Abstract
Ferroptosis, a unique form of regulated necrotic cell death, is caused by excessive iron-dependent lipid peroxidation. However, the underlying mechanisms driving ferroptosis in human cancers remain elusive. In this study, we identified TRIM3, an E3 ubiquitin-protein ligase, as a key regulator of ferroptosis. TRIM3 is downregulated in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), two major types of non-small cell lung cancer (NSCLC). Forced expression of TRIM3 promotes cell death by enhancing the cellular level of ROS and lipid peroxidation. Moreover, our in vivo study determined that TRIM3 overexpression diminishes the tumorigenicity of NSCLC cells, indicating that TRIM3 functions as a tumor suppressor in NSCLC. Mechanistically, TRIM3 directly interacts with SLC7A11/xCT through its NHL domain, leading to SCL7A11 K11-linked ubiquitination at K37, which promotes SLC7A11 proteasome-mediated degradation. Importantly, TRIM3 expression exhibits a negative correlation with SCL7A11 expression in clinical NSCLC samples, and low TRIM3 expression is associated with a worse prognosis. This study reveals that TRIM3 functions as a tumor suppressor that can impede the tumorigenesis of NSCLC by degrading SLC7A11, suggesting a novel therapeutic strategy against NSCLC.
Subject terms: Non-small-cell lung cancer, Ubiquitylation
Introduction
Lung cancer remains one of the most commonly diagnosed malignancies, causing 1.796 million cancer-related mortalities (approximately 18.4% of the total cases) annually on a global scale [1, 2]. Non-small cell lung cancer (NSCLC) represents over 85% of lung cancer cases and its two main subtypes, accounting for approximately 80% of NSCLC cases, are lung adenocarcinoma (LUAD) and lung squamous carcinoma (LUSC) [3, 4]. Despite the availability of chemotherapy, irradiation, and immunotherapy, the 5-year survival rate of patients with advanced-stage lung cancer remains unsatisfactory, dropping to 0-10% compared to 60% for early-stage patients, due to chemo/radioresistance and immunogenic heterogeneity [5–7]. Thus, there is an urgent need to explore novel strategies for NSCLC treatment.
Ferroptosis, a unique modality of programmed cell death driven by iron-dependent lipid peroxidation, has been reported to play a vital role in tumor suppression [8, 9]. Activation of ferroptosis has been reported to contribute to the efficacy of cancer treatments, such as chemotherapy [10], radiotherapy [11], and immune checkpoint blockade [12]. Thus, a better understanding of ferroptosis may lead to new therapeutic strategies for NSCLC. The classic cellular anti-ferroptosis mechanism is achieved by the SLC7A11 (also known as xCT) signaling axis [13, 14]. As a core regulator of ferroptosis, SLC7A11 is capable of exporting intracellular glutamate and importing extracellular cystine, thereby inhibiting ferroptosis [15]. The expression and activity of SLC7A11 are regulated at multiple levels. Upon oxidative stress conditions, transcription of SLC7A11 could be induced by stress-responsive transcription factors, including nuclear factor erythroid 2-related factor 2 (NRF2) [16] and activating transcription factor 4 (ATF4) [17], and the methylation of histone H3 [18]. Apart from transcriptional regulation, SLC7A11 expression is also controlled at the post-translational level. For instance, OTUB1 functions as a deubiquitinase that interacts with SLC7A11 and maintains SLC7A11 protein stability [19]. However, despite these observations, the dysregulation of SLC7A11 in NSCLC progression remains largely unknown.
Tripartite motif containing 3 (TRIM3), a member of the ring type E3 ligases [20], was first identified to associate with myosin and mediate its target protein transportation in cells [21]. Recently, TRIM3 was shown to be involved in a variety of biological processes including cell proliferation [22], cell apoptosis [23], and cell differentiation [24] in several types of human cancers. However, its role in NSCLC has not been characterized.
Here, our study showed a tumor suppressor role of TRIM3 in NSCLC. Overexpression of TRIM3 impeded tumor cell survival by promoting cellular ferroptosis. Molecular biology studies uncovered that TRIM3 interacted with SLC7A11 and catalyzed K11-linked polyubiquitination of SLC7A11 at K37, which led to SLC7A11 proteasome-mediated degradation, suggesting a key post-transcriptional modification mode of SLC7A11. Its therapeutic application holds tantalizing promise in regenerative medicine.
Methods and materials
Cell culture
Human NSCLC cell lines (A549, H1975, H2122, H1573, H1944, and HCC827) and human embryonic kidney (HEK293T) cells were obtained from Beyotime Company (Shanghai, China) and cultured in DMEM supplemented with 10% fetal bovine serum (10099141C; Gibco) and 1% penicillin/streptomycin (C0222, Beyotime). BEAS-2B and 16HBE cell lines were obtained from ATCC and maintained in a special culture medium (CM-0496 and CM-0249, Procell). All cell lines were free from mycoplasma contamination and kept in a humidified atmosphere containing 5% CO2 at 37 °C.
Antibodies and reagents
The following antibodies and reagents were used in this study: β-actin (AF0003, Beyotime), TRIM3 (sc-136363, Santa Cruz Biotechnology), TRIM3 (ab111840, Abcam), Ki-67 (ab15580, Abcam), anti-DDDDK tag (ab18230, Abcam), anti-Myc tag (ab32, Abcam), anti-HA tag (ab9110, Abcam), SLC7A11 (ab307601, Abcam), ACSL4 (ab307601, Abcam), GPX4 (ab125006, Abcam), Nrf2 (ab62352, Abcam), MG132 (S1748, Beyotime), chloroquine (C843545, Macklin), cycloheximide (66-81-9, Sigma-Aldrich), bortezomib (HY-10227, MedChemExpress),
Plasmids, siRNA, and transfection
Fag-TRIM3 WT or its mutant (C22/25S) and Myc-SLC7A11 were generated by cloning their coding regions with the N-terminal FLAG or Myc sequence into the pCDH-CMV-MCS-EF1α-Puro vector. Plasmids encoding HA-ubiquitin WT and its mutants (K6, K11, K27, K29, K33, K48, K63) were obtained from Addgene. The following siRNA targeting TRIM3 were used: siTRIM3#1: 3′-TATGCAGTATCTGCCTGGATCGGTA-5′; siTRIM3#2: 3′-UGAAUACACUGGUUGUUGC-5′. Cells were transfected with the indicated plasmids and siRNAs with Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions.
RNA isolation and real-time q-PCR
Total RNA was extracted by using TRIzol reagent (15596-026, Invitrogen). cDNA synthesis kit (K1622, Thermo Fisher) was used for reverse transcription. The primer sequences were as follows: TRIM3, forward: 5′-GGCTGACTGGGGCAACAGCCGCATC-3′, reverse 5′-ATCTGCAGAACCACTGTATGGTCCA-3′. SLC7A11, forward: 5′- TCTCCA AAGGAGGTTACCTGC-3′, reverse 5′- AGACTCCCCTCAGTAAAGTGAC-3′. GAPDH, forward: 5′-GAAGGTGAAGGTCGGAGTC-3′, reverse: 5′-GAAGATGGTGATGGGATTTC-3′. The relative TRIM3 mRNA levels were normalized to GAPDH and determined using the 2-ΔΔCt method.
Human tissues and immunohistochemistry (IHC)
Human NSCLC tissues were obtained from the Affiliated Taizhou People’s Hospital of Nanjing Medical University and Shanghai Outdo Biotech (Shanghai, China).
IHC staining of human tissues and xenograft tumors was performed by Servicebio Technology (Wuhan, China). A microarray containing 85 primary NSCLC tissues and adjacent normal lung tissues (Shanghai Outdo Biotech) was used to test the expression of TRIM3 and SLC7A11. The proportion score reflected the proportion of positively stained cells (0, 0%; 1, 1–25%; 2, 26–50%; 3, 51–75%; 4, 75–100%), and the intensity score revealed the staining intensity (0, no staining; 1, weak; 2, intermediate; 3, strong). The expression of TRIM3 and SLC7A11 was calculated by multiplying the proportion and intensity score. Due to the relatively low expression of TRIM3 and the relatively high expression of SLC7A11 in tumors, IHC scores ≤3 for TRIM3 were regarded as low expression, and IHC scores >3 were considered high. IHC scores ≤6 for SLC7A11 were regarded as low expression, and scores >6 were considered high.
Colony formation and CCK-8 assay
For the colony formation assay, the cells were plated in six-well plates at a density of 300 cells/well and cultured for 2–3 weeks. Then the culture medium was removed, and the colonies were stained with crystal violet (0.1% in 20% methanol) after being fixed with 4% paraformaldehyde.
The Cell Counting Kit-8 (CK04, DOJINDO) was used to perform the CCK-8 assay. Briefly, lung cancer cells were plated in 96-well plates. Each well was incubated with 100 μl DMEM (Gibco) supplemented with 10 μl CCK-8 reagent at 37 °C for 2 h. The absorbance of each well was measured at 450 nm using a microplate reader (Thermo Fisher Scientific, USA). All experimental data were a result of three replicates.
Invasion, migration, and wound-healing assay
For cell invasion and migration assays, 1 × 104 tumor cells were suspended in 100 μl serum-free medium and then plated in an 8.0-μm, 24-well plate with or without Matrigel-coated chamber inserts (Corning). Then, 500 μl DMEM containing 10% fetal bovine serum was added to the bottom of the insert. After incubation for 24 h, the cells were fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet blue for 30 min. The positively stained cells were captured using a Nikon inverted microscope.
For the wound-healing assay, cells were transplanted into a 6-well plate. Cells at a density of approximately 90% were scraped with a 10-μl pipette tip. After washing with PBS, the cells were kept in serum-free DMEM medium. Photographs were taken at 0 h and 24 h after wounding. Statistical analysis was performed with Image-Pro Plus 6.0 software. Data are shown as the average of three independent experiments.
GST pull-down assay
To generate GST and GST-tagged proteins from bacteria, Escherichia coli BL21 (DE3) cells containing the GST, GST-SLC7A11 WT, and GST-SLC7A11 K37R plasmids were induced for protein expression using 0.5 mM isopropyl-d-1-thiogalactopyranoside at 37 °C for 5 h. The proteins immobilized on glutathione Sepharose beads were verified using SDS-PAGE and aliquoted for storage at ***-80 °C.
For the GST pull-down assay, purified Flag-TRIM3 WT and Flag-TRIM3 C22/25S were incubated with Glutathione Sepharose 4B (Catia) bound to GST or GST-SLC7A11 in binding buffer at 4 °C overnight. After washing three times with the binding buffer, the pull-down samples were analyzed by immunoblotting.
In vivo ubiquitination assay
An in vivo ubiquitination assay was performed using denature-IP (d-IP). Cells were lysed in SDS-denaturing buffer (62.5 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 1.5% β-mercaptoethanol) and boiled for 10 min. Cell lysates were then diluted ten to forty-fold in native lysis buffer (50 mM Tris-HCl pH 7.4, 0.5% Triton X-100, 200 mM NaCl, 10% glycerol). After centrifugation, the supernatants were subjected to immunoprecipitation and analyzed by immunoblotting.
In vitro ubiquitination assay
Purified Flag-TRIM3 or its CS mutant proteins were incubated with E1 UBE1, E2 UbcH5c, His-ubiquitin, and adenosine triphosphate in the absence or presence of purified GST-SLC7A11 WT or GST-SLC7A11 K37R. The mixture was subjected to Ni-NTA pull-down assay and then analyzed by immunoblotting.
Subcutaneous tumor xenograft models
Female 5-week-old nude mice (GemPharmatech Co., Ltd) were used for animal study. A total of 5 × 106 A549 cells transfected as indicated were suspended in 100 µl DMEM and subcutaneously implanted in the right flank of each mouse. The tumor size was determined using digital calipers every three days. Tumor volume = length × width2 × 0.52. At the end of the study, the mice were sacrificed, and the tumors were collected and weighed.
Detection of malondialdehyde (MDA) and 4-hydroxynonenal (4-HNE)
Lipid peroxidation detection kits (ab118970 and ab238538) obtained from Abcam were used to determine the levels of MDA and 4-HNE in cell lysates according to the manufacturer’s instructions. For MDA analysis, 2 × 106 cells were collected with MDA lysis buffer supplemented with 1% BHT. After centrifugation, 200 µl supernatant was added to a 1.5 ml EP tube containing 600 µl of Developer VII/TBA reagent and incubated at 95 °C for 60 min. After cooling to room temperature in an ice bath, 200 µl reaction mix was added into a 96-well microplate, and the absorbance of each well at 532 nm was measured. For 4-HNE analysis, 1 × 106 cells were collected with RIPA buffer and subjected to centrifugation. Then, 50 µl of supernatant was added to the 4-HNE conjugate-coated plate and incubated at room temperature for 10 min. After incubation with anti-4-HNE antibody and secondary antibody-HRP, substrate solution was added and the absorbance at 450 nm was measured.
Measurement of ROS and lipid oxidation levels
The ROS Assay Kit (S0033, Beyotime) was used to determine the levels of ROS in tumor cells. A Lipid Peroxidation Probe (L267, DOJINDO) was used to determine the levels of lipid ROS. In brief, the cells were plated in six-well plates at a density of 2 × 105 cells/well. The next day, cells were harvested by trypsinization and incubated with 10 µM DCFH-DA (for ROS assay) or BDP 581/591 C11 working solution (for lipid ROS assay) at 37 °C for 30 min in the dark. After washing three times, the fluorescence intensity of ROS was tested by flow cytometry (FACSCalibur, BD Biosciences).
Determination of iron content
The FerroOrange Kit (GC19943, Glpbio) was used to determine the iron concentration in cells. Briefly, cells were incubated with 1 µM FerroOrange working solution at 37 °C for 30 min and then incubated with FerroOrange staining solution. Images were captured using a fluorescence microscope (THUNDER DMi8, LEICA) at an excitation wavelength of 532 nm and an emission wavelength of 580 nm.
Statistical analysis
The data are shown as the mean ± SD of at least three independent experiments. Statistical analysis was performed using GraphPad software 7.0. Student’s t-test was used for comparisons between two groups, and one-way analysis of variance was used for comparisons among more than two groups. The log-rank test was used to determine the statistical significance of Kaplan–Meier survival curves. A P value less than 0.05 was considered statistically significant.
Result
TRIM3 is downregulated in non-small cell lung cancer
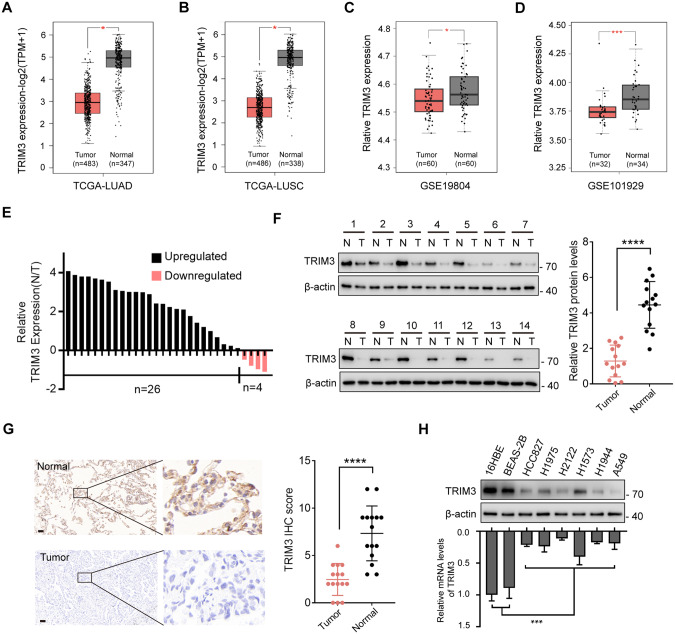
To determine the expression of TRIM3 in non-small cell lung cancer (NSCLC), we performed in silico analysis using the GEPIA2 website [25]. As shown in Fig. 1A and B, the mRNA levels of TRIM3 were significantly lower in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) compared to that in adjacent normal lung tissues. Similar expression trends were observed in three other cohorts of NSCLC datasets (Fig. 1C, D and Fig. S1A). Real-time q-PCR confirmed lower TRIM3 mRNA levels in 30 clinical NSCLC samples compared to that in paired adjacent normal tissues (Fig. 1E). In confirmation of these transcriptomic results, we noticed decreased TRIM3 protein levels in tumor tissues (Fig. 1F and G). Moreover, we also determined TRIM3 expression in a panel of NSCLC cell lines. As expected, the protein and mRNA levels in two normal cell lines, 16HBE cells (human bronchial epithelial cell) and BEAS-2B cells (human lung epithelial cell), were markedly higher than those in tumor cells (Fig. 1H). Taken together, these data indicated a lower expression of TRIM3 in NSCLC.
Fig. 1. TRIM3 is downregulated in non-small cell lung cancer.
A Box plot analysis of TRIM3 mRNA levels in lung adenocarcinoma (LUAD) tissues (n = 483) and normal lung tissues (n = 347) from the TCGA database. B Box plot analysis of TRIM3 mRNA levels in lung squamous cell carcinoma (LUSC) tissues (n = 486) and normal lung tissues (n = 338) from the TCGA database. C, D Box plot analysis of TRIM3 mRNA levels in non-small cell lung cancer (NSCLC) tissues and normal lung tissues from the GSE19804 (C) and GSE101929 (D) datasets. E Expression of TRIM3 in 30 paired clinical samples with NSCLC and adjacent tissues was analyzed by q-PCR. F Western blot analysis for the protein levels of TRIM3 in 14 paired clinical samples with NSCLC and adjacent tissues. G Representative IHC images of TRIM3 expression in normal and tumor tissues. Scale bars: 100 μm. H Protein and relative mRNA levels of TRIM3 in human normal lung epithelial cells, human normal bronchial epithelial cells, and a panel of lung cancers were determined by western blot and q-PCR, respectively. Data are represented as the mean ± SD. Statistical analysis was performed using Student’s t-test (A–D, F, and G) and two-sided t-test (H), *p < 0.05; ***p < 0.001; ****p < 0.0001.
TRIM3 overexpression impedes the proliferation and invasion of lung cancer cells
To investigate the functional significance of TRIM3 in NSCLC, we overexpressed TRIM3 in A549 and H2122 cells in which TRIM3 expression was relatively low and knocked down TRIM3 in H1573 cells in which TRIM3 expression was relatively high (Fig. 2A and B). The CCK-8 assay revealed that enforced expression of TRIM3 significantly impeded NSCLC cell proliferation (Fig. 2C), whereas TRIM3 knockdown promoted tumor cell survival (Fig. 2D), and these results were further confirmed by the colony formation assay (Fig. 2E and F). Moreover, invasion, migration, and wound-healing assays revealed that TRIM3 overexpression significantly impaired the invasion and migration of NSCLC cells (Fig. 2G and H). In contrast, TRIM3 depletion strengthened the invasion and migration of H1573 cells (Fig. 2I and J). In vivo, the TRIM3-overexpressing tumors grew much more slowly than the tumors in the vector control group (Fig. 2K–M). In addition, the IHC assay showed that the expression level of Ki-67 in TRIM3-overexpressing tumors were markedly decreased compared to that in the vector control group (Fig. 2N). Collectively, these results indicate that TRIM3 functions as a tumor suppressor in NSCLC.
Fig. 2. TRIM3 overexpression impedes the proliferation and invasion of lung cancer cells.
A Western blot assay of TRIM3 expression in A549 and H2122 cells transfected with TRIM3 or vector control. B Western blot assay of TRIM3 expression in H1573 cells transfected with siTRIM3 RNA or siRNA control. C, D CCK-8 assay of A549, H2122 (C) and H1573 (D) cells transfected as indicated. E, F Colony formation assay of A549, H2122 (C) and H1573 (D) cells transfected as indicated. Scale bars: 0.5 cm. G, H Invasion (G) and wound-healing assay (H) of A549 and H2122 cells transfected with TRIM3 or vector control. I, J Invasion (I) and wound-healing assay (J) of H1573 cells transfected with siTRIM3 RNA or siRNA control. K Representative tumor images in nude mice bearing A549 cells transfected as indicated. L The tumor volume was monitored every 3 days. M The tumor weight. N The tumors were harvested and the expression of Ki-67 in different groups was determined by IHC. Data are represented as the mean ± SD (n = 3). Statistical analysis was performed using Student’s t-test, *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
TRIM3 facilitates ferroptosis in lung cancer cells
Accumulated evidence has revealed that multiple tumor suppressors inhibit tumor progression via cell death pathways including necrosis [26], apoptosis [27], autophagy [28], and ferroptosis [29]. As we noticed that cells with TRIM3 overexpression exhibited a higher cell death rate compared to that of the control cells (Fig. S2A), we thus wondered whether TRIM3 impedes NSCLC cell survival through these pathways. To this end, we treated A549 and H2122 cells with two ferroptosis inhibitors (Liproxstatin-1 and Ferrostatin-1), an apoptosis inhibitor (Z-VAD-FMK), a necrosis inhibitor (Necrostatin-1), or an autophagy inhibitor (3-MA). As shown in Fig. 3A and B, cell death induced by TRIM3 overexpression was reversed by Liproxstatin-1 and Ferrostatin-1, but not by other inhibitors, indicating that TRIM3 may promote cell death through ferroptosis. Consistent with this hypothesis, we found that TRIM3-knockdown H1573 cells were resistant to Erastin-induced cell death (Fig. 3C). Moreover, forced expression of TRIM3 led to elevated lipid peroxidation, ROS levels and free iron levels in A549 and H2122 cells (Fig. 3D–G and Fig. S2B and C). In contrast, TRIM3-knockdown rescued Erastin-induced lipid peroxidation, ROS levels and free iron levels (Fig. 3H–K and Fig. S2D and E). Furthermore, transmission electron microscopy (TEM) showed a shrunken morphology of mitochondria in cells overexpressing TRIM3 (Fig. S2F). However, Erastin-induced morphological changes in mitochondria were reversed by TRIM3 depletion (Fig. S2G). All these data suggested that TRIM3 strengthens ferroptosis in NSCLC cells.
Fig. 3. TRIM3 facilitates ferroptosis in lung cancer cells.
A, B Trypan blue staining assays using vector control- or TRIM3-overexpressing A549 and H2122 cells treated with two ferroptosis inhibitors (500 nM Lip-1 and 10 μM Fer-1), an apoptosis inhibitor (10 μM Z-VAD-FMK), a necrosis inhibitor (10 μM Nec-1), or an autophagy inhibitor (1 mmol/L 3-MA). C Trypan blue staining assays using H1573 cells transfected with siRNA control or siTRIM3 RNA upon treatment with the ferroptosis inducer (20 μM Erastin). D–G Relative expression levels of MDA (D-E), 4-HNE (F) and ROS (G) in A549 and H2122 cells transfected with TRIM3 or vector control. H–K Relative expression levels of MDA (H, I), 4-HNE (J), and ROS (K) in H1573 cells transfected with siTRIM3 RNA or siRNA control. Data are represented as the mean ± SD (n = 3). Statistical analysis was performed using Student’s t-test, *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
TRIM3 promotes xCT degradation
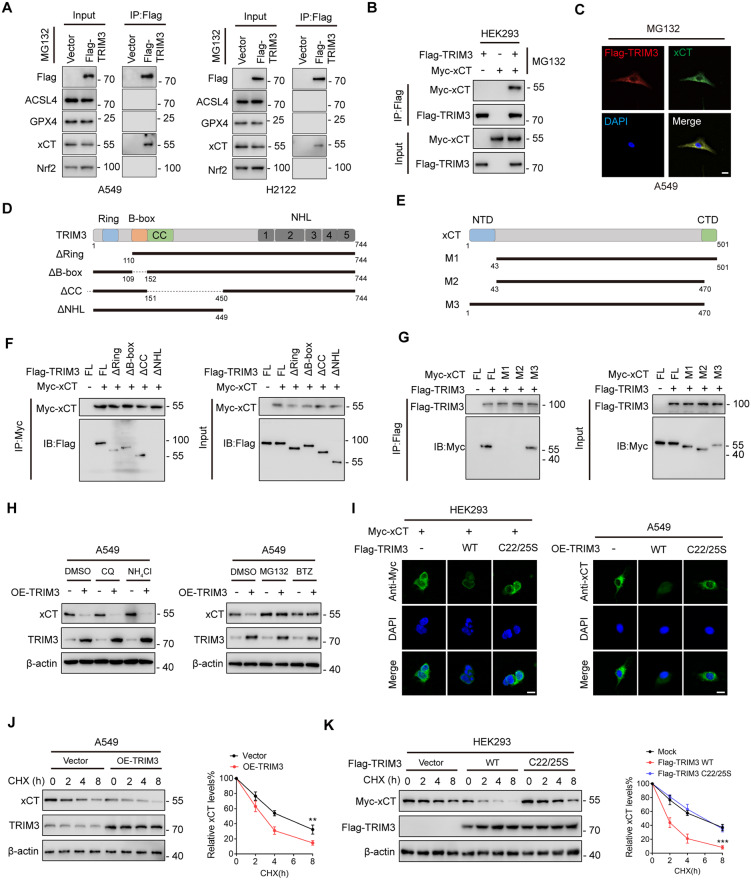
It is well-established that ferroptosis is accurately regulated by several core proteins such as GPX4, SLC7A11 (xCT), Nrf2, and ACSL4 [30]. To further investigate the underlying mechanisms by which TRIM3 regulates ferroptosis, we performed mass spectrometry (MS) analysis (Fig. S3A and Table S1). The result of MS analysis showed that xCT is co-precipitated with Flag-TRIM3 in A549 cells (Fig. S3B). Moreover, after immunoprecipitating ectopically expressed Flag-TRIM3 in A549 and H2122 cells, we found that endogenous xCT, but not other proteins, co-precipitated with TRIM3 (Fig. 4A). Complementarily, we also confirmed an interaction between ectopically expressed Flag-TRIM3 and Myc-xCT (Fig. 4B). Confocal analysis determined the co-localization of these proteins (Fig. 4C and Fig. S3C). Mapping TRIM3-xCT interaction motifs revealed that the NHL domain of TRIM3 as well as the N-terminal sequences of xCT were required for their interaction (Fig. 4D–G).
Fig. 4. TRIM3 promotes xCT degradation.
A A549 and H2122 cells transfected with Flag-TRIM3 were subjected to IP analysis with anti-Flag antibody and then analyzed by IB with the indicated antibodies. B HEK293 cells transfected with Flag-TRIM3 and Myc-xCT were subjected to IP analysis with anti-Flag antibody and then analyzed by IB with the indicated antibodies. C Confocal assay showing co-localization of Flag-TRIM3 (red) and xCT (green) in A549 cells. Nuclei were counterstained with DAPI (blue). Scale bar: 10 μm. D, E Sketch map of full-length (FL) Flag-labeled TRIM3, Myc-labeled xCT, and their various deletion mutants. F HEK293 cells were co-transfected with Myc-xCT and FL Flag-TRIM3 or deletion mutants. After treatment with MG132 (20 μM) for 6 h, cell lysates were subjected to IP followed by western blotting assay with the indicated antibodies. G HEK293 cells were co-transfected with Flag-TRIM3 and FL Myc-xCT or deletion mutants. After treatment with MG132 (20 μM) for 6 h, cell lysates were subjected to IP followed by western blot assay with the indicated antibodies. H A549 cells transfected with TRIM3 or vector control were treated as indicated. The xCT and TRIM3 expression levels were determined by western blotting. I Confocal assay of xCT expression in HEK293 and A549 cells transfected as indicated. J A549 cells transfected with TRIM3 or vector control were treated with CHX for the indicated times. The protein levels of xCT and TRIM3 were determined by western blotting. Quantification of xCT levels relative to β-actin are shown. K Myc-xCT was co-transfected with vector control, Flag-TRIM3 WT or Flag-TRIM3 C22/25S into HEK293 cells. After being treated with CHX for the indicated times, the cells were subjected to western blot analysis. Quantification of xCT levels relative to β-actin is shown. Data are represented as the mean ± SD (n = 3). Statistical analysis was performed using Student’s t-test, **p < 0.01; ***p < 0.001.
TRIM3, as an E3 ligase, has been reported to mediate ubiquitin-proteasomal degradation of its substrates [23]. We thus wondered whether TRIM3 plays a role in xCT stability. After overexpression of TRIM3, we noticed that the protein abundance of xCT was severely decreased, which was reversed by two proteasome inhibitors (MG132 and Bortezomib), but not by the lysosomal inhibitors (NH4Cl and chloroquine) (Fig. 4H). Moreover, confocal analysis revealed that ectopic expression of wild-type (WT) TRIM3, but not its catalytically inactive mutant (C22/25S), decreased Myc-xCT abundance (Fig. 4I). Consistently, overexpression of TRIM3 significantly promoted the degradation of xCT through treatment with CHX (Fig. 4J and Fig. S3D), while TRIM3 C22/25S did not support this effect (Fig. 4K). In addition, neither TRIM3 WT nor TRIM3 C22/25S altered xCT mRNA levels (Fig. S3E). Taken together, these data suggest that TRIM3 is an xCT-interacting partner and regulates the xCT protein level at the post-transcriptional level.
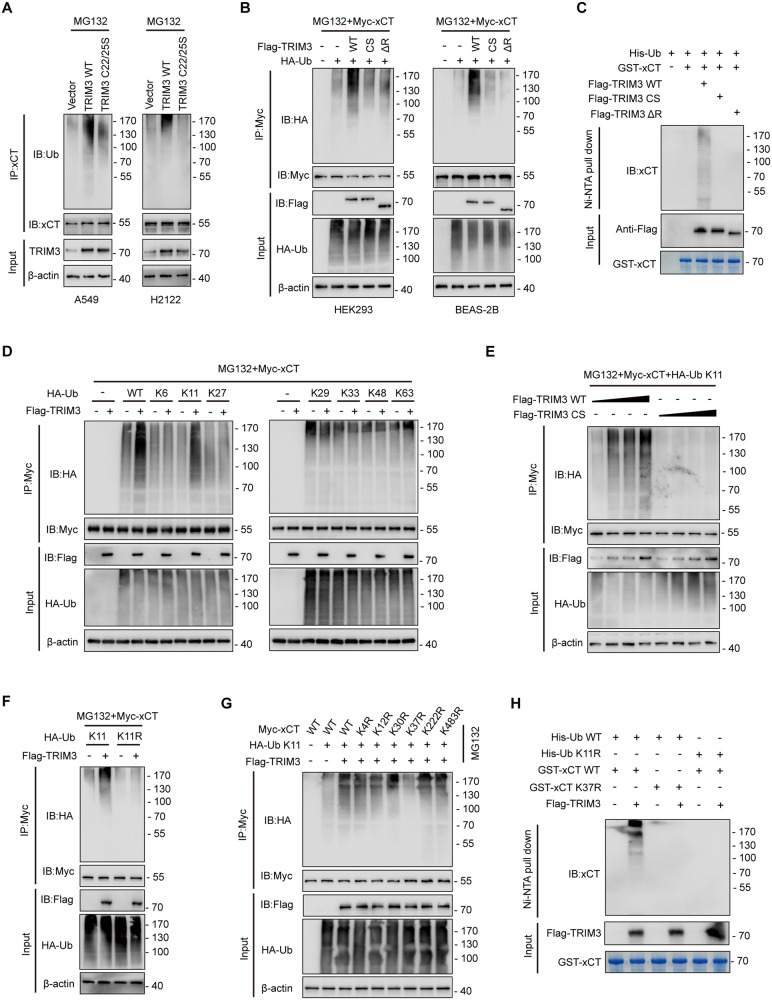
TRIM3 catalyzes K11-linked ubiquitination of xCT at K37
Next, we sought to determine the effect of TRIM3 on the ubiquitination of xCT. Forced expression of TRIM3 WT, but not the TRIM3 C22/25S mutant, enhanced the polyubiquitination levels of endogenous xCT in A549 and H2122 cells (Fig. 5A). Consistently, Myc-xCT was heavily ubiquitinated upon co-expression with TRIM3 WT, but not with TRIM3 C22/25S or the TRIM3 mutant (TRIM3 ΔR) lacking its Ring domain (exerts E3 ligases that mediates the attachment of ubiquitin to their substrates) (Fig. 5B). These results were further validated by an in vitro ubiquitination assay (Fig. 5C). Moreover, co-transfection experiments with linkage-specific ubiquitin showed that TRIM3-mediated K11-linked polyubiquitination of xCT (Fig. 5D), and this effect of TRIM3 was dose-dependent (Fig. 5E). Complementarily, the ubiquitin mutant K11R, in which K11 was replaced with Arg (R) was unable to support xCT this modification (Fig. 5F). Furthermore, we identified six putative ubiquitination sites of xCT that were reported previously using the PhosphosSitePlus website (Fig. S4A) [31]. To investigate whether TRIM3 catalyzes ubiquitination of xCT at these sites, we constructed a panel of xCT mutants and co-expressed them with Flag-TRIM3, respectively, and found that TRIM3 failed to mediate K11-linked polyubiquitination of xCT K37R (Fig. 5G), which was further determined by an in vitro ubiquitin assay (Fig. 5H). In addition, the amino acid sequence around K37 of xCT is highly conserved (Fig. S4B). These data indicate that TRIM3 specifically catalyzes K11-linked polyubiquitination of xCT at K37.
Fig. 5. TRIM3 catalyzes K11-linked ubiquitination of xCT at K37.
A A549 and H2122 cells transfected with vector control, TRIM3 WT or TRIM3 C22/25S were subjected to denaturing-IP (d-IP) with ant-xCT antibody and analyzed by western blotting. Cells were treated with MG132 (20 μM) for 6 h before collection. B Myc-xCT was co-transfected with Flag-TRIM3 WT, Flag-TRIM3 C22/25S or Flag-TRIM3 ΔR into HEK293 cells. After treatment with MG132 (20 μM) for 6 h, the cells were collected and subjected to d-IP and western blot assays. C Purified GST-xCT was incubated with purified Flag-TRIM3 WT, Flag-TRIM3 C22/25S or Flag-TRIM3 ΔR proteins. Then the reaction mixtures were subjected to Ni-NTA pull down and the precipitated proteins were analyzed by western blotting using an anti-xCT antibody. D HEK293 cells were transfected with Myc-xCT and Flag-TRIM3 together with ubiquitin WT or different ubiquitin mutants. After treatment with MG132 (20 μM) for 6 h, the cells were collected and subjected to d-IP and western blot assays. E HEK293 cells transfected with Myc-xCT and increasing amounts of Flag-TRIM3 WT or Flag-TRIM3 C22/25S were subjected to d-IP with anti-Myc antibody and analyzed by western blotting. Cells were treated with MG132 (20 μM) for 6 h before collection. F HEK293 cells were transfected with Myc-xCT and Flag-TRIM3 together with ubiquitin WT or ubiquitin K11R mutant. After treatment with MG132 (20 μM) for 6 h, the cells were collected and subjected to d-IP and western blot assays. G HEK293 cells transfected with Flag-TRIM3, ubiquitin K11, and different Myc-xCT mutants were subjected to d-IP with anti-Myc antibody and analyzed by western blotting. Cells were treated with MG132 (20 μM) for 6 h before collection. H Purified GST-xCT WT or GST-xCT K37R was incubated with purified Flag-TRIM3 proteins and His-ubiquitin WT or His-ubiquitin K11R. Then the reaction mixtures were subjected to Ni-NTA pull down, and the precipitated proteins were analyzed by western blotting using an anti-xCT antibody.
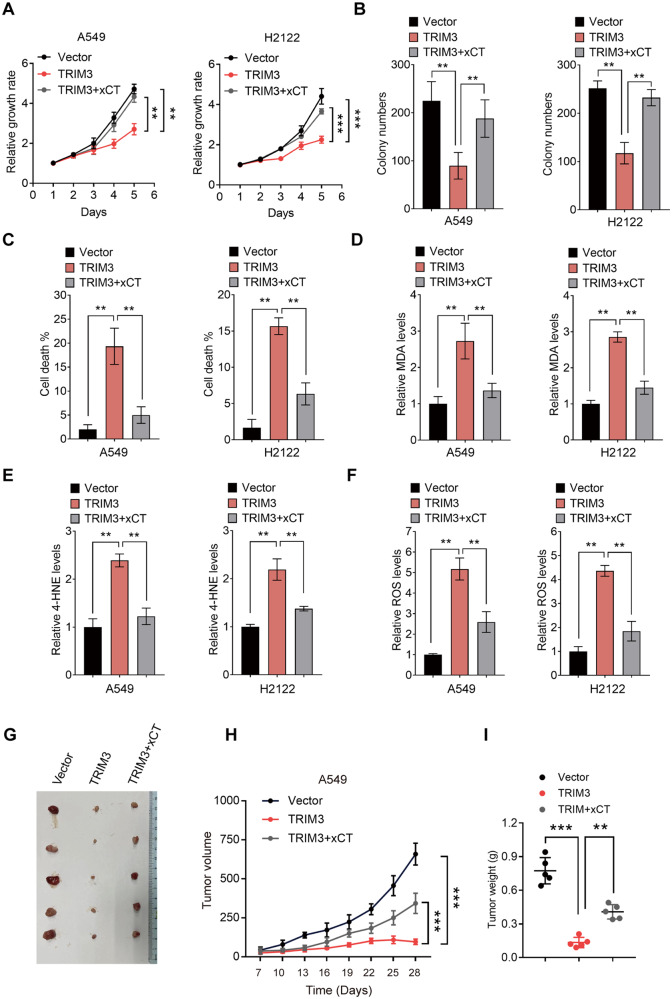
TRIM3 promotes ferroptosis in NSCLC cells through xCT
Coupled with the effect of TRIM3 on regulating xCT stability, we were motivated to determine the role of the TRIM3/xCT axis in the biological function of NSCLC. As expected, the inhibitory effect of TRIM3 overexpression on tumor cell proliferation and survival could be largely reversed by xCT (Fig. 6A–C). Moreover, ferroptosis events triggered by TRIM3 were dramatically reduced by exogenous expression of xCT (Fig. 6D–F and Fig. S5A–C). Using a mouse model, we confirmed that compared with the mice implanted with vector control-transduced A549 cells, those implanted with TRIM3-overexpressing A549 cells displayed a lower rate of tumor growth, which was rescued by xCT (Fig. 6G–I). In summary, these data suggest that TRIM3 causes ferroptosis through xCT, thus limiting the tumorigenesis of NSCLC.
Fig. 6. TRIM3 promotes ferroptosis in NSCLC cells through xCT.
A CCK-8 assay of A549 and H2122 cells transfected as indicated. B Colony formation assay of A549 and H2122 cells transfected as indicated. C The cell death rate in A549 and H2122 cells transfected as indicated was determined by trypan blue staining assay. D–F Relative ferroptosis levels in A549 and H2122 cells transfected as indicated were determined by MDA (D), 4-HNE (E), and ROS (F) levels. G Representative tumor images in nude mice bearing A549 cells transfected as indicated. H The tumor volume was monitored every 3 days. I The tumor weight. Data are represented as the mean ± SD (n = 3). Statistical analysis was performed using Student’s t-test, *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
TRIM3 shows a negative correlation with xCT protein levels in clinical NSCLC samples
To investigate the clinical relevance of our findings, we detected the expression of TRIM3, xCT, and MDA in serial sections of 84 human NSCLC specimens. The expression level of TRIM3 exhibited a negative correlation with that of xCT (Fig. 7A and B). Moreover, survival analysis of the 84 human NSCLC specimens revealed that low TRIM3 expression and high xCT expression were positively associated with worse outcomes for NSCLC patients (Fig. 7C). Finally, pan-cancer analysis of the TCGA database revealed that TRIM3 was downregulated not only in LUAD and LUSC but also in other types of cancers, further supporting the anti-tumor role of TRIM3 (Fig. 7D).
Fig. 7. TRIM3 shows a negative correlation with xCT protein levels in clinical NSCLC samples.
A Representative images of IHC assay of TRIM3, xCT, and MDA in clinical NSCLC samples. Scale bars: 100 µm. B Correlation analysis of TRIM3 and xCT expression in (A); chi-square test. C Kaplan–Meier analyses for patients in (A); long-rank test. D Pan-cancer analysis of relative TRIM3 mRNA levels in the TCGA database.
Discussion
How to effectively eliminate tumor cells while leaving the normal cells intact has always been a challenge in cancer treatment. Multiple studies suggest that ferroptosis, an intracellular iron-dependent form of programmed cell death, exerts a vital role in tumor suppression, due to cancer cells exhibiting an increased iron demand compared with healthy, non-cancer cells [32]. Thus, a better understanding of the underlying mechanisms driving ferroptosis may provide new strategies for cancer therapy.
TRIM3 (tripartite motif containing 3), a member of TRIM family proteins, is a RING finger E3 ligase that was initially identified as a regulator of myosin function which is responsible for protein transport in cells [33]. However, recent studies have shown a decreased expression level of TRIM3 in a few human cancers including hepatocellular carcinoma (HCC) [34] and glioblastoma (GBM) [35]. In this study, we found that TRIM3 was significantly downregulated in NSCLC and determined its tumor-suppressive role in NSCLC. The functional investigation demonstrated that overexpression of TRIM3 severely impeded lung cancer cell proliferation and tumorigenesis by enhancing cellular levels of free iron, lipid peroxidation, and ROS, highlighting its vital role in ferroptosis. In recent years, growing evidence has suggested that ferroptosis is interconnected with apoptosis [36], autophagy [37], and necrosis [26, 38]. For instance, autophagy can induce ferroptosis by producing lysosomal ROS and labile iron via NCOA4-mediated ferritinophagy [39]. Moreover, ferroptotic agents can enhance the function of apoptotic agent tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) in promoting cell apoptosis [40], raising the possibility that TRIM3-mediated ferroptosis may sensitize tumor cells to other types of cell death. Thus, future studies need to further explore the role of TRIM3 in other types of regulated cell death.
It is well-established that ferroptosis is strictly regulated by the cystine/glutamate antiporter SLC7A11 (also known as xCT) [41], which functions to transport cystine for glutathione biosynthesis and ROS clearance. The expression level of SLC7A11 was found to be upregulated in multiple human malignancies [42–44], which is crucial for promoting tumor occurrence, development, and drug resistance [45]. Accumulating evidence has demonstrated that SLC7A11 is precisely regulated at transcriptional and post-translational levels. However, the post-transcriptional modification of SLC7A11 in lung cancer cells remains unknown. In this study, we identified TRIM3 as a key regulator of SLC7A11. TRIM3 interacted with SLC7A11 through its NHL domain and specifically assembled the K11-linked polyubiquitin chain at K37 of SLC7A11, which further led to SLC7A11 proteasome-mediated degradation, suggesting a novel post-translational mechanism of SLC7A11. In recent years, various attempts have been made to identify and characterize pharmacological inhibitors of SLC7A11 [46]. Erastin is the most well-studied SLC7A11 inhibitor, due to its low solubility and poor metabolic stability, erastin is not ideal for animal research [47]. Our work suggests a promising therapeutic strategy that inhibition the SLC7A11 pathway by promoting its degradation rather than directly inhibiting SLC7A11.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Supplementary information
Acknowledgements
The mechanistic scheme of this study was drawn using Figdraw (www.figdraw.com).
Author contributions
Conception and design: ZJW; data acquisition, analysis, and interpretation: ZJW, NS; investigation: LY, SY, YW, and YL; acquisition of patient specimens: QZ and GHH; article drafting and revising: ZJW; and article writing: ZJW. All authors approved the final version of the manuscript.
Funding
This work was supported by the “333 projects” of Jiangsu Province (grant numbers: BRA2020190).
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
All participants provided informed consent. All human tissue research in this study had the approval of ethics committees of the Affiliated Taizhou People’s Hospital of Nanjing Medical University (Taizhou, China) and Shanghai Outdo Biotech (Shanghai, China). All of the animal experiments were performed by the relevant guidelines and regulations and were approved by the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhangjie Wang, Na Shen, Ziao Wang.
Contributor Information
Gaohua Han, Email: danny_75@njmu.edu.cn.
Qi Zhang, Email: zhangqi1225@njmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01239-5.
References
- 1.Brody H. Lung cancer. Nature. 2020;587:S7. doi: 10.1038/d41586-020-03152-0. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Le X, Nilsson M, Goldman J, Reck M, Nakagawa K, Kato T, et al. Dual EGFR-VEGF pathway inhibition: a promising strategy for patients With EGFR-Mutant NSCLC. J Thorac Oncol. 2021;16:205–15. doi: 10.1016/j.jtho.2020.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–54. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 5.Wang M, Herbst RS, Boshoff C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat Med. 2021;27:1345–56. doi: 10.1038/s41591-021-01450-2. [DOI] [PubMed] [Google Scholar]
- 6.Abbosh C, Frankell AM, Harrison T, Kisistok J, Garnett A, Johnson L, et al. Tracking early lung cancer metastatic dissemination in TRACERx using ctDNA. Nature. 2023;616:553–62. doi: 10.1038/s41586-023-05776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 8.Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22:266–82. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Li X, Ge C, Min J, Wang F. The multifaceted role of ferroptosis in liver disease. Cell Death Differ. 2022;29:467–80. doi: 10.1038/s41418-022-00941-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Q, Deng T, Zhang H, Zuo D, Zhu Q, Bai M, et al. Adipocyte-derived exosomal MTTP suppresses ferroptosis and promotes chemoresistance in colorectal cancer. Adv Sci. 2022;9:e2203357. doi: 10.1002/advs.202203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seibt TM, Proneth B, Conrad M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic Biol Med. 2019;133:144–52. doi: 10.1016/j.freeradbiomed.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Deng T, Liu R, Ning T, Yang H, Liu D, et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol Cancer. 2020;19:43. doi: 10.1186/s12943-020-01168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lei G, Zhuang L, Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer. 2022;22:381–96. doi: 10.1038/s41568-022-00459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan HF, Zou T, Tuo QZ, Xu S, Li H, Belaidi AA, et al. Ferroptosis: mechanisms and links with diseases. Signal Transduct Target Ther. 2021;6:49. doi: 10.1038/s41392-020-00428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2021;12:599–620. doi: 10.1007/s13238-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong H, Xia Y, Jin S, Xue C, Wang Y, Hu R, et al. Nrf2 attenuates ferroptosis-mediated IIR-ALI by modulating TERT and SLC7A11. Cell Death Dis. 2021;12:1027. doi: 10.1038/s41419-021-04307-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao R, Kalathur RKR, Coto-Llerena M, Ercan C, Buechel D, Shuang S, et al. YAP/TAZ and ATF4 drive resistance to Sorafenib in hepatocellular carcinoma by preventing ferroptosis. EMBO Mol Med. 2021;13:e14351. doi: 10.15252/emmm.202114351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol. 2013;14:211–24. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu T, Jiang L, Tavana O, Gu W. The deubiquitylase OTUB1 mediates ferroptosis via stabilization of SLC7A11. Cancer Res. 2019;79:1913–24. doi: 10.1158/0008-5472.CAN-18-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li WW, Nie Y, Yang Y, Ran Y, Luo WW, Xiong MG, et al. Ubiquitination of TLR3 by TRIM3 signals its ESCRT-mediated trafficking to the endolysosomes for innate antiviral response. Proc Natl Acad Sci USA. 2020;117:23707–16. doi: 10.1073/pnas.2002472117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan Q, Sun W, Kujala P, Lotfi Y, Vida TA, Bean AJ. CART: an Hrs/actinin-4/BERP/myosin V protein complex required for efficient receptor recycling. Mol Biol Cell. 2005;16:2470–82. doi: 10.1091/mbc.e04-11-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhuang T, Wang B, Tan X, Wu L, Li X, Li Z, et al. TRIM3 facilitates estrogen signaling and modulates breast cancer cell progression. Cell Commun Signal. 2022;20:45. doi: 10.1186/s12964-022-00861-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Zhang Y, Pei X, Guo G, Xue B, Duan X, et al. TRIM3 inhibits P53 signaling in breast cancer cells. Cancer Cell Int. 2020;20:559. doi: 10.1186/s12935-020-01630-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen G, Kong J, Tucker-Burden C, Anand M, Rong Y, Rahman F, et al. Human Brat ortholog TRIM3 is a tumor suppressor that regulates asymmetric cell division in glioblastoma. Cancer Res. 2014;74:4536–48. doi: 10.1158/0008-5472.CAN-13-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C, Tang Z, Zhang W, Ye Z, Liu F. GEPIA2021: integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021;49:W242–W246. doi: 10.1093/nar/gkab418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su Z, Yang Z, Xu Y, Chen Y, Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol Cancer. 2015;14:48. doi: 10.1186/s12943-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emdad L, Bhoopathi P, Talukdar S, Pradhan AK, Sarkar D, Wang XY, et al. Recent insights into apoptosis and toxic autophagy: the roles of MDA-7/IL-24, a multidimensional anti-cancer therapeutic. Semin Cancer Biol. 2020;66:140–54. doi: 10.1016/j.semcancer.2019.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–42. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D, Li Y. The interaction between ferroptosis and lipid metabolism in cancer. Signal Transduct Target Ther. 2020;5:108. doi: 10.1038/s41392-020-00216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie Y, Wang B, Zhao Y, Tao Z, Wang Y, Chen G, et al. Mammary adipocytes protect triple-negative breast cancer cells from ferroptosis. J Hematol Oncol. 2022;15:72. doi: 10.1186/s13045-022-01297-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tong X, Tang R, Xiao M, Xu J, Wang W, Zhang B, et al. Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research. J Hematol Oncol. 2022;15:174. doi: 10.1186/s13045-022-01392-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia X, Zhao C, Zhao W. Emerging roles of MHC class I region-encoded E3 ubiquitin ligases in innate immunity. Front Immunol. 2021;12:687102. doi: 10.3389/fimmu.2021.687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao J, Zhang XF, Pan QZ, Zhao JJ, Jiang SS, Wang Y, et al. Decreased expression of TRIM3 is associated with poor prognosis in patients with primary hepatocellular carcinoma. Med Oncol. 2014;31:102. doi: 10.1007/s12032-014-0102-9. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Raheja R, Yeh N, Ciznadija D, Pedraza AM, Ozawa T, et al. TRIM3, a tumor suppressor linked to regulation of p21(Waf1/Cip1.) Oncogene. 2014;33:308–15. doi: 10.1038/onc.2012.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan J, Wan P, Choksi S, Liu ZG. Necroptosis and tumor progression. Trends Cancer. 2022;8:21–27. doi: 10.1016/j.trecan.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maremonti F, Meyer C, Linkermann A. Mechanisms and models of kidney tubular necrosis and nephron loss. J Am Soc Nephrol. 2022;33:472–86. doi: 10.1681/ASN.2021101293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou B, Liu J, Kang R, Klionsky DJ, Kroemer G, Tang D. Ferroptosis is a type of autophagy-dependent cell death. Semin Cancer Biol. 2020;66:89–100. doi: 10.1016/j.semcancer.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Mou Y, Wang J, Wu J, He D, Zhang C, Duan C, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol. 2019;12:34. doi: 10.1186/s13045-019-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sui S, Xu S, Pang D. Emerging role of ferroptosis in breast cancer: new dawn for overcoming tumor progression. Pharm Ther. 2022;232:107992. doi: 10.1016/j.pharmthera.2021.107992. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Chen Y, Wang X, Tian H, Wang Y, Jin J, et al. Stem cell factor SOX2 confers ferroptosis resistance in lung cancer via upregulation of SLC7A11. Cancer Res. 2021;81:5217–29. doi: 10.1158/0008-5472.CAN-21-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J, Zhou Y, Xie S, Wang J, Li Z, Chen L, et al. Metformin induces ferroptosis by inhibiting UFMylation of SLC7A11 in breast cancer. J Exp Clin Cancer Res. 2021;40:206. doi: 10.1186/s13046-021-02012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouyang S, Li H, Lou L, Huang Q, Zhang Z, Mo J, et al. Inhibition of STAT3-ferroptosis negative regulatory axis suppresses tumor growth and alleviates chemoresistance in gastric cancer. Redox Biol. 2022;52:102317. doi: 10.1016/j.redox.2022.102317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen L, Zhang J, Zheng Z, Yang F, Liu S, Wu Y, et al. PHGDH inhibits ferroptosis and promotes malignant progression by upregulating SLC7A11 in bladder cancer. Int J Biol Sci. 2022;18:5459–74. doi: 10.7150/ijbs.74546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Xia X, Huang P. xCT: a critical molecule that links cancer metabolism to redox signaling. Mol Ther. 2020;28:2358–66. doi: 10.1016/j.ymthe.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X, Kang R, Kroemer G, Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021;18:280–96. doi: 10.1038/s41571-020-00462-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its supplementary information files].