; Data curation, Zhidong Chen, Xinpei Wang; Supervision, Junqing Wang, Zhe Wang, Chenglin Wang; Project administration, Junqing Wang, Zhe Wang; Funding acquisition, Junqing Wang, Zhe Wang;. All authors have read and agreed to the published version of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence this paper.
Acknowledgements
Zhidong Chen and Xinpei Wang contributed equally to this work. The authors thank Yonghui Lv (School of Pharmaceutical Sciences, Shenzhen Campus of Sun Yat-sen University) for the review and editing of the manuscript.
Contributor Information
Junqing Wang, Email: wangjunqing@mail.sysu.edu.cn.
Zhe Wang, Email: wangzh379@mail.sysu.edu.cn.
References
- 1.Carter P.J., Lazar G.A. Next generation antibody drugs: pursuit of the 'high-hanging fruit'. Nat Rev Drug Discov. 2018;17:197–223. doi: 10.1038/nrd.2017.227. [DOI] [PubMed] [Google Scholar]
- 2.Crook Z.R., Nairn N.W., Olson J.M. Miniproteins as a powerful modality in drug development. Trends Biochem Sci. 2020;45:332–346. doi: 10.1016/j.tibs.2019.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies J.A., Ireland S., Harding S., Sharman J.L., Southan C., Dominguez-Monedero A. Inverse pharmacology: Approaches and tools for introducing druggability into engineered proteins. Biotechnol Adv. 2019;37 doi: 10.1016/j.biotechadv.2019.107439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinch M.S. An overview of FDA-approved biologics medicines. Drug Discov Today. 2015;20:393–398. doi: 10.1016/j.drudis.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Meyer B.K., Shameem M. In: Therapeutic Protein Drug Products. Meyer B.K., editor. Woodhead Publishing; 2012. 1 - Commercial therapeutic protein drug products; pp. 1–11. [Google Scholar]
- 6.D. Kevin, The top 20 drugs by worldwide sales in 2021, Fierce, Pharma, 2022.
- 7.Oostindie S.C., Lazar G.A., Schuurman J., Parren P.W.H.I. Avidity in antibody effector functions and biotherapeutic drug design. Nat Rev Drug Discov. 2022;21:715–735. doi: 10.1038/s41573-022-00501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.H. Ausserwöger, M.M. Schneider, T.W. Herling, P. Arosio, G. Invernizzi, T.P.J. Knowles, N. Lorenzen, Non-specificity as the sticky problem in therapeutic antibody development, Nature Reviews Chemistry, 2022. [DOI] [PubMed]
- 9.Frokjaer S., Otzen D.E. Protein drug stability: a formulation challenge. Nat Rev Drug Discov. 2005;4:298–306. doi: 10.1038/nrd1695. [DOI] [PubMed] [Google Scholar]
- 10.Jain T., Sun T., Durand S., Hall A., Houston N.R., Nett J.H., Sharkey B., Bobrowicz B., Caffry I., Yu Y., Cao Y., Lynaugh H., Brown M., Baruah H., Gray L.T., Krauland E.M., Xu Y., Vásquez M., Wittrup K.D. Biophysical properties of the clinical-stage antibody landscape. Proc Natl Acad Sci. 2017;114:944–949. doi: 10.1073/pnas.1616408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabia L.A., Desai A.A., Jhajj H.S., Tessier P.M. Understanding and overcoming trade-offs between antibody affinity, specificity, stability and solubility. Biochem Eng J. 2018;137:365–374. doi: 10.1016/j.bej.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makowski E.K., Kinnunen P.C., Huang J., Wu L., Smith M.D., Wang T., Desai A.A., Streu C.N., Zhang Y., Zupancic J.M., Schardt J.S., Linderman J.J., Tessier P.M. Co-optimization of therapeutic antibody affinity and specificity using machine learning models that generalize to novel mutational space, Nature. Communications. 2022;13:3788. doi: 10.1038/s41467-022-31457-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai P.-K., Gallegos A., Mody N., Sathish H.A., Trout B.L. Machine learning prediction of antibody aggregation and viscosity for high concentration formulation development of protein therapeutics. mAbs. 2022;14:2026208. doi: 10.1080/19420862.2022.2026208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.J.S. Kingsbury, A. Saini, S.M. Auclair, L. Fu, M.M. Lantz, K.T. Halloran, C. Calero-Rubio, W. Schwenger, C.Y. Airiau, J. Zhang, Y.R. Gokarn, A single molecular descriptor to predict solution behavior of therapeutic antibodies, Science Advances, 6 eabb0372. [DOI] [PMC free article] [PubMed]
- 15.Kurtzhals P., Østergaard S., Nishimura E., Kjeldsen T. Derivatization with fatty acids in peptide and protein drug discovery. Nat Rev Drug Discov. 2022 doi: 10.1038/s41573-022-00529-w. [DOI] [PubMed] [Google Scholar]
- 16.Yallapragada V.V.B., Walker S.P., Devoy C., Buckley S., Flores Y., Tangney M. Function2Form Bridge—Toward synthetic protein holistic performance prediction, Proteins: Structure. Funct, Bioinforma. 2020;88:462–475. doi: 10.1002/prot.25825. [DOI] [PubMed] [Google Scholar]
- 17.Owens J. Determining druggability. Nat Rev Drug Discov. 2007;6(187–187) [Google Scholar]
- 18.Oprea T.I., Hasselgren C. In: Comprehensive Medicinal Chemistry III. Chackalamannil S., Rotella D., Ward S.E., editors. Elsevier; Oxford: 2017. 3.17 - Predicting Target and Chemical Druggability; pp. 429–439. [Google Scholar]
- 19.Bailly M., Mieczkowski C., Juan V., Metwally E., Tomazela D., Baker J., Uchida M., Kofman E., Raoufi F., Motlagh S., Yu Y., Park J., Raghava S., Welsh J., Rauscher M., Raghunathan G., Hsieh M., Chen Y.-L., Nguyen H.T., Nguyen N., Cipriano D., Fayadat-Dilman L. Predicting Antibody Developability Profiles Through Early Stage Discovery Screening. mAbs. 2020;12:1743053. doi: 10.1080/19420862.2020.1743053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chauhan V.M., Zhang H., Dalby P.A., Aylott J.W. Advancements in the co-formulation of biologic therapeutics. J Control Release. 2020;327:397–405. doi: 10.1016/j.jconrel.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawat P., Kumar S., Michael M. Gromiha, An in-silico method for identifying aggregation rate enhancer and mitigator mutations in proteins. Int J Biol Macromol. 2018;118:1157–1167. doi: 10.1016/j.ijbiomac.2018.06.102. [DOI] [PubMed] [Google Scholar]
- 22.Karadag M., Arslan M., Kaleli N.E., Kalyoncu S. In: Advances in Protein Chemistry and Structural Biology. Donev R., editor. Academic Press; 2020. Chapter Four - Physicochemical determinants of antibody-protein interactions; pp. 85–114. [DOI] [PubMed] [Google Scholar]
- 23.Hajduk P.J., Huth J.R., Tse C. Predicting protein druggability. Drug Discov Today. 2005;10:1675–1682. doi: 10.1016/S1359-6446(05)03624-X. [DOI] [PubMed] [Google Scholar]
- 24.Benet L.Z., Hosey C.M., Ursu O., Oprea T.I. BDDCS, the Rule of 5 and drugability. Adv Drug Deliv Rev. 2016;101:89–98. doi: 10.1016/j.addr.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amidon G.L., Lennernäs H., Shah V.P., Crison J.R. A Theoretical Basis for a Biopharmaceutic Drug Classification: The Correlation of in Vitro Drug Product Dissolution and in Vivo Bioavailability. Pharm Res. 1995;12:413–420. doi: 10.1023/a:1016212804288. [DOI] [PubMed] [Google Scholar]
- 26.Narayanan H., Dingfelder F., Butté A., Lorenzen N., Sokolov M., Arosio P. Machine learning for biologics: opportunities for protein engineering, developability, and formulation. Trends Pharmacol Sci. 2021;42:151–165. doi: 10.1016/j.tips.2020.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Navarro S., Ventura S. Computational methods to predict protein aggregation. Curr Opin Struct Biol. 2022;73 doi: 10.1016/j.sbi.2022.102343. [DOI] [PubMed] [Google Scholar]
- 28.Norman R.A., Ambrosetti F., Bonvin A.M.J.J., Colwell L.J., Kelm S., Kumar S., Krawczyk K. Computational approaches to therapeutic antibody design: established methods and emerging trends. Brief Bioinforma. 2020;21:1549–1567. doi: 10.1093/bib/bbz095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed L., Gupta P., Martin K.P., Scheer J.M., Nixon A.E., Kumar S. Intrinsic physicochemical profile of marketed antibody-based biotherapeutics. Proc Natl Acad Sci. 2021;118 doi: 10.1073/pnas.2020577118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W., Ye Z., Gao H., Ouyang D. Computational pharmaceutics - A new paradigm of drug delivery. J Control Release. 2021;338:119–136. doi: 10.1016/j.jconrel.2021.08.030. [DOI] [PubMed] [Google Scholar]
- 31.Amaro R.E., Mulholland A.J. Multiscale methods in drug design bridge chemical and biological complexity in the search for cures. Nat Rev Chem. 2018;2:0148. doi: 10.1038/s41570-018-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leaver-Fay A., Tyka M., Lewis S.M., Lange O.F., Thompson J., Jacak R., Kaufman K.W., Renfrew P.D., Smith C.A., Sheffler W., Davis I.W., Cooper S., Treuille A., Mandell D.J., Richter F., Ban Y.-E.A., Fleishman S.J., Corn J.E., Kim D.E., Lyskov S., Berrondo M., Mentzer S., Popović Z., Havranek J.J., Karanicolas J., Das R., Meiler J., Kortemme T., Gray J.J., Kuhlman B., Baker D., Bradley P. In: Methods in Enzymology. Johnson M.L., Brand L., editors. Academic Press; 2011. Chapter nineteen - Rosetta3: An Object-Oriented Software Suite for the Simulation and Design of Macromolecules; pp. 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. Honma, H. Suzuki, Can molecular dynamics facilitate the design of protein–protein-interaction inhibitors?, Nature Reviews Rheumatology, 2022. [DOI] [PubMed]
- 34.Karplus M. Development of Multiscale Models for Complex Chemical Systems: From H+H2 to Biomolecules (Nobel Lecture) Angew Chem Int Ed. 2014;53:9992–10005. doi: 10.1002/anie.201403924. [DOI] [PubMed] [Google Scholar]
- 35.Bender B.J., Gahbauer S., Luttens A., Lyu J., Webb C.M., Stein R.M., Fink E.A., Balius T.E., Carlsson J., Irwin J.J., Shoichet B.K. A practical guide to large-scale docking. Nat Protoc. 2021;16:4799–4832. doi: 10.1038/s41596-021-00597-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider P., Walters W.P., Plowright A.T., Sieroka N., Listgarten J., Goodnow R.A., Fisher J., Jansen J.M., Duca J.S., Rush T.S., Zentgraf M., Hill J.E., Krutoholow E., Kohler M., Blaney J., Funatsu K., Luebkemann C., Schneider G. Rethinking drug design in the artificial intelligence era. Nat Rev Drug Discov. 2020;19:353–364. doi: 10.1038/s41573-019-0050-3. [DOI] [PubMed] [Google Scholar]
- 37.Wainberg M., Merico D., Delong A., Frey B.J. Deep learning in biomedicine. Nat Biotechnol. 2018;36:829–838. doi: 10.1038/nbt.4233. [DOI] [PubMed] [Google Scholar]
- 38.Chen Z.-D., Zhao L., Chen H.-Y., Gong J.-N., Chen X., Chen C.Y.-C. A novel artificial intelligence protocol to investigate potential leads for Parkinson's disease. RSC Adv. 2020;10:22939–22958. doi: 10.1039/d0ra04028b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X., Chen H.-Y., Chen Z.-D., Gong J.-N., Chen C.Y.-C. A novel artificial intelligence protocol for finding potential inhibitors of acute myeloid leukemia. J Mater Chem B. 2020;8:2063–2081. doi: 10.1039/d0tb00061b. [DOI] [PubMed] [Google Scholar]
- 40.Kuriata A., Iglesias V., Pujols J., Kurcinski M., Kmiecik S., Ventura S. Aggrescan3D (A3D) 2.0: prediction and engineering of protein solubility. Nucleic Acids Res. 2019;47:W300–W307. doi: 10.1093/nar/gkz321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dauparas J., Anishchenko I., Bennett N., Bai H., Ragotte R.J., Milles L.F., Wicky B.I.M., Courbet A., de Haas R.J., Bethel N., Leung P.J.Y., Huddy T.F., Pellock S., Tischer D., Chan F., Koepnick B., Nguyen H., Kang A., Sankaran B., Bera A.K., King N.P., Baker D. Robust deep learning–based protein sequence design using ProteinMPNN. Science. 2022;378:49–56. doi: 10.1126/science.add2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smiatek J., Jung A., Bluhmki E. Towards a Digital Bioprocess Replica: Computational Approaches in Biopharmaceutical Development and Manufacturing. Trends Biotechnol. 2020;38:1141–1153. doi: 10.1016/j.tibtech.2020.05.008. [DOI] [PubMed] [Google Scholar]
- 43.Wang G., Haringa C., Noorman H., Chu J., Zhuang Y. Developing a computational framework to advance bioprocess scale-up. Trends Biotechnol. 2020;38:846–856. doi: 10.1016/j.tibtech.2020.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Gierach I., Galiardi J.M., Marshall B., Wood D.W. In: Remington (Twenty-third Edition) Adejare A., editor. Academic Press; 2021. Chapter 26 - Protein drug production and formulation; pp. 489–547. [Google Scholar]
- 45.Hassanin I.A., Elzoghby A.O. Self-assembled non-covalent protein-drug nanoparticles: an emerging delivery platform for anti-cancer drugs. Expert Opin Drug Deliv. 2020;17:1437–1458. doi: 10.1080/17425247.2020.1813713. [DOI] [PubMed] [Google Scholar]
- 46.Hoogenboezem E.N., Duvall C.L. Harnessing albumin as a carrier for cancer therapies. Adv Drug Deliv Rev. 2018;130:73–89. doi: 10.1016/j.addr.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yardley D.A. nab-Paclitaxel mechanisms of action and delivery. J Control Release. 2013;170:365–372. doi: 10.1016/j.jconrel.2013.05.041. [DOI] [PubMed] [Google Scholar]
- 48.Liu X., Mohanty R.P., Maier E.Y., Peng X., Wulfe S., Looney A.P., Aung K.L., Ghosh D. Controlled loading of albumin-drug conjugates ex vivo for enhanced drug delivery and antitumor efficacy. J Control Release. 2020;328:1–12. doi: 10.1016/j.jconrel.2020.08.015. [DOI] [PubMed] [Google Scholar]
- 49.Jabbour E., Paul S., Kantarjian H. The clinical development of antibody–drug conjugates — lessons from leukaemia. Nat Rev Clin Oncol. 2021;18:418–433. doi: 10.1038/s41571-021-00484-2. [DOI] [PubMed] [Google Scholar]
- 50.Drago J.Z., Modi S., Chandarlapaty S. Unlocking the potential of antibody–drug conjugates for cancer therapy. Nat Rev Clin Oncol. 2021;18:327–344. doi: 10.1038/s41571-021-00470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MaHam A., Tang Z., Wu H., Wang J., Lin Y. Protein-based nanomedicine platforms for drug delivery. Small. 2009;5:1706–1721. doi: 10.1002/smll.200801602. [DOI] [PubMed] [Google Scholar]
- 52.Xu D., Chen X., Chen Z., Lv Y., Li Y., Li S., Xu W., Mo Y., Wang X., Chen Z., Chen T., Wang T., Wang Z., Wu M., Wang J. Silico Approach Reveal Nanodisc Formul Doxorubicin. 2022;10 doi: 10.3389/fbioe.2022.859255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Z., Chen X., Huang J., Wang J., Wang Z. Harnessing protein corona for biomimetic nanomedicine design. Biomimetics. 2022 doi: 10.3390/biomimetics7030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Irvine D.J., Dane E.L. Enhancing cancer immunotherapy with nanomedicine. Nat Rev Immunol. 2020;20:321–334. doi: 10.1038/s41577-019-0269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitchell M.J., Billingsley M.M., Haley R.M., Wechsler M.E., Peppas N.A., Langer R. Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov. 2021;20:101–124. doi: 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang Z., Xiao Y., Kong N., Liu C., Chen W., Huang X., Xu D., Ouyang J., Feng C., Wang C., Wang J., Zhang H., Tao W. Nano-bio interfaces effect of two-dimensional nanomaterials and their applications in cancer immunotherapy. Acta Pharm Sin B. 2021;11:3447–3464. doi: 10.1016/j.apsb.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beck H., Härter M., Haß B., Schmeck C., Baerfacker L. Small molecules and their impact in drug discovery: A perspective on the occasion of the 125th anniversary of the Bayer Chemical Research Laboratory. Drug Discov Today. 2022;27:1560–1574. doi: 10.1016/j.drudis.2022.02.015. [DOI] [PubMed] [Google Scholar]
- 59.Leeson P.D., Springthorpe B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat Rev Drug Discov. 2007;6:881–890. doi: 10.1038/nrd2445. [DOI] [PubMed] [Google Scholar]
- 60.Vargason A.M., Anselmo A.C., Mitragotri S. The evolution of commercial drug delivery technologies. Nat Biomed Eng. 2021;5:951–967. doi: 10.1038/s41551-021-00698-w. [DOI] [PubMed] [Google Scholar]
- 61.Ortmayr K., de la Cruz Moreno R., Zampieri M. Expanding the search for small-molecule antibacterials by multidimensional profiling. Nat Chem Biol. 2022;18:584–595. doi: 10.1038/s41589-022-01040-4. [DOI] [PubMed] [Google Scholar]
- 62.Childs-Disney J.L., Yang X., Gibaut Q.M.R., Tong Y., Batey R.T., Disney M.D. Targeting RNA structures with small molecules. Nat Rev Drug Discov. 2022;21:736–762. doi: 10.1038/s41573-022-00521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Offringa R., Kötzner L., Huck B., Urbahns K. The expanding role for small molecules in immuno-oncology. Nat Rev Drug Discov. 2022;21:821–840. doi: 10.1038/s41573-022-00538-9. [DOI] [PubMed] [Google Scholar]
- 64.Drucker D.J. Advances in oral peptide therapeutics. Nat Rev Drug Discov. 2020;19:277–289. doi: 10.1038/s41573-019-0053-0. [DOI] [PubMed] [Google Scholar]
- 65.Crommelin D.J.A., Storm G., Verrijk R., de Leede L., Jiskoot W., Hennink W.E. Shifting paradigms: biopharmaceuticals versus low molecular weight drugs. Int J Pharm. 2003;266:3–16. doi: 10.1016/s0378-5173(03)00376-4. [DOI] [PubMed] [Google Scholar]
- 66.Anselmo A.C., Gokarn Y., Mitragotri S. Non-invasive delivery strategies for biologics. Nat Rev Drug Discov. 2019;18:19–40. doi: 10.1038/nrd.2018.183. [DOI] [PubMed] [Google Scholar]
- 67.Bajracharya R., Song J.G., Back S.Y., Han H.-K. Recent Advancements in Non-Invasive Formulations for Protein Drug Delivery. Comput Struct Biotechnol J. 2019;17:1290–1308. doi: 10.1016/j.csbj.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vishali D.A., Monisha J., Sivakamasundari S.K., Moses J.A., Anandharamakrishnan C. Spray freeze drying: Emerging applications in drug delivery. J Control Release. 2019;300:93–101. doi: 10.1016/j.jconrel.2019.02.044. [DOI] [PubMed] [Google Scholar]
- 69.Falconer R.J. Advances in liquid formulations of parenteral therapeutic proteins. Biotechnol Adv. 2019;37 doi: 10.1016/j.biotechadv.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 70.Viola M., Sequeira J., Seiça R., Veiga F., Serra J., Santos A.C., Ribeiro A.J. Subcutaneous delivery of monoclonal antibodies: How do we get there? J Control Release. 2018;286:301–314. doi: 10.1016/j.jconrel.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 71.Garidel P., Kuhn A.B., Schäfer L.V., Karow-Zwick A.R., Blech M. High-concentration protein formulations: How high is high? Eur J Pharm Biopharm. 2017;119:353–360. doi: 10.1016/j.ejpb.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 72.Elgundi Z., Reslan M., Cruz E., Sifniotis V., Kayser V. The state-of-play and future of antibody therapeutics. Adv Drug Deliv Rev. 2017;122:2–19. doi: 10.1016/j.addr.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 73.Melo G.B., Cruz N.F.Sd, Emerson G.G., Rezende F.A., Meyer C.H., Uchiyama S., Carpenter J., Shiroma H.F., Farah M.E., Maia M., Rodrigues E.B. Critical analysis of techniques and materials used in devices, syringes, and needles used for intravitreal injections. Prog Retin Eye Res. 2021;80 doi: 10.1016/j.preteyeres.2020.100862. [DOI] [PubMed] [Google Scholar]
- 74.Li J., Cheng Y., Chen X., Zheng S. Impact of electroviscous effect on viscosity in developing highly concentrated protein formulations: Lessons from non-protein charged colloids. Int J Pharm: X. 2019;1 doi: 10.1016/j.ijpx.2018.100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang J., Du Y., Zhou P., Ding J., Xia S., Wang Q., Chen F., Zhou M., Zhang X., Wang W., Wu H., Lu L., Zhang S. Predicting unseen antibodies’ neutralizability via adaptive graph neural networks. Nat Mach Intell. 2022;4:964–976. [Google Scholar]
- 76.Parkinson J., Hard R., Wang W. The RESP AI model accelerates the identification of tight-binding antibodies. Nat Commun. 2023;14:454. doi: 10.1038/s41467-023-36028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu Z., Kan S.B.J., Lewis R.D., Wittmann B.J., Arnold F.H. Mach Learn-Assist Dir Protein Evol Comb Libr. 2019;116:8852–8858. doi: 10.1073/pnas.1901979116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu X., Luo Y., Li P., Song S., Peng J. Deep geometric representations for modeling effects of mutations on protein-protein binding affinity. PLoS Comput Biol. 2021;17 doi: 10.1371/journal.pcbi.1009284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen X., Gentili M., Hacohen N., Regev A. A cell-free nanobody engineering platform rapidly generates SARS-CoV-2 neutralizing nanobodies, Nature. Communications. 2021;12:5506. doi: 10.1038/s41467-021-25777-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Biswas S., Khimulya G., Alley E.C., Esvelt K.M., Church G.M. Low-N protein engineering with data-efficient deep learning. Nat Methods. 2021;18:389–396. doi: 10.1038/s41592-021-01100-y. [DOI] [PubMed] [Google Scholar]
- 81.Qiu Y., Hu J., Wei G.-W. Cluster learning-assisted directed evolution. Nat Comput Sci. 2021;1:809–818. doi: 10.1038/s43588-021-00168-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu G., Zeng H., Mueller J., Carter B., Wang Z., Schilz J., Horny G., Birnbaum M.E., Ewert S., Gifford D.K. Antibody complementarity determining region design using high-capacity machine learning. Bioinformatics. 2019;36:2126–2133. doi: 10.1093/bioinformatics/btz895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schneider C., Buchanan A., Taddese B., Deane C.M. DLAB: deep learning methods for structure-based virtual screening of antibodies. Bioinformatics. 2021;38:377–383. doi: 10.1093/bioinformatics/btab660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jemimah S., Sekijima M., Gromiha M.M. ProAffiMuSeq: sequence-based method to predict the binding free energy change of protein–protein complexes upon mutation using functional classification. Bioinformatics. 2019;36:1725–1730. doi: 10.1093/bioinformatics/btz829. [DOI] [PubMed] [Google Scholar]
- 85.Renaud N., Geng C., Georgievska S., Ambrosetti F., Ridder L., Marzella D.F., Réau M.F., Bonvin A.M.J.J., Xue L.C. DeepRank: a deep learning framework for data mining 3D protein-protein interfaces. Nat Commun. 2021;12:7068. doi: 10.1038/s41467-021-27396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rodrigues C.H.M., Myung Y., Pires D.E.V., Ascher D.B. mCSM-PPI2: predicting the effects of mutations on protein–protein interactions. Nucleic Acids Res. 2019;47:W338–W344. doi: 10.1093/nar/gkz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Myung Y., Rodrigues C.H.M., Ascher D.B., Pires D.E.V. mCSM-AB2: guiding rational antibody design using graph-based signatures. Bioinformatics. 2019;36:1453–1459. doi: 10.1093/bioinformatics/btz779. [DOI] [PubMed] [Google Scholar]
- 88.Myung Y., Pires D.E.V., Ascher D.B. mmCSM-AB: guiding rational antibody engineering through multiple point mutations. Nucleic Acids Res. 2020;48:W125–W131. doi: 10.1093/nar/gkaa389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rodrigues C.H.M., Pires D.E.V., Ascher D.B. mmCSM-PPI: predicting the effects of multiple point mutations on protein–protein interactions. Nucleic Acids Res. 2021;49:W417–W424. doi: 10.1093/nar/gkab273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang M., Cang Z., Wei G.-W. A topology-based network tree for the prediction of protein–protein binding affinity changes following mutation. Nat Mach Intell. 2020;2:116–123. doi: 10.1038/s42256-020-0149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robert P.A., Arulraj T., Meyer-Hermann M. Ymir: A 3D structural affinity model for multi-epitope vaccine simulations. iScience. 2021;24 doi: 10.1016/j.isci.2021.102979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang N., Chen Y., Lu H., Zhao F., Alvarez R.V., Goncearenco A., Panchenko A.R., Li M. MutaBind2: Predicting the Impacts of Single and Multiple Mutations on Protein-Protein Interactions. iScience. 2020;23 doi: 10.1016/j.isci.2020.100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang X., Zheng W., Pearce R., Zhang Y. SSIPe: accurately estimating protein–protein binding affinity change upon mutations using evolutionary profiles in combination with an optimized physical energy function. Bioinformatics. 2019;36:2429–2437. doi: 10.1093/bioinformatics/btz926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Viricel C., de Givry S., Schiex T., Barbe S. Cost function network-based design of protein–protein interactions: predicting changes in binding affinity. Bioinformatics. 2018;34:2581–2589. doi: 10.1093/bioinformatics/bty092. [DOI] [PubMed] [Google Scholar]
- 95.Romero-Molina S., Ruiz-Blanco Y.B., Mieres-Perez J., Harms M., Münch J., Ehrmann M., Sanchez-Garcia E. PPI-Affinity: A Web Tool for the Prediction and Optimization of Protein–Peptide and Protein–Protein Binding Affinity. J Proteome Res. 2022;21:1829–1841. doi: 10.1021/acs.jproteome.2c00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dunbrack R.L., Cang Z., Wei G.-W. TopologyNet: Topology based deep convolutional and multi-task neural networks for biomolecular property predictions. PLOS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Akbar R., Robert P.A., Weber C.R., Widrich M., Frank R., Pavlović M., Scheffer L., Chernigovskaya M., Snapkov I., Slabodkin A., Mehta B.B., Miho E., Lund-Johansen F., Andersen J.T., Hochreiter S., Hobæk Haff I., Klambauer G., Sandve G.K., Greiff V. In silico proof of principle of machine learning-based antibody design at unconstrained scale. mAbs. 2022;14:2031482. doi: 10.1080/19420862.2022.2031482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lim Y.W., Adler A.S., Johnson D.S. Predicting antibody binders and generating synthetic antibodies using deep learning. mAbs. 2022;14:2069075. doi: 10.1080/19420862.2022.2069075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cannon D.A., Shan L., Du Q., Shirinian L., Rickert K.W., Rosenthal K.L., Korade M., 3rd, van Vlerken-Ysla L.E., Buchanan A., Vaughan T.J., Damschroder M.M., Popovic B. Experimentally guided computational antibody affinity maturation with de novo docking, modelling and rational design. PLoS Comput Biol. 2019;15 doi: 10.1371/journal.pcbi.1006980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Saksena S.D., Liu G., Banholzer C., Horny G., Ewert S., Gifford D.K. Computational counterselection identifies nonspecific therapeutic biologic candidates, Cell Reports. Methods. 2022;2 doi: 10.1016/j.crmeth.2022.100254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Boughter C.T., Borowska M.T., Guthmiller J.J., Bendelac A., Wilson P.C., Roux B., Adams E.J. Biochemical patterns of antibody polyreactivity revealed through a bioinformatics-based analysis of CDR loops. eLife. 2020;9 doi: 10.7554/eLife.61393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mason D.M., Friedensohn S., Weber C.R., Jordi C., Wagner B., Meng S.M., Ehling R.A., Bonati L., Dahinden J., Gainza P., Correia B.E., Reddy S.T. Optimization of therapeutic antibodies by predicting antigen specificity from antibody sequence via deep learning. Nat Biomed Eng. 2021;5:600–612. doi: 10.1038/s41551-021-00699-9. [DOI] [PubMed] [Google Scholar]
- 103.Harvey E.P., Shin J.-E., Skiba M.A., Nemeth G.R., Hurley J.D., Wellner A., Shaw A.Y., Miranda V.G., Min J.K., Liu C.C., Marks D.S., Kruse A.C. An in silico method to assess antibody fragment polyreactivity. Nat Commun. 2022;13:7554. doi: 10.1038/s41467-022-35276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Alley E.C., Khimulya G., Biswas S., AlQuraishi M., Church G.M. Unified rational protein engineering with sequence-based deep representation learning. Nat Methods. 2019;16:1315–1322. doi: 10.1038/s41592-019-0598-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Linsky T.W., Noble K., Tobin A.R., Crow R., Carter L., Urbauer J.L., Baker D., Strauch E.-M. Sampling of structure and sequence space of small protein folds. Nat Commun. 2022;13:7151. doi: 10.1038/s41467-022-34937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang S., Tang H., Zhao Y., Zuo L. BayeStab: Predict Eff Mutat Protein Stab Uncertain Quantif. 2022;31 [Google Scholar]
- 107.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46:W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rodrigues C.H.M., Pires D.E.V., Ascher D.B. DynaMut2: Assessing changes in stability and flexibility upon single and multiple point missense mutations. 2021;30:60–69. doi: 10.1002/pro.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cao H., Wang J., He L., Qi Y., Zhang J.Z. DeepDDG: Predicting the Stability Change of Protein Point Mutations Using Neural Networks. J Chem Inf Model. 2019;59:1508–1514. doi: 10.1021/acs.jcim.8b00697. [DOI] [PubMed] [Google Scholar]
- 110.Tu H., Han Y., Wang Z., Li J. Clustered tree regression to learn protein energy change with mutated amino acid. Brief Bioinforma. 2022;23 doi: 10.1093/bib/bbac374. [DOI] [PubMed] [Google Scholar]
- 111.Iqbal S., Ge F., Li F., Akutsu T., Zheng Y., Gasser R.B., Yu D.-J., Webb G.I., Song J. PROST: AlphaFold2-aware Sequence-Based Predictor to Estimate Protein Stability Changes upon Missense Mutations. J Chem Inf Model. 2022;62:4270–4282. doi: 10.1021/acs.jcim.2c00799. [DOI] [PubMed] [Google Scholar]
- 112.Keskin O., Chen Y., Lu H., Zhang N., Zhu Z., Wang S., Li M. PremPS: Predicting the impact of missense mutations on protein stability. PLOS Comput Biol. 2020;16 doi: 10.1371/journal.pcbi.1008543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen C.-W., Lin M.-H., Liao C.-C., Chang H.-P., Chu Y.-W. iStable 2.0: Predicting protein thermal stability changes by integrating various characteristic modules. Comput Struct Biotechnol J. 2020;18:622–630. doi: 10.1016/j.csbj.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang Y., Urolagin S., Niroula A., Ding X., Shen B., Vihinen M. PON-tstab: Protein Variant Stability Predictor. Import Train Data Qual. 2018;19:1009. doi: 10.3390/ijms19041009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang Y., Zhao J., Zeng L., Vihinen M. ProTstab2 Predict Protein Therm Stabilities. 2022;23:10798. doi: 10.3390/ijms231810798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Caldararu O., Blundell T.L., Kepp K.P. Three simple properties explain protein stability change upon mutation. J Chem Inf Model. 2021;61:1981–1988. doi: 10.1021/acs.jcim.1c00201. [DOI] [PubMed] [Google Scholar]
- 117.Samaga Y.B.L., Raghunathan S., Priyakumar U.D. SCONES: self-consistent neural network for protein stability prediction upon mutation. J Phys Chem B. 2021;125:10657–10671. doi: 10.1021/acs.jpcb.1c04913. [DOI] [PubMed] [Google Scholar]
- 118.Gopi S., Devanshu D., Krishna P., Naganathan A.N. pStab: prediction of stable mutants, unfolding curves, stability maps and protein electrostatic frustration. Bioinformatics. 2017;34:875–877. doi: 10.1093/bioinformatics/btx697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen J., Zhang S., Wang W., Pang L., Zhang Q., Liu X. Mutation-Induced Impacts on the Switch Transformations of the GDP- and GTP-Bound K-Ras: Insights from Multiple Replica Gaussian Accelerated Molecular Dynamics and Free Energy Analysis. J Chem Inf Model. 2021;61:1954–1969. doi: 10.1021/acs.jcim.0c01470. [DOI] [PubMed] [Google Scholar]
- 120.Scarabelli G., Oloo E.O., Maier J.K.X., Rodriguez-Granillo A. Accurate Prediction of Protein Thermodynamic Stability Changes upon Residue Mutation using Free Energy Perturbation. J Mol Biol. 2022;434 doi: 10.1016/j.jmb.2021.167375. [DOI] [PubMed] [Google Scholar]
- 121.Saurabh S., Kalonia C., Li Z., Hollowell P., Waigh T., Li P., Webster J., Seddon J.M., Lu J.R., Bresme F. Understanding the Stabilizing Effect of Histidine on mAb Aggregation: A Molecular Dynamics Study. Mol Pharm. 2022;19:3288–3303. doi: 10.1021/acs.molpharmaceut.2c00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ding X., Zou Z., Brooks C.L. Iii, Deciphering protein evolution and fitness landscapes with latent space models. Nat Commun. 2019;10:5644. doi: 10.1038/s41467-019-13633-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li Q., Yan Y., Liu X., Zhang Z., Tian J., Wu N. Enhancing thermostability of a psychrophilic alpha-amylase by the structural energy optimization in the trajectories of molecular dynamics simulations. Int J Biol Macromol. 2020;142:624–633. doi: 10.1016/j.ijbiomac.2019.10.004. [DOI] [PubMed] [Google Scholar]
- 124.Bunc M., Hadži S., Graf C., Bončina M., Lah J. Aggregation time machine: a platform for the prediction and optimization of long-term antibody stability using short-term kinetic analysis. J Med Chem. 2022;65:2623–2632. doi: 10.1021/acs.jmedchem.1c02010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Heads J.T., Kelm S., Tyson K., Lawson A.D.G. A computational method for predicting the aggregation propensity of IgG1 and IgG4(P) mAbs in common storage buffers. mAbs. 2022;14:2138092. doi: 10.1080/19420862.2022.2138092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Prabakaran R., Rawat P., Kumar S., Michael Gromiha M. ANuPP: A Versatile Tool to Predict Aggregation Nucleating Regions in Peptides and Proteins. J Mol Biol. 2021;433 doi: 10.1016/j.jmb.2020.11.006. [DOI] [PubMed] [Google Scholar]
- 127.Moreira C.A., Philot E.A., Lima A.N., Scott A.L. Predicting regions prone to protein aggregation based on SVM algorithm. Appl Math Comput. 2019;359:502–511. [Google Scholar]
- 128.Louros N., Orlando G., De Vleeschouwer M., Rousseau F., Schymkowitz J. Structure-based machine-guided mapping of amyloid sequence space reveals uncharted sequence clusters with higher solubilities. Nat Commun. 2020;11:3314. doi: 10.1038/s41467-020-17207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Orlando G., Silva A., Macedo-Ribeiro S., Raimondi D., Vranken W. Accurate prediction of protein beta-aggregation with generalized statistical potentials. Bioinformatics. 2019;36:2076–2081. doi: 10.1093/bioinformatics/btz912. [DOI] [PubMed] [Google Scholar]
- 130.Wen L., Lyu M., Xiao H., Lan H., Zuo Z., Yin Z. Protein Aggregation and Performance Optimization Based on Microconformational Changes of Aromatic Hydrophobic Regions. Mol Pharm. 2018;15:2257–2267. doi: 10.1021/acs.molpharmaceut.8b00115. [DOI] [PubMed] [Google Scholar]
- 131.Feng J., Jiang M., Shih J., Chai Q. Antibody apparent solubility prediction from sequence by transfer learning. iScience. 2022;25 doi: 10.1016/j.isci.2022.105173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Navarro S., Ventura S. Computational re-design of protein structures to improve solubility. Expert Opin Drug Discov. 2019;14:1077–1088. doi: 10.1080/17460441.2019.1637413. [DOI] [PubMed] [Google Scholar]
- 133.Han X., Zhang L., Zhou K., Wang X. ProGAN: Protein solubility generative adversarial nets for data augmentation in DNN framework. Comput Chem Eng. 2019;131 [Google Scholar]
- 134.Raimondi D., Orlando G., Fariselli P., Moreau Y. Insight into the protein solubility driving forces with neural attention. PLoS Comput Biol. 2020;16 doi: 10.1371/journal.pcbi.1007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rawi R., Mall R., Kunji K., Shen C.-H., Kwong P.D., Chuang G.-Y. PaRSnIP: sequence-based protein solubility prediction using gradient boosting machine. Bioinformatics. 2017;34:1092–1098. doi: 10.1093/bioinformatics/btx662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Khurana S., Rawi R., Kunji K., Chuang G.-Y., Bensmail H., Mall R. DeepSol: a deep learning framework for sequence-based protein solubility prediction. Bioinformatics. 2018;34:2605–2613. doi: 10.1093/bioinformatics/bty166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hou Q., Kwasigroch J.M., Rooman M., Pucci F. SOLart: a structure-based method to predict protein solubility and aggregation. Bioinformatics. 2019;36:1445–1452. doi: 10.1093/bioinformatics/btz773. [DOI] [PubMed] [Google Scholar]
- 138.Y. Yang, L. Zeng, M. Vihinen, PON-Sol2: Prediction of Effects of Variants on Protein Solubility, 22, 2021: 8027. [DOI] [PMC free article] [PubMed]
- 139.Hon J., Marusiak M., Martinek T., Kunka A., Zendulka J., Bednar D., Damborsky J. SoluProt: prediction of soluble protein expression in Escherichia coli. Bioinformatics. 2021;37:23–28. doi: 10.1093/bioinformatics/btaa1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Paladin L., Piovesan D., Silvio C.E. Tosatto, SODA: prediction of protein solubility from disorder and aggregation propensity. Nucleic Acids Res. 2017;45:W236–W240. doi: 10.1093/nar/gkx412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Han X., Shih J., Lin Y., Chai Q., Cramer S.M. Development of QSAR models for in silico screening of antibody solubility. mAbs. 2022;14:2062807. doi: 10.1080/19420862.2022.2062807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Han X., Wang X., Zhou K. Develop machine learning-based regression predictive models for engineering protein solubility. Bioinformatics. 2019;35:4640–4646. doi: 10.1093/bioinformatics/btz294. [DOI] [PubMed] [Google Scholar]
- 143.Hawkins-Hooker A., Depardieu F., Baur S., Couairon G., Chen A., Bikard D. Generating functional protein variants with variational autoencoders. PLoS Comput Biol. 2021;17 doi: 10.1371/journal.pcbi.1008736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lai P.-K. DeepSCM: An efficient convolutional neural network surrogate model for the screening of therapeutic antibody viscosity. Comput Struct Biotechnol J. 2022;20:2143–2152. doi: 10.1016/j.csbj.2022.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Apgar J.R., Tam A.S.P., Sorm R., Moesta S., King A.C., Yang H., Kelleher K., Murphy D., D'Antona A.M., Yan G., Zhong X., Rodriguez L., Ma W., Ferguson D.E., Carven G.J., Bennett E.M., Lin L. Modeling and mitigation of high-concentration antibody viscosity through structure-based computer-aided protein design. PLoS One. 2020;15 doi: 10.1371/journal.pone.0232713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lai P.-K., Fernando A., Cloutier T.K., Gokarn Y., Zhang J., Schwenger W., Chari R., Calero-Rubio C., Trout B.L. Machine Learning Applied to Determine the Molecular Descriptors Responsible for the Viscosity Behavior of Concentrated Therapeutic Antibodies. Mol Pharm. 2021;18:1167–1175. doi: 10.1021/acs.molpharmaceut.0c01073. [DOI] [PubMed] [Google Scholar]
- 147.Kingsbury J.S., Saini A., Auclair S.M., Fu L., Lantz M.M., Halloran K.T., Calero-Rubio C., Schwenger W., Airiau C.Y., Zhang J., Gokarn Y.R. A Single Mol Descr Predict Solut Behav Ther antibodies. 2020;6:eabb0372. doi: 10.1126/sciadv.abb0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lai P.-K., Swan J.W., Trout B.L. Calculation of therapeutic antibody viscosity with coarse-grained models, hydrodynamic calculations and machine learning-based parameters. mAbs. 2021;13:1907882. doi: 10.1080/19420862.2021.1907882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tomar D.S., Li L., Broulidakis M.P., Luksha N.G., Burns C.T., Singh S.K., Kumar S. In-silico prediction of concentration-dependent viscosity curves for monoclonal antibody solutions. mAbs. 2017;9:476–489. doi: 10.1080/19420862.2017.1285479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Izadi S., Patapoff T.W., Walters B.T. Multiscale Coarse-Grained Approach to Investigate Self-Association of Antibodies. Biophys J. 2020;118:2741–2754. doi: 10.1016/j.bpj.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Prihoda D., Maamary J., Waight A., Juan V., Fayadat-Dilman L., Svozil D., Bitton D.A. BioPhi: A platform for antibody design, humanization, and humanness evaluation based on natural antibody repertoires and deep learning. mAbs. 2022;14:2020203. doi: 10.1080/19420862.2021.2020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sang Z., Xiang Y., Bahar I., Shi Y. Llamanade: An open-source computational pipeline for robust nanobody humanization. Structure. 2022;30:418–429. doi: 10.1016/j.str.2021.11.006. e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Marks C., Hummer A.M., Chin M., Deane C.M. Humanization of antibodies using a machine learning approach on large-scale repertoire data. Bioinformatics. 2021;37:4041–4047. doi: 10.1093/bioinformatics/btab434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Manavalan B., Govindaraj R.G., Shin T.H., Kim M.O., Lee G. iBCE-EL: A N Ensemble Learn Framew Improv Linear B-Cell Epitope Predict. 2018;9 doi: 10.3389/fimmu.2018.01695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Alghamdi W., Attique M., Alzahrani E., Ullah M.Z., Khan Y.D. LBCEPred: a machine learning model to predict linear B-cell epitopes. Brief Bioinforma. 2022;23 doi: 10.1093/bib/bbac035. [DOI] [PubMed] [Google Scholar]
- 157.Choi Y., Verma D., Griswold K.E., Bailey-Kellogg C. In: Computational Protein Design. Samish I., editor. Springer; New York, New York, NY: 2017. EpiSweep: Computationally Driven Reengineering of Therapeutic Proteins to Reduce Immunogenicity While Maintaining Function; pp. 375–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yachnin B.J., Mulligan V.K., Khare S.D., Bailey-Kellogg C. MHCEpitopeEnergy, a Flexible Rosetta-Based Biotherapeutic Deimmunization Platform. J Chem Inf Model. 2021;61:2368–2382. doi: 10.1021/acs.jcim.1c00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schubert B., Scharfe C., Donnes P., Hopf T., Marks D., Kohlbacher O. Population-specific design of de-immunized protein biotherapeutics. PLoS Comput Biol. 2018;14 doi: 10.1371/journal.pcbi.1005983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jespersen M.C., Mahajan S., Peters B., Nielsen M., Marcatili P. Antib Specif B-Cell Epitope Predict: Leverag-- Inf Antib-Antigen Protein Complex. 2019;10 doi: 10.3389/fimmu.2019.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nelapati A.K., Das B.K., Ponnan Ettiyappan J.B., Chakraborty D. In-silico epitope identification and design of Uricase mutein with reduced immunogenicity. Process Biochem. 2020;92:288–302. [Google Scholar]
- 163.Pennington L.F., Gasser P., Kleinboelting S., Zhang C., Skiniotis G., Eggel A., Jardetzky T.S. Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex. Nat Commun. 2021;12:7069. doi: 10.1038/s41467-021-27397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lu R.-M., Hwang Y.-C., Liu I.J., Lee C.-C., Tsai H.-Z., Li H.-J., Wu H.-C. Development of therapeutic antibodies for the treatment of diseases. J Biomed Sci. 2020;27:1. doi: 10.1186/s12929-019-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Laustsen A.H., Greiff V., Karatt-Vellatt A., Muyldermans S., Jenkins T.P. Animal Immunization, in Vitro Display Technologies, and Machine Learning for Antibody Discovery. Trends Biotechnol. 2021;39:1263–1273. doi: 10.1016/j.tibtech.2021.03.003. [DOI] [PubMed] [Google Scholar]
- 166.Hoogenboom H.R. Selecting and screening recombinant antibody libraries. Nat Biotechnol. 2005;23:1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 167.Hanes J., Schaffitzel C., Knappik A., Plückthun A. Picomolar affinity antibodies from a fully synthetic naive library selected and evolved by ribosome display. Nat Biotechnol. 2000;18:1287–1292. doi: 10.1038/82407. [DOI] [PubMed] [Google Scholar]
- 168.Markel U., Essani K.D., Besirlioglu V., Schiffels J., Streit W.R., Schwaneberg U. Advances in ultrahigh-throughput screening for directed enzyme evolution. Chem Soc Rev. 2020;49:233–262. doi: 10.1039/c8cs00981c. [DOI] [PubMed] [Google Scholar]
- 169.Maute R.L., Gordon S.R., Mayer A.T., McCracken M.N., Natarajan A., Ring N.G., Kimura R., Tsai J.M., Manglik A., Kruse A.C., Gambhir S.S., Weissman I.L., Ring A.M. Engineering high-affinity PD-1 variants for optimized immunotherapy and immuno-PET imaging. Proc Natl Acad Sci. 2015;112:E6506–E6514. doi: 10.1073/pnas.1519623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Taft J.M., Weber C.R., Gao B., Ehling R.A., Han J., Frei L., Metcalfe S.W., Overath M.D., Yermanos A., Kelton W., Reddy S.T. Deep mutational learning predicts ACE2 binding and antibody escape to combinatorial mutations in the SARS-CoV-2 receptor-binding domain. Cell. 2022;185:4008–4022. doi: 10.1016/j.cell.2022.08.024. e4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nimrod G., Fischman S., Austin M., Herman A., Keyes F., Leiderman O., Hargreaves D., Strajbl M., Breed J., Klompus S., Minton K., Spooner J., Buchanan A., Vaughan T.J., Ofran Y. Computational Design of Epitope-Specific Functional Antibodies. Cell Rep. 2018;25:2121–2131. doi: 10.1016/j.celrep.2018.10.081. e2125. [DOI] [PubMed] [Google Scholar]
- 172.Yang K.K., Wu Z., Arnold F.H. Machine-learning-guided directed evolution for protein engineering. Nat Methods. 2019;16:687–694. doi: 10.1038/s41592-019-0496-6. [DOI] [PubMed] [Google Scholar]
- 173.Wu Z., Kan S.B.J., Lewis R.D., Wittmann B.J., Arnold F.H. Machine learning-assisted directed protein evolution with combinatorial libraries. Proc Natl Acad Sci. 2019;116:8852–8858. doi: 10.1073/pnas.1901979116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Huang P.-S., Boyken S.E., Baker D. The coming of age of de novo protein design. Nature. 2016;537:320–327. doi: 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- 175.Walker S.P., Yallapragada V.V.B., Tangney M. Arming Yourself for The In Silico Protein Design Revolution. Trends Biotechnol. 2021;39:651–664. doi: 10.1016/j.tibtech.2020.10.003. [DOI] [PubMed] [Google Scholar]
- 176.Hummer A.M., Abanades B., Deane C.M. Advances in computational structure-based antibody design. Curr Opin Struct Biol. 2022;74 doi: 10.1016/j.sbi.2022.102379. [DOI] [PubMed] [Google Scholar]
- 177.Schissel C.K., Mohapatra S., Wolfe J.M., Fadzen C.M., Bellovoda K., Wu C.-L., Wood J.A., Malmberg A.B., Loas A., Gómez-Bombarelli R., Pentelute B.L. Deep learning to design nuclear-targeting abiotic miniproteins. Nat Chem. 2021;13:992–1000. doi: 10.1038/s41557-021-00766-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ovchinnikov S., Huang P.-S. Structure-based protein design with deep learning. Curr Opin Chem Biol. 2021;65:136–144. doi: 10.1016/j.cbpa.2021.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pan X., Kortemme T. Recent advances in de novo protein design: Principles, methods, and applications. J Biol Chem. 2021;296 doi: 10.1016/j.jbc.2021.100558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Woolfson D.N., Brief A. History of De Novo Protein Design: Minimal, Rational, and Computational. J Mol Biol. 2021;433 doi: 10.1016/j.jmb.2021.167160. [DOI] [PubMed] [Google Scholar]
- 181.Ferruz N., Höcker B. Controllable protein design with language models. Nat Mach Intell. 2022;4:521–532. [Google Scholar]
- 182.Leman J.K., Weitzner B.D., Lewis S.M., Adolf-Bryfogle J., Alam N., Alford R.F., Aprahamian M., Baker D., Barlow K.A., Barth P., Basanta B., Bender B.J., Blacklock K., Bonet J., Boyken S.E., Bradley P., Bystroff C., Conway P., Cooper S., Correia B.E., Coventry B., Das R., De Jong R.M., DiMaio F., Dsilva L., Dunbrack R., Ford A.S., Frenz B., Fu D.Y., Geniesse C., Goldschmidt L., Gowthaman R., Gray J.J., Gront D., Guffy S., Horowitz S., Huang P.-S., Huber T., Jacobs T.M., Jeliazkov J.R., Johnson D.K., Kappel K., Karanicolas J., Khakzad H., Khar K.R., Khare S.D., Khatib F., Khramushin A., King I.C., Kleffner R., Koepnick B., Kortemme T., Kuenze G., Kuhlman B., Kuroda D., Labonte J.W., Lai J.K., Lapidoth G., Leaver-Fay A., Lindert S., Linsky T., London N., Lubin J.H., Lyskov S., Maguire J., Malmström L., Marcos E., Marcu O., Marze N.A., Meiler J., Moretti R., Mulligan V.K., Nerli S., Norn C., Ó’Conchúir S., Ollikainen N., Ovchinnikov S., Pacella M.S., Pan X., Park H., Pavlovicz R.E., Pethe M., Pierce B.G., Pilla K.B., Raveh B., Renfrew P.D., Burman S.S.R., Rubenstein A., Sauer M.F., Scheck A., Schief W., Schueler-Furman O., Sedan Y., Sevy A.M., Sgourakis N.G., Shi L., Siegel J.B., Silva D.-A., Smith S., Song Y., Stein A., Szegedy M., Teets F.D., Thyme S.B., Wang R.Y.-R., Watkins A., Zimmerman L., Bonneau R. Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nat Methods. 2020;17:665–680. doi: 10.1038/s41592-020-0848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ding W., Nakai K., Gong H. Protein design via deep learning. Brief Bioinforma. 2022;(23) doi: 10.1093/bib/bbac102. bbac102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Frappier V., Keating A.E. Data-driven computational protein design. Curr Opin Struct Biol. 2021;69:63–69. doi: 10.1016/j.sbi.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Dilliard S.A., Cheng Q., Siegwart D.J. On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles. Proc Natl Acad Sci. 2021;118 doi: 10.1073/pnas.2109256118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ban Z., Yuan P., Yu F., Peng T., Zhou Q., Hu X. Machine learning predicts the functional composition of the protein corona and the cellular recognition of nanoparticles. Proc Natl Acad Sci. 2020;117:10492–10499. doi: 10.1073/pnas.1919755117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lazarovits J., Sindhwani S., Tavares A.J., Zhang Y., Song F., Audet J., Krieger J.R., Syed A.M., Stordy B., Chan W.C.W. Supervised Learning and Mass Spectrometry Predicts the in Vivo Fate of Nanomaterials. ACS Nano. 2019;13:8023–8034. doi: 10.1021/acsnano.9b02774. [DOI] [PubMed] [Google Scholar]
- 188.Haddad Y., Charousova M., Zivotska H., Splichal Z., Merlos Rodrigo M.A., Michalkova H., Krizkova S., Tesarova B., Richtera L., Vitek P., Stokowa-Soltys K., Hynek D., Milosavljevic V., Rex S., Heger Z. Norepinephrine transporter-derived homing peptides enable rapid endocytosis of drug delivery nanovehicles into neuroblastoma cells. J Nanobiotechnol. 2020;18:95. doi: 10.1186/s12951-020-00654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Song M., Fu W., Liu Y., Yao H., Zheng K., Liu L., Xue J., Xu P., Chen Y., Huang M., Li J. Unveiling the molecular mechanism of pH-dependent interactions of human serum albumin with chemotherapeutic agent doxorubicin: A combined spectroscopic and constant-pH molecular dynamics study. J Mol Liq. 2021;333 [Google Scholar]
- 190.Starr C.G., Tessier P.M. Selecting and engineering monoclonal antibodies with drug-like specificity. Curr Opin Biotechnol. 2019;60:119–127. doi: 10.1016/j.copbio.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wang W., Ohtake S. Science and art of protein formulation development. Int J Pharm. 2019;568 doi: 10.1016/j.ijpharm.2019.118505. [DOI] [PubMed] [Google Scholar]
- 192.Muralidhara B.K., Wong M. Critical considerations in the formulation development of parenteral biologic drugs. Drug Discov Today. 2020;25:574–581. doi: 10.1016/j.drudis.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 193.Krause M.E., Sahin E. Chemical and physical instabilities in manufacturing and storage of therapeutic proteins. Curr Opin Biotechnol. 2019;60:159–167. doi: 10.1016/j.copbio.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 194.Marabotti A., Scafuri B., Facchiano A. Predicting the stability of mutant proteins by computational approaches: an overview. Brief Bioinforma. 2021;22 doi: 10.1093/bib/bbaa074. bbaa074. [DOI] [PubMed] [Google Scholar]
- 195.Magliery T.J. Protein stability: computation, sequence statistics, and new experimental methods. Curr Opin Struct Biol. 2015;33:161–168. doi: 10.1016/j.sbi.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Dobson C.M. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 197.Goldenzweig A., Fleishman S.J. Principles of Protein Stability and Their Application in Computational Design. Annu Rev Biochem. 2018;87:105–129. doi: 10.1146/annurev-biochem-062917-012102. [DOI] [PubMed] [Google Scholar]
- 198.Smith A.M., Borkovec M., Trefalt G. Forces between solid surfaces in aqueous electrolyte solutions. Adv Colloid Interface Sci. 2020;275 doi: 10.1016/j.cis.2019.102078. [DOI] [PubMed] [Google Scholar]
- 199.Ni H., Amme R.C. Ion redistribution in an electric double layer. J Colloid Interface Sci. 2003;260:344–348. doi: 10.1016/s0021-9797(02)00229-1. [DOI] [PubMed] [Google Scholar]
- 200.Xu A.Y., Castellanos M.M., Mattison K., Krueger S., Curtis J.E. Studying Excipient Modulated Physical Stability and Viscosity of Monoclonal Antibody Formulations Using Small-Angle Scattering. Mol Pharm. 2019;16:4319–4338. doi: 10.1021/acs.molpharmaceut.9b00687. [DOI] [PubMed] [Google Scholar]
- 201.Wang S., Tang H., Zhao Y., Zuo L. BayeStab: Predicting effects of mutations on protein stability with uncertainty quantification. Protein Sci. 2022;31 doi: 10.1002/pro.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nisthal A., Wang C.Y., Ary M.L., Mayo S.L. Protein stability engineering insights revealed by domain-wide comprehensive mutagenesis. Proc Natl Acad Sci. 2019;116:16367–16377. doi: 10.1073/pnas.1903888116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tu H., Han Y., Wang Z., Li J. Clustered tree regression to learn protein energy change with mutated amino acid. Brief Bioinforma. 2022;23 doi: 10.1093/bib/bbac374. bbac374. [DOI] [PubMed] [Google Scholar]
- 204.Kuai R., Ochyl L.J., Bahjat K.S., Schwendeman A., Moon J.J. Designer vaccine nanodiscs for personalized cancer immunotherapy. Nat Mater. 2017;16:489–496. doi: 10.1038/nmat4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chen L., Yu C., Xu W., Xiong Y., Cheng P., Lin Z., Zhang Z., Knoedler L., Panayi A.C., Knoedler S., Wang J., Mi B., Liu G. Dual-Targeted Nanodiscs Revealing the Cross-Talk between Osteogenic Differentiation of Mesenchymal Stem Cells and Macrophages. ACS Nano. 2023;17:3153–3167. doi: 10.1021/acsnano.2c12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kadiyala P., Li D., Nuñez F.M., Altshuler D., Doherty R., Kuai R., Yu M., Kamran N., Edwards M., Moon J.J., Lowenstein P.R., Castro M.G., Schwendeman A. High-density lipoprotein-mimicking nanodiscs for chemo-immunotherapy against glioblastoma multiforme. ACS Nano. 2019;13:1365–1384. doi: 10.1021/acsnano.8b06842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wang J., Wang A.Z., Lv P., Tao W., Liu G. Advancing the pharmaceutical potential of bioinorganic hybrid lipid-based assemblies. Adv Sci. 2018;5:1800564. doi: 10.1002/advs.201800564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lv Y., Chen X., Chen Z., Shang Z., Li Y., Xu W., Mo Y., Wang X., Xu D., Li S., Wang Z., Wu M., Wang J. Melittin Tryptophan Substitution with a Fluorescent Amino Acid Reveals the Structural Basis of Selective Antitumor Effect and Subcellular Localization in Tumor Cells. Toxins. 2022 doi: 10.3390/toxins14070428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Xu D., Chen X., Li Y., Chen Z., Xu W., Wang X., Lv Y., Wang Z., Wu M., Liu G., Wang J. Reconfigurable Peptide Analogs of Apolipoprotein A-I Reveal Tunable Features of Nanodisc Assembly. Langmuir. 2023;39:1262–1276. doi: 10.1021/acs.langmuir.2c03082. [DOI] [PubMed] [Google Scholar]
- 210.Gupta N., Ansari A., Dhoke G.V., Chilamari M., Sivaccumar J., Kumari S., Chatterjee S., Goyal R., Dutta P.K., Samarla M., Mukherjee M., Sarkar A., Mandal S.K., Rai V., Biswas G., Sengupta A., Roy S., Roy M., Sengupta S. Computationally designed antibody–drug conjugates self-assembled via affinity ligands. Nat Biomed Eng. 2019;3:917–929. doi: 10.1038/s41551-019-0470-8. [DOI] [PubMed] [Google Scholar]
- 211.Shah M. Commentary: New perspectives on protein aggregation during Biopharmaceutical development. Int J Pharm. 2018;552:1–6. doi: 10.1016/j.ijpharm.2018.09.049. [DOI] [PubMed] [Google Scholar]
- 212.Ebo J.S., Guthertz N., Radford S.E., Brockwell D.J. Using protein engineering to understand and modulate aggregation. Curr Opin Struct Biol. 2020;60:157–166. doi: 10.1016/j.sbi.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Meric G., Robinson A.S., Roberts C.J. Driving Forces for Nonnative Protein Aggregation and Approaches to Predict Aggregation-Prone Regions. Annu Rev Chem Biomol Eng. 2017;8:139–159. doi: 10.1146/annurev-chembioeng-060816-101404. [DOI] [PubMed] [Google Scholar]
- 214.Roberts C.J. Therapeutic protein aggregation: mechanisms, design, and control. Trends Biotechnol. 2014;32:372–380. doi: 10.1016/j.tibtech.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Santos J., Pujols J., Pallarès I., Iglesias V., Ventura S. Computational prediction of protein aggregation: Advances in proteomics, conformation-specific algorithms and biotechnological applications. Comput Struct Biotechnol J. 2020;18:1403–1413. doi: 10.1016/j.csbj.2020.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Prabakaran R., Rawat P., Thangakani A.M., Kumar S., Gromiha M.M. Protein aggregation: in silico algorithms and applications. Biophys Rev. 2021;13:71–89. doi: 10.1007/s12551-021-00778-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chennamsetty N., Voynov V., Kayser V., Helk B., Trout B.L. Design of therapeutic proteins with enhanced stability. Proc Natl Acad Sci. 2009;106:11937–11942. doi: 10.1073/pnas.0904191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sankar K., Krystek S.R., Jr, Carl S.M., Day T., Maier J.K.X. AggScore: Prediction of aggregation-prone regions in proteins based on the distribution of surface patches. Protein: Struct, Funct, Bioinforma. 2018;86:1147–1156. doi: 10.1002/prot.25594. [DOI] [PubMed] [Google Scholar]
- 219.Prabakaran R., Rawat P., Kumar S., Gromiha M.M. Evaluation of in silico tools for the prediction of protein and peptide aggregation on diverse datasets. Brief Bioinforma. 2021;(22) doi: 10.1093/bib/bbab240. bbab240. [DOI] [PubMed] [Google Scholar]
- 220.d’Arcy R., El Mohtadi F., Francini N., DeJulius C.R., Back H., Gennari A., Geven M., Lopez-Cavestany M., Turhan Z.Y., Yu F., Lee J.B., King M.R., Kagan L., Duvall C.L., Tirelli N. A Reactive Oxygen Species-Scavenging ‘Stealth’ Polymer, Poly(thioglycidyl glycerol), Outperforms Poly(ethylene glycol) in Protein Conjugates and Nanocarriers and Enhances Protein Stability to Environmental and Biological Stressors. J Am Chem Soc. 2022;144:21304–21317. doi: 10.1021/jacs.2c09232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Thakral S., Sonje J., Munjal B., Suryanarayanan R. Stabilizers and their interaction with formulation components in frozen and freeze-dried protein formulations. Adv Drug Deliv Rev. 2021;173:1–19. doi: 10.1016/j.addr.2021.03.003. [DOI] [PubMed] [Google Scholar]
- 222.Ohtake S., Kita Y., Arakawa T. Interactions of formulation excipients with proteins in solution and in the dried state. Adv Drug Deliv Rev. 2011;63:1053–1073. doi: 10.1016/j.addr.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 223.Tamasi M.J., Patel R.A., Borca C.H., Kosuri S., Mugnier H., Upadhya R., Murthy N.S., Webb M.A., Gormley A.J. Machine Learning on a Robotic Platform for the Design of Polymer–Protein Hybrids. Adv Mater. 2022;34:2201809. doi: 10.1002/adma.202201809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Jo S., Xu A., Curtis J.E., Somani S., MacKerell A.D., Jr. Computational Characterization of Antibody–Excipient Interactions for Rational Excipient Selection Using the Site Identification by Ligand Competitive Saturation-Biologics Approach. Mol Pharm. 2020;17:4323–4333. doi: 10.1021/acs.molpharmaceut.0c00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Chang C.C.H., Song J., Tey B.T., Ramanan R.N. Bioinformatics approaches for improved recombinant protein production in Escherichia coli: protein solubility prediction. Brief Bioinforma. 2014;15:953–962. doi: 10.1093/bib/bbt057. [DOI] [PubMed] [Google Scholar]
- 226.González-Montalbán N., García-Fruitós E., Villaverde A. Recombinant protein solubility—does more mean better? Nat Biotechnol. 2007;25:718–720. doi: 10.1038/nbt0707-718. [DOI] [PubMed] [Google Scholar]
- 227.Waldo G.S. Genetic screens and directed evolution for protein solubility. Curr Opin Chem Biol. 2003;7:33–38. doi: 10.1016/s1367-5931(02)00017-0. [DOI] [PubMed] [Google Scholar]
- 228.Cabantous S., Waldo G.S. In vivo and in vitro protein solubility assays using split GFP. Nat Methods. 2006;3:845–854. doi: 10.1038/nmeth932. [DOI] [PubMed] [Google Scholar]
- 229.de Marco A. Protocol for preparing proteins with improved solubility by co-expressing with molecular chaperones in Escherichia coli. Nat Protoc. 2007;2:2632–2639. doi: 10.1038/nprot.2007.400. [DOI] [PubMed] [Google Scholar]
- 230.Chai Q., Shih J., Weldon C., Phan S., Jones B.E. Development of a high-throughput solubility screening assay for use in antibody discovery. mAbs. 2019;11:747–756. doi: 10.1080/19420862.2019.1589851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Klesmith J.R., Bacik J.-P., Wrenbeck E.E., Michalczyk R., Whitehead T.A. Trade-offs between enzyme fitness and solubility illuminated by deep mutational scanning. Proc Natl Acad Sci. 2017;114:2265–2270. doi: 10.1073/pnas.1614437114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Jezek J., Rides M., Derham B., Moore J., Cerasoli E., Simler R., Perez-Ramirez B. Viscosity of concentrated therapeutic protein compositions. Adv Drug Deliv Rev. 2011;63:1107–1117. doi: 10.1016/j.addr.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 233.Watt R.P., Khatri H., Dibble A.R.G. Injectability as a function of viscosity and dosing materials for subcutaneous administration. Int J Pharm. 2019;554:376–386. doi: 10.1016/j.ijpharm.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 234.Daugherty A.L., Mrsny R.J. Formulation and delivery issues for monoclonal antibody therapeutics. Adv Drug Deliv Rev. 2006;58:686–706. doi: 10.1016/j.addr.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 235.Mitragotri S., Burke P.A., Langer R. Overcoming the challenges in administering biopharmaceuticals: formulation and delivery strategies. Nat Rev Drug Discov. 2014;13:655–672. doi: 10.1038/nrd4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Shire S.J. Formulation and manufacturability of biologics. Curr Opin Biotechnol. 2009;20:708–714. doi: 10.1016/j.copbio.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 237.Zhang Z., Liu Y. Recent progresses of understanding the viscosity of concentrated protein solutions. Curr Opin Chem Eng. 2017;16:48–55. [Google Scholar]
- 238.Yearley Eric J., Godfrin Paul D., Perevozchikova T., Zhang H., Falus P., Porcar L., Nagao M., Curtis Joseph E., Gawande P., Taing R., Zarraga Isidro E., Wagner Norman J., Liu Y. Observation of Small Cluster Formation in Concentrated Monoclonal Antibody Solutions and Its Implications to Solution Viscosity. Biophys J. 2014;106:1763–1770. doi: 10.1016/j.bpj.2014.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zidar M., Rozman P., Belko-Parkel K., Ravnik M. Control of viscosity in biopharmaceutical protein formulations. J Colloid Interface Sci. 2020;580:308–317. doi: 10.1016/j.jcis.2020.06.105. [DOI] [PubMed] [Google Scholar]
- 240.Agrawal N.J., Helk B., Kumar S., Mody N., Sathish H.A., Samra H.S., Buck P.M., Li L., Trout B.L. Computational tool for the early screening of monoclonal antibodies for their viscosities. mAbs. 2016;8:43–48. doi: 10.1080/19420862.2015.1099773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Banik N., Braun S., Gerit Brandenburg J., Fricker G., Kalonia D.S., Rosenkranz T. Technology development to evaluate the effectiveness of viscosity reducing excipients. Int J Pharm. 2022;626 doi: 10.1016/j.ijpharm.2022.122204. [DOI] [PubMed] [Google Scholar]
- 242.Proj M., Zidar M., Lebar B., Strašek N., Miličić G., Žula A., Gobec S. Discovery of compounds with viscosity-reducing effects on biopharmaceutical formulations with monoclonal antibodies. Comput Struct Biotechnol J. 2022;20:5420–5429. doi: 10.1016/j.csbj.2022.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Sauna Z.E., Lagassé D., Pedras-Vasconcelos J., Golding B., Rosenberg A.S. Evaluating and Mitigating the Immunogenicity of Therapeutic Proteins. Trends Biotechnol. 2018;36:1068–1084. doi: 10.1016/j.tibtech.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 244.Harris J.M., Chess R.B. Effect of pegylation on pharmaceuticals. Nat Rev Drug Discov. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 245.Schellenberger V., Wang C.-w, Geething N.C., Spink B.J., Campbell A., To W., Scholle M.D., Yin Y., Yao Y., Bogin O., Cleland J.L., Silverman J., Stemmer W.P.C. A recombinant polypeptide extends the in vivo half-life of peptides and proteins in a tunable manner. Nat Biotechnol. 2009;27:1186–1190. doi: 10.1038/nbt.1588. [DOI] [PubMed] [Google Scholar]
- 246.Binder U., Skerra A. PASylation®: A versatile technology to extend drug delivery. Curr Opin Colloid Interface Sci. 2017;31:10–17. [Google Scholar]
- 247.Lee E.C., Liang Q., Ali H., Bayliss L., Beasley A., Bloomfield-Gerdes T., Bonoli L., Brown R., Campbell J., Carpenter A., Chalk S., Davis A., England N., Fane-Dremucheva A., Franz B., Germaschewski V., Holmes H., Holmes S., Kirby I., Kosmac M., Legent A., Lui H., Manin A., O'Leary S., Paterson J., Sciarrillo R., Speak A., Spensberger D., Tuffery L., Waddell N., Wang W., Wells S., Wong V., Wood A., Owen M.J., Friedrich G.A., Bradley A. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nat Biotechnol. 2014;32:356–363. doi: 10.1038/nbt.2825. [DOI] [PubMed] [Google Scholar]
- 248.Peters B., Nielsen M., Sette A. T Cell Epitope Predictions. Annu Rev Immunol. 2020;38:123–145. doi: 10.1146/annurev-immunol-082119-124838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Zinsli L.V., Stierlin N., Loessner M.J., Schmelcher M. Deimmunization of protein therapeutics – Recent advances in experimental and computational epitope prediction and deletion. Comput Struct Biotechnol J. 2021;19:315–329. doi: 10.1016/j.csbj.2020.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Griswold K.E., Bailey-Kellogg C. Design and engineering of deimmunized biotherapeutics. Curr Opin Struct Biol. 2016;39:79–88. doi: 10.1016/j.sbi.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Nagata S., Pastan I. Removal of B cell epitopes as a practical approach for reducing the immunogenicity of foreign protein-based therapeutics. Adv Drug Deliv Rev. 2009;61:977–985. doi: 10.1016/j.addr.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Sela-Culang I., Ofran Y., Peters B. Antibody specific epitope prediction—emergence of a new paradigm. Curr Opin Virol. 2015;11:98–102. doi: 10.1016/j.coviro.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Gustafsson E., Rosén A., Barchan K., van Kessel K.P.M., Haraldsson K., Lindman S., Forsberg C., Ljung L., Bryder K., Walse B., Haas P.-J., van Strijp J.A.G., Furebring C. Directed evolution of chemotaxis inhibitory protein of Staphylococcus aureus generates biologically functional variants with reduced interaction with human antibodies. Protein Eng, Des Sel. 2010;23:91–101. doi: 10.1093/protein/gzp062. [DOI] [PubMed] [Google Scholar]
- 254.Liu W., Onda M., Lee B., Kreitman R.J., Hassan R., Xiang L., Pastan I. Recombinant immunotoxin engineered for low immunogenicity and antigenicity by identifying and silencing human B-cell epitopes. Proc Natl Acad Sci. 2012;109:11782–11787. doi: 10.1073/pnas.1209292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Lin J.C., Ettinger R.A., Schuman J.T., Zhang A.H., Wamiq-Adhami M., Nguyen P.C., Nakaya-Fletcher S.M., Puranik K., Thompson A.R., Pratt K.P. Six amino acid residues in a 1200 Å2 interface mediate binding of factor VIII to an IgG4κ inhibitory antibody. PloS One. 2015;10 doi: 10.1371/journal.pone.0116577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Khetan R., Curtis R., Deane C.M., Hadsund J.T., Kar U., Krawczyk K., Kuroda D., Robinson S.A., Sormanni P., Tsumoto K., Warwicker J., Martin A.C.R. Current advances in biopharmaceutical informatics: guidelines, impact and challenges in the computational developability assessment of antibody therapeutics. mAbs. 2022;14:2020082. doi: 10.1080/19420862.2021.2020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 258.Bender A., Cortes-Ciriano I. Artificial intelligence in drug discovery: what is realistic, what are illusions? Part 2: a discussion of chemical and biological data. Drug Discov Today. 2021;26:1040–1052. doi: 10.1016/j.drudis.2020.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Steinwandter V., Borchert D., Herwig C. Data science tools and applications on the way to Pharma 4.0. Drug Discov Today. 2019;24:1795–1805. doi: 10.1016/j.drudis.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 260.Bender A., Cortés-Ciriano I. Artificial intelligence in drug discovery: what is realistic, what are illusions? Part 1: Ways to make an impact, and why we are not there yet. Drug Discov Today. 2021;26:511–524. doi: 10.1016/j.drudis.2020.12.009. [DOI] [PubMed] [Google Scholar]
- 261.Kmiecik S., Gront D., Kolinski M., Wieteska L., Dawid A.E., Kolinski A. Coarse-Grained Protein Models and Their Applications. Chem Rev. 2016;116:7898–7936. doi: 10.1021/acs.chemrev.6b00163. [DOI] [PubMed] [Google Scholar]
- 262.Polêto M.D., Lemkul J.A. Integration of experimental data and use of automated fitting methods in developing protein force fields. Commun Chem. 2022;5:38. doi: 10.1038/s42004-022-00653-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Bhatia H., Carpenter T.S., Ingólfsson H.I., Dharuman G., Karande P., Liu S., Oppelstrup T., Neale C., Lightstone F.C., Van Essen B., Glosli J.N., Bremer P.-T. Machine-learning-based dynamic-importance sampling for adaptive multiscale simulations. Nat Mach Intell. 2021;3:401–409. [Google Scholar]
- 264.Vlachas P.R., Arampatzis G., Uhler C., Koumoutsakos P. Multiscale simulations of complex systems by learning their effective dynamics. Nat Mach Intell. 2022;4:359–366. [Google Scholar]
- 265.Lazim R., Suh D., Choi S. Advances in molecular dynamics simulations and enhanced sampling methods for the study of protein systems. Int J Mol Sci. 2020 doi: 10.3390/ijms21176339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Schlick T., Portillo-Ledesma S. Biomolecular modeling thrives in the age of technology, Nature Computational. Science. 2021;1:321–331. doi: 10.1038/s43588-021-00060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Noé F., Tkatchenko A., Müller K.-R., Clementi C. Machine learning for molecular simulation. Annu Rev Phys Chem. 2020;71:361–390. doi: 10.1146/annurev-physchem-042018-052331. [DOI] [PubMed] [Google Scholar]
- 268.Zhu J., Wang J., Han W., Xu D. Neural relational inference to learn long-range allosteric interactions in proteins from molecular dynamics simulations. Nat Commun. 2022;13:1661. doi: 10.1038/s41467-022-29331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Gentile F., Yaacoub J.C., Gleave J., Fernandez M., Ton A.-T., Ban F., Stern A., Cherkasov A. Artificial intelligence–enabled virtual screening of ultra-large chemical libraries with deep docking. Nat Protoc. 2022;17:672–697. doi: 10.1038/s41596-021-00659-2. [DOI] [PubMed] [Google Scholar]
- 270.Patel V., Shah M. Artificial intelligence and machine learning in drug discovery and development. Intell Med. 2022;2:134–140. [Google Scholar]