Abstract
Vibrio parahaemolyticus is an organism well adapted to communal life on surfaces. When grown on a surface or in a viscous layer, the bacterium induces a large gene system and differentiates to swarmer cells capable of movement over and colonization of surfaces. V. parahaemolyticus displays additional phenotypic versatility manifested as variable colony morphology, switching between translucent and opaque colony types. Although not itself luminescent, V. parahaemolyticus produces autoinducer molecules capable of inducing luminescence in Vibrio harveyi. To examine the role of quorum signaling in the lifestyles of V. parahaemolyticus, the functional homolog of the gene encoding the V. harveyi autoinducer-controlled transcriptional regulatory protein LuxR was cloned. Sequence analysis of the clone predicted an open reading frame with a deduced product 96% identical to LuxR. Introduction of the clone carrying the luxR-like locus into V. parahaemolyticus dramatically affected colony morphology, converting a translucent strain to an opaque one. When the coding sequence for the luxR homolog was placed under the control of the Ptac promoter, conversion to the opaque phenotype became inducible by isopropyl-β-d-thiogalactopyranoside. Allelic disruption of the luxR-like gene on the chromosome of an opaque strain produced a translucent strain proficient in swarming ability. Primer extension mapping demonstrated opaR transcription in opaque but not translucent cell types. It is postulated that this gene, which has been named opaR, encodes a transcription factor controlling cell type. The underlying genetic basis for opaque-translucent variation may be the consequence of a genomic alteration detected in the opaR locus of opaque and translucent strains.
In order to survive in changing environments, bacteria possess enormous adaptive capabilities that allow them to modulate their behavior and reprogram gene expression in response to environmental cues. Vibrio parahaemolyticus, a ubiquitous marine bacterium and human pathogen, seems particularly adapted to growth on surfaces or in biofilms. In response to its physical environment, V. parahaemolyticus induces the expression of a large gene system that allows differentiation to swarmer cells. This cell type is adapted for movement on surfaces or through highly viscous environments and thus for colonization of surfaces (20).
V. parahaemolyticus displays another adaptive mechanism. The organism switches between a translucent colony type and an opaque colony type. Opaque-translucent variation has been observed in other bacteria. For example, in Neisseria gonorrhoeae, phase-variable opacity is associated with the presence of specific outer membrane proteins (35). Furthermore, the expression of opacity proteins is correlated with adherence (17, 18). It has been postulated that cell surface components, e.g., the N. gonorrhoeae opacity proteins, lead to autoaggregation and differential cell packing. The organization presumably translates into differences in the transmission of light by the bacterial colony (34). Opacity has also been associated with the production of extracellular polysaccharide. The closely related bacterium Vibrio vulnificus undergoes opaque-translucent switching (26). For this bacterium, virulence and opacity have been correlated with the presence of capsular polysaccharide (38).
Density-dependent sensing has been postulated to be an important component of bacterial colonization and growth in communities (6, 23). Small-molecule signaling and intercellular communication may provide the bacterial cell methods for discrimination between a free-living, low-cell-density state and an attached, high-density environment (13, 36). Intriguingly, N-acyl homoserine lactones (AHLs) have been demonstrated to be involved in the swarming behavior of the bacterium Serratia liquefaciens (10). This observation, i.e., that there is a cell-density-dependent component of the swarming of S. liquefaciens, coupled with the knowledge that V. parahaemolyticus produces autoinducer molecules (14), provoked examination of the role of density-dependent signaling in V. parahaemolyticus.
Nonluminescent V. parahaemolyticus produces autoinducer molecules that stimulate light production of luminous Vibrio harveyi (14, 30). The molecular scheme of density-dependent signaling in V. harveyi contrasts with the luxI-luxR system found in many bacteria, the best example being that of the regulation of luminescence in Vibrio fischeri. In V. fischeri, luxI encodes a protein responsible for the synthesis of an AHL autoinducer, and luxR encodes a protein that activates transcription of the luminescence operon in response to the level of autoinducer (13). When the level of autoinducer exceeds a threshold concentration, LuxR binds autoinducer (15) and activates the expression of luminescence genes. A variety of bacteria produce AHL signaling molecules, and a number of homologs of the V. fischeri luxR gene product have been demonstrated to be involved in intercellular communication controlling diverse gene systems (13, 36). These proteins contain signature motifs for DNA binding and autoinducer binding.
Luminescence in V. harveyi is also controlled in a cell-density-dependent manner; however, the model for quorum sensing in V. harveyi is more elaborate than the luxI-luxR paradigm of V. fischeri in that there are additional layers of gene control. V. harveyi LuxR is the transcriptional activator of the luminescence operon (29), although it is not homologous with members of the V. fischeri LuxR family. LuxR of V. harveyi does not interact with an autoinducer. V. harveyi produces two autoinducer molecules. It is postulated that the two autoinducers each interact with cognate regulatory proteins to control the transcription of luxR. In this cascade manner, the synthesis of LuxR and the ultimate output of light are controlled by the density of the culture and the autoinducer levels (5, 25).
V. parahaemolyticus BB22 produces the two autoinducer molecules which are capable of stimulating the dual systems found in V. harveyi (4). Since luxR transcription is modulated by multiple density-sensing inputs, a loss of function of one signaling system, for example, in the gene encoding one of the autoinducer synthases, does not eliminate luminescence or the regulation of luminescence in V. harveyi because the second system remains functional. So, in order to investigate the role of density-dependent signaling in V. parahaemolyticus, it seemed logical to begin by examining the ultimate transcriptional activator component, i.e., LuxR. A gene was cloned from V. parahaemolyticus which substituted for the V. harveyi luxR gene to activate the expression of the V. harveyi luminescence operon.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this work are described in Table 1. For propagation of V. parahaemolyticus strains, the following media were used: HI (25 g of heart infusion broth [Difco] and 20 g of NaCl per liter) and the minimal medium of Broach et al. (8) supplemented with 0.4% galactose, 20 mM NH4Cl, and 2% NaCl (final concentration). Casamino Acids (Difco) were added to the minimal medium, as appropriate, at a final concentration of 0.25%. Solidified media were prepared by use of 2% Bacto Agar (Difco) for HI Swarm Minus, 1.5% Bacto Agar for HI Swarm Plus, and 1.4% Bacto Agar for minimal Swarm Plus plates. Medium that supports movement over the surface is designated Swarm Plus, while medium that does not allow swarming is designated Swarm Minus. Vibrio strains were grown at 30°C with overnight incubation on complex media, 5 to 6 days on minimal swarm medium, and 2 to 3 days on minimal swarm medium with Casamino Acids. Minimal swarm plates were wrapped in Parafilm to prevent desiccation. Luria broth was prepared as described by Miller (24). Ampicillin, chloramphenicol, and tetracycline were used at final concentrations of 75, 10, and 10 μg/ml, respectively. The final isopropyl-β-d-thiogalactopyranoside (IPTG; Gold BioTechnology, Inc., St. Louis, Mo.) concentration was 0.5 mM.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| V. parahaemolyticus | ||
| BB22TR | Wild type; translucent variant | R. Belas (7) |
| BB22OP | Wild type; opaque variant | BB22TR (this work) |
| LM1017 | lfgE313::lux (formerly lafX313::lux) | RS313 (22) |
| LM4449 | Opaque phenotype; BB22TR/pLM1950 | BB22TR (this work) |
| LM4437 | Translucent phenotype; opaR::Tn5 | BB22TR (this work) |
| LM4462 | Translucent phenotype; opaR::Tn5 | BB22OP (this work) |
| LM4565 | IPTG-inducible opaque phenotype; LM4462/pLM2035 | LM4462 (this work) |
| LM4572 | Translucent phenotype; LM4462/pLM1835 | LM4462 (this work) |
| LM4582 | Translucent phenotype; BB22TR/pLAFRII | BB22TR (this work) |
| E. coli | ||
| DH5α | F−endA1 hsdR17 (rK−) supE44 thi-1 λ− recA1 deoR gyrA96 relA1 Δ(argF-lacZYA)U169 φ80dlacZΔM15 | Bethesda Research Laboratories |
| LLM1956 | DH5α/pRS205 (dark) | This work |
| LLM1957 | DH5α/pRS205/pLM1950 (bright) | This work |
| LLM1958 | DH5α/pRS205/pLM1952 (dark) | This work |
| Plasmids | ||
| pLAFRII | Tetr broad-host-range cosmid vector | 11 |
| pRS205 | Apr; luxCDABE+ operon (V. harveyi) | R. Showalter (29) |
| pLM1835 | Camr Apr broad-host-range expression vector carrying lacIq and Ptac | pMMB66EH (12, 33) |
| pLM1950 | TetropaR+ | pLAFRII V. parahaemolyticus bank (21) |
| pLM1952 | Tetr KanropaR294::Tn5 | pLM1950 (this work) |
| pLM1980 | Apr; pUC19 with 2.0-kb PstI fragment from pLM1952 containing part of Tn5 insertion in opaR | pUC19 and pLM1952 |
| pLM2035 | Camr ApropaR+ under Ptac control | pLM1835 (this work) |
Opaque-translucent variation is slow, so it is possible to obtain essentially “pure” cultures of opaque or translucent colony types having less than 1 alternate form per 1,000 colonies. Cultures for RNA and DNA preparations were checked for variation by diluting and plating on HI Swarm Minus medium to observe single colonies.
Bioluminescence measurements.
Bioluminescence was quantified with a TD-20/22 luminometer (Turner Designs, Sunnyvale, Calif.) by measuring 0.1- and 0.2-ml samples of cultures appropriately diluted to give a linear response. Luminescence was reported as specific light units, which are relative light units per minute per milliliter per unit of optical density at 600 nm (OD600). Bioluminescence was also monitored by exposing plates to Kodak XAR5 X-ray film.
Genetic and molecular techniques.
Transposon mutagenesis with Tn5 has been described elsewhere (29). Procedures for the transfer of clones from Escherichia coli to V. parahaemolyticus via conjugation and gene replacement techniques have also been described elsewhere (31). General DNA manipulations were adapted from the methods of Sambrook et al. (27). Chromosomal DNA was prepared according to the protocol of Woo et al. (37). Southern blot analysis of restricted genomic DNA (19) was performed on Hybond-NX membranes (Amersham Life Science, Buckinghamshire, England). The cosmid bank was constructed by ligating 15 to 20 kb of size-fractionated V. parahaemolyticus DNA with vector pLAFRII (21). DNA for the bank, which was constructed in 1987, was prepared from a translucent strain; however, the culture from which the DNA was prepared was not monitored for opaque variants.
The primers for the PCRs that revealed differences between opaque and translucent colony types were HPT (also used for primer extension; see below) and LUXUR1 (5′-GAACTGACGGACAACATGGTTGAG-3′); they were used at an annealing temperature of 43°C. These primers should prime in the sequences beginning at bp 919 and bp 1539, respectively. Thirty cycles of amplification were performed by use of Taq polymerase (Perkin-Elmer, Foster City, Calif.) with extension times of 50 s. The molecular size standard on agarose gels was the 1-kb ladder obtained from GIBCO BRL (Gaithersburg, Md.).
For the construction of pLM2035, a gel-purified 660-bp PCR product was ligated into the BamHI and SalI restriction sites of pLM1835. The following primers were used at an annealing temperature of 49°C: LUXRF (5′-CGCGGATCCATGGCAAGGAAAATGGATATGGAC-3′) and LUXRR (5′-GGACGTCGACGCTTTAGTGTTCGCGATTGTAGATGC-3′). Forty amplification cycles were performed with 1-min extension times. After ligation, clones were transformed into E. coli LLM1956 and screened for IPTG-inducible light production.
DNA sequencing analysis.
The sequence of interest was obtained for both strands from cosmid clone pLM1950 by the DNA Core Facility of the University of Iowa by use of a fluorescence automated sequencer (model 373A; Applied Biosystems, Division of Perkin-Elmer). Sequence information precisely localizing the opaR294::Tn5 insertion was obtained from clone pLM1980 by use of the oligonucleotide TnR (5′-CCGCACGATGAAGAGCAG-3′), which primed to the end of Tn5. Synthetic oligonucleotides were prepared by Integrated DNA Technologies, Inc. (Coralville, Iowa). Sequence assembly was performed by use of the Genetics Computer Group (GCG) software package. Searches for homology were performed at the National Center for Biotechnology Information (NCBI) with the BLAST network service (1, 2).
Primer extension analysis.
RNA was prepared with Trizol reagent (GIBCO BRL Life Technologies, Grand Island, N.Y.) according to the manufacturer’s protocol. Cells grown overnight on HI Swarm Plus plates were resuspended to an OD600 of 2.0 units per ml, and 25 μl was used to inoculate each HI Swarm Plus plate by spreading. Multiple plates were incubated for 5 to 6 h, and cells were harvested by resuspension in 0.3 M sucrose. Approximately 1 ml of Trizol reagent was used per 2.0 OD600 units for RNA extraction. Primer extension analysis was performed by use of the avian myeloblastosis virus reverse transcriptase primer extension system (Promega, Madison, Wis.) according to the manufacturer’s protocol. Reaction mixtures contained approximately 1 to 2 μg of total RNA per final reaction volume of 13 μl. Two oligonucleotide primers were used: 5′-GTTGCTTACGTTTAAGAGGAG-3′ (OPAR) and 5′-CTTCACTGCCTTGGTAAC-3′ (HPT). Annealing reactions were performed at 48°C. Cosmid pLM1950 was primed with OPAR and HPT oligonucleotides for the sequencing reactions by the dideoxy chain termination procedure of Sanger et al. (28) with a Sequenase 2.0 kit (United States Biochemicals) and [α-35S]dATP (Amersham).
Nucleotide sequence accession number.
The nucleotide sequence studied here has been deposited in GenBank under accession no. AF035967.
RESULTS
The LuxR homolog of V. parahaemolyticus.
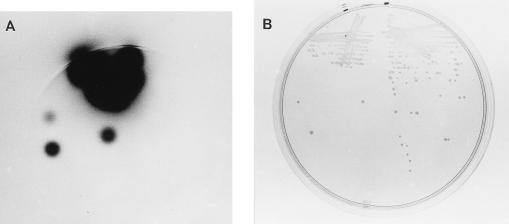
The homolog of the V. harveyi luxR gene was retrieved from a V. parahaemolyticus cosmid gene bank by selecting in E. coli for clones encoding a transcriptional regulator that activated the expression of the V. harveyi luminescence operon luxCDABE carried on plasmid pRS205. The expression in E. coli of the lux operon from V. harveyi requires the product of the luxR gene; thus, E. coli LLM1956 carrying pRS205 is nonluminescent. The cosmid bank was conjugated into LLM1956, and exconjugants were screened for light production. Bright clones were obtained. One clone, pLM1950, was mutagenized with transposon Tn5 to obtain insertions inactivating and thus localizing the gene responsible for the induction of luminescence. Figure 1 contrasts the light produced by two E. coli strains harboring the lux plasmid and either the activating cosmid pLM1950 (on left side of each plate) or the mutated cosmid pLM1952 (on right side of each plate). Heterologous trans activation by the V. parahaemolyticus clone of the V. harveyi promoter was approximately 25,000-fold (994,000.0 versus 3.0 specific light units for wild-type and mutated cosmids, respectively) and was of the same order of magnitude as that obtained for the homologous V. harveyi luxR-luxCDABE pair (29).
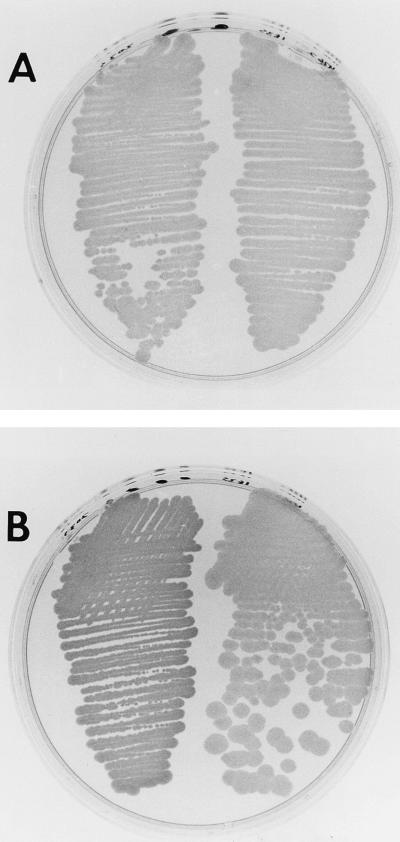
FIG. 1.
Activation of luminescence by a V. parahaemolyticus clone. Strain LLM1957 (left side of each plate) is E. coli DH5α containing two plasmids, pRS205 (V. harveyi lux operon) and pLM1950 (V. parahaemolyticus luxR-like locus). Strain LLM1958 (right side of each plate) is E. coli DH5α containing two plasmids, pRS205 and pLM1952 (a mutated form of pLM1950 containing Tn5). Strains were grown overnight on a Luria broth-ampicillin-tetracycline plate and exposed to X-ray film for 15 min (A) or photographed directly in incident light (B).
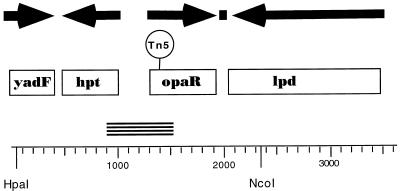
The Tn5 insertion targeted a 2-kb PstI restriction fragment that was subcloned and used for initial sequencing and gene localization. The entire sequenced region, containing three complete open reading frames and one partial coding sequence, is depicted in Fig. 2. The four deduced protein sequences showed homology to E. coli YadF (and other members of a carbonic anhydrase family), a number of hypoxanthine ribosyltransferases (Hpt), V. harveyi LuxR, and E. coli dihydrolipoamide dehydrogenase (Lpd). The position of the transposon that eliminated the activation of luminescence is shown in Fig. 2. It mapped within the open reading frame encoding a deduced protein found 96% identical to V. harveyi LuxR by GCG BestFit analysis. The predicted polypeptide also showed strong similarity (79% similarity and 72% identity by GCG BestFit analysis) to one other protein in the NCBI database, HapR, a positive regulator of the hemagglutinin/protease gene of Vibrio cholerae (16). The hpt genes are linked to the luxR-like genes in V. parahaemolyticus, V. harveyi, and V. cholerae.
FIG. 2.
Organization of the luxR-like locus of V. parahaemolyticus. The complete double-stranded nucleotide sequence of the region from bp 1 to 3605 was obtained from cosmid pLM1950. Boxes indicate open reading frames (ORFs). Arrows point in the direction of transcription of the deduced coding regions. The solid square shows the location of a potential rho-independent transcriptional terminator. The ORF for yadF (from bp 3 to 411) is incomplete; however, over 136 amino acids its product shows 72% similarity and 63% identity to E. coli YadF by GCG BestFit analysis. The predicted product of hpt (coding region from bp 488 to 1018) is 176 amino acids long and is 99% similar and 95% identical to V. harveyi Hpt. The ORF from bp 1338 to 1952 codes for a 204-amino-acid polypeptide showing 98% similarity and 96% identity to V. harveyi LuxR. The 475-amino-acid deduced product of the ORF from bp 2080 to 3507 exhibits 92% similarity and 87% identity to E. coli Lpd. Tn5 in pLM1952 is contained on a 2.3-kb HpaI-NcoI restriction fragment, and DNA sequencing localized the insertion to bp 1392, which is within the coding region for the LuxR homolog. This gene is designated opaR in V. parahaemolyticus. The PCR product produced with the HPT and LUXUR1 primers is represented by the stacked short lines.
Disruption of the luxR-like gene fails to affect swarming.
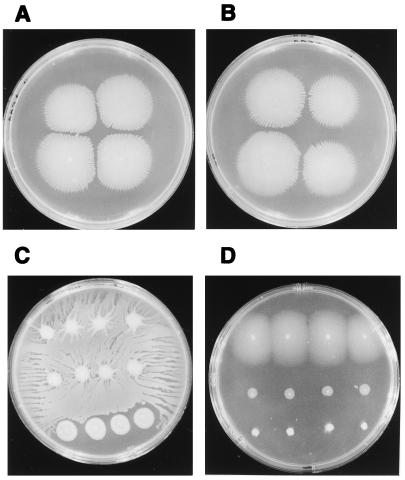
Swarming motility of S. liquefaciens has been shown to be controlled by quorum sensing (10). An S. liquefaciens mutant strain unable to synthesize autoinducer failed to swarm on minimal medium supplemented with Casamino Acids. On rich medium, initiation of swarming by this mutant strain was severely delayed compared to that of the wild type. Swarming could be restored to wild-type patterns by the addition of exogenous autoinducer. To examine the role of quorum sensing in V. parahaemolyticus, the transposon insertion which abrogated luxCDABE transcriptional activation in E. coli and interrupted the coding region for the LuxR-like protein was introduced into the V. parahaemolyticus chromosome via allelic replacement. Construction of the correct gene replacement was confirmed by Southern blot analysis. The resulting strain, LM4437, was examined for swarming motility on a variety of media and showed no demonstrable defect in swarming. The swarming phenotype on minimal medium with Casamino Acids is shown in Fig. 3: the mutant strain (Fig. 3B) swarmed as well as the wild-type strain (Fig. 3A). It is important to note that in contrast to S. liquefaciens, V. parahaemolyticus swarmed on unsupplemented minimal medium. There was no demonstrable difference in swarming on this type of medium between the wild-type strain and a strain with a defect in the luxR-like gene (Fig. 3C, rows 1 and 2, respectively). For comparison, strain LM1017, a nonswarming mutant, was also inoculated on this plate (Fig. 3C, row 3). Thus, the gene encoding the LuxR homolog is not required for swarming in V. parahaemolyticus.
FIG. 3.
Swarming patterns of wild-type and mutant strains with defects in the luxR-like gene opaR. (A and B) Wild-type BB22TR (A) and LM4437 (opaR::Tn5 in BB22TR) (B) on minimal Swarm Plus plates supplemented with Casamino Acids. (C) BB22TR, LM4462 (opaR::Tn5 in BB22OP), and LM1017 (a swarm-negative mutant) (top to bottom) on a minimal Swarm Plus plate with no Casamino Acids supplementation. (D) BB22TR, LM1017, and BB22OP (top to bottom) on a complex medium (HI) Swarm Plus plate. Four single colonies of each strain were inoculated into each row on each plate.
The V. parahaemolyticus LuxR homolog affects the opaque and translucent phenotypes.
Descendants of a single colony of V. parahaemolyticus can have multiple colony morphologies. The variants are described as opaque and translucent colony types as a result of differences in the transmission of light by the colonies (Fig. 4A). The properties of opaque and translucent colony types are distinct. For example, when an opaque colony is transferred from a plate with a toothpick, the entire colony adheres to the tip, sometimes lifting off the plate as a long string. In certain kinds of liquid medium, an opaque colony looks particulate due to small aggregates or clumps of cells, and on swarm plates an opaque colony performs very poorly, exhibiting little or no movement across the surface. A comparison of translucent (top row) and opaque (bottom row) swarming colonies is shown in Fig. 3D. Inoculated in the middle row of the plate is the nonswarming mutant strain LM1017.
FIG. 4.
Comparison of opaque and translucent phenotypes. In photographs, opaque strains appear darker than translucent strains. (A) BB22TR (left) and BB22OP (right). (B) LM4582 (BB22TR with pLAFRII; translucent phenotype) (left) and LM4449 (BB22TR with pLM1950; opaque phenotype) (right). (C) LM4462 (opaque strain with a transposon in opaR; translucent phenotype) (left) and BB22OP (right). Strains were grown overnight at 30°C on HI Swarm Minus plates with antibiotic as appropriate.
Introduction of pLM1950, the cosmid carrying the luxR-like gene, into the translucent strain BB22TR converted the strain to an opaque colony type. Figure 4B shows the opaque phenotype of strain BB22TR carrying cosmid pLM1950 compared to the translucent phenotype of strain BB22TR carrying the control cosmid pLAFRII. To test whether the LuxR homolog was directly involved in the opaque-translucent switch, the transposon in the luxR-like gene was used for allelic replacement in an opaque strain. Introduction of the Tn5 mutation carried by pLM1952 into the chromosome of an opaque strain converted the opaque strain to a translucent colony type (strain LM4462; Fig. 4C). Furthermore, the introduction of the Tn5 mutation resulted in the concomitant gain of swarming ability. Strain LM4462 swarmed as well as strain BB22TR (Fig. 3C). Thus, the V. parahaemolyticus gene encoding the LuxR homolog seems to be implicated in the control of opacity and is designated opaR.
Expression of opaR controls opacity.
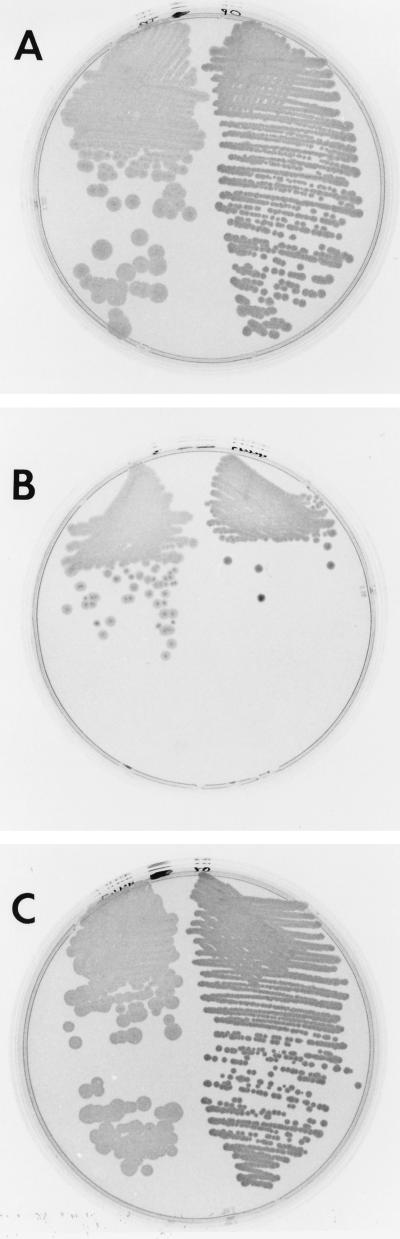
In order to confirm the role of opaR in regulating opacity, a promoterless opaR coding region was cloned into the IPTG-inducible expression vector pLM1835. This vector also contains the lacIq gene. Clone pLM2035 was transferred by conjugation to translucent strain LM4462, which contains the opaR::Tn5 insertion. The strain remained translucent when either pLM1835 or pLM2035 was introduced (Fig. 5A). The addition of IPTG to the plates induced the opaque morphology for the strain carrying pLM2035, but the control strain carrying pLM1835 remained translucent (Fig. 5B). A comparison of the IPTG-induced opaque phenotype with the wild-type translucent and opaque phenotypes is shown in Fig. 6. The plates were illuminated from above rather than below (as was done for the other figures) in order to show the contrast between the opaque and translucent colony types in another manner. The colony morphology of the wild-type opaque strain (Fig. 6B, right side) was identical to the IPTG-induced morphology (Fig. 6A, right side). Thus, induction of the expression of opaR resulted in transformation to the opaque phenotype. The introduction of pLM2035 into BB22TR also produced a translucent strain that could be converted to an opaque colony type by use of IPTG.
FIG. 5.
IPTG-inducible opacity. On the right half of each plate is LM4572 (opaR::Tn5 with pLM1835), and on the left is LM4565 (opaR::Tn5 with pLM2035). Plasmid pLM2035 contains opaR under the control of the Ptac promoter in the vector pLM1835. The media used were HI Swarm MInus with chloramphenicol (A) and HI Swarm Minus with chloramphenicol and IPTG (B).
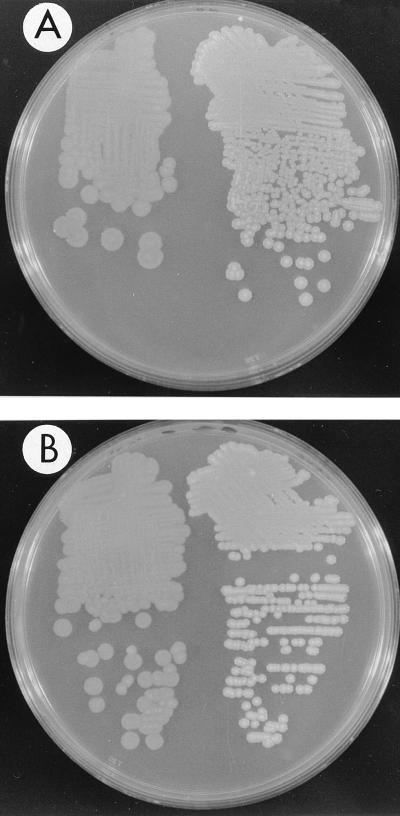
FIG. 6.
Comparison of OpaR-induced opacity in opaque and translucent wild-type strains. (A) LM4572 (opaR::Tn5 with pLM1835) (left) and LM4565 (opaR::Tn5 with pLM2035) (right) were grown on HI Swarm Minus with chloramphenicol and IPTG. Plasmid pLM2035 contains opaR under the control of the Ptac promoter in the vector pLM1835. (B) Translucent (left) and opaque (right) wild-type strains were grown on HI Swarm Minus. Plates were photographed with illumination from above, in contrast to the plates in other figures, which were photographed with illumination from below.
IPTG-induced expression of opaR induced the opaque phenotype. This finding suggests that the translucent phenotype is the result of the lack of expression of opaR. To test such an idea, as well as to establish the basis for examining the possibility of autoinducer-mediated regulation of opaR expression, primer extension mapping was used to define the promoter region and to examine the expression of opaR in opaque and translucent strains. RNA was prepared from opaque and translucent strains that had been grown on plates for 5 h. The labeled primer extension products for both the hpt and the opaR genes are indicated by the arrows next to the sequencing lanes in Fig. 7. The intergenic region between hpt and opaR is very similar to the equivalent V. harveyi region, and a comparison of the two sequences is shown below the autoradiograms of the primer extension gels in Fig. 7. The promoter regions for opaR and luxR seem well conserved with respect to potential ς70 promoter sequences and the transcription start sites. The expression of opaR correlated with the opaque phenotype. The production of opaR mRNA was detected only in the opaque strain and not in the translucent strain (Fig. 7, left panel, lane 1 versus lane 2). As a control, primer extension was performed with identical RNA preparations and conditions and with an hpt-derived primer. In this case, transcripts were identified for both opaque and translucent strain mRNAs (Fig. 7, right panel, lanes 1 and 2). In fact, slightly more total RNA was used in the translucent strain than in the opaque strain primer extension reactions, and this finding is reflected in more product in lane 2 than in lane 1 of Fig. 7 with the hpt-derived primer. The expression of hpt was observed in both cell types, while opaR-specific RNA could be detected only in the opaque strain and was not be observed in the translucent strain.
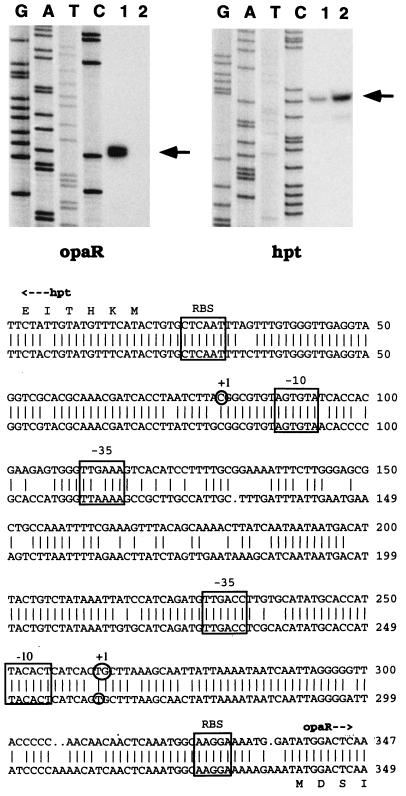
FIG. 7.
Primer extension mapping of opaR and hpt transcripts. RNA was prepared from BB22OP (lane 1) and BB22TR (lane 2). Lanes G, A, T, and C correspond to the sequencing reactions generated with opaR- or hpt-specific primers for pLM1950 template DNA. The positions of the primer extension products are indicated by arrows. Below the autoradiogram is a GCG BestFit comparison of the V. parahaemolyticus (top) and V. harveyi (bottom) intergenic regions spanning the hpt and the opaR (or luxR) genes. Features of the sequence include potential ribosome binding sites identified by homology with the Shine-Dalgarno sequence (RBS), potential ς70 promoters (boxed −10 and −35 regions), and transcription start sites (circled nucleotides labeled +1). For V. harveyi, the luxR start site was determined by Miyamoto et al. (25), and the promoter boxes were proposed by Showalter et al. (29) and Miyamoto et al. (25). For V. parahaemolyticus, a strong doublet was observed for the opaR primer extension product (circle including two bases). The doublet was very clear in autoradiogram exposures shorter than the one shown here. It should be noted that there are multiple potential ATG start codons for opaR, and the translation depicted in this figure represents the particular N terminus having optimal spacing with respect to a potential Shine-Dalgarno sequence and corresponding to the predicted V. harveyi N terminus.
Opaque-translucent variation involves genomic rearrangements.
Southern analysis of opaque and translucent variant strains revealed a difference in the chromosome structure in the opaR locus. Figure 8A shows the physical rearrangement observed on a Southern blot containing digested chromosomal DNA prepared from opaque and translucent strains. Also on this blot is digested DNA prepared from the opaR::Tn5 mutant strain LM4462. The blot was probed with cosmid pLM1950, which contains approximately 20 kb of recombinant V. parahaemolyticus DNA encompassing the opaR locus. Locations of transposon mutations can be mapped on a Southern blot because insertion of a transposon results in a large perturbation of the sequence. In Fig. 8A, lanes 5 and 6, which contained LM4462 DNA, a 2.3-kb fragment was missing. This finding is consistent with the sequence data, which precisely determined the insertion to be at bp 1392 (Fig. 2) within the coding region for opaR (from bp 1338 to bp 1952) and on a 2.3-kb HpaI-NcoI restriction fragment. Thus, the opaR gene can be mapped to this fragment. This same fragment showed a size aberration in opaque and translucent variants, while the other restriction fragments remained relatively constant in size among the strains (except for the new bands appearing in LM4462, which were the consequence of the transposon insertion). When a blot identical to that shown in Fig. 8A was reprobed with only a structural coding sequence (with plasmid pLM2035), the fluctuating band in the 2.3-kb region was the only band detected (data not shown). The rearrangement in the chromosome structure observed for the opaque and translucent strains appeared to be closely linked to the opaR gene.
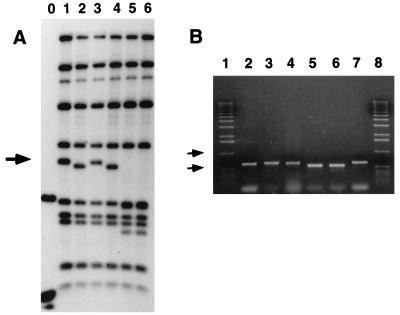
FIG. 8.
Genomic organization of the opaR locus in opaque and translucent strains. (A) Southern blot of chromosomal DNAs prepared from the following strains: lanes 1 and 3, BB22OP; lanes 2 and 4, BB22TR; and lanes 5 and 6, LM4462 (opaR::Tn5). DNAs were digested with HpaI and NcoI, loaded in the indicated lanes, electrophoresed on an 0.8% agarose gel, Southern blotted, and probed with 32P-labeled pLM1950. The 2.3-kb band marked with an arrow shows perturbations, while the other bands remained constant in size among the lanes. Molecular size markers were loaded in lane 0, and the two bands visualized correspond to 1.6 and 0.5 kb. (B) PCR amplification of opaR. Products were displayed on a 0.6% ethidium bromide-stained agarose gel. Lanes correspond to PCR products containing DNAs from the following sources: 2 and 7, cosmid pLM1950; 3 and 4, BB22OP; and 5 and 6, BB22TR. Molecular size markers were loaded in lanes 1 and 8. Arrows indicate 1- and 0.5-kb markers. The predicted size of the PCR product derived from the sequence of pLM1950 is 620 bp. The specific primers used were designed to amplify the sequence between bp 919 and 1539 depicted in Fig. 2.
The rearrangement was analyzed in another manner (Fig. 8B). Two oligonucleotides were used to prime a PCR amplification between bp 919 and 1539 on the map shown in Fig. 2. This region contains the intergenic region between hpt and opaR and part of the N-terminal coding region for opaR. The sizes of the amplified products were different when template DNAs from opaque and translucent strains were used (two strains of each). The size of the product amplified from opaque strains (Fig. 8B, lanes 3 and 4) corresponded well with the size of the product produced when the template DNA was the sequenced clone pLM1950 (lanes 2 and 7), while the size of the translucent strain product was smaller by approximately 100 bp (lanes 5 and 6). Cosmid pLM1950 was cloned by selecting for the expression of opaR in E. coli, and the size of the PCR product amplified with pLM1950 corresponded to the size of the product produced with chromosomal DNA prepared from the opaque form, which is the state observed to express opaR mRNA. The size of the PCR product was different for translucent strains, and the translucent form failed to produce an opaR transcript. The PCR results in combination with the transcription data are consistent with the hypothesis that opaR is not expressed in translucent strains due to chromosomal organization.
DISCUSSION
V. parahaemolyticus is a bacterium with multiple personalities, each adapted for survival in a particular environment. The swimmer cell possesses a sheathed polar flagellum that is capable of rotating very fast and propelling the bacterium in liquid medium at speeds averaging 60 μm per s (3). However, in viscous medium or on surfaces, the polar flagellum fails to function, leading to the formation of the swarmer cell (20). The swarmer cell can be very long and multinucleate and can possess numerous lateral flagella in addition to the polar flagellum (7, 20). Induction of the lateral flagellar system endows the bacterium with the ability to move through viscous layers or over surfaces. The result of swarmer cell differentiation is colonization of surfaces. In addition to the swimmer-swarmer cell dimorphism, V. parahaemolyticus exhibits another kind of phenotypic switching, described as opaque-translucent variation in colony morphology. The molecular basis for the difference in colony morphology is not known. For other organisms it has been associated with differences in cell surface characteristics, e.g., outer membrane proteins and encapsulation (35, 38). It has been postulated that differences in colony structure or packing result in differential light transmission by opaque and translucent forms (34).
This work describes the identification of a gene, opaR, that encodes a transcriptional regulatory protein controlling opacity. The product of the opaR gene is 96% identical to and functionally exchangeable with the LuxR protein of V. harveyi. The gene was cloned by selecting for activation of the V. harveyi luxCDABE operon in E. coli. V. parahaemolyticus produces autoinducer molecules capable of inducing luminescence in V. harveyi (4, 14). It has been a puzzle why nonluminescent V. parahaemolyticus makes signaling molecules, and one hypothesis has postulated a role in swarming behavior. Precedent for such an idea derives from S. liquefaciens. In this organism, loss of function of a gene encoding an autoinducer synthase abolishes swarming motility on certain types of media (10). To test this hypothesis in V. parahaemolyticus, the gene encoding the LuxR homolog was targeted for introduction of a knockout mutation. Strains carrying such a defect were demonstrated to display no swarming defect. In the course of these experiments, swarming was examined on minimal medium and, contrary to observations for other swarming organisms, such as S. liquefaciens, wild-type V. parahaemolyticus was observed to swarm on minimal medium without supplementation with Casamino Acids. Mutants with opaR defects also swarmed well on this medium.
Evidence suggests that opaR controls opacity. Introduction of a clone containing the opaR locus into a translucent strain transformed the colony morphology to opaque. Transposon-mediated disruption of opaR in an opaque genetic background converted the strain to a translucent phenotype. Expression of the gene from an exogenously controlled promoter determined opacity; i.e., opacity could be manipulated by use of an IPTG-inducible promoter controlling opaR expression.
Expression of opaR may be ultimately governed by the particular state of the DNA. The opaR gene is expressed in opaque strains but not in translucent strains. DNA rearrangements in the opaR locus were detected in opaque and translucent strains. The locus was cloned from a cosmid bank by use of positive selection for expression, i.e., activation of the V. harveyi lux operon in E. coli. Thus, both clone pLM1950 and opaque strains are expression competent for opaR. PCR analysis demonstrated that the physical state of the DNA on the clone appears to correspond to that found in opaque strains. Interestingly, the bank from which the clone was retrieved was constructed from a translucent strain (21). It is possible that the culture from which the chromosomal DNA was prepared contained some opaque variants. Current work involves analyzing other clones retrieved directly from the same bank by DNA hybridization as well as examining lineages of opaque-translucent variants in order to define the molecular basis for the rearrangement. It is possible that switching is the result of a gene or promoter inversion similar to Salmonella typhimurium flagellar phase variation (32) or some kind of phase-shifting event involving the loss of DNA. It is intriguing that the expression of luminescence in V. harveyi is also an unstable phenotype. Colonies show striking sectoring and segregate into bright and dim phenotypes with respect to light production (32).
What regulates opacity? Opaque-translucent switching may alternate between expression-competent and expression-incompetent states; however, the transcription of opaR may also be regulated by intercellular or environmental signaling mechanisms. Although V. parahaemolyticus and V. harveyi are phylogenetically closely related (9) and have functionally exchangeable components with respect to autoinducer and LuxR functions, it remains to be established whether opaR expression in V. parahaemolyticus is actually density dependent. It is provocative that the intergenic region and the transcription initiation sites are conserved between the organisms. Furthermore, the gene organization and the sequence conservation extend to the one other described homolog of V. harveyi LuxR, HapR, a positive regulator found in V. cholerae (16). HapR controls the expression of a soluble hemagglutinin. Interestingly, hapR mutants of El Tor strains have a rugose colony morphology.
What is the role of opaR? opaR is a master regulatory gene controlling cell type. Opaque and translucent strains form distinct colony structures. The colony organization is most probably determined by cell surface characteristics. Particular cell surface components, such as proteins or capsules, may aid survival under specific circumstances. For example, at times it may be beneficial to be adhesive. At other times, perhaps during the development of a biofilm, it may be critical to have mobility. Once colonization is established, the highly motile swarmer lifestyle may not always be advantageous. Also, an ability to detach, perhaps autoagglutinate, and take up a planktonic existence may play an important role in survival. These possibilities, plus the potential role of autoinducer signaling in the growth of communities, will be interesting to pursue. The alternating personalities of V. parahaemolyticus may have important significance for the development of ideas with respect to attachment, detachment, and how bacterial populations adapt to growth on surfaces, form structured communities, and develop in biofilms.
ACKNOWLEDGMENTS
I especially thank Mike Silverman for introducing me to opaque and translucent Vibrio and Rich Showalter for cloning and mutagenizing opaR. Also, acknowledgments are appreciatively due to Bill Elliot, who worked on some of the gene replacements, Todd Ontl, who cloned the Tn nub from the opaR::Tn5 plasmid, Y. K. Kim for expert technical assistance, Jodi Enos-Berlage for insightful discussion, and the DNA Core Facility and Medical Photography and Graphics at the University of Iowa for excellent support.
This research was supported by Public Health Service grant GM43196 from the National Institutes of Health.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atsumi T, McCarter L, Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion membrane forces. Nature (London) 1992;355:182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- 4.Bassler B L, Greenberg E P, Stevens A M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassler B L, Wright M, Silverman M R. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 6.Batchelor S E, Cooper M, Chhabra S R, Glover L A, Stewart G S A B, Williams P, Prossner J I. Cell density-regulated recovery of starved biofilm populations of ammonia-oxidizing bacteria. Appl Environ Microbiol. 1997;63:2281–2286. doi: 10.1128/aem.63.6.2281-2286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belas R, Simon M, Silverman M. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J Bacteriol. 1986;167:210–218. doi: 10.1128/jb.167.1.210-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broach J, Neumann C, Kustu S. Mutant strains (nit) of Salmonella typhimurium with a pleiotropic defect in nitrogen metabolism. J Bacteriol. 1976;128:86–98. doi: 10.1128/jb.128.1.86-98.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorsch M, Lane D, Stackebrandt E. Towards a phylogeny of the genus Vibrio based on 16S rRNA sequences. Int J Syst Bacteriol. 1992;42:58–63. doi: 10.1099/00207713-42-1-58. [DOI] [PubMed] [Google Scholar]
- 10.Eberl L, Winson M K, Sternberg C, Stewart G S A B, Christiansen G, Chhabra S R, Bycroft B, Williams P, Molin S, Givskov M. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behavior of Serratia liquefaciens. Mol Microbiol. 1996;20:127–136. doi: 10.1111/j.1365-2958.1996.tb02495.x. [DOI] [PubMed] [Google Scholar]
- 11.Friedman A, Long S R, Brown S E, Buikema W J, Ausubel F. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 12.Fuerste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 13.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 14.Greenberg E P, Hastings J W, Ulitzur S. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch Microbiol. 1979;120:87–91. [Google Scholar]
- 15.Hanselka B L, Greenberg E P. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J Bacteriol. 1995;177:815–817. doi: 10.1128/jb.177.3.815-817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jobling M G, Holmes R K. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol. 1997;26:1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 17.Kupsch E M, Knepper B, Kuroki T, Heuer I, Meyer T F. Variable opacity (Opa) outer membrane proteins account for the cell tropisms displayed by Neisseria gonorrhoeae for human leukocytes and epithelial cells. EMBO J. 1993;12:641–650. doi: 10.1002/j.1460-2075.1993.tb05697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambden P R, Heckels J E, James L T, Watt P J. Variations in surface protein composition associated with virulence properties in opacity types of Neisseria gonorrhoeae. J Gen Microbiol. 1979;114:305–312. doi: 10.1099/00221287-114-2-305. [DOI] [PubMed] [Google Scholar]
- 19.Martin M, Showalter R, Silverman M. Identification of a locus controlling expression of luminescence genes in Vibrio harveyi. J Bacteriol. 1989;171:2406–2414. doi: 10.1128/jb.171.5.2406-2414.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarter L, Silverman M. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol Microbiol. 1990;4:1057–1062. doi: 10.1111/j.1365-2958.1990.tb00678.x. [DOI] [PubMed] [Google Scholar]
- 21.McCarter L L, Silverman M. Phosphate regulation of gene expression in Vibrio parahaemolyticus. J Bacteriol. 1987;169:3441–3449. doi: 10.1128/jb.169.8.3441-3449.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarter L L, Wright M E. Identification of genes encoding components of the swarmer cell flagellar motor and propeller and a sigma factor controlling differentiation of Vibrio parahaemolyticus. J Bacteriol. 1993;175:3361–3371. doi: 10.1128/jb.175.11.3361-3371.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLean R J C, Whitely M, Stickler D J, Fuqua W C. Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol Lett. 1997;154:259–263. doi: 10.1111/j.1574-6968.1997.tb12653.x. [DOI] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 25.Miyamoto C M, Chatterjee J, Swartzman E, Szittner R, Meighen E A. The role of the lux autoinducer in regulating luminescence in Vibrio harveyi: control of luxR expression. Mol Microbiol. 1996;19:767–775. doi: 10.1046/j.1365-2958.1996.417948.x. [DOI] [PubMed] [Google Scholar]
- 26.Reddy G P, Hayat U, Abeygunawardana C, Fox C, Wright A C, Maneval D R, Jr, Bush C A, Morris J G., Jr Purification and determination of the structure of capsular polysaccharide of Vibrio vulnificus M106-24. J Bacteriol. 1992;174:2620–2630. doi: 10.1128/jb.174.8.2620-2630.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Showalter R, Martin M O, Silverman M R. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J Bacteriol. 1990;172:2946–2954. doi: 10.1128/jb.172.6.2946-2954.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Showalter, R., M. Silverman, and L. McCarter. Unpublished observation.
- 31.Silverman M, Showalter R, McCarter L. Genetic analysis in Vibrio. Methods Enzymol. 1991;204:515–536. doi: 10.1016/0076-6879(91)04026-k. [DOI] [PubMed] [Google Scholar]
- 32.Simon M I, Silverman M. Recombinational regulation of gene expression in bacteria. In: Beckwith J, Davies J, Gallant J A, editors. Gene function in prokaryotes. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 211–227. [Google Scholar]
- 33.Stewart B J, Enos-Berlage J L, McCarter L L. The lonS gene regulates swarmer cell differentiation of Vibrio parahaemolyticus. J Bacteriol. 1997;179:107–114. doi: 10.1128/jb.179.1.107-114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swanson J. Studies on gonococcus infection. XII. Colony color and opacity variants of gonococci. Infect Immun. 1977;19:320–331. doi: 10.1128/iai.19.1.320-331.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swanson J. Studies on gonococcus infection. XIV. Cell wall protein differences among color/opacity colony variants of Neisseria gonorrhoeae. Infect Immun. 1978;21:292–302. doi: 10.1128/iai.21.1.292-302.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swift S, Throup J P, Williams P, Salmond G P C, Stewart G S A B. Quorum sensing: a population-density component in the determination of bacterial phenotype. Trends Biochem Sci. 1996;21:214–219. [PubMed] [Google Scholar]
- 37.Woo T H S, Cheng A F, Ling J M. An application of a simple method for the preparation of bacterial DNA. BioTechniques. 1992;13:696–697. [PubMed] [Google Scholar]
- 38.Wright L, Simpson L M, Oliver J D, Morris J G., Jr Relation of capsular materials and colony opacity to virulence of Vibrio vulnificus. Infect Immun. 1990;47:446–451. doi: 10.1128/iai.47.2.446-451.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]