Abstract
Purpose
The advent of proteomics provides new opportunities to investigate the molecular mechanisms underlying male infertility. The selection of relevant targets based on a single analysis is not always feasible, due to the growing number of proteomic studies with conflicting results. Thus, this study aimed to systematically review investigations comparing the sperm proteome of normozoospermic and infertile men to define a panel of proteins with the potential to be used to evaluate sperm quality.
Materials and Methods
A literature search was conducted on PubMed, Web of Science, and Scopus databases following the PRISMA guidelines. To identify proteins systematically reported, first the studies were divided by condition into four groups (asthenozoospermia, low motility, unexplained infertility, and infertility related to risk factors) and then, all studies were analysed simultaneously (poor sperm quality). To gain molecular insights regarding identified proteins, additional searches were performed within the Human Protein Atlas, Mouse Genome Informatics, UniProt, and PubMed databases.
Results
Thirty-two studies were included and divided into 4 sub-analysis groups. A total of 2752 proteins were collected, of which 38, 1, 3 and 2 were indicated as potential markers for asthenozoospermia, low motility, unexplained infertility and infertility related to risk factors, respectively, and 58 for poor sperm quality. Among the identified proteins, ACR, ACRBP, ACRV1, ACTL9, AKAP4, ATG3, CCT2, CFAP276, CFAP52, FAM209A, GGH, HPRT1, LYZL4, PRDX6, PRSS37, REEP6, ROPN1B, SPACA3, SOD1, SPEM1, SPESP1, SPINK2, TEKT5, and ZPBP were highlighted due to their roles in male reproductive tissues, association with infertility phenotypes or participation in specific biological functions in spermatozoa.
Conclusions
Sperm proteomics allows the identification of protein markers with the potential to overcome limitations in male infertility diagnosis and to understand changes in sperm function at the molecular level. This study provides a reliable list of systematically reported proteins that could be potential targets for further basic and clinical studies.
Keywords: Biological processes, Male infertility, Proteome, Proteomics, Spermatozoa
INTRODUCTION
On a global scale, infertility affects 8%–12% of reproductive-age couples. Of these, the male factor is responsible for 20-30% of the total, yet contributes to 50% of overall infertility cases [1]. A complex set of aetiologies and risk factors are known to correlate with male infertility. Nevertheless, the precise mechanisms responsible for male infertility remain unknown in 30%–50% of patients [2]. The cornerstone tool to diagnose male infertility is semen analysis, facilitating the clinical evaluation of both macro and microscopic aspects of semen samples [3,4]. These parameters can crudely distinguish fertile from infertile men according to the World Health Organization (WHO) guidelines [5]. However, the reality is that semen analysis has a relatively poor diagnostic value; this technique does not identify defects associated with the functional and molecular aspects of human sperm, and it fails to reliably predict fertilization potential and pregnancy success [6,7,8]. To improve infertility diagnosis, additional sperm functional tests have emerged to further evaluate key features of spermatozoa such as DNA fragmentation, assessment of reactive oxygen species and oxidative stress, membrane ion channels, acrosome reaction and mitochondrial function [4,9]. Nonetheless, these tests generally lack accuracy, often require a subjective interpretation of data, and also fail to explain underlying molecular causes of infertility [6,10].
Currently, proteomics is a powerful source of information to further understand and characterize the molecular mechanisms underlying both physiological and pathological conditions. In the reproductive field, several analyses have sought to characterize the proteome of ejaculated spermatozoa [11,12]. Indeed, spermatozoa are an ideal cell type for such studies as they can be isolated as a highly purified and relatively homogeneous material source. Additionally, since spermatozoa are considered quiescent cells presumably lacking active transcription and translation, the sperm proteome reflects the protein content of mature cells [13]. Regarding male infertility, proteomics has been applied to further understand changes in the sperm proteome and identify dysregulated pathways in certain clinical conditions [14,15]. In these proteomic studies, ejaculated spermatozoa from normozoospermic and infertile men were compared to identify differentially expressed proteins (DEPs). Through bioinformatic workflows, the DEPs are analysed to unveil altered pathways and defective biological processes allowing the molecular understanding of deregulated mechanisms [16,17]. Such bioinformatics analyses are also useful to identify potential biomarkers and therapeutic targets for subsequent investigation [17,18].
Considering the high number of available proteomic studies performed on human sperm, it is not feasible to draw robust conclusions based on single studies. The main objective of this systematic review was to compare human sperm proteomes from infertile and normozoospermic men and to identify a panel of candidate protein that could serve as potential biomarkers to evaluate male infertility. Also, using the available information related to those proteins in spermatozoa, we intended to highlight the most relevant examples.
MATERIALS AND METHODS
1. Protocol and registration
This review was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) [19,20]. The review protocol was registered on the international prospective register for systematic reviews – PROSPERO – before data extraction was completed (CRD42021257114).
2. Search strategy
An extensive search of the literature, published before 1st January 2023, was performed using the PubMed, Web of Science, and Scopus databases. Four classes of terms were defined and combined using Boolean operators to direct searchers to proteomic studies that analysed sperm samples from donors with different conditions related to male infertility. The terms included in each class are summarized in Supplement Table 1. Relevant articles referenced in the included studies were further evaluated for potential inclusion in this review.
3. Selection criteria
Among the identified studies, only those conforming to six inclusion criteria were selected: 1) publication in an indexed journal with full text in English or Portuguese; 2) freshly ejaculated human sperm samples were evaluated according to WHO guidelines [5]; 3) studies comparing a minimum of two experimental classes: (a) a group composed of men with poor-sperm quality related to significant changes in sperm parameters (concentration, motility, morphology, DNA fragmentation, ROS levels); male infertility conditions; poor reproductive outcomes; or conditions affecting fertility potential (lifestyle, systemic diseases, other relevant biological factors); (b) a control group composed of normozoospermic healthy donors; 4) spermatozoa isolated from whole semen samples using washing protocols (simple washing gradient or density washing gradient); 5) quantitative proteomic analyses; and 6) proteins were described using the gene name and/or UniProt/SwissProt ID. Review articles, metanalyses, commentaries, and proteomic analyses performed in other species were positively excluded and recorded, with descriptive exclusion criteria, in Supplement Table 2.
4. Data collection
Literature searches and study screens were independently performed by two reviewers (POC and JM). Disagreements between the judgments of the two primary reviewers were resolved by a third individual (JVS). From each included proteomic study, information related to the number of participants (n), participants’ age, clinical condition evaluated, proteomic methodologies and parameters used, and the number of DEPs were collected. Additionally, DEPs were retrieved and recorded with the respective fold-change (increased or decreased). To avoid redundancy, all DEPs were mapped in the UniProt database and annotated with UniProtKB/Swiss-Prot accession number (downloaded on 5th January 2023) (Supplement Table 3).
5. Quality of evidence assessment
To evaluate the quality of the included proteomic studies, the risk of bias assessment QUADAS-2 tool was employed [21]. Reviewers’ judgments were aided by available signalling questions. Each question was answered with “yes”, “no” or “unclear”, where “yes” indicates a low risk of bias. The risk of bias was classified as “low”, “high”, or “some concern”. The plot and graph of the risk of bias were created through the Robvis application [22]. These evaluations were performed independently, by two reviewers, as described above.
6. Identification of protein candidates
To identify a list of potential protein candidates as sperm quality markers, two complementary approaches were used. Firstly, to identify protein candidates for specific male infertility conditions, four subgroups were defined: (i) asthenozoospermia (AZS); (ii) low sperm motility; (iii) unexplained infertility; and (iv) infertility related to risk factors. Proteins were considered candidates if they were reported in a minimum of three independent analyses. Secondly, to unveil protein candidates associated with poor sperm quality under different conditions, all included studies were cross compared. Proteins were highlighted as candidates if they were identified in at least two distinct conditions and at least four independent analyses. In both approaches, a Venn diagram analysis was performed to cross-compare increased and decreased DEPs using the JVenn tool [23]. In both analyses, proteins with inconsistent expression patterns in different studies were excluded from further evaluation.
7. Detailed molecular insights of identified DEPs
Data regarding the selective expression of identified DEPs in the male reproductive tract were collected from the Human Protein Atlas (HPA) (version 22.0, downloaded on 5th January 2023) [24]. To predict any potential associations between the identified DEPs and male infertility defects and phenotypes, the Mouse Genome Informatics (MGI) database was employed (downloaded on 5th January 2023) [25]. The UniProt database provided information regarding gene ontology (downloaded on 5th January 2023). From the information collected, only terms related to sperm physiology and processes were retrieved. Finally, all DEPs were searched within the PubMed database to identify studies that report the role of these proteins in mammalian spermatozoa (until 10th January 2023).
RESULTS
1. Description of the included studies
A total of 1,133 articles were identified by a literature search of the PubMed, Web of Science, and Scopus databases and screening of references associated with such publications. After duplicate exclusion and initial screening, 423 articles were fully evaluated. A total of 32 studies [26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57] were selected for inclusion in this review (Table 1, Fig. 1). A majority of these studies used sperm samples from independent men as a control group and/or condition group (Table 1). Wu et al [52] employed samples collected from the same men after testicular-heat exposure. Most investigations used comparison groups composed of men of similar ages (data not shown). Only Liu et al [32] used two male groups of different ages, since the major objective of this study was to determine the influence of aging on the sperm proteome. Concerning proteomic strategies, gel-based methods (one-dimensional [1D] or two-dimensional polyacrylamide gel electrophoresis [2DE or 2DPAGE]; differential gel electrophoresis [DIGE]) and gelfree methods (tandem mass tag [TMT]; isobaric tags for relative and absolute quantitation [iTRAQ]; and free label) were the most common techniques used for protein separation, coupled with tandem mass spectrometry (MS/MS) or time-of-flight (TOF) analyses (Table 1). From the included studies, a total of 4,579 DEPs were collected, corresponding to 2,752 unique proteins according to the UniProt database (Supplement Table 3). Considering the evaluated conditions, studies were categorized into four subgroups as detailed in Table 1.
Table 1. Summary of the included proteomic studies.
| No | Reference | Condition | Participants | Proteomic method | Number of DEPs | Analysis group | ||
|---|---|---|---|---|---|---|---|---|
| Controls (#) | Cases (#) | Reported | Mapped | |||||
| 1 | Chhikara et al, 2023 [48] | AZS | 3 normozoospermic men | 12 asthenozoospermic men | 62 DEPs: ↑7 and ↓55 | 62 DEPs: ↑7 and ↓55 | (i) AZS | |
| 2 | Grande et al, 2022 [50] | Secondary hypogonadism | 5 normogonadic normozoospermic men | 5 hypogonadic men | TMT labelling and LC-MS/MS | 43 DEPs: ↑11 and ↓32 | 43 DEPs: ↑11 and ↓32 | Other sperm alterations |
| 3 | Liu et al, 2022 [27] | Low motility | 20 normozoospermic men (high motile sperm fraction) | Same 20 normozoospermic men (low motile sperm fraction) | 2DE and MALDI-TOF/TOF-MS | 21 DEPs: ↑13 and ↓8 | 21 DEPs: ↑13 and ↓8 | (ii) Low motility |
| 4 | Yang et al, 2022 [26] | AZS | 7 normozoospermic men | 11 asthenozoospermic men | UHPLC | 1,430 DEPs: ↑701 and ↓729 | 1,430 DEPs: ↑701 and ↓729 | (i) AZS |
| 5 | Liang et al, 2021 [38] | Severe oligoasthenoteratozoospermia | 12 normozoospermic men | 12 severe oligoasthenoteratozoospermic men | iTRAQ labelling and 2D-LC-MS/ MS | 938 DEPs: ↑226 and ↓712 | 938 DEPs: ↑226 and ↓712 | Other sperm alterations |
| 6 | Naglot et al, 2021 [51] | Recurrent pregnancy loss | 3 normozoospermic men | 3 normozoospermic men whose spouse had suffered three or more miscarriages | Label free and LC-MS/MS | 27 DEPs: ↑22 and ↓5 | 27 DEPs: ↑22 and ↓5 | (iii) Unexplained infertility |
| 7 | Mohanty et al, 2020 [49] | Recurrent pregnancy loss | 20 normozoospermic men | 16 normozoospermic men whose spouse had suffered two miscarriages | Nano-UHPLC-MS/MS | 23 DEPs: ↑8 and ↓15 | 23 DEPs: ↑8 and ↓15 | (iii) Unexplained infertility |
| 8 | Wu et al, 2020 [52] | Hyperthermia | 10 normozoospermic men | Same 10 normozoospermic men (exposed to heat) | iTRAQ labelling and 2D-LC-MS/ MS | 61 DEPs: ↑28 and ↓33 | 61 DEPs: ↑28 and ↓33 | (iv) Risk factor |
| 9 | Guo et al, 2019 [53] | Globozoospermia | 3 normozoospermic men | 3 globozoospermic men | TMT labelling and LC-MS/MS | 491 DEPs: ↑370 and ↓121 | 462 DEPs: ↑347 and ↓114 | Other sperm alterations |
| 10 | Guo et al, 2019 [54] | AZS | 5 normozoospermic men | 5 asthenozoospermic men | TMT labelling and LC-MS/MS | 152 DEPs: ↑84 and ↓68 | 152 DEPs: ↑84 and ↓68 | (i) AZS |
| 11a | Moscatelli et al, 2019 [55] | AZS | 3 normozoospermic men | 3 asthenozoospermic men | Label free and LC-MS/MS | 86 DEPs: ↑35 and ↓51 | 84 DEPs ↑34 and ↓50 | (i) AZS |
| 11b | Moscatelli et al, 2019 [55] | Severe AZS | 3 severe asthenozoospermic men | 88 DEPs: ↑41 and ↓47 | 88 DEPs: ↑41 and ↓47 | (i) AZS | ||
| 12 | Sinha et al, 2019 [56] | AZS | 5 normozoospermic men | 5 asthenozoospermic men | 2D-DIGE and MALDI-TOF/MS | 7 DEPs: all ↓ | 7 DEPs: all ↓ | (i) AZS |
| 13 | Xue et al, 2019 [57] | Recurrent pregnancy loss | 7 fertile normozoospermic men | 10 normozoospermic men whose spouse had suffered two miscarriages | iTRAQ labelling and HPLC-MS/ MS | 38 DEPs: ↑25 and ↓13 | 37 DEPs: ↑25 and ↓12 | (iii) Unexplained infertility |
| 14 | Liu et al, 2018 [28] | ZP binding failure | 20 normozoospermic men with IVF pregnancy | 20 normozoospermic men with R-ICSI pregnancy | iTRAQ labelling and MALDI-TOF/TOF-MS | 56 DEPs: ↑36 and ↓20 | 56 DEPs: ↑36 and ↓20 | (iii) Unexplained infertility |
| 15 | Nowicka-Bauer et al, 2018 [29] | AZS | 10 normozoospermic men | 4 asthenozoospermic men | 2DE and MALDI-TOF/MS | 25 DEPs: ↑18 and ↓7 | 23 DEPs: ↑16 and ↓7 | (i) AZS |
| 16 | Wang et al, 2018 [30] | Low PR | 10 normozoospermic men with highly fecundity (PR>50%) | 10 normozoospermic men with low fecundity (PR=0) | 2D-DIGE and MALDI-TOF/MS | 25 DEPs: ↑8 and ↓17 | 25 DEPs: ↑8 and ↓17 | (iii) Unexplained infertility |
| 17 | Saraswat et al, 2017 [31] | AZS | 5 normozoospermic men | 8 asthenozoospermic men | Label free and UHPLC-MS | 341 DEPs: ↑167 and ↓174 | 341 DEPs: ↑167 and ↓174 | (i) AZS |
| 18 | Liu et al, 2015 [32] | Ageing | 60 healthy young men | 60 old men | 2DE and MALDI-TOF/MS | 22 DEPs: ↑9 and ↓13 | 20 DEPs: ↑8 and ↓12 | (iv) Risk factors |
| 19 | Liu et al, 2015 [33] | Obesity and severe AZS | 3 normozoospermic men | 3 obese men with/severe AZS | HPLC-MS/MS | 127 DEPs: ↑22 and ↓105 | 125 DEPs: ↑22 and ↓103 | (i) AZS (iv) Risk factors |
| 20a | Amaral et al, 2014 [34] | AZS | 5 normozoospermic men | 5 asthenozoospermic men | TMT labelling and LC-MS/MS | 80 DEPs: ↑30 and ↓50 | 80 DEPs: ↑30 and ↓50 | (i) AZS |
| 20b | Amaral et al, 2014 [34] | Low motility | 5 normozoospermic men (high motile sperm fraction) | Same 5 normozoospermic men (high motile sperm fraction) | TMT labelling and LC-MS/MS | 93 DEPs: ↑47 and ↓46 | 93 DEPs: ↑47 and ↓46 | (ii) Low motility |
| 21 | Frapsauce et al, 2014 [35] | ZP binding failure | 3 normozoospermic men | 3 normozoospermic men with complete ZP binding failure | 2D-DIGE and MALDI-TOF/MS | 12 DEPs: ↑3 and ↓9 | 12 DEPs: ↑3 and ↓9 | (iii) Unexplained infertility |
| 22a | Légaré et al, 2014 [36] | Idiopathic infertility | 3 normozoospermic men | 6 infertile men | iTRAQ labelling and LC-MS/MS | 18 DEPS: ↑7 and ↓11 | 18 DEPS: ↑7 and ↓11 | Other sperm alterations |
| 22b | Légaré et al, 2014 [36] | IVF failure | 3 normozoospermic men | 4 men with a history of IVF failure | iTRAQ labelling and LC-MS/MS | 33 DEPs: ↑15 and ↓18 | 33 DEPs: ↑15 and ↓18 | (iii) Unexplained infertility |
| 23 | McReynolds et al, 2014 [43] | Blastocyst development | 6 normozoospermic men and good blastocyst development | 6 normozoospermic men and bad blastocyst development | 1D-PAGE and LC-MS/MS | 49 DEPs: ↑20 and ↓29 | 49 DEPs: ↑20 and ↓29 | (iii) Unexplained infertility |
| 24 | Hosseinifar et al, 2013 [44] | Varicocele and oligozoospermia | 20 normozoospermic men | 20 oligozoospermic men with varicocele | 2DE and MALDI-TOF/TOF-MS | 10 DEPs: ↑1 and ↓9 | 9 DEPs: ↑1 and ↓8 | (iv) Risk factors Other sperm alterations |
| 25 | Intasqui et al, 2013 [45] | DF | 11 normozoospermic men with low DF | 6 normozoospermic men with high DF | 2DE and Nano-UHPLC-ESI-MS | 94 DEPs: ↑23 and ↓71 | 94 DEPs: ↑23 and ↓71 | Other sperm alterations |
| 26 | Shen et al, 2013 [46] | AZS | 30 normozoospermic men | 30 asthenozoospermic men | 2DE and MALDI-TOF/TOF-MS | 15 DEPs: ↑4 and ↓11 | 15 DEPs: ↑4 and ↓11 | (i) AZS |
| 27 | Zhu et al, 2013 [47] | IVF failure | 3 normozoospermic men with successful IVF | 3 normozoospermic men without successful IVF | TMT labelling and HPLC-MS/MS | 21 DEPs: ↑5 and ↓16 | 21 DEPs: ↑5 and ↓16 | (iii) Unexplained infertility |
| 28 | Xu et al, 2012 [42] | Null pregnancy | 10 fertile normozoospermic men | 10 infertile normozoospermic men | 2DE and MALDI-TOF/TOF-MS | 24 DEPs: ↑15 and ↓9 | 18 DEPs: ↑11 and ↓7 | (iii) Unexplained infertility |
| 29a | Paasch et al, 2011 [37] | DM 1 | 21 normozoospermic men | 8 DM1 men | DIGE and MALDI-TOF/TOF-MS | 8 DEPs: ↑6 and ↓2 | 8 DEPs: ↑6 and ↓2 | (iv) Risk factors |
| 29b | Paasch et al, 2011 [37] | DM 2 | 7 DM2 men | 10 DEPs: ↑7 and ↓3 | 10 DEPs: ↑7 and ↓3 | (iv) Risk factors | ||
| 29c | Paasch et al, 2011 [37] | Obesity | 13 non-diabetic obese men | 7 DEPs: ↑6 and ↓1 | 7 DEPs: ↑6 and ↓1 | (iv) Risk factors | ||
| 30 | Siva et al, 2010 [39] | AZS | 20 normozoospermic men | 17 asthenozoospermic men | 2DE and MALDI-MS/MS | 8 DEPs: ↑3 and ↓5 | 7 DEPs: ↑3 and ↓4 | (i) AZS |
| 31a | Kriegel et al, 2009 [40] | Diabetes type 1 | 5 normozoospermic men | 2 DM1 men | 2D-DIGE and MALDI-TOF/MS | 8 DEPs: ↑5 and ↓3 | 8 DEPs: ↑5 and ↓3 | (iv) Risk factors |
| 31b | Kriegel et al, 2009 [40] | Obesity | 2 non-diabetic obese men | 9 DEPs: ↑3 and ↓6 | 9 DEPs: ↑3 and ↓6 | (iv) Risk factors | ||
| 32 | Martínez-Heredia et al, 2008 [41] | AZS | 10 normozoospermic men | 20 asthenozoospermic men | 2DE and PDQuest | 17 DEPs: ↑10 and ↓7 | 16 DEPs: ↑10 and ↓6 | (i) AZS |
For each study, the evaluated condition, the number of individuals included in each group, the proteomic method, the number of DEPs (reported and mapped in the UniProt database), and the analysis group are indicated.
1D: one-dimensional, 2DE: two-dimensional, AZS: asthenozoospermia, DEP: differentially expressed protein, DF: DNA fragmentation, DIGE: differential gel electrophoresis, DM1: diabetes mellitus type 1, DM2: diabetes mellitus type 2, ESI: electrospray ionization, HPLC: high-performance liquid chromatography, iTRAQ: isobaric tags for relative and absolute quantitation, IVF: in vitro fertilization, LC: liquid chromatography, MALDI: matrix-assisted laser desorption/ionization, MS/MS: tandem mass spectrometry, PAGE: polyacrylamide gel electrophoresis, PR: pregnancy rate, R-ICSI: rescue intracytoplasmic sperm injection, TMT: tandem mass tag, TOF: time-of-flight, UHPLC: ultra-high-performance liquid chromatography, ZP: zona pellucida, #: number of patients.
Fig. 1. Study selection. Flow diagram illustrating the selection process of proteomic studies employed in this systematic review. Systematic results search and study selection process are represented according to the PRISMA guidelines (2020). MS: mass spectrometry.
2. Quality appraisal
The bias risk of 30 of the included studies was evaluated as “low”, whilst two studies [38,47] were appraised with “some concerns” (Supplement Fig. 1). As determined by the QUADAS-2 tool, the studies included in this systematic review were classified as good quality.
3. Candidate protein markers for AZS
AZS is one of the most common causes of male infertility (~20% of cases) and is characterized by a reduction (total motility <40%) or total lack of motility [5]. Among the focused conditions, the sperm proteome related to AZS was the most often analysed (n=12) (Table 1). One of the included studies presented two independent analyses performed in samples from asthenozoospermic and severe asthenozoospermic (total motility <12%) patients [55]. Another investigation included samples from obese men with severe AZS (total motility <20%) [33]. The study conducted by Chhikara et al [48] contains several sub-analyses, but only DEPs identified among ejaculated samples of normozoospermic and asthenozoospermic men were considered. A total of 1,998 unique DEPs were identified in AZS sperm, of which 878 were exclusively increased and 992 were exclusively decreased (Fig. 2A). As potential protein candidates, 12 increased and 26 decreased proteins were identified (Table 2). Of these 38 proteins, ACRBP, TEKT5, ACR, ACRV1, CCIN, IZUMO4, AKAP4, SPACA3, GARIN3, ACTRT2, LYZL4, and ZPBP have enriched expression in testis, REEP6 is enhanced in testis, SPINK2 is enriched in epididymis, and ACP3 is a prostate-enriched protein (Table 2). Furthermore, ACP3, ACRBP, CA2, REEP6, ACR, CD46, SPINK2, AKAP4 and ZPBP were associated with fertility defects in mice (Table 2). Although not mentioned in the MGI database, two proteins (PRDX6 and SOD1) were also associated with male infertility phenotypes in mice. Prdx6 null mice presented spermatozoa with abnormal chromatin structure, low motility, impaired capacitation, decreased sperm/oocyte interaction, and fertility competence (reviewed by O’Flaherty [58]). Sod1 null mice had altered spermatogenesis, presenting spermatozoa with low motility, impaired capacitation, inability to penetrate ZP, and reduced fertilization potential [59,60,61,62].
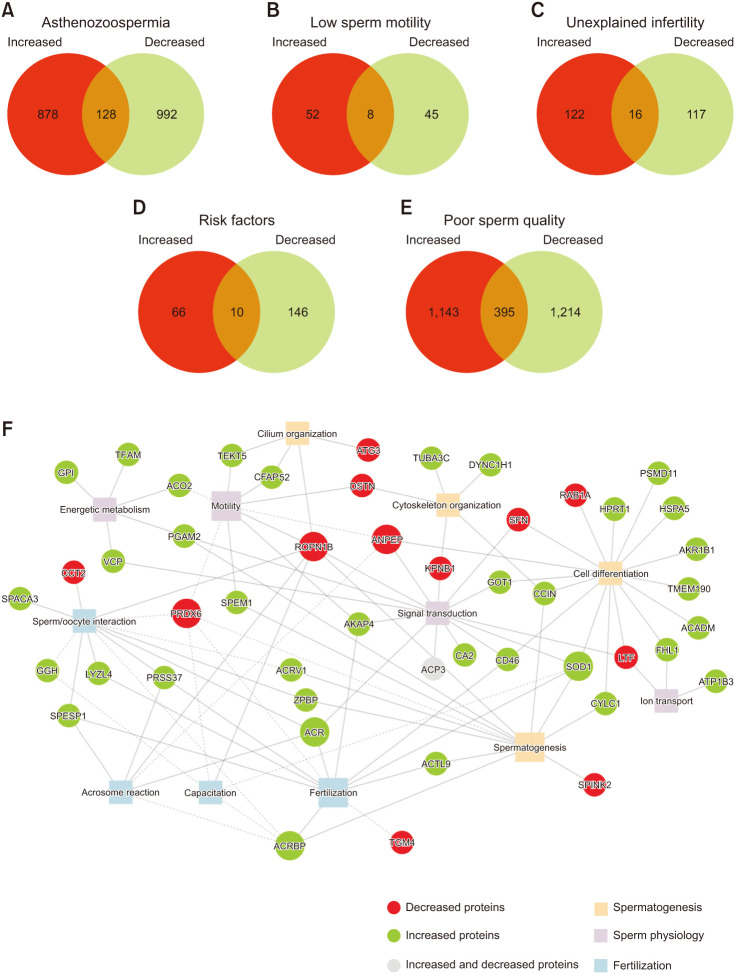
Fig. 2. Differentially expressed proteins (DEPs) in male infertility-related conditions and overall poor-sperm quality. Number of DEPs (increased and decreased) in (A) asthenozoospermia, (B) low sperm motility, (C) unexplained infertility, (D) infertility related to risk factors, (E) poor sperm quality. (F) DEPs related to the retrieved biological processes in the UniProt database (solid line) and PubMed search (dash line). Larger circles represent proteins associated with a higher number of biological processes. Larger squares represent biological processes with a higher number of associated proteins.
Table 2. Candidate protein markers for male infertility-related conditions.
| UniProtKB | Gene name | Protein name | # | Expression level | Reference | Tissue expression | Male fertility defects | Biological process | |
|---|---|---|---|---|---|---|---|---|---|
| (i) Asthenozoospermia | |||||||||
| P15309 | ACP3 | Prostatic acid phosphatase | 4 | ↑ | [26,31,34,54] | Prostate (enriched) | Abnormal prostate gland | Signal transduction | |
| P60981 | DSTN | Destrin | 3 | ↑ | [34,55] | - | - | Cytoskeleton organization; Motility | |
| O00231 | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11 | 3 | ↑ | [26,55] | - | - | Cell differentiation | |
| P15144 | ANPEP | Aminopeptidase N | 3 | ↑ | [26,31,55] | - | - | Cell differentiation; Signal transduction | |
| P26640 | VARS1 | Valine-tRNA ligase | 3 | ↑ | [26,31,54] | - | - | - | |
| P30041 | PRDX6 | Peroxiredoxin-6 | 3 | ↑ | [26,31,34] | - | - | - | |
| P62820 | RAB1A | Ras-related protein Rab-1A | 3 | ↑ | [26,31,54] | - | - | Cell differentiation | |
| P78371 | CCT2 | T-complex protein 1 subunit beta | 3 | ↑ | [26,29,31] | - | - | Sperm/oocyte interaction | |
| Q14974 | KPNB1 | Importin subunit beta 1 | 3 | ↑ | [26,31,33] | - | - | Cytoskeleton organization; Signal transduction | |
| Q96FW1 | OTUB1 | Ubiquitin thioesterase OTUB1 | 3 | ↑ | [26,55] | - | - | - | |
| Q9H0I9 | TKTL2 | Transketolase-like protein 2 | 3 | ↑ | [26,31,54] | - | - | - | |
| Q9HB40 | SCPEP1 | Retinoid-inducible serine carboxypeptidase | 3 | ↑ | [26,54,55] | - | - | - | |
| P17174 | GOT1 | Aspartate aminotransferase, cytoplasmic | 4 | ↓ | [26,54,55] | - | - | Cell differentiation; Signal transduction | |
| Q8NEB7 | ACRBP | Acrosin-binding protein | 4 | ↓ | [26,31,48,54] | Testis (enriched) | Abnormal acrosome and nucleus, decreased sperm motility | Fertilization; Spermatogenesis | |
| Q92820 | GGH | Gamma-glutamyl hydrolase | 4 | ↓ | [26,54,55] | - | - | - | |
| Q96M29 | TEKT5 | Tektin-5 | 4 | ↓ | [26,31,33,34] | Testis (enriched) | - | Cilium organization; Motility | |
| Q99798 | ACO2 | Aconitate hydratase, mitochondrial | 4 | ↓ | [26,34,55] | - | - | Energy metabolism | |
| P00441 | SOD1 | Superoxide dismutase [Cu-Zn] | 3 | ↓ | [26,48,54] | - | - | Fertilization; Cell differentiation; Spermatogenesis; Signal transduction | |
| P00918 | CA2 | Carbonic anhydrase 2 | 3 | ↓ | [26,48,54] | - | Reduced male fertility; dilated efferent testis ductules and rete testis | Signal transduction | |
| Q96HR9 | REEP6 | Receptor expression-enhancing protein 6 | 3 | ↓ | [31,55] | Testis (enhanced) | OZS, TZS, AZS, impaired acrosome reaction | - | |
| P06744 | GPI | Glucose-6-phosphate isomerase | 3 | ↓ | [26,34,55] | - | - | Energy metabolism | |
| P10323 | ACR | Acrosin | 3 | ↓ | [26,54,55] | Testis enriched | Delayed fertilization | Acrosome reaction; Fertilization; Signal transduction; Sperm/oocyte interaction | |
| P15121 | AKR1B1 | Aldo-keto reductase family 1 member B1 | 3 | ↓ | [26,33,54] | - | - | Cell differentiation | |
| P15529 | CD46 | Membrane cofactor protein | 3 | ↓ | [26,55] | - | Abnormal fertilization | Cell differentiation; Fertilization; Signal transduction | |
| P20155 | SPINK2 | Serine protease inhibitor Kazal-type 2 | 3 | ↓ | [26,31,48] | Epididymis (enriched) | Azoospermia | Spermatogenesis | |
| P26436 | ACRV1 | Acrosomal protein SP-10 | 3 | ↓ | [26,31,54] | Testis (enriched) | - | Spermatogenesis | |
| Q13642 | FHL1 | Four and a half LIM domains protein 1 | 3 | ↓ | [26,31,34] | - | - | Cell differentiation; Ion transport | |
| Q13939 | CCIN | Calicin | 3 | ↓ | [26,31,33] | Testis (enriched) | - | Cell differentiation; Cytoskeleton organization; Spermatogenesis | |
| Q1ZYL8 | IZUMO4 | Izumo sperm-egg fusion protein 4 | 3 | ↓ | [26,48,54] | Testis (enriched) | - | - | |
| Q5JQC9 | AKAP4 | A-kinase anchor protein 4 | 3 | ↓ | [26,29,31] | Testis (enriched) | Impaired sperm motility | Fertilization; Motility; Signal transduction | |
| Q5JRX3 | PITRM1 | Presequence protease, mitochondrial | 3 | ↓ | [26,33,55] | - | - | - | |
| Q8IXA5 | SPACA3 | Sperm acrosome membrane-associated protein 3 | 3 | ↓ | [26,31,54] | Testis (enriched) | - | Sperm/oocyte interaction | |
| Q8IYQ7 | THNSL1 | Threonine synthase-like 1 | 3 | ↓ | [26,55] | - | - | - | |
| Q8TC56 | GARIN3 | Golgi-associated RAB2 interactor protein 3 | 3 | ↓ | [26,31,48] | Testis (enriched) | - | - | |
| Q8TDY3 | ACTRT2 | Actin-related protein T2 | 3 | ↓ | [26,33,48] | Testis (enriched) | - | - | |
| Q96KX0 | LYZL4 | Lysozyme-like protein 4 | 3 | ↓ | [26,34,54] | Testis (enriched) | Normal fertility | Fertilization; Sperm/oocyte interaction | |
| Q9BS86 | ZPBP | Zona pellucida-binding protein 1 | 3 | ↓ | [26,48,54] | Testis (enriched) | Abnormal sperm morphology | Sperm/oocyte interaction; Spermatogenesis | |
| Q9H3G5 | CPVL | Probable serine carboxypeptidase CPVL | 3 | ↓ | [26,54,55] | - | - | - | |
| (ii) Low motility | |||||||||
| P11021 | HSPA5 | Endoplasmic reticulum chaperone BiP | 2 | ↑ | [27,34] | - | - | Cell differentiation | |
| (iii) Unexplained infertility | |||||||||
| Q5JQC9 | AKAP4 | A-kinase anchor protein 4 | 4 | ↓ | [30,35,42,57] | Testis (enriched) | Impaired sperm motility | Fertilization; Motility; Signal transduction | |
| P02788 | LTF | Lactotransferrin | 3 | ↓ | [43,49,51] | - | - | Cell differentiation; Ion transport; Signal transduction | |
| P0DPH7 | TUBA3C | Tubulin alpha-3C chain | 3 | ↓ | [28,30,35] | - | - | Cytoskeleton organzitation | |
| (iv) Infertility related to risk factors | |||||||||
| P02788 | LT F | Lactotransferrin | 3 | ↑ | [37] | - | - | Cell differentiation; Ion transport; Signal transduction | |
| Q6UWU2 | GLB1L | Beta-galactosidase-1-like protein | 3 | ↓ | [37] | Testis (enriched) | - | - | |
Highlighted protein candidates in (i) asthenozoospermia, (ii) low sperm motility, (iii) unexplained infertility, and (iv) infertility related to lifestyle factors. For each protein, this table indicates selective expression in male reproductive tissues, the male infertility defects/phenotypes observed in knock-out mice, and the relevant biological processes (retrieved from UniProt database).
AZS: asthenozoospermia, OZS: oligozoospermia, TZS: teratozoospermia, #: number of counts.
4. Candidate protein markers for low sperm motility
Regarding semen analysis, motility is one of the major predictors of sperm function [5]. After spermatogenesis and epididymal maturation, a fraction of spermatozoa still presents low motility, or even immotility, providing a useful biological sample to determine the molecular mechanisms responsible for sperm (im)motility. Two studies analysed the proteome of the low motility sperm fraction of normozoospermic men (Table 1). A total of 105 unique proteins were identified, of which 52 were increased and 45 were decreased (Fig. 2B). Only one protein (HSPA5) was described in both studies as decreased in sperm with low motility (Table 2).
5. Candidate protein markers for unexplained male infertility and poor reproductive outcomes
In unexplained male infertility, men present normal semen analysis and physical and endocrine abnormalities related to infertility were excluded. Yet, men with these conditions cannot establish a successful pregnancy [6,63]. In these cases, the female infertility factor has been excluded a priori either by clinical evaluation or oocyte morphological analysis. The ten included studies in this category were related to recurrent pregnancy loss (n=3), ZP binding failure (n=2), low pregnancy rate (n=1), null pregnancy (n=1), in vitro fertilization (IVF) failure (n=2), and inappropriate blastocyst development (n=1) (Table 1). In these proteomic analyses both comparison groups were composed of normozoospermic men. From the included studies, 255 unique DEPs were identified, 122 being exclusively increased and 117 exclusively decreased (Fig. 2C). Three decreased DEPs (AKAP4, LTF, and TUBA3C) were systematically reported in these studies (Table 2). Although not mentioned in the HPA, TUBA3C is a testis-specificα-tubulin isoform, amenable to acetylation, present in human sperm [64].
6. Candidate protein markers for infertility associated with risk factors
A multitude of causes and risk factors have been associated with the increasing male infertility incidence; these include genetic defects (diabetes mellitus [DM] type 1), acquired factors (for example varicocele, obesity, and DM type 2), and biological factors (ageing) [2]. Although those causes and risk factors are biologically distinct, their presence is correlated with a significant decrease in sperm quality [65]. In this section, six studies were included to unveil common DEPs in infertility associated with risk factors: hyperthermia (n=1), DM type 1 (n=2), DM type 2 (n=1), obesity (n=3), varicocele (n=1), and ageing (n=1) (Table 1). A total of 222 unique proteins were identified, of which 66 were increased, 146 were decreased and 10 were both increased and decreased (Fig. 2D). Two proteins (LTF and GLB1L) were-positively identified as potential markers (Table 2). GLB1L expression is enhanced in the testis (Table 2).
7. Candidate protein markers of poor sperm quality
From the 2,752 unique proteins identified in all the studies included in this review (Supplement Table 3), 1,143 and 1,214 proteins were exclusively increased and decreased, respectively (Fig. 2E). Amongst these, 11 increased and 47 decreased DEPs were highlighted as potential protein markers of poor sperm quality (Table 3). Twenty-four proteins (ACO2, ACRBP, ACRV1, ACTRT2, ANPEP, CA2, CCIN, CD46, FHL1, GARIN3, GGH, GLB1L, GOT1, IZUMO4, KPNB1, LYZL4, OTUB1, PITRM1, RAB1A, SPACA3, SPINK2, TEKT5, THNSL1, and ZPBP) were previously identified in the subgroup analyses (Table 2, 3). Among the 58 highlighted proteins, 22 (H2AC1, ACRBP, ZPBP, CCIN, GARIN3, ACRV1, TEKT5, FAM205A, LYZL4, ACTRT2, PRSS37, CYLC1, IZUMO4, FAM209A, ACTL9, FNDC8, TMEM190, GARIN4, SPESP1, LYZL1, SPACA3, and SPEM1) are testis-enriched, two (GLB1L and CFAP276) are testis-enhanced, one is enriched epididymis (SPINK2), and one (TGM4) is prostate-enriched (Table 3). Fifteen proteins (TGM4, DYNC1H1, ACRBP, ZPBP, CD46, PRSS37, CA2, FAM209A, ACTL9, HPRT1, SPINK2, CFAP276, SPESP1, SPEM1, and ROPN1B) were associated with defects in male fertility such as abnormal sperm morphology, impaired/reduced fertilization, and abnormal male reproductive tissues morphology (Table 3).
Table 3. Potential protein markers for poor sperm quality.
| UniProtKB | Gene name | Protein name | # | Conditions (references) | Tissue expression | Male fertility defects | Biological process | |
|---|---|---|---|---|---|---|---|---|
| Increased proteins | ||||||||
| P31947 | SFN | 14-3-3 protein sigma | 5 | AZS [26,31]; LM [34]; RPL [51,57] | - | - | Cell differentiation; Signal transduction | |
| P15144 | ANPEP | Aminopeptidase N | 4 | AZS [26,31,55]; Idiopathic Infertility [36] | - | - | Cell differentiation; Signal transduction | |
| P49221 | TGM4 | Protein-glutamine gamma-glutamyltransferase 4 | 4 | AZS [31]; DF [45]; IVF failure [36]; RPL [57] | Prostate (enriched) | Impaired copulatory plug formation, reduced fertilization | - | |
| P52597 | HNRNPF | Heterogeneous nuclear ribonucleoprotein F | 4 | AZS [26,54]; Hyperthermia [52]; Severe OAT [38] | - | - | - | |
| P56192 | MARS1 | Methionine--tRNA ligase, cytoplasmic | 4 | AZS [26,54]; GZS [53]; LM [34] | - | - | - | |
| P62820 | RAB1A | Ras-related protein Rab-1A | 4 | AZS [26,31,54]; Severe OAT [38] | - | - | Cell differentiation | |
| Q14204 | DYNC1H1 | Cytoplasmic dynein 1 heavy chain 1 | 4 | AZS [26,55]; GZS [54]; RPL [51] | - | Male infertility | Cytoskeleton organization | |
| Q14974 | KPNB1 | Importin subunit beta 1 | 4 | AZS [26,31]; GZS [53]; AZS and obesity [33] | - | - | Cytoskeleton organization; Signal transduction | |
| Q96FW1 | OTUB1 | Ubiquitin thioesterase OTUB1 | 4 | AZS [26,55]; GZS [53] | - | - | - | |
| Q9NT62 | ATG3 | Ubiquitin-like-conjugating enzyme ATG3 | 4 | AZS [26,54]; GZS [53]; Severe OAT [38] | - | Cilium organization | - | |
| Q96QV6 | H2AC1 | Histone H2A type 1-A | 4 | GZS [54]; AZS [31,48]; ZP binding failure [28] | Testis (enriched) | - | - | |
| Decreased proteins | ||||||||
| Q8NEB7 | ACRBP | Acrosin-binding protein | 8 | AZS [26,31,54]; RPL [57]; GZS [53]; ZP binding failure [28]; Severe OAT [38] | Testis (enriched) | Abnormal acrosome and nucleus, and decreased sperm motility | Fertilization; Spermatogenesis | |
| Q92820 | GGH | Gamma-glutamyl hydrolase | 8 | AZS [26,54,55]; GZS [53]; LM [34]; DM-1 and 2 [37] | - | - | - | |
| Q9BS86 | ZPBP | Zona pellucida-binding protein 1 | 8 | Hyperthermia [52]; AZS [26,48,54]; GZS [53]; ZP binding failure [28]; Idiopathic infertility [36]; Severe OAT [38] | Testis (enriched) | Abnormal sperm morphology | Sperm/oocyte interaction; Spermatogenesis | |
| Q13939 | CCIN | Calicin | 7 | AZS [26,31,54]; GZS [53]; AZS and obesity [33]; DF [45]; Hyperthermia [52]; Severe OAT [38] | Testis (enriched) | - | Cell differentiation; Cytoskeleton organization; Spermatogenesis | |
| Q6UWU2 | GLB1L | Beta-galactosidase-1-like protein | 7 | AZS [26,54] GZS [53]; DM-1 and 2, obesity [37]; Severe OAT [38] | Testis (enhanced) | - | - | |
| Q8TC56 | GARIN3 | Golgi-associated RAB2 interactor protein 3 | 7 | GZS [53]; RPL [57]; Hyperthermia [52]; AZS [26,31,48] Severe OAT [38] | Testis (enriched) | - | - | |
| P26436 | ACRV1 | Acrosomal protein SP-10 | 6 | AZS [26,31,54]; GZS [53]; IVF failure [47]; DF[45] | Testis (enriched) | - | Spermatogenesis | |
| Q96M29 | TEKT5 | Tektin-5 | 6 | AZS [26,31Protein FAM205A ref-type="bibr" rid="B34">34]; AZS and obesity [33]; Blastocyst development [43]; Hyperthermia [52] | Testis (enriched) | - | Cilium organization; Motility | |
| P15529 | CD46 | Membrane cofactor protein | 6 | Hyperthermia [52]; GZS [53]; AZS [26,55]; Severe OAT [38] | - | Abnormal fertilization | Cell differentiation; Fertilization; Signal transduction | |
| P17174 | GOT1 | Aspartate aminotransferase, cytoplasmic | 6 | LM [34]; AZS [26,54,55]; Severe OAT [38] | - | - | Cell differentiation; Signal transduction | |
| Q6ZU69 | FAM205A | Protein FAM205A | 6 | GZS [53]; AZS [26,54]; DF [45]; RPL [57]; Severe OAT [38] | Testis (enriched) | - | - | |
| Q96KX0 | LYZL4 | Lysozyme-like protein 4 | 6 | Blastocyst development [43]; GZS [53]; AZS [26,34,54]; Severe OAT [38] | Testis (enriched) | - | Fertilization; Sperm/oocyte interaction | |
| Q8TDY3 | ACTRT2 | Actin-related protein T2 | 6 | AZS [26,48]; GZS [53]; AZS and obesity [33]; Hyperthermia [52]; | Testis (enriched) | - | - | |
| A4D1T9 | PRSS37 | Probable inactive serine protease 37 | 5 | AZS [26,54]; GZS [53]; Severe OAT [38]; Secondary hypogonadism [50] | Testis (enriched) | Impaired ZP binding and migration from the uterus to oviduct | Acrosome reaction; Fertilization; Sperm/oocyte interaction | |
| P00918 | CA2 | Carbonic anhydrase 2 | 5 | AZS [26,48,54]; Idiopathic Infertility [36]; Severe OAT [38] | - | Reduced male fertility, dilated rete testis and efferent ductulus | Signal transduction | |
| P15259 | PGAM2 | Phosphoglycerate mutase 2 | 5 | IVF failure [36]; AZS [26,54]; DF [45]; Severe OAT [38] | - | - | Energetic metabolism; Signal transduction; Spermatogenesis | |
| P35663 | CYLC1 | Cylicin-1 | 5 | AZS [26]; GZS [53]; AZS and obesity [33]; Hyperthermia [52]; Severe OAT [38] | Testis (enriched) | - | Cell differentiation; Spermatogenesis | |
| P54709 | ATP1B3 | Sodium/potassium-transporting ATPase subunit beta-3 | 5 | Hyperthermia [52]; AZS [26,55]; IVF failure [47]; Severe OAT [38] | - | - | Ion transport | |
| P55072 | VCP | Transitional endoplasmic reticulum ATPase | 5 | LM [34]; DF [45]; IVF failure [36]; ZP binding failure [35]; Severe OAT [38] | - | - | Energetic metabolism; Signal transduction | |
| Q13642 | FHL1 | Four and a half LIM domains protein 1 | 5 | AZS [31,34][26]; ZP binding failure [28]; Severe OAT [38] | - | - | Cell differentiation; Ion transport | |
| Q1ZYL8 | IZUMO4 | Izumo sperm-egg fusion protein 4 | 5 | GZS [53]; ZP binding failure [28]; AZS [26,48,54] | Testis (enriched) | - | - | |
| Q5JX71 | FAM209A | Protein FAM209A | 5 | DF [45]; Blastocyst development [43]; Severe OAT [38]; AZS [26,48] | Testis (enriched) | AZS, GZS, and abnormal acrosome morphology | - | |
| Q8TC94 | ACTL9 | Actin-like protein 9 | 5 | AZS [26]; LM [34]; GZS [53]; Ageing [32]; Hyperthermia [52] | Testis (enriched) | Abnormal sperm morphology, impaired fertilization | Fertilization; Spermatogenesis | |
| Q8TC99 | FNDC8 | Fibronectin type III domain-containing protein 8 | 5 | GZS [53]; Hypertermia [52]; Severe OAT [38]; AZS [26,48] | Testis (enriched) | - | - | |
| Q8WZ59 | TMEM190 | Transmembrane protein 190 | 5 | GZS [53]; Hyperthermia [52]; DF [45]; AZS [31]; Severe OAT [38] | Testis (enriched) | - | Cell differentiation | |
| Q8IYT1 | GARIN4 | Golgi-associated RAB2 interactor protein 4 | 5 | GZS [53]; Hypertermia [52]; Severe OAT [38]; AZS [26,48] | Testis (enriched) | - | - | |
| Q96QE4 | LRRC37B | Leucine-rich repeat-containing protein 37B | 5 | GZS [53]; AZS [26,31]; Hyperthermia [52]; Severe OAT [38] | - | - | - | |
| Q99798 | ACO2 | Aconitate hydratase, mitochondrial | 5 | AZS [26,34,55]; Severe OAT [38] | - | - | Energy metabolism | |
| P00492 | HPRT1 | Hypoxanthine-guanine phosphoribosyltransferase | 4 | Blastocyst development [43]; LM [34]; AZS [55]; Severe OAT [38] | - | Testicular atrophy | Cell differentiation | |
| P04066 | FUCA1 | Tissue alpha-L-fucosidase | 4 | AZS [26,54]; GZS [53]; Severe OAT [38] | - | - | - | |
| P11310 | ACADM | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | 4 | LM [34]; AZS [26,55]; Severe OAT [38] | - | - | Cell differentiation | |
| P20155 | SPINK2 | Serine protease inhibitor Kazal-type 2 | 4 | DF [45]; AZS [26,31,48] | Epididymis (enriched) | Azoospermia | Spermatogenesis | |
| P23786 | CPT2 | Carnitine O-palmitoyltransferase 2, mitochondrial | 4 | ZP binding failure [35]; AZS [26,55]; Severe OAT [38] | - | - | - | |
| P28838 | LAP3 | Cytosol aminopeptidase | 4 | DF [45]; IVF failure [36]; Severe OAT [38]; AZS [26] | - | - | - | |
| P56730 | PRSS12 | Neurotrypsin | 4 | AZS [26,54]; GZS [53]; Severe OAT [38] | - | - | - | |
| Q00059 | TFAM | Transcription factor A, mitochondrial | 4 | GZS [53]; Severe OAT [38]; IVF failure [36]; AZS [26] | - | - | Energy metabolism | |
| Q5JRX3 | PITRM1 | Presequence protease, mitochondrial | 4 | Obesity and AZS [33]; Severe OAT [38]; AZS [26,55] | - | - | - | |
| Q5T5A4 | CFAP276 | Protein CFAP276 | 4 | AZS and obesity [33]; LM [34]; Blastocyst development [43]; Severe OAT [38] | Testis enhanced | Impaired embryo implantation | - | |
| Q6UW49 | SPESP1 | Sperm equatorial segment protein 1 | 4 | Hyperthermia [52]; GZS [53]; AZS [54]; DF [45] | Testis enriched | Decreased fertilization frequency and delayed fertilization | Acrosome reaction; Fertilization; Sperm/oocyte interaction | |
| Q6UWQ5 | LYZL1 | Lysozyme-like protein 1 | 4 | Obesity [40]; GZS [53]; AZS [26,34] | Testis enriched | - | ||
| Q8IXA5 | SPACA3 | Sperm acrosome membrane-associated protein 3 | 4 | GZS [53]; AZS [26,31,54] | Testis enriched | - | Sperm/oocyte interaction | |
| Q8IYQ7 | THNSL1 | Threonine synthase-like 1 | 4 | AZS [26,55]; Severe OAT [38] | - | - | - | |
| Q8N1V2 | CFAP52 | Cilia- and flagella-associated protein 52 | 4 | AZS [26,34]; LM [34]; Severe OAT [38] | - | - | Cilium organization; Motility | |
| Q8N4L4 | SPEM1 | Spermatid maturation protein 1 | 4 | GZS [53]; ZP binding failure [28]; Severe OAT [38]; AZS [26] | Testis (enriched) | AZS, TZS, and impaired spermiogenesis | Spermatogenesis; Acrosome reaction; Fertilization; Sperm/oocyte interaction; Motility | |
| Q9BZX4 | ROPN1B | Ropporin-1B | 4 | Severe OAT [38]; AZS [26,31]; Secondary Hypogonadism [50] | Testis (enriched) | AZS, reduced male fertility | Acrosome reaction; Motility; Sperm/oocyte interaction; Spermatogenesis; Capacitation; Cilium organization; Signal transduction | |
| Q9NR28 | DIABLO | Diablo IAP-binding mitochondrial protein | 4 | AZS [26,34]; Blastocyst development [43]; Severe OAT [38] | - | - | - | |
| Q9UBX1 | CTSF | Cathepsin F | 4 | AZS [48,54]; GZS [53]; Severe OAT [38] | - | - | - | |
After cross-comparison of the studies included in this review, relevant protein candidates were identified. For each protein is indicated selective expression in male reproductive tissues, the male infertility defects/phenotypes observed in knock-out (KO) mice, and the relevant biological processes (retrieved from UniProt database).
AZS: asthenozoospermia, DF: DNA fragmentation, DM: diabetes mellitus, GZS: globozoospermia, IVF: in vitro fertilization, LM: low motility, OAT: oligoasthenoteratozoospermia, RPL: recurrent pregnancy loss, TZS: teratozoospermia, ZP: zona pellucida, #: number of counts.
8. Biological processes associated with the identified DEPs
A total of 50 different biological processes were retrieved from the UniProt database associated with the previously identified protein candidates (Supplement Table 4). Among these, 13 were highlighted as being relevant for sperm physiology: ion transport, signal transduction, energy metabolism (including glycolysis and aerobic respiration), spermatogenesis, cell differentiation, cytoskeleton organization, cilium organization, motility, capacitation, sperm/oocyte interaction, acrosome reaction, and fertilization (Fig. 2F, squares). From the 76 highlighted DEPs, 44 are associated with those processes (Tables 2, 3, Fig. 2F).
Additionally, the search performed on PubMed allowed the identification of additional associations between ACRV1, ACRBP, CD46, TGM4, ANPEP, GGH, ACO2, PRDX6, and SOD1 and sperm-related processes. ACRV1 is involved in sperm-oocyte interaction [66,67,68]. Studies performed in mice and boar spermatozoa showed ACRBP involvement in the acrosome reaction, capacitation, and sperm/oocyte interaction [69,70]. TGM4 is involved in the formation of copulatory plugs [71,72]. ANPEP activity is associated with both sperm motility and acrosome reaction in mice and humans [73,74,75,76]. A recent study reported that GGH forms a complex with T-complex protein 1 subunit beta (CCT2) and glutathione S-transferase Mu 3 protein (GSTM-3), both involved in sperm capacitation and sperm/oocyte interaction [77], suggesting the involvement of GGH in these processes. Low ACO2 levels correlate with the reduced motility of human sperm [78]. CD46 blockaded with antibodies decreased sperm/oocyte interactions [79,80]. PRDX6 activity is associated with spermatogenesis, motility, capacitation, and fertility competence [58,81,82]. Several studies have reported that SOD1 provides protection against oxidative stress during epididymal maturation and capacitation of mammalian sperm [83,84,85]. Moreover, increased levels of SOD1 were associated with a high fertility rate in cattle [86].
Taken together, the retrieved biological processes can be grouped into three main categories related to the spermatozoon life journey: 1) spermatogenesis (cell differentiation, spermatogenesis, cilium organization, and cytoskeleton organization) (Fig. 2F, orange squares), 2) sperm physiology (signal transduction, ion transport, energy metabolism, motility, and capacitation) (Fig. 2F, lilac squares), and 3) fertilization (sperm/oocyte interaction, acrosome reaction and fertilization) (Fig. 2F, blue squares). The most common processes associated with the identified DEPs were cell differentiation (n=16), signal transduction (n=14), sperm/oocyte interaction (n=12), spermatogenesis (n=12), fertilization (n=11), and motility (n=9) (highlighted in Fig. 2F with bigger squares). Among the DEPs, ROPN1B (n=7), PRDX6 (n=5) ACRBP (n=5), SOD1 (n=5), ANPEP (n=4), and ACR (n=4) were DEPs associated with most processes (highlighted in Fig. 2F with bigger circles).
DISCUSSION
Male infertility conditions have a plethora of underlying causes resulting in the dysregulation of various biochemical pathways and cellular processes. These altered molecular mechanisms are reflected in qualitative and quantitative differences in the sperm proteome [12]. Although there are some literature reviews focused on sperm proteomic studies, there is a general lack of analyses to indicate which DEPs are systematically reported in male infertility conditions. We believe this present study is to be the first systematic review to critically analyse proteomic data from the spermatozoa of infertile men. As a consequence of this analysis, it was possible to identify a set of relevant DEPs which are likely to prove viable targets for further basic and clinical research to unveil male infertility biomarkers.
One of the inherent challenges of this systematic analysis was the effective combination of proteomic studies to identify candidate proteins without losing relevant targets. As indicated by this work, a profitable strategy was to combine two approaches: 1) group the studies by infertility condition; and 2) concurrently analyse all data. Thus, it was possible to establish panels of protein candidates for specific conditions (such as AZS, low motility, unexplained infertility, and infertility related to risk factors) and for overall poor sperm quality. Evidence that this strategy was successful is provided by the identification of the following proteins: ACP3, PSMD11, VARS1, PRDX6, CCT2, SOD1, REEP6, GPI, ACR, AKAP4, CPVL, HSPA5, LTF, and TUBA3C. These proteins were highlighted in subgroup analyses but would be excluded for overall poor sperm quality because they presented inconsistent expression among the different infertility conditions.
For the highlighted proteins, it was important to understand their molecular role(s) in spermatozoa, which could be helpful to string the candidates' list. Currently, biomarkers research is focused on the discovery of tissue-specific candidates which facilitate a greater understanding of specific molecular mechanisms underlying clinical conditions [87]. From the 72 identified proteins (Table 2, 3), 28 have a selective expression in the testis and 3 in other male reproductive tissues. TGM4, ACP3, and LTF are components of the seminal fluid and bind to the sperm surface through membrane proteins [88,89]. In physiological conditions, these proteins are involved in sperm processes such as motility, capacitation, and fertilization [88,89]. However, in male infertility, it is not fully understood whether these proteins are abnormally expressed or whether the spermatozoa present molecular changes on their surface to differentially bind them. Additional proteomic studies to analyse, in parallel, altered proteins in spermatozoa and seminal fluid could clarify this point. Among the testis-selective expressed proteins, ACRBP, SPESP1, ACTL9, PRSS37, HPRT1, CFAP276, SPESP1, FAM209A, SPEM1, ACR, REEP6, AKAP4, ZPBP, and ROPN1B were associated with male infertility phenotypes in KO mice models. Considering their expression and impact upon mouse fertility, they must represent strong candidates for further investigation. Conversely, 49 identified proteins were associated with the 13 relevant processes for spermatogenesis [90,91], sperm survival, maintenance, maturation, and fertilization [92,93,94,95], suggesting that these processes may be dysregulated in patients with poor sperm quality. Two mitochondrial proteins (ACO2 and TFAM) were associated with energetic metabolism. Besides the contribution of mitochondria role to energy production, mitochondrial integrity loss has been associated with increased ROS production, oxidative stress, and apoptosis-like events in spermatozoa [96]. Thus, ACO2 and TFAM may also be indicative of mitochondrial dysfunction in poor sperm quality. ACO2, ACRBP, ACR, ACRV1, ACTL9, AKAP4, ANPEP, CCT2, CFAP52, DSTN, GGH, LYZL4, PRDX6, PRSS37, ROPN1B, SOD1, SPACA3, SPEM1, SPESP1, TEKT5, and ZPBP were associated with sperm-specific processes (motility, capacitation, sperm/oocyte interaction, acrosome reaction, and fertilization). These proteins are also strong candidates for further research as potential biomarkers.
In the included studies, some limitations were identified and can be taken into consideration for further study design. Firstly, the inconsistencies observed in the expression levels of some proteins under the same conditions may result from the proteomic methods employed. Though gel-based methods remain viable in proteomic analyses, they present certain limitations compared to gel-free methods which are more sensitive and accurate in protein quantification [97]. Since proteomics is a constantly evolving field, the emergence of new software and instrumentation may be useful for protein identification and quantification in male infertility. A pertinent example is the study performed by Yang et al [26]; these authors used a 4D proteomic strategy to study AZS allowing the identification of a higher number of DEPs (n=1,430) compared to similar studies [31,34,54,55]. Secondly, there is a limited number (or even a lack) of proteomic studies focused on the analysis of the sperm proteome in some male infertility conditions such as idiopathic infertility, globozoospermia, and ageing. Another factor that may be overlooked in proteomics analyses is participant heterogeneity. Indeed, confounders (such as age match, ethnic group, lifestyle, and region of residence) are poorly addressed in sperm proteomic analysis. Detailed participant questionnaires may help to establish more homogeneous comparison groups and so improve the identification of reliable protein candidates. In unexplained male infertility analyses, the female infertility factor was excluded by clinical evaluation or by oocyte morphological selection. Nevertheless, the molecular dysfunctions associated with the female factor were not considered and may also contribute to poor reproductive outcomes. Final relevant comment is the limited data regarding the role of the identified DEPs in mammalian spermatozoa. Most of these are exhaustively investigated in somatic cells but remain poorly characterized in this highly differentiated cell type. Consequently, the identified proteins in this analysis are obvious candidates to be further explored in future investigations.
CONCLUSIONS
Proteomics, a valuable tool to study the molecular mechanisms of male infertility and poor sperm quality, can provide large datasets of DEPs. These datasets further reflect the complexity of male infertility since the same clinical condition may originate from the dysregulation of different molecular pathways. The present review identified, for the first time, DEPs systematically reported in proteomic studies focused on male infertility. Herein, we provide a list of 76 potential protein candidates to assess sperm quality in specific conditions or which are more generally related to poor sperm quality. Some of these proteins (ACR, ACRBP, ACRV1, ACTL9, AKAP4, ATG3, CCT2, CFAP276, CFAP52, FAM209A, GGH, HPRT1, LYZL4, PRDX6, PRSS37, REEP6, ROPN1B, SPACA3, SOD1, SPEM1, SPESP1, SPINK2, TEKT5, and ZPBP) have a selective expression in male tissues or have been associated with male infertility phenotypes or biological processes relevant to sperm function. These DEPs are obvious targets for further basic and clinical research and may be validated for screening purposes, potentiating an improved clinical diagnosis of the origins of male infertility.
Footnotes
Conflict of Interest: The authors have nothing to disclose.
Funding: This study was supported by the Institute of Biomedicine - iBiMED (UIDB/04501/2020 and UIDP/04501/2020) and by an individual grant from the Foundation for Science Technology (FCT) of the Portuguese Ministry of Science and Higher Education to PC (2020.10111.BD).
- Conceptualization: POC, JVS, MF.
- Data curation: POC, JVS.
- Formal analysis: POC.
- Funding Acquisition: MF, JVS.
- Investigation: POC, JM.
- Methodology: POC, JVS.
- Project administration: MF.
- Supervision: PFO, MF.
- Writing – original draft: POC.
- Writing – review & editing: POC, JH, MF, JVS.
Supplementary Materials
Supplementary materials can be found via https://doi.org/10.5534/wjmh.220262.
Quality assessment of the included studies. (A) Risk of bias plot representing the domain-level judgments for each included study. (B) Distribution of the risk of bias per domain. The plot and graph were generated in the Robvis application.
Search terms used to identify proteomic studies performed in human spermatozoa
Excluded studies after screening and full-text assessment. Each excluded study was recorded with the DOI number, year, title, and respective exclusion motive
Differentially expressed proteins in male infertility-related conditions. All accession numbers identified in included studies were collected and mapped in the UniProt database. For each mapped protein is indicated the UniProtKB number, gene name, protein name, fold-change, and the review status according to the UniProt database.
Gene ontology terms for biological processes. For each protein candidate, the gene ontology terms were retrieved from the UniProt database and were grouped according to their relationship.
References
- 1.Vander Borght M, Wyns C. Fertility and infertility: definition and epidemiology. Clin Biochem. 2018;62:2–10. doi: 10.1016/j.clinbiochem.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal A, Baskaran S, Parekh N, Cho CL, Henkel R, Vij S, et al. Male infertility. Lancet. 2021;397:319–333. doi: 10.1016/S0140-6736(20)32667-2. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal A, Majzoub A, Parekh N, Henkel R. A schematic overview of the current status of male infertility practice. World J Mens Health. 2020;38:308–322. doi: 10.5534/wjmh.190068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khatun A, Rahman MS, Pang MG. Clinical assessment of the male fertility. Obstet Gynecol Sci. 2018;61:179. doi: 10.5468/ogs.2018.61.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization (WHO) WHO laboratory manual for the examination and processing of human semen. 5th ed. Geneva: WHO; 2010. [Google Scholar]
- 6.Pandruvada S, Royfman R, Shah TA, Sindhwani P, Dupree JM, Schon S, et al. Lack of trusted diagnostic tools for undetermined male infertility. J Assist Reprod Genet. 2021;38:265–276. doi: 10.1007/s10815-020-02037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C, Swerdloff RS. Limitations of semen analysis as a test of male fertility and anticipated needs from newer tests. Fertil Steril. 2014;102:1502–1507. doi: 10.1016/j.fertnstert.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alshahrani S, Aldossari K, Al-Zahrani J, Gabr AH, Henkel R, Ahmad G. Interpretation of semen analysis using WHO 1999 and WHO 2010 reference values: abnormal becoming normal. Andrologia. 2018;50:e12838. doi: 10.1111/and.12838. [DOI] [PubMed] [Google Scholar]
- 9.Oehninger S, Franken DR, Ombelet W. Sperm functional tests. Fertil Steril. 2014;102:1528–1533. doi: 10.1016/j.fertnstert.2014.09.044. [DOI] [PubMed] [Google Scholar]
- 10.Esteves SC. Are specialized sperm function tests clinically useful in planning assisted reproductive technology? Int Braz J Urol. 2020;46:116–123. doi: 10.1590/S1677-5538.IBJU.2020.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Rose MB, Piccolomini MM, Soares Belo AS, Borges E, Jr, Filho FF. Proteomics in human reproduction. Protein Pept Lett. 2018;25:420–423. doi: 10.2174/0929866525666180412164602. [DOI] [PubMed] [Google Scholar]
- 12.Agarwal A, Panner Selvam MK, Baskaran S. Proteomic analyses of human sperm cells: understanding the role of proteins and molecular pathways affecting male reproductive health. Int J Mol Sci. 2020;21:1621. doi: 10.3390/ijms21051621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilany K, Minai-Tehrani A, Amini M, Agharezaee N, Arjmand B. The challenge of human spermatozoa proteome: a systematic review. J Reprod Infertil. 2017;18:267–279. [PMC free article] [PubMed] [Google Scholar]
- 14.Panner Selvam MK, Agarwal A. Update on the proteomics of male infertility: a systematic review. Arab J Urol. 2018;16:103–112. doi: 10.1016/j.aju.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Intasqui P, Agarwal A, Sharma R, Samanta L, Bertolla RP. Towards the identification of reliable sperm biomarkers for male infertility: a sperm proteomic approach. Andrologia. 2018;50:e12919. doi: 10.1111/and.12919. [DOI] [PubMed] [Google Scholar]
- 16.Trindade F, Ferreira R, Magalhães B, Leite-Moreira A, Falcão-Pires I, Vitorino R. How to use and integrate bioinformatics tools to compare proteomic data from distinct conditions? A tutorial using the pathological similarities between aortic valve stenosis and coronary artery disease as a case-study. J Proteomics. 2018;171:37–52. doi: 10.1016/j.jprot.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 17.Corda PO, Silva JV, Pereira SC, Barros A, Alves MG, Fardilha M. Bioinformatic approach to unveil key differentially expressed proteins in human sperm after slow and rapid cryopreservation. Front Cell Dev Biol. 2022;9:759354. doi: 10.3389/fcell.2021.759354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panner Selvam MK, Baskaran S, Sikka SC. Telomere signaling and maintenance pathways in spermatozoa of infertile men treated with antioxidants: an in silico approach using bioinformatic analysis. Front Cell Dev Biol. 2021;9:768510. doi: 10.3389/fcell.2021.768510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6:e1000100. doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2 Group. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 22.McGuinness LA, Higgins JPT. Risk-of-bias VISualization (robvis): an R package and Shiny web app for visualizing risk-of-bias assessments. Res Synth Methods. 2021;12:55–61. doi: 10.1002/jrsm.1411. [DOI] [PubMed] [Google Scholar]
- 23.Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics. 2014;15:293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 25.Bult CJ, Blake JA, Smith CL, Kadin JA, Richardson JE Mouse Genome Database Group. Mouse Genome Database (MGD) Nucleic Acids Res. 2019;47:D801–D806. doi: 10.1093/nar/gky1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Liu Q, Yu B, Han B, Yang B. 4D-quantitative proteomics signature of asthenozoospermia and identification of extracellular matrix protein 1 as a novel biomarker for sperm motility. Mol Omics. 2022;18:83–91. doi: 10.1039/d1mo00257k. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Teng Z, Wang Z, Zhu P, Song Z, Liu F. Expressions of HSPA1L and HSPA9 are associated with poor sperm quality of low-motility spermatozoa in fertile men. Andrologia. 2022;54:e14321. doi: 10.1111/and.14321. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Liu G, Liu J, Zhu P, Wang J, Wang Y, et al. iTRAQbased analysis of sperm proteome from normozoospermic men achieving the rescue-ICSI pregnancy after the IVF failure. Clin Proteomics. 2018;15:27. doi: 10.1186/s12014-018-9203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowicka-Bauer K, Lepczynski A, Ozgo M, Kamieniczna M, Fraczek M, Stanski L, et al. Sperm mitochondrial dysfunction and oxidative stress as possible reasons for isolated asthenozoospermia. J Physiol Pharmacol. 2018;69:403–417. doi: 10.26402/jpp.2018.3.05. [DOI] [PubMed] [Google Scholar]
- 30.Wang XM, Xiang Z, Fu Y, Wu HL, Zhu WB, Fan LQ. Comparative proteomics reveal the association between SPANX proteins and clinical outcomes of artificial insemination with donor sperm. Sci Rep. 2018;8:6850. doi: 10.1038/s41598-018-25032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saraswat M, Joenväärä S, Jain T, Tomar AK, Sinha A, Singh S, et al. Human spermatozoa quantitative proteomic signature classifies normo- and asthenozoospermia. Mol Cell Proteomics. 2017;16:57–72. doi: 10.1074/mcp.M116.061028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu FJ, Liu X, Han JL, Wang YW, Jin SH, Liu XX, et al. Aged men share the sperm protein PATE1 defect with young asthenozoospermia patients. Hum Reprod. 2015;30:861–869. doi: 10.1093/humrep/dev003. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Guo Y, Song N, Fan Y, Li K, Teng X, et al. Proteomic pattern changes associated with obesity-induced asthenozoospermia. Andrology. 2015;3:247–259. doi: 10.1111/andr.289. [DOI] [PubMed] [Google Scholar]
- 34.Amaral A, Paiva C, Attardo Parrinello C, Estanyol JM, Ballescà JL, Ramalho-Santos J, et al. Identification of proteins involved in human sperm motility using high-throughput differential proteomics. J Proteome Res. 2014;13:5670–5684. doi: 10.1021/pr500652y. [DOI] [PubMed] [Google Scholar]
- 35.Frapsauce C, Pionneau C, Bouley J, Delarouziere V, Berthaut I, Ravel C, et al. Proteomic identification of target proteins in normal but nonfertilizing sperm. Fertil Steril. 2014;102:372–380. doi: 10.1016/j.fertnstert.2014.04.039. [DOI] [PubMed] [Google Scholar]
- 36.Légaré C, Droit A, Fournier F, Bourassa S, Force A, Cloutier F, et al. Investigation of male infertility using quantitative comparative proteomics. J Proteome Res. 2014;13:5403–5414. doi: 10.1021/pr501031x. [DOI] [PubMed] [Google Scholar]
- 37.Paasch U, Heidenreich F, Pursche T, Kuhlisch E, Kettner K, Grunewald S, et al. Identification of increased amounts of eppin protein complex components in sperm cells of diabetic and obese individuals by difference gel electrophoresis. Mol Cell Proteomics. 2011;10:M110.007187. doi: 10.1074/mcp.M110.007187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang J, Zheng Y, Zeng W, Chen L, Yang S, Du P, et al. Proteomic profile of sperm in infertile males reveals changes in metabolic pathways. Protein J. 2021;40:929–939. doi: 10.1007/s10930-021-10013-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siva AB, Kameshwari DB, Singh V, Pavani K, Sundaram CS, Rangaraj N, et al. Proteomics-based study on asthenozoospermia: differential expression of proteasome alpha complex. Mol Hum Reprod. 2010;16:452–462. doi: 10.1093/molehr/gaq009. [DOI] [PubMed] [Google Scholar]
- 40.Kriegel TM, Heidenreich F, Kettner K, Pursche T, Hoflack B, Grunewald S, et al. Identification of diabetes- and obesity-associated proteomic changes in human spermatozoa by difference gel electrophoresis. Reprod Biomed Online. 2009;19:660–670. doi: 10.1016/j.rbmo.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Martínez-Heredia J, de Mateo S, Vidal-Taboada JM, Ballescà JL, Oliva R. Identification of proteomic differences in asthenozoospermic sperm samples. Hum Reprod. 2008;23:783–791. doi: 10.1093/humrep/den024. [DOI] [PubMed] [Google Scholar]
- 42.Xu W, Hu H, Wang Z, Chen X, Yang F, Zhu Z, et al. Proteomic characteristics of spermatozoa in normozoospermic patients with infertility. J Proteomics. 2012;75:5426–5436. doi: 10.1016/j.jprot.2012.06.021. [DOI] [PubMed] [Google Scholar]
- 43.McReynolds S, Dzieciatkowska M, Stevens J, Hansen KC, Schoolcraft WB, Katz-Jaffe MG. Toward the identification of a subset of unexplained infertility: a sperm proteomic approach. Fertil Steril. 2014;102:692–699. doi: 10.1016/j.fertnstert.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 44.Hosseinifar H, Gourabi H, Salekdeh GH, Alikhani M, Mirshahvaladi S, Sabbaghian M, et al. Study of sperm protein profile in men with and without varicocele using two-dimensional gel electrophoresis. Urology. 2013;81:293–300. doi: 10.1016/j.urology.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 45.Intasqui P, Camargo M, Del Giudice PT, Spaine DM, Carvalho VM, Cardozo KH, et al. Unraveling the sperm proteome and post-genomic pathways associated with sperm nuclear DNA fragmentation. J Assist Reprod Genet. 2013;30:1187–1202. doi: 10.1007/s10815-013-0054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen S, Wang J, Liang J, He D. Comparative proteomic study between human normal motility sperm and idiopathic asthenozoospermia. World J Urol. 2013;31:1395–1401. doi: 10.1007/s00345-013-1023-5. [DOI] [PubMed] [Google Scholar]
- 47.Zhu Y, Wu Y, Jin K, Lu H, Liu F, Guo Y, et al. Differential proteomic profiling in human spermatozoa that did or did not result in pregnancy via IVF and AID. Proteomics Clin Appl. 2013;7:850–858. doi: 10.1002/prca.201200078. [DOI] [PubMed] [Google Scholar]
- 48.Chhikara N, Tomar AK, Datta SK, Yadav S. Proteomic changes in human spermatozoa during in vitro capacitation and acrosome reaction in normozoospermia and asthenozoospermia. Andrology. 2023;11:73–85. doi: 10.1111/andr.13289. [DOI] [PubMed] [Google Scholar]
- 49.Mohanty G, Jena SR, Nayak J, Kar S, Samanta L. Quantitative proteomics decodes clusterin as a critical regulator of paternal factors responsible for impaired compensatory metabolic reprogramming in recurrent pregnancy loss. Andrologia. 2020;52:e13498. doi: 10.1111/and.13498. [DOI] [PubMed] [Google Scholar]
- 50.Grande G, Barrachina F, Soler-Ventura A, Jodar M, Mancini F, Marana R, et al. The role of testosterone in spermatogenesis: lessons from proteome profiling of human spermatozoa in testosterone deficiency. Front Endocrinol (Lausanne) 2022;13:852661. doi: 10.3389/fendo.2022.852661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naglot S, Tomar AK, Singh N, Yadav S. Label-free proteomics of spermatozoa identifies candidate protein markers of idiopathic recurrent pregnancy loss. Reprod Biol. 2021;21:100539. doi: 10.1016/j.repbio.2021.100539. [DOI] [PubMed] [Google Scholar]
- 52.Wu YQ, Rao M, Hu SF, Ke DD, Zhu CH, Xia W. Effect of transient scrotal hyperthermia on human sperm: an iTRAQ-based proteomic analysis. Reprod Biol Endocrinol. 2020;18:83. doi: 10.1186/s12958-020-00640-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo Y, Jiang J, Zhang H, Wen Y, Zhang H, Cui Y, et al. Proteomic analysis of Dpy19l2-deficient human globozoospermia reveals multiple molecular defects. Proteomics Clin Appl. 2019;13:e1900007. doi: 10.1002/prca.201900007. [DOI] [PubMed] [Google Scholar]
- 54.Guo Y, Jiang W, Yu W, Niu X, Liu F, Zhou T, et al. Proteomics analysis of asthenozoospermia and identification of glucose-6-phosphate isomerase as an important enzyme for sperm motility. J Proteomics. 2019;208:103478. doi: 10.1016/j.jprot.2019.103478. [DOI] [PubMed] [Google Scholar]
- 55.Moscatelli N, Lunetti P, Braccia C, Armirotti A, Pisanello F, De Vittorio M, et al. Comparative proteomic analysis of proteins involved in bioenergetics pathways associated with human sperm motility. Int J Mol Sci. 2019;20:3000. doi: 10.3390/ijms20123000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sinha A, Singh V, Singh S, Yadav S. Proteomic analyses reveal lower expression of TEX40 and ATP6V0A2 proteins related to calcium ion entry and acrosomal acidification in asthenozoospermic males. Life Sci. 2019;218:81–88. doi: 10.1016/j.lfs.2018.12.016. [DOI] [PubMed] [Google Scholar]
- 57.Xue D, Zhang Y, Wang Y, Wang J, An F, Sun X, et al. Quantitative proteomic analysis of sperm in unexplained recurrent pregnancy loss. Reprod Biol Endocrinol. 2019;17:52. doi: 10.1186/s12958-019-0496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O'Flaherty C. Peroxiredoxin 6: the protector of male fertility. Antioxidants (Basel) 2018;7:173. doi: 10.3390/antiox7120173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsunoda S, Kawano N, Miyado K, Kimura N, Fujii J. Impaired fertilizing ability of superoxide dismutase 1-deficient mouse sperm during in vitro fertilization. Biol Reprod. 2012;87:121. doi: 10.1095/biolreprod.112.102129. [DOI] [PubMed] [Google Scholar]
- 60.Selvaratnam JS, Robaire B. Effects of aging and oxidative stress on spermatozoa of superoxide-dismutase 1- and catalase-null mice. Biol Reprod. 2016;95:60. doi: 10.1095/biolreprod.116.141671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garratt M, Bathgate R, de Graaf SP, Brooks RC. Copper-zinc superoxide dismutase deficiency impairs sperm motility and in vivo fertility. Reproduction. 2013;146:297–304. doi: 10.1530/REP-13-0229. [DOI] [PubMed] [Google Scholar]
- 62.Nguyen-Powanda P, Robaire B. Aging and oxidative stress alter DNA repair mechanisms in male germ cells of superoxide dismutase-1 null mice. Biol Reprod. 2021;105:944–957. doi: 10.1093/biolre/ioab114. [DOI] [PubMed] [Google Scholar]
- 63.Hamada A, Esteves SC, Nizza M, Agarwal A. Unexplained male infertility: diagnosis and management. Int Braz J Urol. 2012;38:576–594. doi: 10.1590/s1677-55382012000500002. [DOI] [PubMed] [Google Scholar]
- 64.Bhagwat S, Dalvi V, Chandrasekhar D, Matthew T, Acharya K, Gajbhiye R, et al. Acetylated α-tubulin is reduced in individuals with poor sperm motility. Fertil Steril. 2014;101:95–104.e3. doi: 10.1016/j.fertnstert.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 65.Okonofua FE, Ntoimo LFC, Omonkhua A, Ayodeji O, Olafusi C, Unuabonah E, et al. Causes and risk factors for male infertility: a scoping review of published studies. Int J Gen Med. 2022;15:5985–5997. doi: 10.2147/IJGM.S363959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cruz A, Sullivan DB, Doty KF, Hess RA, Canisso IF, Reddi PP. Acrosomal marker SP-10 (gene name Acrv1) for staging of the cycle of seminiferous epithelium in the stallion. Theriogenology. 2020;156:214–221. doi: 10.1016/j.theriogenology.2020.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coonrod SA, Herr JC, Westhusin ME. Inhibition of bovine fertilization in vitro by antibodies to SP-10. J Reprod Fertil. 1996;107:287–297. doi: 10.1530/jrf.0.1070287. [DOI] [PubMed] [Google Scholar]
- 68.Hamatani T, Tanabe K, Kamei K, Sakai N, Yamamoto Y, Yoshimura Y. A monoclonal antibody to human SP-10 inhibits in vitro the binding of human sperm to hamster oolemma but not to human Zona pellucida. Biol Reprod. 2000;62:1201–1208. doi: 10.1095/biolreprod62.5.1201. [DOI] [PubMed] [Google Scholar]
- 69.Baba T, Niida Y, Michikawa Y, Kashiwabara S, Kodaira K, Takenaka M, et al. An acrosomal protein, sp32, in mammalian sperm is a binding protein specific for two proacrosins and an acrosin intermediate. J Biol Chem. 1994;269:10133–10140. [PubMed] [Google Scholar]
- 70.Kato Y, Kumar S, Lessard C, Bailey JL. ACRBP (Sp32) is involved in priming sperm for the acrosome reaction and the binding of sperm to the zona pellucida in a porcine model. PLoS One. 2021;16:e0251973. doi: 10.1371/journal.pone.0251973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tseng HC, Tang JB, Gandhi PS, Luo CW, Ou CM, Tseng CJ, et al. Mutual adaptation between mouse transglutaminase 4 and its native substrates in the formation of copulatory plug. Amino Acids. 2012;42:951–960. doi: 10.1007/s00726-011-1009-9. [DOI] [PubMed] [Google Scholar]
- 72.Dean MD. Genetic disruption of the copulatory plug in mice leads to severely reduced fertility. PLoS Genet. 2013;9:e1003185. doi: 10.1371/journal.pgen.1003185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khatun A, Rahman MS, Ryu DY, Kwon WS, Pang MG. Elevated aminopeptidase N affects sperm motility and early embryo development. PLoS One. 2017;12:e0184294. doi: 10.1371/journal.pone.0184294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khatun A, Kang KH, Ryu DY, Rahman MS, Kwon WS, Pang MG. Effect of aminopeptidase N on functions and fertility of mouse spermatozoa in vitro. Theriogenology. 2018;118:182–189. doi: 10.1016/j.theriogenology.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 75.Subirán N, Agirregoitia E, Valdivia A, Ochoa C, Casis L, Irazusta J. Expression of enkephalin-degrading enzymes in human semen and implications for sperm motility. Fertil Steril. 2008;89(5 Suppl):1571–1577. doi: 10.1016/j.fertnstert.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 76.Subirán N, Pinto FM, Agirregoitia E, Candenas L, Irazusta J. Control of APN/CD13 and NEP/CD10 on sperm motility. Asian J Androl. 2010;12:899–902. doi: 10.1038/aja.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castillo J, Bogle OA, Jodar M, Torabi F, Delgado-Dueñas D, Estanyol JM, et al. Proteomic changes in human sperm during sequential in vitro capacitation and acrosome reaction. Front Cell Dev Biol. 2019;7:295. doi: 10.3389/fcell.2019.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang M, Liu BJ, Wang SQ, Xu Y, Han P, Li PC, et al. The role of mitochondrial aconitate (ACO2) in human sperm motility. Syst Biol Reprod Med. 2014;60:251–256. doi: 10.3109/19396368.2014.915360. [DOI] [PubMed] [Google Scholar]
- 79.Anderson DJ, Abbott AF, Jack RM. The role of complement component C3b and its receptors in sperm-oocyte interaction. Proc Natl Acad Sci U S A. 1993;90:10051–10055. doi: 10.1073/pnas.90.21.10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taylor CT, Biljan MM, Kingsland CR, Johnson PM. Inhibition of human spermatozoon-oocyte interaction in vitro by monoclonal antibodies to CD46 (membrane cofactor protein) Hum Reprod. 1994;9:907–911. doi: 10.1093/oxfordjournals.humrep.a138615. [DOI] [PubMed] [Google Scholar]
- 81.Moawad AR, Fernandez MC, Scarlata E, Dodia C, Feinstein SI, Fisher AB, et al. Deficiency of peroxiredoxin 6 or inhibition of its phospholipase A2 activity impair the in vitro sperm fertilizing competence in mice. Sci Rep. 2017;7:12994. doi: 10.1038/s41598-017-13411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee D, Moawad AR, Morielli T, Fernandez MC, O'Flaherty C. Peroxiredoxins prevent oxidative stress during human sperm capacitation. Mol Hum Reprod. 2017;23:106–115. doi: 10.1093/molehr/gaw081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Park K, Jeon S, Song YJ, Yi LS. Proteomic analysis of boar spermatozoa and quantity changes of superoxide dismutase 1, glutathione peroxidase, and peroxiredoxin 5 during epididymal maturation. Anim Reprod Sci. 2012;135:53–61. doi: 10.1016/j.anireprosci.2012.08.027. [DOI] [PubMed] [Google Scholar]
- 84.Ge W, Xiao L, Duan H, Zhao X, Li J, Hu J. Proteomic analysis of iTRAQ in melatonin-treated sheep epididymal epithelial cells. Reprod Domest Anim. 2022;57:1406–1417. doi: 10.1111/rda.14217. [DOI] [PubMed] [Google Scholar]
- 85.Zhang R, Guo X, Liang C, Pei J, Bao P, Yin M, et al. Identification and validation of yak (Bos grunniens) frozen-thawed sperm proteins associated with capacitation and the acrosome reaction. J Proteome Res. 2022;21:2754–2770. doi: 10.1021/acs.jproteome.2c00528. [DOI] [PubMed] [Google Scholar]
- 86.Bromfield EG, Aitken RJ, Anderson AL, McLaughlin EA, Nixon B. The impact of oxidative stress on chaperone-mediated human sperm-egg interaction. Hum Reprod. 2015;30:2597–2613. doi: 10.1093/humrep/dev214. [DOI] [PubMed] [Google Scholar]
- 87.Hekselman I, Yeger-Lotem E. Mechanisms of tissue and cell-type specificity in heritable traits and diseases. Nat Rev Genet. 2020;21:137–150. doi: 10.1038/s41576-019-0200-9. [DOI] [PubMed] [Google Scholar]
- 88.Hamada A, Sharma R, du Plessis SS, Willard B, Yadav SP, Sabanegh E, et al. Two-dimensional differential in-gel electrophoresis-based proteomics of male gametes in relation to oxidative stress. Fertil Steril. 2013;99:1216–1226.e2. doi: 10.1016/j.fertnstert.2012.11.046. [DOI] [PubMed] [Google Scholar]
- 89.O'Rand MG, Widgren EE, Hamil KG, Silva EJ, Richardson RT. Functional studies of eppin. Biochem Soc Trans. 2011;39:1447–1449. doi: 10.1042/BST0391447. [DOI] [PubMed] [Google Scholar]
- 90.Azhar M, Altaf S, Uddin I, Cheng J, Wu L, Tong X, et al. Towards post-meiotic sperm production: genetic insight into human infertility from mouse models. Int J Biol Sci. 2021;17:2487–2503. doi: 10.7150/ijbs.60384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cannarella R, Condorelli RA, Mongioì LM, La Vignera S, Calogero AE. Molecular biology of spermatogenesis: novel targets of apparently idiopathic male infertility. Int J Mol Sci. 2020;21:1728. doi: 10.3390/ijms21051728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Finkelstein M, Etkovitz N, Breitbart H. Ca2+ signaling in mammalian spermatozoa. Mol Cell Endocrinol. 2020;516:110953. doi: 10.1016/j.mce.2020.110953. [DOI] [PubMed] [Google Scholar]
- 93.Nowicka-Bauer K, Szymczak-Cendlak M. Structure and function of ion channels regulating sperm motility-an overview. Int J Mol Sci. 2021;22:3259. doi: 10.3390/ijms22063259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Syeda SS, Sánchez G, McDermott JP, Hong KH, Blanco G, Georg GI. The Na+ and K+ transport system of sperm (ATP1A4) is essential for male fertility and an attractive target for male contraception. Biol Reprod. 2020;103:343–356. doi: 10.1093/biolre/ioaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gervasi MG, Visconti PE. Molecular changes and signaling events occurring in spermatozoa during epididymal maturation. Andrology. 2017;5:204–218. doi: 10.1111/andr.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Castellini C, D'Andrea S, Cordeschi G, Totaro M, Parisi A, Di Emidio G, et al. Pathophysiology of mitochondrial dysfunction in human spermatozoa: focus on energetic metabolism, oxidative stress and apoptosis. Antioxidants (Basel) 2021;10:695. doi: 10.3390/antiox10050695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aslam B, Basit M, Nisar MA, Khurshid M, Rasool MH. Proteomics: technologies and their applications. J Chromatogr Sci. 2017;55:182–196. doi: 10.1093/chromsci/bmw167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quality assessment of the included studies. (A) Risk of bias plot representing the domain-level judgments for each included study. (B) Distribution of the risk of bias per domain. The plot and graph were generated in the Robvis application.
Search terms used to identify proteomic studies performed in human spermatozoa
Excluded studies after screening and full-text assessment. Each excluded study was recorded with the DOI number, year, title, and respective exclusion motive
Differentially expressed proteins in male infertility-related conditions. All accession numbers identified in included studies were collected and mapped in the UniProt database. For each mapped protein is indicated the UniProtKB number, gene name, protein name, fold-change, and the review status according to the UniProt database.
Gene ontology terms for biological processes. For each protein candidate, the gene ontology terms were retrieved from the UniProt database and were grouped according to their relationship.