Abstract
Expression of the genes for the membrane-bound Fo sector of the Escherichia coli F1Fo proton-translocating ATPase can respond to changes in metabolic conditions, and these changes are reflected in alterations in the subunit stoichiometry of the oligomeric Fo proton channel. Transcriptional and translational lacZ fusions to the promoter and to two Fo genes show that, during growth on the nonfermentable carbon source succinate, transcription of the operon and translation of uncB, encoding the a subunit of Fo, are higher than during growth on glucose. In contrast, translation of the uncE gene, encoding the c subunit of Fo, is higher during growth on glucose than during growth on succinate. Translation rates of both uncB and uncE change as culture density increases, but transcription rates do not. Quantitation of the c stoichiometry shows that more c subunits are assembled into the F1Fo ATPase in cells grown on glucose than in cells grown on succinate. E. coli therefore appears to have a mechanism for regulating the composition and, presumably, the function of the ATPase in response to metabolic circumstances.
F1Fo ATPases, or F-type ATPases, consist of two large sectors. The Fo sector forms a proton channel across the membrane. The F1 sector is the catalytic sector containing the sites for ATP synthesis or hydrolysis. These enzymes utilize the energy in a transmembrane electrochemical gradient of protons to drive the synthesis of ATP from ADP and Pi. In plants (chloroplasts) and animals (mitochondria), these enzymes operate primarily as ATP synthases. In facultative bacteria such as Escherichia coli, the enzymes can act as either ATP synthases or ATPases. The proton gradient generated by the electron transport chain can be used to drive transport and ATP synthesis. However, the enzyme can also hydrolyze ATP generated from glycolysis and pump protons across the membrane, restoring the proton gradient, which is used for a variety of transport processes (for a review, see reference 6). The enzyme therefore catalyzes a reversible reaction whose direction presumably depends upon the metabolic circumstances. Exactly how the direction of the reaction is determined is not known. There are no known allosteric or covalent activators or inhibitors of the enzyme, which might favor one direction of this reaction over the other. The activity of the mitochondrial F1Fo ATPase is inhibited by a specific inhibitor protein which prevents the enzyme from acting as an ATPase (7), but E. coli does not have an analogous protein, presumably because both ATP hydrolysis and ATP synthesis are essential, depending upon the metabolic conditions.
We have previously demonstrated that the stoichiometry of the c subunit in the purified F1Fo ATPase can vary, depending upon the expression of uncE, the gene encoding the c subunit of the Fo sector (15). We therefore assayed transcription of the unc operon and translation of the first two Fo genes, uncB and uncE, under conditions of growth on glucose minimal and succinate minimal media to determine if the expression of either gene changes significantly when cells are growing on a fermentable or a nonfermentable carbon source. We also measured the effects of those growth conditions on the stoichiometry of the c subunit assembled into the F1Fo complex, to determine if the expression of ATPase genes and the structure of the ATPase can both respond to different growth conditions.
MATERIALS AND METHODS
unc-lac fusion constructions.
The translational uncB′-′lacZ fusions in pDKWH107 and pKS104 and the translational uncE′-′lacZ fusion in pKS105 have been described previously (9, 19). The transcriptional fusion in pWSB56 was constructed for this study by cloning the SspI-BamHI fragment from pKS105 into the promoter-detection plasmid pRZ5255, which carries a trp-lac fusion containing the entire lacZ gene (14). β-Galactosidase activity produced by this construction is dependent upon a cloned promoter, but lacZ translation is initiated from the translational initiation region of lacZ, not the uncB gene.
Assays of β-galactosidase produced by fusion genes.
The fusions were transferred from plasmids into λRZ5 and integrated into the λ att site of MC1000 Unc+, as described previously (19). The resultant single-copy lysogens were grown at 37°C on minimal medium containing 100 mM KPi (pH 7), 2 g of (NH4)2SO4 per liter, 250 mg of l-leucine per liter, 2 mM MgCl2, 200 mg of B1 per liter, and either 4 g of glucose per liter or 8 g of Na-succinate per liter. Samples were removed at values for optical density at 600 nm between 0.2 and 1.2 and assayed for β-galactosidase activity by the Miller assay (12).
Immunoblots.
F1Fo complexes were purified from cells grown on either minimal-glucose medium or minimal-succinate medium, as described by Foster and Fillingame (4), and immunoprecipitated with anti-F1, as described previously (15). This procedure has been shown to precipitate Fo subunits which are associated with F1 subunits, but not Fo subunits alone. Immunoblots were treated overnight with antibody raised against E. coli F1 and Fo subunits and then with secondary antibody consisting of anti-rabbit antibody conjugated with either alkaline phosphatase or horseradish peroxidase. Final development was with the GIBCO/BRL (bromo-chloro-indolylphosphate-p-toluidine salt (BCIP)–nitroblue tetrazolium chloride reagents for colorimetric determinations or with the Amersham ECL chemiluminescent reagents followed by exposure to X-ray film. Both types of blots were photographed, and the bands in the black-and-white photographs were scanned and quantitated with a Hewlett-Packard ScanJet Plus scanner with Scanning Gallery Plus 5.0 densitometry software version 1 from Stratagene. Intensities were adjusted for the different backgrounds apparent in different sections of the photograph. The previous study showed that this method of quantitation produced results linear with amounts of protein loaded in each lane (15).
RESULTS AND DISCUSSION
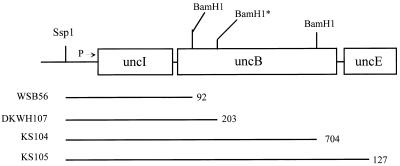
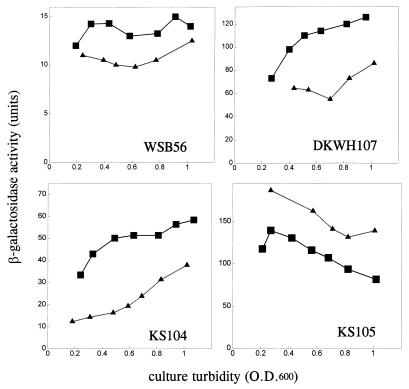
Figure 1 shows the beginning of the unc operon and indicates the sites at which lacZ was fused to unc genes to create either a transcriptional fusion (WSB56) or translational fusions to either uncB (DKWH107 and KS104) or uncE (KS105). Unc+ cells lysogenized with λ phage carrying each of these fusions in single copy were grown on minimal medium containing either glucose or succinate as the sole carbon and energy source. Samples taken at various times were assayed for β-galactosidase activities. Figure 2 shows the β-galactosidase activities produced by these fusions as a function of culture turbidity. A single-copy lysogen carrying the WSB56 transcriptional fusion produced approximately 25% more activity when grown on minimal-succinate medium than when grown on minimal-glucose medium. Lysogens carrying either of the translational fusions to uncB also produced more β-galactosidase activity when grown on succinate than when grown on glucose. A single-copy lysogen carrying the fusion to uncE, however, produced significantly less β-galactosidase activity when cells were grown on minimal-succinate medium than when they were grown on minimal-glucose medium. The expression of both the uncB′-′lacZ fusions increased during growth. The expression of the uncE′-′lacZ fusion gene, however, decreased significantly as culture turbidity increased. The activity of the transcriptional fusion did not change as culture turbidity increased in these experiments. Since the effect of growth on succinate on expression of uncE was the opposite of its effect on both translation of uncB and transcription of the unc operon, these results suggest the existence of some carbon source-dependent regulatory mechanism specific for the expression of uncE.
FIG. 1.
The start of the unc operon indicating the locations of fusions to lacZ. The promoter (P) and the first three genes of the unc operon are indicated with restriction enzyme recognition sites used to make these fusions in plasmids. The translational unB′-′lacZ fusions in pDKWH107 and pKS104 and the translational uncE′-′lacZ fusion in pKS105 have been described previously (9, 19). The transcriptional fusion in pWSB56 was constructed for this study by cloning the SspI-BamHI fragment from the pKS105 fusion into the promoter detection plasmid pRZ5255, which carries a trp-lac fusion containing the entire lacZ gene (14). β-Galactosidase activity produced by this construction is dependent upon a cloned promoter, but lacZ translation is initiated from the translational initiation region of lacZ, not the uncB gene. The horizontal lines indicate the amount of unc DNA cloned either into the transcriptional fusion vector to make the WSB56 fusion or into translational fusion vectors to make the DKWH107, KS104, and KS105 fusions, and the number following each line indicates exactly how many bases of each gene are present in each fusion construction. The BamHI* site is not normally present in uncB but was constructed for the purpose of making the fusion in DKWH107 (9).
FIG. 2.
The fusions described for Fig. 1 were transferred from plasmids into λRZ5, integrated into the λ att site of MC1000 Unc+, and assayed as described in Materials and Methods. The points on these plots are the averages of duplicate assays, which typically varied by less than 5%. The effects of glucose and succinate on promoter activity are shown in the graph of the WSB56 lysogen, effects of carbon source on translation of uncB (a subunit) are shown in the graphs of the DKWH107 and KS104 fusions, and effects of carbon source on translation of uncE (c subunit) are shown in the graph of the KS105 fusion. ▪, succinate-grown cultures; ▴, glucose-grown cultures.
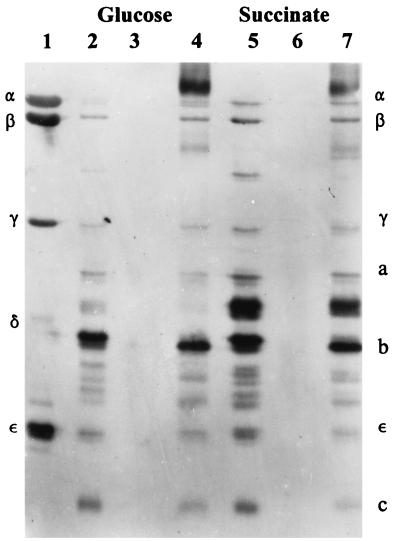
Previous studies showed that a change in the expression of uncE relative to that of other ATPase genes produced a change in the amount of the c subunit assembled into the F1Fo complex (15). We attempted to determine if this carbon source-dependent change in gene expression was also reflected in a change in the composition of the ATPase, specifically in a change in the relative stoichiometry of the c subunit assembled into the ATPase of cells grown on these different carbon sources. We purified the F1Fo complex and immunoprecipitated it with anti-F1. This procedure immunoprecipitates only those Fo subunits assembled into the F1Fo complex (15). Figure 3 shows a blot developed with the Amersham ECL chemiluminescence detection system. The amounts of β, b, ɛ, and c subunits could be easily quantitated from this blot by densitometry. Although the γ and a subunits are visible, they could not be accurately measured because the transfer to nitrocellulose was poor near the center of this blot. The relative amounts of precipitating β, b, and ɛ subunits were the same in these two immunoprecipitates (compare lanes 4 and 7 in Fig. 3), but the relative amount of precipitating c subunit in the F1Fo prepared from glucose-grown cells (lane 4) was significantly higher than the relative amount of precipitating c subunit in the F1Fo prepared from succinate-grown cells (lane 7). In most experiments, we developed the immunoblots with antibody preparations which reacted more strongly to the c subunit than did that used for the blot in Fig. 3, and we could routinely measure only the amounts of the b and c subunits, as we reported previously (15). Table 1 shows the results of three such experiments in which three preparations of F1Fo were prepared from cells grown on Luria-Bertani (LB) medium or on minimal medium containing either glucose or succinate. The amounts of b and c subunits precipitated by anti-F1 antiserum were quantitated, and the c/b ratio was calculated. In all cases, the c/b ratio was higher in F1Fo purified from cells grown on glucose than in F1Fo purified from cells grown on succinate. The b/ɛ ratio did not change in F1Fo preparations purified from cells grown on different media (data not shown), indicating that the change in c/b ratio is probably caused by changes in the c stoichiometry. The relative amount of c assembled into F1Fo is highest in cells grown on minimal-glucose medium, lower in cells grown on minimal-succinate medium, and lowest in cells grown on LB medium, in which the primary source of carbon and energy is amino acids.
FIG. 3.
Immunoblot of F1Fo complexes purified from cells grown on either minimal-glucose medium or minimal-succinate medium and immunoprecipitated with anti-F1. This procedure has been shown to precipitate Fo subunits which are associated with F1 subunits but not Fo subunits alone (15). Lanes: 1, purified F1; 2, F1Fo purified from cells grown on minimal-glucose medium; 3, control (with control serum) immunoprecipitation of glucose F1Fo; 4, immunoprecipitation (with anti-F1 antiserum) of glucose F1Fo; 5, F1Fo purified from cells grown on minimal-succinate medium; 6, control (with control serum) immunoprecipitation of succinate F1Fo; 7, immunoprecipitation (with anti-F1 antiserum) of succinate F1Fo. This blot was developed with dilute preparations of antiserum in order to minimize the background. The overall reactivity of the c subunit is therefore much less than in the experiments described for Table 1.
TABLE 1.
Quantitation of the c/b ratio in immunoprecipitates of F1Fo preparations purified from cells grown on LB, minimal-glucose, or minimal-succinate mediuma
| Expt | Ratio for medium
|
||||
|---|---|---|---|---|---|
| c/b
|
Glucose/succinate | Glucose/LB | |||
| LB | Glucose | Succinate | |||
| 1 | 0.8 | 1.4 | 1.1 | 1.3 | 1.8 |
| 2 | 0.4 | 1.1 | 0.6 | 1.8 | 2.8 |
| 3 | 0.4 | 0.9 | 0.5 | 1.8 | 2.3 |
These data are from three different sets of three different F1Fo preparations immunoprecipitated, immunoblotted, and quantitated as described for Fig. 3. In the first two experiments, the blots were developed with alkaline phosphatase-conjugated secondary antibody and GIBCO/BRL nitroblue tetrazolium-BCIP reagents. In the third experiment, the blot was developed with horseradish peroxidase-conjugated secondary antibody followed by the Amersham ECL chemiluminescence reagents. The first three columns show the c/b ratios calculated from these quantitations. The variations in the absolute numbers result from different dilutions of antibody and different incubation times for antibody treatments. These differ from experiment to experiment but are the same for the three preparations from any given experiment. The last two columns show the ratio of c/b ratios from the glucose and succinate preparations and from the glucose and LB preparations. Other data (see Results) indicate that the relative amount of b is constant in each preparation; these last two columns, therefore, show the differences in c stoichiometries between F1Fo preparations.
These measurements demonstrate that both gene expression and subunit stoichiometry of Fo can change depending on carbon source. Although the exact mechanism responsible for altering the synthesis and assembly of c is not addressed in these studies, the uncE gene is preceded by an unusual translational enhancer sequence which could serve as a target for regulation (11). It appears as if function as an ATP synthase is correlated with a lower c stoichiometry, and function as a proton pump is correlated with a higher c stoichiometry. In our previous studies demonstrating that a change in expression of uncE produces a change in c stoichiometry, the complexes containing more c subunits were poor ATP synthases compared to the complexes containing fewer c subunits, despite the fact that membranes containing either type of complex had nearly identical ATPase activities and ATP-dependent proton pumping activities (15, 18).
In mitochondria and chloroplasts, organelles involved in aerobic metabolism, the stoichiometry of the c subunit has been shown elsewhere to be 6 (13, 16, 17). Presumably, these ATPases function primarily, if not exclusively, as ATP synthases. In the vacuolar ATPase of Saccharomyces cerevisiae, which acts exclusively as a proton pump, the stoichiometry of the N,N′-dicyclohexylcarbodiimide-reactive membrane sector component is also 6 (1), but the subunit size is doubled. In bacteria, plants, and animals, the c subunit consists of two transmembrane helices. For the vacuolar ATPase, the homologous protein is twice the size and consists of four transmembrane helices. Therefore, the vacuolar ATPase proton pump has twice the number of transmembrane helices in its “c subunit” as do the ATPases which act as ATP synthases. Additionally, in certain systems, the membrane-bound sector of the vacuolar ATPase appears to change size in response to salinity stress, and this size change is accompanied by increases in the mRNA and protein levels of subunit c. Lüttge and Ratajczak (10) have interpreted these results as indication that the c stoichiometry of these vacuolar ATPases increases under stress. In E. coli, the two most careful studies on c stoichiometry in the ATPase purified from cells grown on glucose produced values of 10 ± 1 (5) or a range of 8 to 14 (8). It may be that these F1Fo preparations consist of a mixture of complexes which contain a range of c stoichiometries. Our data on gene expression, together with the biochemical studies on how much c is actually assembled into Fo, demonstrate that the c stoichiometry in the ATPase purified from cells grown on succinate is lower than that found for cells grown on glucose.
The possible advantages to a facultative organism of being able to change the c stoichiometry of the ATPase.
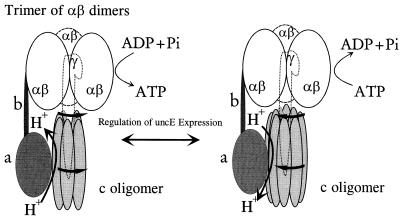
Figure 4 shows the model for rotational catalysis described by Duncan et al. (3). The proton motive force drives a flow of protons through an interface in Fo between the a subunit and the c oligomer and produces rotation of the c oligomer. As each c subunit moves into and out of functional contact with the a subunit to form the proton channel, this rotation is transmitted by the γ subunit to the αβ hexamer, illustrated as a trimer of catalytic αβ dimers. In this model, the energy of the proton motive force is transmitted into the cellular phosphorylation potential ΔGp in a stepwise cogged fashion. Since each full turn of the complex produces three molecules of ATP, the H+/ATP ratio will depend upon the ratio of c subunits to catalytic sites (i.e., c stoichiometry/3). The presence of more c subunits would produce a higher H+/ATP stoichiometry and the presence of fewer c subunits would produce a lower H+/ATP stoichiometry.
FIG. 4.
Possible effect of a variable c stoichiometry. This figure shows the model for rotational catalysis as described by Duncan et al. (3). The Fo subunits a, b, and c are shown, as is the F1 γ subunit. The α and β subunits are depicted as a trimer of αβ dimers. The rotation of the c oligomer in response to a proton motive force produces rotation of the γ subunit within the trimer of αβ dimers. If the cogging of the c oligomer into and out of contact with the a subunit to form the actual proton channel is rate limiting, then, for a given proton motive force, the rate of ATP synthesis or hydrolysis will depend on the size of the c oligomer. Lower stoichiometries would produce higher rates.
If this model is correct, the rate-limiting step in energy coupling is probably the time it takes one c subunit to move one such cog during rotation, since physical rotation of the c oligomer is probably slower than either transmembrane movement of a proton through the channel or substrate binding, catalysis, or release. If each step in rotation is rate limiting, there might not be a significant difference in proton translocation rates of membranes carrying ATPases with different c stoichiometries. For a given proton motive force, however, the rate of rotation of the entire c oligomer, and therefore of the γ subunit, would increase for smaller c stoichiometries and decrease for larger c stoichiometries. During growth on succinate, a decrease in the number of c subunits would decrease the H+/ATP ratio and therefore decrease the extent of ΔGp which could be created by a given Δp, but because the c oligomer would be smaller, rotation would be faster and the rate of ATP synthesis would be increased. A higher c stoichiometry and H+/ATP ratio would theoretically produce a higher ΔGp for a given Δp, but the speed of rotation and therefore of ATP synthesis might then be too slow to keep up with cellular energy demands. We have shown that the enzyme carrying more c subunits synthesizes ATP more slowly than the enzyme carrying fewer c subunits (15, 18). When the enzyme is hydrolyzing ATP to pump protons, an increase in the number of c subunits, and the resulting increase in H+/ATP ratio, would decrease the magnitude of Δp which could be generated by a given ΔGp but might then minimize the rate of ATP hydrolysis and the resultant ATP depletion. Therefore, if this model is accurate, E. coli adjusts the c stoichiometry of the ATPase to maximize the rate of ATP synthesis during oxidative phosphorylation and to minimize the rate of ATP hydrolysis during proton pumping, at the expense of overall energy coupling efficiency. In 1990, Cross and Taiz (2) proposed that the structure of ATPases had evolved to adjust H+/ATPase ratios in order to maximize the efficiency of energy coupling. That model describes evolutionary changes, and our model describes metabolic regulation. The two conclusions are therefore not necessarily incompatible.
ACKNOWLEDGMENT
This research was supported by American Heart Association grant-in-aid 93007630.
REFERENCES
- 1.Arai H, Terres G, Pink S, Forgac M J. Topography and subunit stoichiometry of the coated vesicle proton pump. J Biol Chem. 1988;263:8796–8802. [PubMed] [Google Scholar]
- 2.Cross R L, Taiz L. Gene duplication as a means for altering H+/ATP ratios during the evolution of FoF1 ATPases and synthases. FEBS Lett. 1990;259:227–229. doi: 10.1016/0014-5793(90)80014-a. [DOI] [PubMed] [Google Scholar]
- 3.Duncan T M, Bulygin V V, Zhou Y, Hutcheon M L, Cross R L. Rotation of subunits during catalysis by Escherichia coli F1 ATPase. Proc Natl Acad Sci USA. 1995;9:10964–10968. doi: 10.1073/pnas.92.24.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster D L, Fillingame R H. Energy-transducing H+-ATPase of Escherichia coli. J Biol Chem. 1979;254:8230–8236. [PubMed] [Google Scholar]
- 5.Foster D L, Fillingame R H. Stoichiometry of subunits in the H+-ATPase complex of Escherichia coli. J Biol Chem. 1982;257:2009–2015. [PubMed] [Google Scholar]
- 6.Harold F M, Maloney P C. Energy transduction by ion currents. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 283–306. [Google Scholar]
- 7.Harris D A, Das A M. Control of mitochondrial ATP synthesis in the heart. Biochem J. 1991;280:561–573. doi: 10.1042/bj2800561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hermolin J, Fillingame R H. H+-ATPase activity of Escherichia coli F1Fo is blocked after reaction of dicyclohexylcarbodiimide with a single proteolipid (subunit c) of the Fo complex. J Biol Chem. 1989;264:3896–3903. [PubMed] [Google Scholar]
- 9.Hsu D K W, Brusilow W S A. Effects of the uncI gene on expression of uncB, the gene coding for the a subunit of the F1Fo ATPase of Escherichia coli. FEBS Lett. 1995;371:127–151. doi: 10.1016/0014-5793(95)00867-9. [DOI] [PubMed] [Google Scholar]
- 10.Lüttge U, Ratajczak R. The physiology, biochemistry, and molecular biology of the plant vacuolar ATPase. Adv Bot Res. 1997;25:253–296. [Google Scholar]
- 11.McCarthy J E G, Schairer H U, Sebald W. Translational initiation frequency of atp genes from Escherichia coli: identification of an intercistronic sequence that enhances translation. EMBO J. 1985;4:519–526. doi: 10.1002/j.1460-2075.1985.tb03659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 13.Nelson N, Eytan E, Notsani B, Sigrist H, Sigrist-Nelson K, Gitler C. Isolation of a chloroplast N,N′-dicyclohexylcarbodiimide-binding proteolipid, active in proton translocation. Proc Natl Acad Sci USA. 1977;74:2375–2378. doi: 10.1073/pnas.74.6.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porter A C G, Brusilow W S A, Simoni R D. Promoter for the unc operon of Escherichia coli. J Bacteriol. 1983;155:1271–1278. doi: 10.1128/jb.155.3.1271-1278.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schemidt T A, Hsu D K W, Deckers-Hebestreit G, Altendorf K, Brusilow W S A. The effects of an atpE ribosome-binding site mutation on the stoichiometry of the c subunit in the F1Fo ATPase of Escherichia coli. Arch Biochem Biophys. 1995;323:423–428. doi: 10.1006/abbi.1995.0063. [DOI] [PubMed] [Google Scholar]
- 16.Sebald W, Graf T, Lukins H B. The dicyclohexylcarbodiimide-binding protein of the mitochondrial ATPase complex from Neurospora crassa and Saccharomyces cerevisiae. Eur J Biochem. 1979;93:587–599. doi: 10.1111/j.1432-1033.1979.tb12859.x. [DOI] [PubMed] [Google Scholar]
- 17.Sigrist-Nelson K, Sigrist H, Azzi A. Characterization of the dicyclohexylcarbodiimide-binding protein isolated from chloroplast membranes. Eur J Biochem. 1978;92:9–14. doi: 10.1111/j.1432-1033.1978.tb12717.x. [DOI] [PubMed] [Google Scholar]
- 18.Solomon K A, Brusilow W S A. Effect of an uncE ribosome-binding site mutation on the synthesis and assembly of the Escherichia coli proton-translocating ATPase. J Biol Chem. 1988;263:5402–5407. [PubMed] [Google Scholar]
- 19.Solomon K A, Hsu D K W, Brusilow W S A. Use of lacZ fusions to measure in vivo expression of the first three genes of the Escherichia coli unc operon. J Bacteriol. 1989;171:3039–3045. doi: 10.1128/jb.171.6.3039-3045.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]