Abstract
In the present study, next generation sequencing was employed to identify and explore the differential expression profiles of microRNAs (miRNAs) in peripheral blood mononuclear cells (PBMCs) of crossbred (B. taurus x B. indicus) and Vechur (B. indicus) cattle in response to the bacterial endotoxin-lipopolysaccharide (LPS). The PBMCs from adult apparently healthy female crossbred cows and Vechur cattle, a native cattle breed of Kerala, India were stimulated with 10 μg/mL of LPS for 6 h. Among the differentially expressed miRNAs, the expression of 13 miRNAs showed statistically significant up regulation while, significant decrease in the expression of 15 miRNAs was noticed in LPS treated PBMCs of Vechur cattle compared to crossbred cows. The expression profiling of miRNA, bta-miR-375, expression of which was found to be significantly down regulated in LPS treated PBMCs of Vechur cattle with respect to crossbred cattle by the NGS studies, is presented in the present manuscript. The decrease in expression of bta-miR-375 noticed by NGS was in accordance with the results of quantitative real time PCR assay. Functional gene enrichment analysis and pathway analysis revealed significant enrichment of predicted targets of bta-miR-375 in many immune related and cell signalling mechanisms. In addition, over representation of targets of bta-miR-375 was also noticed in pathogenesis of many of the bovine diseases. The study could also identify differences in the expression of cytokines, viz. Tumour Necrosis Factor Alpha (TNFα), Interleukin 4 (IL-4) and Interferon-γ (IFNγ) between LPS treated and untreated PBMCs of crossbred and Vechur cattle.
Keywords: Vechur cattle, Peripheral blood mononuclear cells, Lipopolysaccharide, microRNA, Cytokines
1. Introduction
Inflammatory responses towards bacterial endotoxin; lipopolysaccharide (LPS) and studying the mechanisms behind LPS mediated toll-like receptor 4 (TLR-4) signalling are critical in understanding the impact of gram-negative bacterial sepsis. Being a potent activator of innate immune system, LPS can activate the cells of immune system and thereby regulates the synthesis of various pro-inflammatory and anti-inflammatory cytokines. Studies on post-transcriptional regulation by microRNAs over LPS mediated TLR-4 signalling are gaining importance nowadays. These small endogenous non-coding RNA molecules (19–24 nt in length) can modulate a wide array of biological processes including fine tuning of vital cell signalling pathways and regulation of both innate and adaptive immune responses [1]. They mediate development, differentiation and activation of immune cells, regulation of immune related signalling pathways and synthesis and release of pro and anti-inflammatory cytokines. Distinct expression patterns of miRNAs are seen at different stages of inflammation and identification of miRNA based biomarkers are promising approaches for the diagnosis as well as prognosis of many of the important diseases including cancer [2].
Zebu cattle are well-known for their disease resistance, thermo tolerance and general adaptability to the wavering climatic conditions compared to the existing crossbred cattle population [3]. Native cattle breeds are low producers as compared to exotic breeds and the crossbreeding program tremendously increased milk production among livestock population. But higher susceptibility to diseases and poor adaptability to the prevailing agro-climatic conditions pose challenge to successful animal agriculture as a result of the intensive crossbreeding programmes. Exploring the reasons of the immunological sturdiness noted in native cattle breeds is an area of intense research as scientific studies substantiating these claims are largely lacking. Considering the role of miRNAs in regulating immune responses, the current research work was envisaged to identify and unravel the differences in the expression of miRNAs in crossbred (Bos indicus × Bos taurus) and Vechur cattle (Bos indicus) in response to LPS and thereby assessing the significance of miRNAs in conferring a higher disease resistance potential to Vechur cattle; a native cattle breed of Kerala, India. To date, miRNA expression analysis in LPS induced bovine PBMCs has not been well characterised. The present manuscript mainly focuses on the differential expression analysis of the miRNA; bta-miR-375, the expression of which was found to be significantly down regulated in LPS treated PBMCs of Vechur cattle when compared to that of crossbred cattle by miRNAome analysis. The study laid emphasis on validation of the results of NGS, prediction of target genes of bta-miR-375, analysis of cellular pathways associated to the target genes and assessment of differences in the level of cytokines whose biosynthetic pathways were found to be influenced by target genes of bta-miR-375.
Significant down regulation noticed in the expression of bta-miR-375 in Vechur cattle with respect to crossbred cattle identified by NGS was consistent with the results of quantitative real time PCR assay. In-silico target prediction, gene ontology and pathway analysis revealed significant enrichment of targets of bta-miR-375 in multiple immune related mechanisms. Variations observed in the expression patterns of cytokines analysed also points towards the differences between the two genetic groups in immune responses. The study forms the basis for future functional studies as well as challenge experiments to unravel the contribution of miRNAs towards the differences noticed in immune responses among the two genetic groups. This findings of the study contributes significantly to the current knowledge of miRNAs in Gram negative bacterial infection and pathogenesis.
2. Materials and methods
2.1. Animals
All the experimental procedures adopted in the present study were approved by the statutory forums of Kerala Veterinary and Animal Sciences University. The research work did not require any approval from the Institutional Animal Ethics Committee of University. The study was conducted in six each of adult, apparently healthy, age and sex matched crossbred and Vechur cattle maintained at University Livestock Farm and Fodder Research and Development Scheme, and Vechur Conservation Unit, Kerala Veterinary and Animal Sciences University, India, respectively. All the animals were maintained under similar environment and nutritional status. The animals were free from illness or injury during the entire study period, and were confirmed healthy by clinical examination and complete blood count. Animals did not receive any vaccinations or other medications for at least one week prior to sample collection.
2.2. Cell isolation, culture and stimulation
Eighty mL blood was collected aseptically from each animal into EDTA coated tubes and PBMCs were isolated by gradient density centrifugation using Hisep 1077 (Himedia, Mumbai, India) as per manufacturer's instructions, within 2 h of sample collection. Cell viability was assessed by Trypan blue exclusion assay and samples having a viability of ≥95% were selected for the subsequent steps. Cells from respective groups were re-suspended in RPMI-1640 medium with 25 mmol/L HEPES buffer (Himedia), 10% low-endotoxin heat-inactivated foetal calf serum (Himedia), 2 mM l-glutamine (Gibco, Thermo Fisher Scientific), 1 mM sodium pyruvate (Sigma-Aldrich, St. Louis, MO), 50 u/mL penicillin and 50 μg/mL streptomycin to a concentration of 2 × 106 viable cells per mL and cultured in a 24-well plate at 37 °C and 5 % CO2 for a period of 6 h, with and without LPS from Escherichia coli O111:B4 (Sigma Aldrich) at a concentration of 10 μg/mL. The cells were then harvested by centrifugation and stored in RNAlater at 2–8 °C overnight followed by storing at −80 °C for miRNA sequencing. The experiment was replicated to harvest cells for qRT-PCR and supernatant for cytokine analysis. All samples were stored at −80 °C till analysis. Cytokine assay was performed within 48 h of collection of supernatants.
2.3. Total RNA isolation and next-generation sequencing for differential expression analysis of miRNAs
Total RNA was isolated from the respective PBMC cultures using Trizol method. Short-read Illumina Next Generation Sequencing (NGS) was employed for the study of miRNA transcriptome. Small RNA library construction was performed using NEBNext small RNA sample preparation protocol. The constructed small RNA libraries were sequenced on HiSeq 2500 with a 1 × 50bp reads to obtain 40-50 million reads. After the sequencing run, Illumina small RNA-Seq data were processed to generate FASTQ files. Adapter sequences, excessively short reads and low quality reads were trimmed from the raw sequence data using cut adapt tool.
All the unique read sequences with a length of 17–35 bp were aligned to miRBase22 (http://www.mirbase.org) to identify previously known miRNAs of Bos taurus species. Differential expression analysis of miRNAs among the two study groups was carried out by counting the aligned reads with reference genome of Bos taurus using the statistical package DESeq. Mean read count of samples, fold change and p-values were calculated.
2.4. Quantitative real time PCR analysis
Expression profile of the miRNA, bta-miR-375 retrieved from NGS was validated by qRT-PCR assay in Biorad CFX Opus 96 real time PCR system. MicroRNA, bta-miR-191 was selected as the endogenous control [4] and relative quantification was performed. Polyadenylation of the total RNA isolated from the respective experimental groups was performed in Biorad T100 thermal cycler in 10 μL reaction mixture containing 1 μg RNA, 1 mM ATP, 1000 units of E. coli Poly (A) Polymerase (New England Biolabs) and 1x buffer. Polyadenylated RNA samples were reverse transcribed using RT primers designed [5] and were custom synthesised (Sigma-Aldrich). The 10 μL reaction mixture contained 1 μL of polyadenylation product, 1 μM RT primer, 0.5 mM dNTP mix, 2500 units RevertAid M-MuLV Reverse Transcriptase (Thermo Scientific) and 2x reaction buffer. Details of RT primers used are given in Table 1.
Table 1.
Primer sequences for first strand cDNA synthesis of bta-miR-375 and bta-miR-191.
|
Sl. No. |
miRNA | Primer sequences |
|---|---|---|
| 1. | bta-miR-375 | CAGTGCAGGGTCCGAGGTCAGAGCCACCTGGGCAATTTTTTTTTTTTCACGC |
| 2. | bta-miR-191 | CAGTGCAGGGTCCGAGGTCAGAGCCACCTGGGCAATTTTTTTTTTTCAGCTG |
The forward primers used for qRT PCR assay of both the microRNAs and the universal the reverse primer were designed [6] and custom synthesised (Sigma-Aldrich) (Table 2).
Table 2.
qRT PCR primers for bta-miR-375 and internal control miRNA.
| Sl.No | miRNA | qRT PCR primers |
|
|---|---|---|---|
| Forward primer | Universal reverse primer | ||
| 1. | bta-miR-375 | TTTTGTTCGTTCGGCTC | CAGTGCAGGGTCCGAGGT |
| 2. | bta-miR-191 | CAACGGAATCCCAAAAG | |
Quantitative RT-PCR was done using Biorad Sso Advanced Universal SYBR green supermix according to the manufacturer's instructions. After an initial denaturation at 95 °C for 10 min, 40 cycles of denaturation (95 °C for 15 s) and annealing for 60 s at 62.5 °C (bta-miR-375) or 60 °C (bta-miR-191) were programmed. Melt curve analysis was carried out at 65 °C–95 °C with 0.5 °C increment. All the data were analysed using 2-ΔΔCt method [7] and all the results were expressed as Mean ± SE. The ΔCt values were subjected to t-test to determine significant differences between the study groups.
2.5. Target prediction and functional annotation
Targets of bta-miR-375 were predicted using TargetScan (https://www.targetscan.org/), miRWalk (http://mirwalk.umm.uni-heidelberg.de/) and miRanda (http://www.microrna.org/microrna/getDownloads.do). Redundant targets were identified and removed using Venny 2.1.0. (https://bioinfogp.cnb.csic.es/tools/venny). Functional annotation and pathway analysis of predicted targets were studied through DAVID accessible at https://david.ncifcrf.gov/.
2.6. Cytokine analysis
The key pro-inflammatory cytokines TNFα and IFNγ and the anti-inflammatory cytokine IL-4 were measured in the culture supernatants. TNFα was measured after 6 h of incubation while IFNγ and IL-4 levels were measured after an overnight incubation of cells with LPS. The assays were conducted according to manufacturer's instructions using ELISA kits for bovine TNFα, IL-4(Invitrogen, Thermo Fischer Scientific) and IFNγ (Bio-Rad Medical laboratories). All the three cytokines were measured within 48 h of collection of supernatants.
3. Results
3.1. Characterization of the microRNA transcriptome in LPS treated PBMCs of crossbred and Vechur cattle
All RNA samples had 260/280 ratio between 1.8 and 2.0 and RNA Integrity Number of above 8.0. From small RNA libraries prepared from LPS challenged samples that yielded a total of 7.4 and 12.9 million raw reads, respectively, for crossbred and Vechur samples a total of 0.47 and 0.64 million clean reads, with an average Phred score of 34.92 and 34.75, respectively, were obtained (un published data-data will be shared on request). This corresponded to 55.3% and 62.1% of the adapter trimmed reads of mean length of 21 nucleotides, respectively for crossbred and Vechur samples. Known and novel miRNAs were identified (un published data) by aligning all clean reads to bovine genome (Bos taurus) with miRBase-22 mature and miRBase-22 precursor miRNA database.
3.2. Differential expression analysis
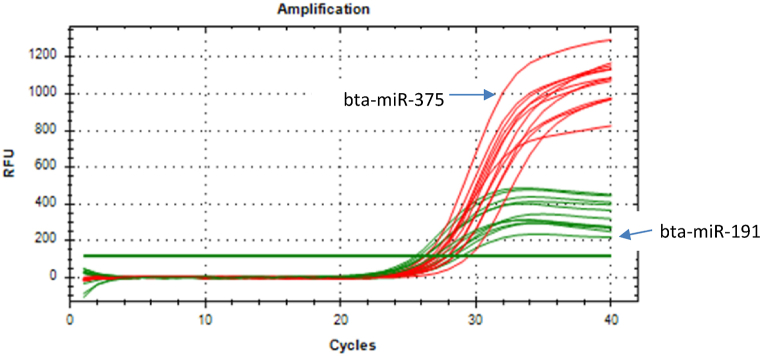
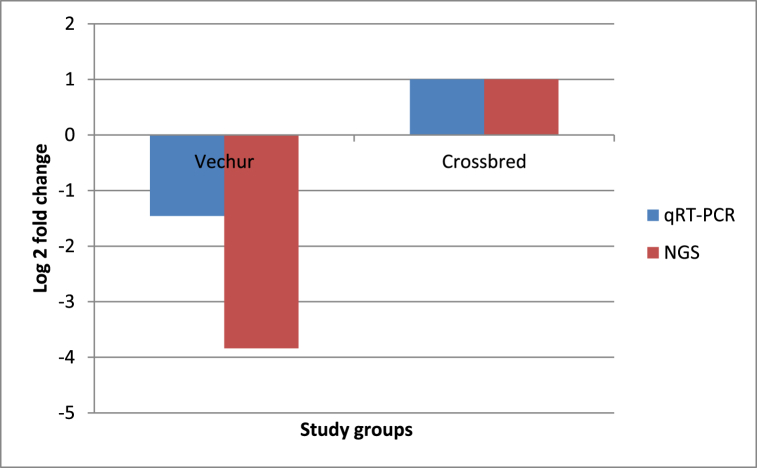
After normalisation of the raw reads, the expression of 13 miRNAs were found to be significantly up-regulated and expression of 15 miRNAs were significantly down-regulated (p < 0.05) in the LPS stimulated PBMCs of Vechur cattle compared to that of crossbred cattle. Among the differentially expressed miRNAs, expression of bta-miR-375 was found to be significantly down regulated with a fold change of 0.069 (p value < 0.05). Validation of the results of NGS of bta-miR-375 by qRT-PCR confirmed the NGS results revealing a statistically significant (p value < 0.05) down regulation with a fold change of 0.364. Fig. 1, Fig. 2 depict the amplification plot for the relative expression of bta-miR-375and log2 fold change for qRT-PCR assay and NGS of bta-miR-375, respectively.
Fig. 1.
Amplification plot for the relative expression of bta-miR-375.
Fig. 2.
Log2 fold change for qRT-PCR assay and NGS of bta-miR-375.
3.3. Prediction of targets
Target analysis of bta-miR-375 using online tools, TargetScan, miRWalk and miRanda revealed 5164, 504 and 256 targets, respectively, with the current data. The total number of predicted targets for bta-miR-375 predicted by the target prediction tools, after removal of redundant ones, was 5573.
3.4. GO and functional classification
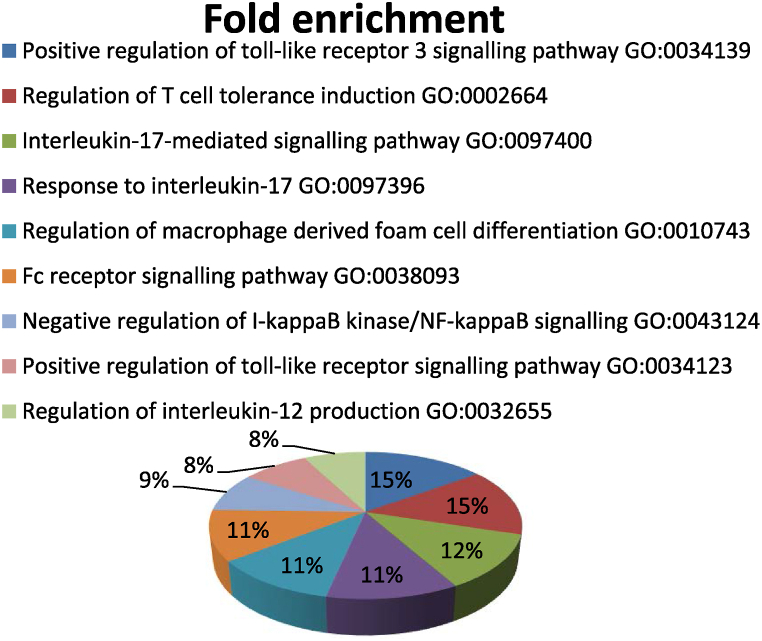
With the default parameters of DAVID data base 1137, 128 and 116 GO terms were identified in the biological process, cellular component and molecular function categories, respectively, by gene ontology analysis of the targets of bta-miR-375. In the biological process category, most of the annotated targets were involved in numerous immune related mechanisms (Fig. 3), mRNA destabilization, posttranscriptional regulation of gene expression, negative regulation of epidermal growth factor receptor signalling pathway, histone modification, negative regulation of histone modification, axon guidance and its regulation, positive regulation of Wnt signalling pathway, regulation of I-kappaB kinase/NF-kappaB signalling, regulation of MAPK cascade, SMAD protein signal transduction, regulation of transforming growth factor beta receptor signalling pathway etc. Coding region instability determinant (CRD) -mediated mRNA stability complex, RNAi effector complex and RISC complex were found to be the highly enriched GO terms in the cellular component category. In the molecular function category, targets of bta-miR-375 were assigned to N-acetyl glucosamine 6-O-sulfotransferase activity, S-methyl transferase activity, translation repressor activity, nucleic acid binding acetyl transferase activity, cytokine receptor activity, miRNA binding etc.
Fig. 3.
Immune related GO terms associated with targets of bt-miR-375.
3.5. Pathway analysis
The KEGG pathway analysis through DAVID demonstrated an overrepresentation of targets of bta-miR-375 in several cell signalling and immune related pathways (Table 3). Glycosaminoglycan biosynthesis, pathways associated to the functioning of adherens junction, axon guidance were the other pathways significantly enriched with the targets of bta-miR-375. In addition, significant enrichment of targets was noted in pathways associated to various types of cancers, hepatitis B, yersiniosis etc.
Table 3.
Pathway analysis of targets of bta-miR-375.
| Sl.No. | Pathway affected | No. of target genes involved | Fold enrichment |
|---|---|---|---|
| 1. | NF-kappa B signalling pathway | 6 | 2.5 |
| 2. | TGF-beta signalling pathway | 40 | 2.0 |
| 3. | PI3K-Akt signalling pathway | 14 | 1.7 |
| 4. | T cell receptor signalling pathway | 41 | 1.7 |
| 5. | HIF-1 signalling pathway | 41 | 1.7 |
| 6. | C-type lectin receptor signalling pathway | 39 | 1.7 |
| 7. | Fc epsilon RI signalling pathway | 25 | 1.6 |
| 8. | Inflammatory mediator regulation of TRP channels | 39 | 1.6 |
| 9. | IL-17 signalling pathway | 31 | 1.5 |
| 10. | Bacterial invasion of epithelial cells | 24 | 1.4 |
| 11. | TNF signalling pathway | 37 | 1.4 |
| 12. | Chemokine signalling pathway | 53 | 1.3 |
3.6. Cytokine analysis
Significantly higher levels of TNFα were observed in the LPS treated PBMCs of crossbred cows compared to that of Vechur cattle, whereas a higher level of IL-4 was noticed in Vechur cattle (p=<0.05). Among the LPS untreated cells also, TNFα level was higher in crossbred cattle and IL-4 level was higher in Vechur. The IFN γ level in LPS treated crossbred samples was higher but the difference was not statistically significant. Results of cytokine analysis are presented in Table 4.
Table 4.
Concentration of cytokines in LPS challenged and unchallenged PBMC cultures.
| TNFα | IL-4 | IFNγ | ||||
|---|---|---|---|---|---|---|
| Groups | Crossbred (pg/mL) | Vechur (pg/mL) | Crossbred (pg/mL) | Vechur (pg/mL) | Crossbred (pg/mL) | Vechur (pg/mL) |
| Test | 39.16 ± 2.82aA | 19.21 ± 0.6aB | 198.98 ± 10.53aB | 713.07 ± 15.62bA | 43.27 ± 1.12aA | 42.01 ± 0.58bA |
| Control | 27.79 ± 0.66bA | 15.1 ± 0.58bB | 301.57 ± 20.06bB | 423.4 ± 18.20aA | 41.77 ± 0.78aB | 45.66 ± 1.30aA |
| F-value for interaction between treatment vs breed = 5.805b; P-value = 0.026 | F-value for interaction between treatment vs breed = 141.405a; P-value <0.001 | F-value for Interaction between treatment vs breed = 6.821b; P-value = 0.017 | ||||
Means having different small letters as superscript differ significantly within a column; Means having different capital letters as superscript differ significantly within a row.
Significant at 0.01 level.
Significant at 0.05 level.
4. Discussion
The present study focussed on the significance of the miRNA, bta-miR-375 in regulation of immune responses among the two genetic groups studied, on post LPS exposure. To our knowledge, this study is the first to analyse miRNA expression profiles between LPS challenged PBMCs of crossbred/exotic and native cattle breeds.
Lipopolysaccharide, the major outer membrane component present in almost all Gram-negative bacteria acts as an extremely potent stimulator of innate immunity in diverse eukaryotic species [8]. Through the activation of TLR4 signalling, LPS can induce the expression of several inflammatory cytokines, viz. TNFα, IL-1β and IL-6 in immune cells [9]. Differential expressions of miRNAs have been identified in primary human macrophages [9], PBMCs of equine [10] and porcine [11] species in response to LPS challenge. Studies have shown that LPS mediated alteration in the expression of miRNAs induced acute inflammation thereby triggering the production of TNFα, IL-1β and IL-6 [12,9,13].
4.1. Characterization of the microRNA transcriptome in LPS treated PBMCs
In our study a total of 7.4 and 12.9 million raw reads and 0.47 and 0.64 million clean reads were identified with an average Phred score of 34.92 and 34.75, respectively, in PBMCs of crossbred and Vechur samples after LPS challenge. On miRNAome profiling, several known and novel miRNAs with potential roles in LPS mediated immune signalling and immune responses were detected in native cattle compared to crossbred cattle (unpublished data). The functions of these novel miRNAs are yet to be explored. We could find out statistically significant differences in the expression of 28 known miRNAs (unpublished data) in LPS treated PBMCs of Vechur cattle compared to crossbred cows (p value < 0.05). Among these differentially expressed miRNAs, we focussed on bta-miR-375 as it exhibited significant decrease in its expression in LPS treated PBMCs of Vechur cattle with respect to the corresponding samples from crossbred cattle. Moreover, recent studies have reported on the crucial roles of miR-375 in modulating immune responses and regulating pathogenesis of a plethora of human diseases [14,6,15,16]. Decreased expression of miR-375 was reported in the intestinal mucosa of patients with Crohn's disease and ulcerative colitis [15]. It has been reported that miR-375 regulate neuronal dendrite formation and overexpression of miR-375 in mouse hippocampus potently reduced dendrite density by repressing the expression of neuronal RNA-binding protein HuD [14]. Similarly, an over expression of miR-375 was noticed in C cells of thyroid gland in medullary thyroid cancer than in reactive C-cell hyperplasia [16].
4.2. qRT-PCR assay for the validation of expression of bta-miR-375
We could identify a statistically significant decrease (p value < 0.05, t-test) in the expression of bta-miR-375 in PBMCs from Vechur cattle on comparison to that of crossbred cows by qRT-PCR assay and the results were in consistent with NGS data. Minor variations noticed between the two assays might be related to sensitivity differences between the two methods regarding profiling miRNA expression. Studies on differential expression of miRNAs in LPS treated PBMCs of equine [10] and porcine [11] species, also revealed discrepancies in the level of miRNA expression between qRT-PCR assay and miRNAome analysis.
4.3. Prediction of targets of bta-miR-375
The number of targets predicted for bta-miR-375 showed differences in the predicted targets among the online tools used for the present study. The reasons for these differences between the target prediction tools may be because of the difference in the features incorporated in each tool for the prediction of target genes even though most tools use some common criteria like seed match, conservation, free energy, and site accessibility etc [17]. All the non-redundant targets retrieved from the online target prediction tools were subjected for gene ontology and pathway analysis.
4.4. Gene ontology and pathway analysis
On gene ontology analysis, it was found that most of the annotated targets of bta-miR-375 were associated with immune related GO terms, viz. regulation of TLR signalling pathway, post-transcriptional regulation of gene expression, epidermal growth factor receptor signalling pathway, histone modification, axon guidance and its regulation, I-kappaB kinase/NF-kappaB signalling, MAPK cascade, SMAD protein signal transduction, TGF-β receptor signalling pathway etc. Analysis of targets of bta-miR-375 also revealed that pathways associated to cell signalling and immune mechanisms to be significantly enriched with bta-miR-375 targets. In addition, pathways associated with the functioning of adherens junction and glycosaminoglycan biosynthesis was also found to be highly enriched with the targets of bta-miR-375. These pathways were reported to have crucial roles in regulation of immune responses [18,19]. Among the immune related pathways the highest fold enrichment was noticed in NF-kappa B signalling pathway followed by TGF-β signalling pathway.
Among the cell surface expressing pattern recognition receptors (PRRs), Toll-like receptors-4 (TLR4), are reported to have key roles in host defense towards LPS, a pathogen-associated molecular pattern (PAMP) seen on outer surface membrane of Gram negative bacteria. TLR4 signalling triggered by LPS is mediated by the cytoplasmic adaptor molecule, MyD88, and serine/threonine kinases of the IL-1R–associated kinase family, followed by phosphorylation of the inhibitory protein IκB-α resulting in the translocation of NF-κB to the nucleus and activation of transcription of many genes encoding production of type I interferons and pro-inflammatory cytokines [20].
Signalling pathway involving transforming growth factor β (TGF-β; a pleiotropic cytokine secreted predominantly by immune cells) are having potent regulatory and anti-inflammatory activity have been reported for its association with lipopolysaccharide mediated TLR4 signalling and thereby influences NF-kappa B signalling [21]. Both NF-kappa B signalling and TGF-β signalling along with LPS mediated TLR4 signalling are central in regulation of immune responses and release of cytokines from immune cells [22,23,24]. Findings of our study, suggest that immune related pathways preferably, Toll-like receptor signalling pathway, I-kappa B kinase/NF-kappa B signalling, SMAD protein signal transduction and TGF-β receptor signalling are inter related and these pathways are significantly influenced by the target genes of bta-miR-375. In support of our findings, the association of these pathways and their importance in regulation of immune responses with respect to the pro inflammatory milieu are reported in recent immunological studies [25]. Controversial reports are there regarding the association of TGF-β signalling pathway and LPS mediated TLR4 signalling. It has been reported that LPS mediated TLR4 signalling is having an inhibitory effect on TGF-β receptor signalling through the coordinated action of SMAD proteins [23] even though, a stimulatory effect of the same pathway on TGF-β signalling has also been reported [22]. LPS-induced NF-kappa B activation was found to be inhibited in murine macrophages by TGF-β1 via Smad3 and Smad6 proteins thereby inhibiting the release of pro-inflammatory cytokines [23]. On the contrary, it was noticed that IL-1β/LPS induces suppression of TGF-β signalling through TRAF6 signalling a myD88 independent mechanism through interaction of IL-1β/LPS with the type III TGF-β receptor [24]. But this inhibitory effect of TGF-β1on NF-kappa B activation can be overcome by triggering Smad3 linker phosphorylation via TLR4–IRAK1-activated ERK1/2 and subsequent blocking Smad6 expression by LPS [23]. Lipopolysaccharide acting via TLR4, transactivating the TGF-β receptor in human vascular smooth muscle cells has been reported but the authors could not detect any LPS mediated inhibition of SMAD 6 expression in these cells [22]. Similar studies showed that LPS enhances Smad2/3-dependent TGF-β1 transcriptional responses in normal fibroblasts through TLR4 signalling [25,26]. The differences in LPS responses noticed might be related to cell specificity, with positive cross talk between TLR4 and TGF-β in non-immune cells, but an inhibitory cross talk in inflammatory cells [25].
In the present study, we could identify that targets of bta-miR-375 were significantly overrepresented in the aforementioned signalling mechanisms. Since, the expression of this miRNA was significantly down regulated in PBMCs of Vechur cattle; the expression of target genes associated with the respective signalling pathways might be up regulated. Significant enrichment of targets of bta-miR-375 observed in mRNA stability complex, RNAi effector complex, RISC complex, translation repressor activity and miRNA binding also indicates the regulatory role of this miRNA in gene expression at post-transcriptional and post translational levels. All these findings suggest that differences exist between the two genetic groups in mechanisms operating during an acute inflammatory response.
These findings were supported by the results of cytokine assay which were involved in the critical immune signalling pathways associated with targets of bta-miR-375.
4.5. Cytokine analysis
The physiological effects of LPS are mainly based on the biosynthesis and activation of molecular mediators like cytokines [12]. We found significantly lower TNFα level in LPS treated PBMC cultures of Vechur cattle in comparison to crossbred cows. It has been also revealed that TNFα level was higher in LPS treated cells when compared to the respective LPS untreated cells of both genetic groups. The significantly higher level of TNFα noticed in LPS untreated PBMCs of crossbred cattle with respect to that of Vechur cattle show the influence of breed in the regulation of LPS mediated immune responses. These differences noticed in the TNFα level may be because of the differences in the activation of NF-κB signalling pathway through the LPS mediated TLR4 signalling and subsequent release of TNFα. Role of LPS in the activation of NF-κB signalling pathway mediated via TLR4 signalling and the enhanced release of pro inflammatory cytokines like TNFα from the activated T cells are reported in recent studies [10,11].
With respect to IL-4 assay, the present study showed a significantly higher IL-4 level in LPS treated and untreated Vechur PBMCs compared to the respective groups from crossbred cattle. It was interesting to note that IL-4 level lowered significantly on LPS treatment in crossbred PBMCs whereas it increased significantly in Vechur cattle. This difference in response to LPS warrants further exploration on the mechanisms involved. Studies on LPS treated equine PBMCs revealed an increase in IL-4 level with time but a statistically significant difference could not be detected between the LPS treated and untreated cells [10].
We could not find out any statistically significant difference in the IFN γ level between the LPS stimulated crossbred and Vechur PBMCs even though a slight rise in their levels was noticed in crossbred samples. Changes noticed in IFN γ level between other study groups were all negligible and did not produce a statistically significant difference. Though, LPS was reported to be a potent stimulator of IFN γ secretion from activated immune cells, the present study could not reveal any significant difference in the IFN γ levels between the two study groups. In contrast to our findings significantly greater level of IFN γ in LPS challenged equine PBMCs compared to the control cells after 8 h of incubation [10].
As the key mediators for orchestrating physiological responses, cytokines are having paramount significance in coordinating immune responses of an organism at the time of a pathological insult by controlling activation, differentiation, chemotaxis and apoptosis of immune cells [12]. Cytokine analysis of our study revealed significant differences between the two genetic groups which could probably be mediated through bta-miR-375 also. Results of cytokine assay was supporting the findings deduced from the results of differential expression profiling of bta-miR-375 and the associated in -silico analysis. It has been also noticed that the major pathways associated to LPS mediated signalling pathways, viz; TLR4 signalling, TGF-β signalling and NF-κB signalling were observed to be significantly enriched with the targets of the miRNA of our study. Recent research findings have reported that TGF-β signalling pathway activates the release of anti-inflammatory cytokines viz; IL-4, IL-10, IL-11, IL-13 etc. whereas, inhibits the release of pro inflammatory cytokines like TNFα, IFNγ, IL-1β, IL-6 etc [27]. In the present study, significant down regulation observed in the expression of bta-miR-375 in PBMCs of Vechur cattle compared to crossbred animals after LPS challenge suggests activation of pathways associated with the expression of corresponding target genes of major signalling pathways like TGF-β signalling. The decrease in the level of TNFα noticed in the LPS treated PBMCs of Vechur cattle may be attributable to the activated TGF-β signalling pathway and subsequent inhibition of TLR4 mediated activation of NF-κB signalling and resulting decrease in the release of pro –inflammatory cytokines.
Similarly, the activated TGF-β signalling pathway might be activating the release of anti-inflammatory cytokine, IL-4 and this might be leading to the high IL-4 level in Vechur PBMC cultures in response to LPS challenge. Likewise, the cytokine, IFN γ secreted primarily by Th1 cell lineage are found to be unaffected by LPS treatment, which suggest that either the target genes associated with this pathway are not affected by miRNA mediated regulation or it might be due to the inhibitory effect of IL-4 on activation of Th1 cell lineage [28,29]. The higher IFN γ noticed in LPS untreated Vechur PBMCs might be due to contamination from any unknown source. The role of miRNAs in regulation of gene expression through the post transcriptional gene silencing mechanisms or by regulation of translational repression might be contributing to the differences noticed in the expression of the aforementioned signalling pathways between the crossbred and Vechur cattle [30]. In contrast to the our findings, there are reports suggesting LPS mediated suppression of TGF-β signalling which in turn overcomes the inhibitory effect of TGFβ on NF-kappa B activation followed by stimulation of the release of pro inflammatory cytokines [23,24]. But supporting the findings of the current study, there are reports explaining activation of TGF-β signalling through LPS mediated TLR4 signalling resulting in inhibition of the release of pro inflammatory cytokines from the activated T cell [22].
5. Conclusion
MicroRNAome analysis is one of the most promising and recent approaches in monitoring the physiological or pathological status of an animal while comparing other transcriptomics biomarkers. These tiny key modulators of gene expression are also reported to have pivotal roles in regulating the complex immune mechanisms of the cells too. The present study suggests involvement of miRNAs in the alleged difference between crossbred and indigenous cattle breeds in response to immune challenges. Though clinical evidences suggesting immune sturdiness have been reported for Vechur cattle, scientific evidences confirming the same are largely lacking. Significant decrease in the expression of bta-miR-375 noticed in LPS treated PBMCs of Vechur cattle when compared to crossbred cattle by NGS, and qRT-PCR assay, and the enrichment of targets of bta-miR-375 in many immune related mechanisms points towards an underlying difference in miRNA mediated immune regulation in the two genetic groups studied. The results of cytokine assay of the present study confirms that the mechanisms behind the LPS induced triggering of immune responses are different in crossbred and Vechur cattle though detailed studies are needed in this regard.
6. Limitations of the study
There are benefits and limitations to the current and latest technologies which all have to be considered while designing miRNA studies. Results of different miRNA assays may exhibit minor variations across the platforms, pointing towards the fact that careful and critical evaluation of the results are needed while interpreting the findings. In the present study too, minor variations were noticed in the expression of miRNA studied; bta-miR-375 between the two technologies used viz. qRT-PCR assay and NGS though both techniques showed a decrease in the expression of miRNA. The high cost for next-generation sequencing is another limitation of these area of research which could be otherwise a fast and more accurate miRNA profiling technique that could greatly augment miRNA research.
Data availability statement
Data will be made available on request.
Additional information
No additional information is available for this paper.
CRediT authorship contribution statement
Divya P. D.: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft. Shynu M.: Conceptualization, Data curation, Formal analysis, Project administration. Jayavardhanan K.K.: Conceptualization, Data curation, Funding acquisition. Uma R.: Conceptualization, Data curation, Formal analysis. Aravindakshan T.V.: Conceptualization, Data curation, Formal analysis. Radhika G.: Conceptualization, Data curation, Formal analysis. Sameer kumar V.B.: Conceptualization, Data curation, Formal analysis. Muhasin Asaf: Resources, Software, Validation. Renjith Sebastian: Resources, Validation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank Kerala Veterinary and Animal Sciences university for providing the financial support for the conduct of research work.
Footnotes
#Part of PhD thesis of first author submitted to Kerala Veterinary & Animal Sciences University, Pookode, Wayanad, Kerala.
References
- 1.Lawless N., Vegh P., Farrelly C.O., Lynn D.J. The role of microRNAs in bovine infection and immunity. Front. Immunol. 2014;5:1–7. doi: 10.3389/fimmu.2014.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Do D.N., Dudemaine P.L., Mathur M., Suravajhala P., Zhao X., Ibeagha-Awemu E.M. miRNA regulatory functions in farm animal diseases, and biomarker potentials for effective therapies. Int. J. Mol. Sci. 2021;22:3080. doi: 10.3390/ijms22063080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pal A., Sharma A., Bhattacharya T.K., Chatterjee P.N., Chakravarty A.K. Molecular characterization and SNP detection of CD14 gene of crossbred cattle. Mol. Biol. Int. 2011:1–13. doi: 10.4061/2011/507346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li D., Liu H., Li Y., Yang M., Qu C., Zhang Y., Liu Y., Zhang X. Identification of suitable endogenous control genes for quantitative RT-PCR analysis of miRNA in bovine solid tissues. Mol. Biol. Rep. 2014;41(10):6475–6480. doi: 10.1007/s11033-014-3530-x. [DOI] [PubMed] [Google Scholar]
- 5.Kang K., Zhang X., Liu H., Wang Z., Zhong J., Huang Z., Peng X., Zeng Y., Wang Y., Yang Y., Luo J., Gou D. A novel real-time PCR assay of microRNAs using S-Poly(T), a specific oligo(dT) reverse transcription primer with excellent sensitivity and specificity. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0048536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin Y., Liu Y., Zhang J., Huang W., Jiang H., Hou Y., Xu C., Zhai C., Gao X., Wang S., Wu Y., Zhu H., Lu S. The expression of miR-375 is associated with carcinogenesis in three subtypes of lung cancer. PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0144187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 8.Alexander C., Rietschel E.T. Bacterial lipopolysaccharides and innate immunity. J. Endotoxin Res. 2001;7(3):167–202. [PubMed] [Google Scholar]
- 9.Naqvi A.R., Zhong S., Dang H., Fordham J.B., Nares S., Khan A. Expression profiling of LPS responsive miRNA in primary human macrophages. J. Microb. Biochem. Technol. 2016;8(2):136–143. doi: 10.4172/1948-5948.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkinson N.J., Buechner-Maxwell V.A., Witonsky S.G., Pleasant R.S., Werre S.R., Ahmed S.A. Characterization of basal and lipopolysaccharide-induced microRNA expression in equine peripheral blood mononuclear cells using Next-Generation Sequencing. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0177664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J., Xu X., Huang X., Zhu H., Chen H., Wang W., Liu Y. Analysis of microRNA expression profiles in porcine PBMCs after LPS stimulation. Innate Immun. 2020;26(5):435–446. doi: 10.1177/1753425920901560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chemonges S., Tung J.P., Fraser J.F. Proteogenomics of selective susceptibility to endotoxin using circulating acute phase biomarkers and bioassay development in sheep: a review. Proteome Sci. 2014;12(1):12. doi: 10.1186/1477-5956-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh J., Mukhopadhyay C.S., Kaur S., Malhotra P., Sethi R.S., Choudhary R.K. Identification of the MicroRNA Repertoire in TLRLig and challenged bubaline PBMCs as a model of bacterial and viral infection. PLoS One. 2016;11(6) doi: 10.1371/journal.pone.0156598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdelmohsen K., Hutchison E.R., Lee E.K., Kuwano Y., Kim M.M., Masuda K., Srikantan S., Subaran S.S., Marasa B.S., Mattson M.P., Gorospe M. miR-375 inhibits differentiation of neurites by lowering HuD levels. Mol. Cell Biol. 2010;17:4197–4210. doi: 10.1128/MCB.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raisch J., Darfeuille-Michaud A., Nguyen H.T. Role of microRNAs in the immune system, inflammation and cancer. World J. Gastroenterol. 2013;19(20):2985–2996. doi: 10.3748/wjg.v19.i20.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romeo P., Colombo C., Granata R., Calareso G., Gualeni A.V., Dugo M., De Cecco L., Rizzetti M.G., Zanframundo A., Aiello A., Carcangiu M.L., Gloghini A., Ferrero S., Licitra L., Greco A., Fugazzola L., Locati L.D., Borrello M.G. Circulating miR-375 as a novel prognostic marker for metastatic medullary thyroid cancer patients. Endocr. Relat. Cancer. 2018;25:217–231. doi: 10.1530/ERC-17-0389. [DOI] [PubMed] [Google Scholar]
- 17.Peterson S.M., Thompson J.A., Ufkin M.L., Sathyanarayana P., Liaw L., Congdon C.B. Common features of microRNA target prediction tools. Front. Genet. 2014;5(23):1–10. doi: 10.3389/fgene.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baum L.G., Cobb B.A. The direct and indirect effects of glycans on immune function. Glycobiology. 2017;27(7):619–624. doi: 10.1093/glycob/cwx036. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y.L., Malik A.B., Sun Y., Hu S., Reynolds A.B., Minshall R.D., Hu G. Innate immune function of the adherens junction protein p120-catenin in endothelial response to endotoxin. J. Immunol. 2011;186(5):3180–3187. doi: 10.4049/jimmunol.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 21.Derynck R., Feng X.H. TGF-beta receptor signaling. Biochim. Biophys. Acta. 1997;1333(2):105–150. doi: 10.1016/s0304-419x(97)00017-6. [DOI] [PubMed] [Google Scholar]
- 22.Afroz R., Kumarapperuma H., Nguyen Q.V.N., Mohamed R., Little P.J., Kamato D. Lipopolysaccharide acting via toll-like receptor 4 transactivates the TGF-β receptor in vascular smooth muscle cells. Cell. Mol. Life Sci. 2022;79(2):121. doi: 10.1007/s00018-022-04159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim E.Y., Kim B.C. Lipopolysaccharide inhibits transforming growth factor-beta1-stimulated Smad6 expression by inducing phosphorylation of the linker region of Smad3 through a TLR4-IRAK1-ERK1/2 pathway. FEBS Lett. 2011;585(5):779–785. doi: 10.1016/j.febslet.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 24.Lim S., Bae E., Kim H.S., Kim T.A., Byun K., Kim B., Hong S., Im J., Yun C., Lee B., Lee B., Hee S., Park S., Letterio J., Kim S. TRAF6 mediates IL-1β/LPS-induced suppression of TGF-β signaling through its interaction with the type III TGF-β receptor. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0032705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharyya S., Kelley K., Melichian D.S., Tamaki Z., Fang F., Su Y., Feng G., Pope R.M., Budinger G.R., Mutlu G.M., Lafyatis R., Radstake T., Feghali-Bostwick C., Varga J. Toll-like receptor 4 signaling augments transforming growth factor-β responses: a novel mechanism for maintaining and amplifying fibrosis in scleroderma. Am. J. Pathol. 2013;182(1):192–205. doi: 10.1016/j.ajpath.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seki E., De Minicis S., Osterreicher C.H., Kluwe J., Osawa Y., Brenner D.A., Schwabe R.F. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat. Med. 2007;13(11):1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- 27.Sanjabi S., Zenewicz L.A., Kamanaka M., Flavell R.A. Anti-inflammatory and pro-inflammatory roles of TGF-beta, IL-10, and IL-22 in immunity and autoimmunity. Curr. Opin. Pharmacol. 2009;9(4):447–453. doi: 10.1016/j.coph.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong C., Flavell R.A. Cell fate decision: T-helper 1 and 2 subsets in immune responses. Arthritis Res. 2000;2(3):179–188. doi: 10.1186/ar85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouyang W., Ranganath S.H., Weindel K., Bhattacharya D., Murphy T.L., Sha W.C., Murphy K.M. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9(5):745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 30.O'Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.