Abstract
Preservatives are essential components in cosmetic products, but their safety issues have attracted widespread attention. There is an urgent need for safe and effective alternatives. Antimicrobial peptides (AMPs) are part of the innate immune system and have potent antimicrobial properties. Using machine learning-assisted rational design, we obtained a novel antibacterial peptide, IK-16-1, with significant antibacterial activity and maintaining safety based on β-defensins. IK-16-1 has broad-spectrum antimicrobial properties against Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, and Candida albicans, and has no haemolytic activity. The use of IK-16-1 holds promise in the cosmetics industry, since it can serve as a preservative synergist to reduce the amount of other preservatives in cosmetics. This study verified the feasibility of combining computational design with artificial intelligence prediction to design AMPs, achieving rapid screening and reducing development costs.
Subject terms: Biotechnology, Computational biology and bioinformatics, Microbiology
Introduction
The cosmetics industry is one of the most rapidly developing sectors worldwide1. Preservatives are essential ingredients in cosmetic products that inhibit microbial growth and prevent spoilage2. Common preservatives include parabens, formaldehyde releasers, isothiazolinones, phenoxyethanol, and organic acids3. However, preservatives are among the most common allergens in cosmetics4. Some preservatives can cause cosmetic contact dermatitis5, such as methylisothiazolinone (MI)6, methylchloroisothiazolinone/methylisothiazolinone (MCI/MI)7, parabens8, and formaldehyde releasers9. Furthermore, certain preservatives in leave-on cosmetics may disrupt skin microbiota balance10. Cosmetic products with low or no preservatives have gained popularity. While combinations of various preservatives represents a current method of improvement11, it cannot fundamentally solve this problem. Therefore, there is an urgent need for safe, potent, broad-spectrum antimicrobial agents.
Other substances present in cosmetic products that also have antibacterial effects are known as ‘preservative synergists’8. Mixing preservatives and preservative synergists effectively decreases the concentration of allergenic preservatives. Certain antibacterial substances, such as glycolipids12 and new antimicrobial nanomaterials13–15, may be used as preservative synergists. However, there are few reports on the development of biomimetic antimicrobial peptide derived from human endogenous proteins for use as cosmetic preservatives.
Antimicrobial peptides (AMPs) are oligopeptides with varying numbers of amino acids (from 5 units to > 100 units)16. AMPs are important components of the innate immune system in many organisms and provide the first line of defense against various pathogens17. AMPs exhibit various activities against Gram-negative and Gram-positive bacteria, fungi, mycobacteria, and some enveloped viruses18,19. Most AMPs can be classified into four groups according to their secondary structure: α-helical, β-sheet, loop, and extended peptides16. Among these structural groups, α-helix and β-sheet structures are more common20. α-helical peptides are the most studied AMPs to date. In α-helix structures, the distance between two adjacent amino acids is around 0.15 nm, and the angle between them with respect to the centre is around 100° from the top view21. Human β defensin type 3 (hBD-3) is a cysteine-rich small antibacterial peptide with broad-spectrum antimicrobial activity and may play an important role in innate epithelial defense. Because hBD-3 kills bacteria by disrupting the cell membrane, which is relatively stable in chemical composition, its antibacterial activity will not cause drug resistance22. Thus, hBD-3 could serve as a potential AMP for designing novel preservatives. However, natural AMPs are large protein molecules that are unstable in various in vitro environments, which limits their applications. Therefore, developing biomimetic AMPs with improved efficacy, greater stability, and lower costs is challenging. Machine learning (ML) is described as the capacity of a system to learn from problem-specific training data to automate the process of analytical model building and solving associated tasks23. ML greatly improves work efficiency and accuracy. Driven by increasing computer power and algorithmic advances, ML has become a powerful tool for finding data patterns24, and it is widely applied in various fields, such as biology25,26, medicine27,28, healthcare29, environment30, fuel cells31 and energy32. ML methods can also be used to develop new AMPs. As more insights are gained into the antimicrobial mechanism, sequence, and structure, ML can gather information to expedite AMP screening and design33–35.
The primary objective of this study was to develop safe and potent broad-spectrum antimicrobial agents to reduce the use of preservatives in cosmetics. In this study, the artificial antibacterial peptide was designed based on hBDs through computational simulation and artificial intelligence (AI) based ML tools, and their antibacterial and haemolytic properties were predicted.
Methods
Designing antimicrobial peptides
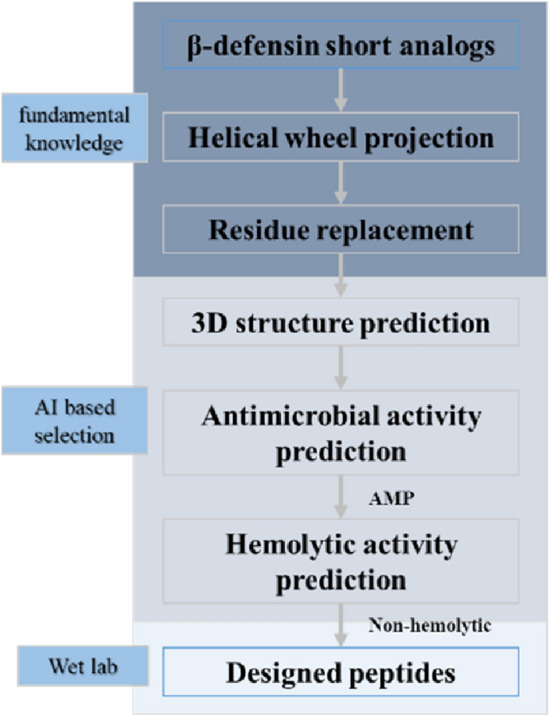
AI deep learning based on prediction algorithms and rational molecular design has been used to develop new biomimetic AMPs based on endogenous human proteins36–38. Specifically, short analogues of β-defensin were used as template peptides and helical wheel projection was conducted. Negatively charged amino acid residues, including Asp (D) and Glu (E), were substituted with positively charged amino acid residues, including Lys (K) and Arg (R), to increase the net positive charge. Several residues were adjusted such that the hydrophobic and hydrophilic residues were located on opposite sides of the helical surface to improve the pore-forming ability of the microbial membrane33. The ab-initio three-dimensional structures of peptides were studied to support the α-helical conformations. Peptides were predicted for their antimicrobial ability by some AI programs39–42. For safety reasons, the haemolytic activity was also predicted using an available AI algorithm43. Antimicrobial activity was predicted using the Antimicrobial Peptide Scanner version 2 (https://www.dveltri.com/ascan/v2/ascan.html), and haemolytic activity was predicted using HAPPENN (http://research.timmons.eu/happenn). All the new top-scoring biomimetic peptides were synthesised and screened for their microbial inhibitory activity.
Disk diffusion test
Four types of bacteria and fungi, namely, Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, and Candida albicans (ATCC, Manassas, VA, USA), were cultured under standard conditions. Candida albicans was cultured with Sabouraud glucose broth medium (Solarbio Life Science, Beijing, China) at 30 °C and 200 rpm; Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa were cultured with Luria Broth (LB) liquid medium (Solarbio Life Science, Beijing, China) at 37 °C and 200 rpm. The paper disks containing 30 μg Kanamycin (Beyotime Biotechnology, Shanghai, China) and 30 μg Amphotericin (Beyotime Biotechnology) were assigned as the positive control, and the medium was assigned as the negative control. The paper disks incorporated with 30 μg of AMP were deposited on the surface of the agar plates pre-inoculated with the bacterial strain to be tested. Specifically, 60 μg AMPs were used to test against Candida albicans. The agar plates were incubated for 24 h under appropriate conditions. Candida albicans was cultured in Sabouraud glucose agar medium in an inverted incubator at 30 °C for 12 h, and the remainder was cultured in LB solid medium in an inverted incubator at 37 °C for 12 h. The diameter of the inhibition zone was measured for statistical analysis. Each test was performed in triplicates.
Minimum inhibitory concentration
Four types of bacteria and fungi–Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, and Candida albicans–were cultured under standard conditions. Candida albicans was cultured in Sabouraud glucose broth medium (Solarbio Life Science, Beijing, China) in a static incubator at 30 °C for 12 h, and the remainder were cultured with LB liquid medium (Solarbio Life Science, Beijing, China) in a static incubator at 37 °C for 12 h. The bacterial order of magnitude was 104/mL–105/mL. A series of two-fold dilutions were performed from the stock solution to obtain different concentrations of peptides from 250 to 1.95 μg/mL, with a total volume of 200 μL per well in a 96-well plate, and then assayed for absorbance at 540 nm. This medium was used as the solvent and was designated as the negative control. Approximately 1 × 104 cells were incubated in a medium containing peptides. After incubating the cells at the appropriate temperature for 24 h, the minimum inhibitory concentration was the lowest peptide concentration that limited visible microbial growth.
Hemolytic activity
For the haemolysis studies, rabbit erythrocytes (Fenghui Biology, Hunan, China) were obtained from healthy blood donors. Blood samples mixed with 0.25% potassium oxalate were immediately separated by centrifugation at 1500 rpm for 10 min. Subsequently, the cells were washed three times with phosphate buffered saline (PBS, Yuanpei Biology, Shanghai, China) and diluted to a 5% suspension with PBS. 250 μL of erythrocyte stock dispersion was mixed with 250 μL of PBS containing different concentrations of AMPs and incubated at 37 °C for 1 h. Intact erythrocytes were removed by centrifugation at 10,000 rpm for 5 min. The absorbance of the resulting supernatant was measured at 540 nm. PBS was used as a negative control (0% lysis), and 0.1% SDS (Solarbio Life Sciences) was used as a positive control (100% lysis). The haemolysis rate was determined using Eq. (1).
| 1 |
where Dtest, Dnc, and Dpc are the 540 nm absorbance of the tested sample, negative control, and positive control, respectively.
Cytotoxicity
HaCaT cells (Fenghui Biology, Hunan, China), a long-lived, spontaneously immortalised human keratinocyte line, was grown in Dulbecco's Modified Eagle Medium (Wisent Bioproducts, Quebec, Canada) supplemented with 10% heat-inactivated foetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA) and 1% penicillin–streptomycin (Langke Biology, Suzhou, China) at 37 ℃ and 5% CO2 until 80–90% of fusion was measured. The experiments were performed in 96-well plates with a final volume of 100 μL of medium per well. HaCaT cell proliferation was tested using the cell counting kit-8 assay (Dojindo Laboratories, Tokyo, Japan). The cells were seeded at a density of 0.5 × 105–1.0 × 105 cells/mL and grown at 37 ℃ until 80–90% fusion was measured. Subsequently, samples at specific concentrations were added to the medium. After incubating the cells for 24 h, all media were removed, and the cells were washed three times with PBS. Next, the cells were incubated for 4 h with fresh medium (100 µL/well) containing 10 µL of the probe. The absorbance was read at 450 nm using a microplate reader.
Antimicrobial assay in products
The basic preservation system contained 1.8% 1,3-propanediol, 0.09% 1,2-Hexanediol, and 0.036% capryl hydroxamic acid. In the synergy test, the 5 μg/mL antimicrobial peptide was mixed with the preservative system and compared with the preservative system without antimicrobial peptide. Four types of bacteria and fungi–Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, and Candida albicans–were cultured under standard conditions. Candida albicans was cultured with Sabouraud glucose broth medium (Solarbio Life Science, Beijing, China) at 30 °C and 200 rpm; Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa were cultured with LB liquid medium (Solarbio Life Science, Beijing, China) at 37 °C and 200 rpm. Following culturing, 1.0 × 106 CFU/mL was added to the cosmetic products and mixed thoroughly. At hour 2 (day 0), day 3, day 7, day 14, and day 22, 1.0 mL of the product was collected and neutralised for 30 min using a 9 mL neutraliser. The 1.0 mL of the neutralised product was centrifuged at 12,000 rpm for 5 min to remove the suspension. The product was resuspended in 0.2 mL of fresh medium, and the CFUs were calculated using the spreading plate method. CFU refers to the number of individual colonies of any microorganism that can grow on a plate of media. The standard unit of measure for CFU is the number of culturable microorganisms present per 1 mL of culture (CFU/mL). The colony forming units per mL (CFU/mL) was determined using Eq. (2).
| 2 |
where N is the number of colonies on the counting plate; d is the sampling volume (mL); V is the dilution multiple.
Results
The design of antimicrobial peptides
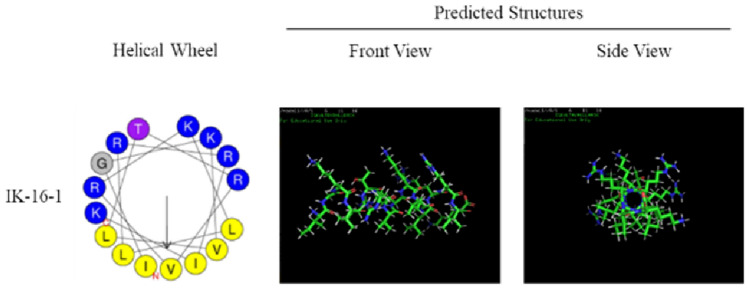
The AMP was selected from the mesenchymal stem cell secretome and used as a template. Figure 1 illustrates the design process of the antibacterial peptides. We selected the amino acid sequences from the 30th to 45th position of beta-defensin 103 as the template and optimised it to determine the amino acid sequence of the target AMP IGKVLTRVKLLRRIK, named IK-16-1. As depicted in Fig. 2, the predicted structure was an α-helix. The probability score for AMPs was 1, indicating that IK-16-1 is highly likely to become an AMP. The probability of haemolysis was 0.006, indicating that IK-16-1 was less likely to be haemolytic.
Figure 1.
Workflow of antimicrobial peptide design.
Figure 2.
Results of Helical Wheel Projection and predicted structures of beta-defensin analogs.
The activities of peptides
To test the inhibitory activity of the peptides, a disk diffusion test and the minimum inhibitory concentration assay was performed. As shown in Table 1, The results showed that IK-16-1 influenced the growth of the four types of microbes. The minimum inhibitory concentration was determined to further test the effects of IK-16-1. As shown in Table 2, IK-16-1 exhibited a broad-spectrum inhibitory effect, although its antifungal activity was not as high as that of antibacterial. Based on the zone of inhibition and minimum inhibitory concentration values, IK-16-1 was found to have broad-spectrum antimicrobial properties against Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, and Candida albicans, which are common microbes that cause cosmetic spoilage.
Table 1.
Zone of inhibition of antimicrobial peptides (AMPs) against different species.
| Species | Samples | Diameter (mm) | Mean ± SD (mm) | ||
|---|---|---|---|---|---|
| Staphylococcus aureus (18 h, 30 μg) | Kanamycin | 26.4 | 25.5 | 26.1 | 26.0 ± 0.4 |
| IK-16-1 | 12.3 | 11.2 | 12.0 | 11.8 ± 0.5 | |
| Escherichia coli (18 h, 30 μg) | Kanamycin | 14.6 | 15.6 | 15.0 | 15.1 ± 0.4 |
| IK-16-1 | 11.2 | 11.3 | 11.5 | 11.3 ± 0.1 | |
| Pseudomonas aeruginosa (18 h, 30 μg) | Kanamycin | 16.3 | 15.1 | 16.0 | 15.8 ± 0.5 |
| IK-16-1 | 11.5 | 10.7 | 10.6 | 10.9 ± 0.4 | |
| Candida albicans (24 h) | Amphotericin (30 μg) | 15.0 | 15.4 | 15.2 | 15.2 ± 0.2 |
| IK-16-1 (60 μg) | 9.6 | 10.3 | 8.7 | 9.5 ± 0.7 | |
SD standard deviation.
Table 2.
Minimum inhibitory concentration (MIC) of IK-16-1 against different species.
| Species | MIC (μg/mL, 18 h) | Mean ± SD (μg/mL, 18 h) | ||
|---|---|---|---|---|
| Candida albicans | 125.0 | 125.0 | 125.0 | 125.0 ± 0.0 |
| Escherichia coli | 125.0 | 125.0 | 62.5 | 104.2 ± 36.1 |
| Pseudomonas aeruginosa | 125.0 | 62.5 | 62.5 | 83.3 ± 36.1 |
| Staphylococcus aureus | 125.0 | 62.5 | 62.5 | 83.3 ± 36.1 |
The safety of peptides
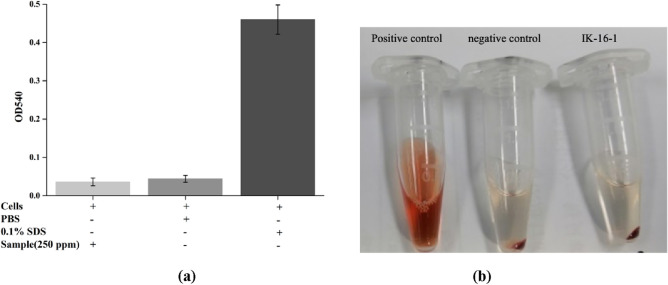
The cytotoxicity and haemolytic activity of IK-16-1 were evaluated to determine its safety. Compared to the blank group, 10 ppm IK-16-1, a common concentration used in cell models, was found to be non-cytotoxic to keratinocytes. As depicted in Fig. 3, no haemolytic activity was observed even at the highest concentration of the peptide solution used.
Figure 3.
The hemolytic results of IK-16-1 at a concentration of 250 μg/mL (positive control: 0.1% SDS, negative control: PBS).
The application of peptides in cosmetic products
To validate the use of IK-16-1 as a synergistic preservative in cosmetics, a common preservative formulation was used in combination with IK-16-1. As shown in Table 3, IK-16-1 significantly inhibited microbial growth at an early stage.
Table 3.
The colony forming unit (CFU) calculation in cosmetic products at different time.
| Species | Day 0 (CFU/mL) | Day3 (CFU/mL) | Day 7 (CFU/mL) | Day 14 (CFU/mL) | Day 22 (CFU/mL) |
|---|---|---|---|---|---|
| Escherichia coli | < 10 | < 10 | < 10 | < 10 | < 10 |
| Staphylococcus aureus | < 10 | < 10 | < 10 | < 10 | < 10 |
| Pseudomonas aeruginosa | < 10 | < 10 | < 10 | < 10 | < 10 |
| Candida albicans | 1.95 × 10 | < 10 | < 10 | < 10 | < 10 |
Discussion
In the present study, we obtained a novel antibacterial peptide IK-16-1 designed based on β-defensin 103 with significant antibacterial activity and maintaining safety by using machine learning. These results confirmed our hypothesis and verified that the workflow for designing AMPs is feasible. Based on the zone of inhibition and minimum inhibitory concentration values, IK-16-1 was found to have broad-spectrum antimicrobial properties against Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, and Candida albicans, which are common microbes that cause cosmetic spoilage. IK-16-1 has a higher antibacterial activity than that of antifungal. IK-16-1 shows good safety, has no cytotoxic to keratinocytes and no haemolytic activity. Notably, the number of preservatives added to cosmetics in the market may exceed the actual requirements. As shown in Table 3, only 30% of the preservatives were used in this study; however, the anticorrosion effect was comparable to that of the normal formulation. The results showed that IK-16-1 effectively killed the microbes in cosmetic products at low concentrations. Therefore, IK-16-1 can be used as a synergistic preservative to reduce the use of preservatives in cosmetics.
AMPs are characterised by both hydrophobic and hydrophilic domains. Most are cationic, and their positive net charge allows them to interact with negatively charged bacterial membranes44. Gram-negative and Gram-positive bacteria have molecules on their outer membranes that confer a negative net charge, allowing electrostatic interactions with cationic peptides45. AMPs can cause bacterial cell death through both membranolytic and non-membranolytic mechanisms by interacting with intracellular targets such as DNA, RNA, and proteins19,46–52. Most AMPs have broad-spectrum antimicrobial activity and are less prone to trigger resistance, which has a good prospect. In this study, peptides with helical structures were adjusted to separate hydrophobic and hydrophilic residues on the surface. Theoretically, the ability of AMPs to form pores could be improved.
ML uses mathematical, statistical, and computational processes to learn from data53. Developing new AMPs through ML is an emerging topic. Lee et al. developed a support vector machine (SVM) based classifier to investigate ⍺-helical AMPs and the interrelated nature of their functional commonality and sequence homology54. Capecchi et al. trained recurrent neural networks (RNN) using data from the Database of Antimicrobial Activity and Structure of Peptides to design short non-haemolytic AMPs55. A recent study used in silico methods to investigate potential AMPs against predatory myxobacteria56. The sources of AMPs are diverse. In this study, we designed AMPs based on endogenous human proteins. Leon et al. used computer-aided tools to identify multifunctional peptides with antimicrobial, antibiofilm, and antioxidant potential from plant peptides57. Tachapuripunya et al. used biochemical, mass spectrometric, and bioinformatics approaches to comprehensively identify putative bioactive peptides from the mucus proteomes of gastropods58. Besides,Ng et al. reported biosurfactants also show antimicrobial properties59, which can be considered for cosmetic applications.
Due to the lack of fundamental knowledge of antibacterial mechanisms, the characteristics generated by different ML programs cannot fully characterise the properties of AMPs. However, with the rapid development of screening methods60 and the continuous updating of calculation algorithms61, challenges in discovering, classifying, and designing AMPs will gradually be solved to provide more alternatives for preventing accidental overgrowth and drug resistance of microorganisms. Notably, wet experiments are the only way to determine the antimicrobial activity of peptides. Integrating ML with targeted experimentation can guide both AMP discovery and design and provide a new understanding of the properties and mechanisms underlying their modes of action62.
This study has some limitations. The exact mechanism by which IK-16-1 combines with preservatives to inhibit bacterial growth was not elucidated, and the mechanism through which IK-16-1 interacts with bacteria and other compounds in emulsions remains unclear. Furthermore, whether IK-16-1 has a synergistic effect with other preservatives, such as phenoxyethanol, requires further investigation. Because the performance of IK-16-1 was tested only under emulsion conditions to form its theoretical helical structure, the effects of other dosage forms should be considered in future applications. In future research, we intend to investigate the synergistic effects of IK-16-1 in relation to other preservatives.
Notwithstanding these limitations, this study verified the feasibility of combining computational design with artificial intelligence prediction to design AMPs derived from human endogenous proteins. IK-16-1 inhibits the growth of common spoilage-causing bacteria. Therefore, it can be used synergistically to effectively preserve cosmetics while containing lower concentrations of allergenic preservatives.
Conclusion
Preservatives, essential ingredients in cosmetic products, are one of the most common allergens. The irritation caused by preservatives has underscore an urgent need for the development of safe and effective broad-spectrum antimicrobial agents. Using machine learning-assisted rational design, we obtained a novel antibacterial peptide IK-16-1 with significant antibacterial activity and maintaining safety based on β-defensins, which could be a promising preservative synergist to reduce the use of allergenic preservatives in cosmetics. This study demonstrated that ML can significantly improve antibacterial activity and safety, shorten development cycles, and reduce development costs.
Acknowledgements
This work was supported by China National Light Industry, Beijing Technology and Business University (Grant Number KLC-2021-YB3). Any opinions, findings, and conclusions are those of the authors and do not necessarily reflect the views of the above agency. We would like to thank Editage (www.editage.cn) for English language editing.
Author contributions
L.Y. wrote the main manuscript text. L.S. and X.F. conducted antibacterial experiments. S.Z. designed artificial AMPs. X.L. reviewed the manuscript. C.H. proposed the framework for this study. J.L. supervised and guided the study. All the authors contributed to the manuscript and approved the submitted version.
Data availability
All data generated or analyzed during this study are available from the corresponding author (Junxiang Li: lijunxiang@acrdc.cn) on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Congfen He, Email: congfenhe@126.com.
Junxiang Li, Email: lijunxiang@acrdc.cn.
References
- 1.Ratajczak P, et al. The growing market for natural cosmetics in Poland: Consumer preferences and industry trends. Clin. Cosmet. Investig. Dermatol. 2023;16:1877–1892. doi: 10.2147/CCID.S411032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glaz P, et al. Effect of commonly used cosmetic preservatives on healthy human skin cells. Cells. 2023;12:1076. doi: 10.3390/cells12071076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halla N, et al. Cosmetics preservation: A review on present strategies. Molecules. 2018;23:1571. doi: 10.3390/molecules23071571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruusgaard-Mouritsen MA, Garvey LH, Johansen JD. Facial contact dermatitis caused by cosmetic-relevant allergens. Contact Dermat. 2021;85:650–659. doi: 10.1111/cod.13966. [DOI] [PubMed] [Google Scholar]
- 5.Uter W, Yazar K, Kratz EM, Mildau G, Lidén C. Coupled exposure to ingredients of cosmetic products: II. Preservatives. Contact Dermat. 2014;70:219–226. doi: 10.1111/cod.12165. [DOI] [PubMed] [Google Scholar]
- 6.Burnett CL, et al. Amended safety assessment of methylisothiazolinone as used in cosmetics. Int. J. Toxicol. 2021;40:5S–19S. doi: 10.1177/10915818211015795. [DOI] [PubMed] [Google Scholar]
- 7.Qin O, et al. Patch test in Chinese in Shanghai with cosmetic allergy to cosmetic series and products. J. Cosmet. Dermatol. 2020;19:2086–2092. doi: 10.1111/jocd.13249. [DOI] [PubMed] [Google Scholar]
- 8.Nowak K, Jabłońska E, Ratajczak-Wrona W. Controversy around parabens: Alternative strategies for preservative use in cosmetics and personal care products. Environ. Res. 2021;198:110488. doi: 10.1016/j.envres.2020.110488. [DOI] [PubMed] [Google Scholar]
- 9.Goossens A, Aerts O. Contact allergy to and allergic contact dermatitis from formaldehyde and formaldehyde releasers: A clinical review and update. Contact Dermat. 2022;87:20–27. doi: 10.1111/cod.14089. [DOI] [PubMed] [Google Scholar]
- 10.Zhang W, et al. Effect of leave-on cosmetic antimicrobial preservatives on healthy skin resident Staphylococcus epidermidis. J. Cosmet. Dermatol. 2023;22:2115–2121. doi: 10.1111/jocd.15690. [DOI] [PubMed] [Google Scholar]
- 11.Lundov MD, Johansen JD, Zachariae C, Moesby L. Low-level efficacy of cosmetic preservatives. Int. J. Cosmet. Sci. 2011;33:190–196. doi: 10.1111/j.1468-2494.2010.00619.x. [DOI] [PubMed] [Google Scholar]
- 12.Ng YJ, et al. Recent advances and discoveries of microbial-based glycolipids: Prospective alternative for remediation activities. Biotechnol. Adv. 2023;68:108198. doi: 10.1016/j.biotechadv.2023.108198. [DOI] [PubMed] [Google Scholar]
- 13.Meftahi A, et al. Nanocelluloses as skin biocompatible materials for skincare, cosmetics, and healthcare: Formulations, regulations, and emerging applications. Carbohydr. Polym. 2022;278:118956. doi: 10.1016/j.carbpol.2021.118956. [DOI] [PubMed] [Google Scholar]
- 14.Moradi N, et al. Synthesis of mesoporous antimicrobial herbal nanomaterial-carrier for silver nanoparticles and antimicrobial sensing. Food Chem. Toxicol. 2022;165:113077. doi: 10.1016/j.fct.2022.113077. [DOI] [PubMed] [Google Scholar]
- 15.Bindhu MR, Umadevi M, Esmail GA, Al-Dhabi NA, Arasu MV. Green synthesis and characterization of silver nanoparticles from Moringa oleifera flower and assessment of antimicrobial and sensing properties. J. Photochem. Photobiol. B. 2020;205:111836. doi: 10.1016/j.jphotobiol.2020.111836. [DOI] [PubMed] [Google Scholar]
- 16.Bahar AA, Ren D. Antimicrobial peptides. Pharmaceuticals (Basel) 2013;6:1543–1575. doi: 10.3390/ph6121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu QH, Patočka J, Kuča K. Insect antimicrobial peptides, a mini review. Toxins. 2018;10:461. doi: 10.3390/toxins10110461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 19.Di Somma A, Moretta A, Canè C, Cirillo A, Duilio A. Antimicrobial and antibiofilm peptides. Biomolecules. 2020;10:652. doi: 10.3390/biom10040652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powers JP, Hancock RE. The relationship between peptide structure and antibacterial activity. Peptides. 2003;24:1681–1691. doi: 10.1016/j.peptides.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 21.Bulet P, Stöcklin R, Menin L. Anti-microbial peptides: From invertebrates to vertebrates. Immunol. Rev. 2004;198:169–184. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L. Interaction of human beta defensin type 3 (hBD-3) with different PIP2-containing membranes, a molecular dynamics simulation study. J. Chem. Inf. Model. 2021;61:4670–4686. doi: 10.1021/acs.jcim.1c00805. [DOI] [PubMed] [Google Scholar]
- 23.Janiesch C, Zschech P, Heinrich K. Machine learning and deep learning. Electron. Mark. 2021;31:685–695. doi: 10.1007/s12525-021-00475-2. [DOI] [Google Scholar]
- 24.Biamonte J, et al. Quantum machine learning. Nature. 2017;549:195–202. doi: 10.1038/nature23474. [DOI] [PubMed] [Google Scholar]
- 25.Crampon K, Giorkallos A, Deldossi M, Baud S, Steffenel LA. Machine-learning methods for ligand-protein molecular docking. Drug Discov. Today. 2022;27:151–164. doi: 10.1016/j.drudis.2021.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Greener JG, Kandathil SM, Moffat L, Jones DT. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022;23:40–55. doi: 10.1038/s41580-021-00407-0. [DOI] [PubMed] [Google Scholar]
- 27.Kang J, Ullah Z, Gwak J, Brain M-B. Tumor classification using ensemble of deep features and machine learning classifiers. Sensors. 2021;21:2222. doi: 10.3390/s21062222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dehkharghanian T, Mu YQ, Tizhoosh HR, Campbell CJV. Applied machine learning in hematopathology. Int. J. Lab. Hematol. 2023;45(Supplement 2):87–94. doi: 10.1111/ijlh.14110. [DOI] [PubMed] [Google Scholar]
- 29.An Q, Rahman S, Zhou JW, Kang JJ. A comprehensive review on machine learning in healthcare industry: Classification, restrictions, opportunities and challenges. Sensors (Basel) 2023;23:4178. doi: 10.3390/s23094178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmad Sobri MZ, et al. Kinetic model derived from machine learning for accurate prediction of microalgal hydrogen production via conversion from low thermally pre-treated palm kernel expeller waste. Chemosphere. 2023;338:139526. doi: 10.1016/j.chemosphere.2023.139526. [DOI] [PubMed] [Google Scholar]
- 31.Su DQ, et al. Application of machine learning in fuel cell research. Energies. 2023;16:4390. doi: 10.3390/en16114390. [DOI] [Google Scholar]
- 32.Wang Z, et al. The role of machine learning to boost the bioenergy and biofuels conversion. Bioresour. Technol. 2022;343:126099. doi: 10.1016/j.biortech.2021.126099. [DOI] [PubMed] [Google Scholar]
- 33.Torres MDT, Sothiselvam S, Lu TK, de la Fuente-Nunez C. Peptide design principles for antimicrobial applications. J. Mol. Biol. 2019;431:3547–3567. doi: 10.1016/j.jmb.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 34.Baindara P, Korpole S, Grover V. Bacteriocins: Perspective for the development of novel anticancer drugs. Appl. Microbiol. Biotechnol. 2018;102:10393–10408. doi: 10.1007/s00253-018-9420-8. [DOI] [PubMed] [Google Scholar]
- 35.Wang G. The antimicrobial peptide database provides a platform for decoding the design principles of naturally occurring antimicrobial peptides. Protein Sci. 2020;29:8–18. doi: 10.1002/pro.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang M, Zhang C, Zhang MZ, Zhang S. Beta-defensin derived cationic antimicrobial peptides with potent killing activity against gram negative and gram positive bacteria. BMC Microbiol. 2018;18:54. doi: 10.1186/s12866-018-1190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gunasekera S, Muhammad T, Strömstedt AA, Rosengren KJ, Göransson U. Alanine and lysine scans of the LL-37-Derived peptide fragment KR-12 reveal key residues for antimicrobial activity. ChemBioChem. 2018;19:931–939. doi: 10.1002/cbic.201700599. [DOI] [PubMed] [Google Scholar]
- 38.Esfandiyari R, et al. Performance evaluation of antimicrobial peptide ll-37 and hepcidin and beta-defensin-2 secreted by mesenchymal stem cells. Heliyon. 2019;5:e02652. doi: 10.1016/j.heliyon.2019.e02652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veltri D, Kamath U, Shehu A. Deep learning improves antimicrobial peptide recognition. Bioinformatics. 2018;34:2740–2747. doi: 10.1093/bioinformatics/bty179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan J, et al. Deep-AmPEP30: Improve short antimicrobial peptides prediction with deep learning. Mol. Ther. Nucleic Acids. 2020;20:882–894. doi: 10.1016/j.omtn.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao X, Wang P, Lin WZ, Jia JH, Chou KC. iAMP-2L: A two-level multi-label classifier for identifying antimicrobial peptides and their functional types. Anal. Biochem. 2013;436:168–177. doi: 10.1016/j.ab.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 42.Thomas S, Karnik S, Barai RS, Jayaraman VK, Idicula-Thomas S. CAMP: A useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2010;38:D774–D780. doi: 10.1093/nar/gkp1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plisson F, Ramírez-Sánchez O, Martínez-Hernández C. Machine learning-guided discovery and design of non-hemolytic peptides. Sci. Rep. 2020;10:16581. doi: 10.1038/s41598-020-73644-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wieprecht T, Apostolov O, Seelig J. Binding of the antibacterial peptide magainin 2 amide to small and large unilamellar vesicles. Biophys. Chem. 2000;85:187–198. doi: 10.1016/S0301-4622(00)00120-4. [DOI] [PubMed] [Google Scholar]
- 45.Pasupuleti M, Schmidtchen A, Malmsten M. Antimicrobial peptides: Key components of the innate immune system. Crit. Rev. Biotechnol. 2012;32:143–171. doi: 10.3109/07388551.2011.594423. [DOI] [PubMed] [Google Scholar]
- 46.Haney EF, Straus SK, Hancock REW. Reassessing the host defense peptide landscape. Front. Chem. 2019;7:43. doi: 10.3389/fchem.2019.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mankoci S, et al. Bacterial membrane selective antimicrobial peptide-mimetic polyurethanes: Structure-property correlations and mechanisms of action. Biomacromolecules. 2019;20:4096–4106. doi: 10.1021/acs.biomac.9b00939. [DOI] [PubMed] [Google Scholar]
- 48.Alghalayini A, Garcia A, Berry T, Cranfield CG. The use of tethered bilayer lipid membranes to identify the mechanisms of antimicrobial peptide interactions with lipid bilayers. Antibiotics (Basel) 2019;8:12. doi: 10.3390/antibiotics8010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brogden KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 50.Cassone M, Frith N, Vogiatzi P, Wade JD, Otvos L. Induced resistance to the designer proline-rich antimicrobial peptide A3-APO does not involve changes in the intracellular target DnaK. Int. J. Pept. Res. Ther. 2009;15:121–128. doi: 10.1007/s10989-009-9176-1. [DOI] [Google Scholar]
- 51.Shah P, Hsiao FS, Ho YH, Chen CS. The proteome targets of intracellular targeting antimicrobial peptides. Proteomics. 2016;16:1225–1237. doi: 10.1002/pmic.201500380. [DOI] [PubMed] [Google Scholar]
- 52.Graf M, Wilson DN. Intracellular antimicrobial peptides targeting the protein synthesis machinery. Adv. Exp. Med. Biol. 2019;1117:73–89. doi: 10.1007/978-981-13-3588-4_6. [DOI] [PubMed] [Google Scholar]
- 53.Lee EY, Lee MW, Fulan BM, Ferguson AL, Wong GCL. What can machine learning do for antimicrobial peptides, and what can antimicrobial peptides do for machine learning? Interface Focus. 2017;7:20160153. doi: 10.1098/rsfs.2016.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee EY, Fulan BM, Wong GC, Ferguson AL. Mapping membrane activity in undiscovered peptide sequence space using machine learning. Proc. Natl Acad. Sci. USA. 2016;113:13588–13593. doi: 10.1073/pnas.1609893113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Capecchi A, et al. Machine learning designs non-hemolytic antimicrobial peptides. Chem. Sci. 2021;12:9221–9232. doi: 10.1039/D1SC01713F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arakal BS, et al. In silico and in vitro analyses reveal promising antimicrobial peptides from myxobacteria. Probiotics Antimicrob. Proteins. 2023;15:202–214. doi: 10.1007/s12602-022-10036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.León Madrazo A, Segura Campos MR. In silico prediction of peptide variants from chia (S. hispanica L.) with antimicrobial, antibiofilm, and antioxidant potential. Comput. Biol. Chem. 2022;98:107695. doi: 10.1016/j.compbiolchem.2022.107695. [DOI] [PubMed] [Google Scholar]
- 58.Tachapuripunya V, Roytrakul S, Chumnanpuen P, E-Kobon T. Unveiling putative functions of mucus proteins and their tryptic peptides in seven gastropod species using comparative proteomics and machine learning-based bioinformatics predictions. Molecules. 2021;26:3475. doi: 10.3390/molecules26113475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ng YJ, et al. Recent advances of biosurfactant for waste and pollution bioremediation: Substitutions of petroleum-based surfactants. Environ. Res. 2022;212:113126. doi: 10.1016/j.envres.2022.113126. [DOI] [PubMed] [Google Scholar]
- 60.Tucker AT, et al. Discovery of next-generation antimicrobials through bacterial self-screening of surface-displayed peptide libraries. Cell. 2018;172:618–628.e13 e613. doi: 10.1016/j.cell.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Das P, et al. Accelerated antimicrobial discovery via deep generative models and molecular dynamics simulations. Nat. Biomed. Eng. 2021;5:613–623. doi: 10.1038/s41551-021-00689-x. [DOI] [PubMed] [Google Scholar]
- 62.Lee EY, Wong GCL, Ferguson AL. Machine learning-enabled discovery and design of membrane-active peptides. Bioorg. Med. Chem. 2018;26:2708–2718. doi: 10.1016/j.bmc.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are available from the corresponding author (Junxiang Li: lijunxiang@acrdc.cn) on reasonable request.