Abstract
We introduce a strategy for learning image registration without acquired imaging data, producing powerful networks agnostic to magnetic resonance imaging (MRI) contrast. While classical methods accurately estimate the spatial correspondence between images, they solve an optimization problem for every new image pair. Learning methods are fast at test time but limited to images with contrasts and geometric content similar to those seen during training. We propose to remove this dependency using a generative strategy that exposes networks to a wide range of images synthesized from segmentations during training, forcing them to generalize across contrasts. We show that networks trained within this framework generalize to a broad array of unseen MRI contrasts and surpass classical state-of-the-art brain registration accuracy by up to 12.4 Dice points for a variety of tested contrast combinations. Critically, training on arbitrary shapes synthesized from noise distributions results in competitive performance, removing the dependency on acquired data of any kind. Additionally, since anatomical label maps are often available for the anatomy of interest, we show that synthesizing images from these dramatically boosts performance, while still avoiding the need for real intensity images during training.
Keywords: Deformable registration, MRI-contrast independence, deep learning without data, image synthesis
1. INTRODUCTION
Image registration estimates spatial correspondences between images and is a fundamental component of many neuroimaging pipelines [1-3]. This work focuses on networks agnostic to magnetic resonance imaging (MRI) contrast that excel both at same-contrast registration (e.g. between two T1-weighted scans, T1w), as well as across contrasts (e.g. T1w to T2-weighted, T2w). Both are important in neuroimaging, where different contrasts are commonly acquired, such as T1w for visualizing anatomy or T2w contrast for detecting abnormal fluids [4]. These scans yield images of different appearance for the same anatomy. For morphometric analyses [1-3], images often need to be registered to an existing atlas, which is typically of a different contrast [5]. Classical methods estimate a deformation field by optimizing an objective that balances image similarity with field regularity [6-10]. While these methods provide strong theory and good results, the optimization needs to be repeated for each image pair, and the objective and optimization have to be adapted to the image type. In contrast, learning methods learn a function that maps an image pair to a deformation field from datasets [11-16]. These methods are fast and have the potential to improve accuracy and robustness to local minima. Unfortunately, learning approaches do not generalize well to new image types unobserved at training. For example, a model trained on T1w-T1w pairs will not accurately register proton-density weighted (PDw) to T1w scans.
Contribution
We propose SynthMorph: a general strategy for learning contrast agnostic registration that can handle unseen MRI contrasts at test time. SynthMorph enables registration within and across contrasts, without the need for real imaging data at training. We first introduce a generative model for label maps of random geometric shape. Second, we generate images of arbitrary contrast conditioned on these maps. Third, the strategy enables us to use a contrast-agnostic loss that measures label overlap. This leads to two network variants with unprecedented generalizability, that register any contrast combination tested without retraining: sm-shapes trains without acquired data of any kind and matches state-of-the-art registration of neuroanatomical MRI, sm-brains trains on images synthesized from brain segmentations only and substantially outperforms all other methods tested. We emphasize that neither variant requires real intensity images.
2. METHOD
Background
Let and be a moving and a fixed 3D image, respectively. We build on unsupervised learning frameworks for non-linear registration: a U-Net-style [17] CNN with parameters implementing the VoxelMorph architecture [11,18] outputs the deformation for image pair {, }. At each training iteration, is given images {, }, and network parameters are updated to minimize a loss that contains a dissimilarity term and a regularization term , where is the network output, and is a constant.
Proposed method overview
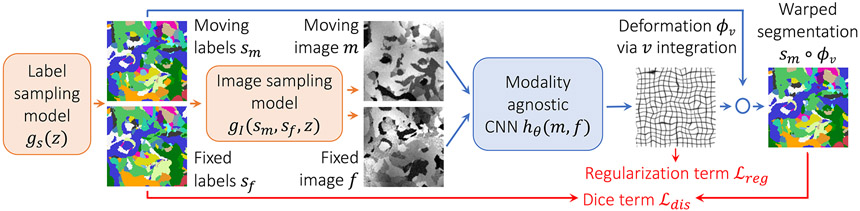
We eliminate the need for acquired training data by synthesizing arbitrary shapes and contrasts from scratch (Figure 1). We generate two paired 3D label maps {, } using a function described below given random seed . However, if anatomical labels are available, generating images from these can further improve accuracy, and we can use these instead of synthesizing segmentations. We then define another function described below that synthesizes two intensity volumes {, } from {, } and . Crucially, synthesizing images from label maps enables us to obviate the dependency of the loss on image contrast and instead use a similarity function that measures label overlap using (soft) Dice [19]:
| (1) |
where is the one-hot encoded label of label map , and and denote voxel-wise multiplication and addition, respectively. We parameterize the deformation with a stationary velocity field (SVF) , which we integrate within the network to obtain a diffeomorphism [6,18,20]. We regularize using , where is the displacement of deformation field .
Fig. 1.
Strategy for learning contrast-agnostic deformable registration. At every mini batch, we synthesize a pair of 3D label maps {, } and then the corresponding 3D images {, } with random contrast. The images are used to train a U-Net-style network, and the label maps are incorporated into a loss that is independent of image contrast.
Label maps
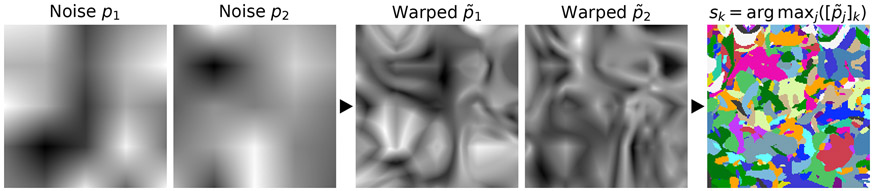
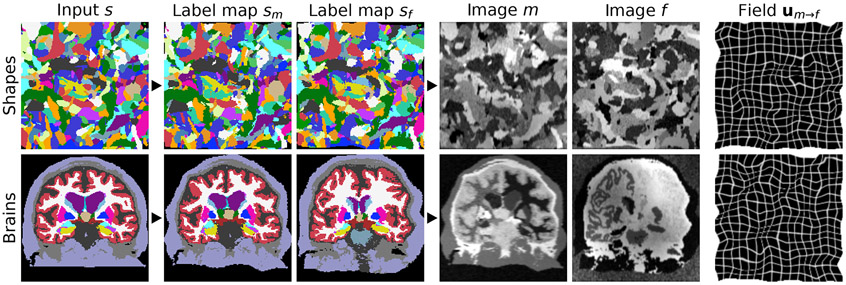
To generate random input label maps with labels, we first draw smooth noise images by sampling voxels from a normal distribution at lower resolution and upsampling (Figure 2). Second, we warp each image with a random deformation field (see below) to obtain images . Third, we create an input label map by assigning, for each voxel of , the label with the highest intensity among the images , i.e. . Given label map , we generate two new label maps by deforming with a random smooth deformation (see below) to produce the moving map and similarly the fixed map . Alternatively, if segmentations are available for the anatomy of interest, such as the brain, we deform two label maps from separate subjects, selected at random, and only synthesize the images. We emphasize that no acquired images (e.g. a T1w MRI) are used during training (Figure 3).
Fig. 2.
Generation of input label maps. A set of smooth 3D noise images is sampled from a standard distribution, then warped by random deformation fields . The label map is synthesized from the warped images : for each voxel of , we determine in which that voxel has the highest intensity and assign the corresponding label , i.e. . The example uses .
Fig. 3.
Data synthesis. Top: from random shapes. Bottom: if available, the synthesis can be initialized with anatomical labels. We generate a pair of label maps {, } and from them images {, } with arbitrary contrast. The registration network then predicts the displacement . If anatomical labels are used, we generate {, } from separate subjects.
Synthetic images
From the label maps {, }, we synthesize gray-scale images {, } [21-23], building on our recent work on segmentation [24]. Given a label map , we draw the intensities of all image voxels for label from the normal distribution . We sample the mean and standard deviation (SD) from continuous distributions and , respectively, where , , , and are hyperparameters. We convolve the image with a Gaussian kernel where . We further corrupt the image with an intensity bias field [25,26], whose voxels we draw from a normal distribution at lower resolution , with . We upsample and take the exponential of each voxel to yield positive values before applying via voxel-wise multiplication. We obtain the images {, } after min-max normalization and voxel-wise contrast exponentiation using parameter , where is the normalized moving image (similarly for the fixed image).
Random transforms
We obtain the transforms for noise image by integrating random SVFs [6,18,20,27]. We draw each voxel of as an independent sample of a normal distribution at reduced resolution , where , and each SVF is integrated and upsampled to full size. We obtain the transforms and similarly, based on hyperparameters and .
3. EXPERIMENTS
We evaluate network variants trained with the proposed strategy and compare to several baselines. SynthMorph achieves unprecedented generalizability among neural networks, matching or exceeding the accuracy of all classical and learning baselines tested.
Data
We test cross-subject registration of 3D brain-MRI scans compiled from the HCP-A [28, 29] and OASIS [30] datasets, which include T1w and T2w acquisitions at 1.5T and 3T with resolution. We also use BIRN [31] PDw scans from 8 subjects. For training sm-brains, we obtain 40 distinct-subject segmentations from the Buckner40 dataset [32], a subset of the fMRIDC structural data [33]. We derive brain and non-brain labels using SAMSEG [3], except for the PDw data, which include manual brain label maps. As we focus on deformable registration, we map all images to a common affine space [1,34] at 1 mm isotropic resolution.
Setup
For each contrast, we test the networks on 30 image pairs, except for PDw, of which we have only 8. We test registration within and across datasets, with and without skull-stripping, using held-out datasets of the same size. To measure registration accuracy, we propagate the moving labels using the predicted warps and compute the Dice metric across the largest brain labels.
Baselines
We test classical registration with ANTs (SyN) [7] using recommended parameters [35] since these methods are already optimized for brain MRI, with the NCC metric within and MI across contrasts. We test NiftyReg [8] with NMI and diffeomorphic transforms as in our approach. We also run deedsBCV [10], where we reduce the grid spacing, search radius and quantization step to improve accuracy in our experiments. As a learning baseline, we train VoxelMorph (vm), using an NCC-based loss and the same architecture as SynthMorph, on 100 skull-stripped T1w images from HCP-A that do not overlap with the validation set. Similarly, we train a model with an NMI-based loss on combinations of 100 T1w and 100 T2w images.
SynthMorph variants
For contrast and shape agnostic training, we generate {} from one of 100 random-shape segmentations at each iteration (sm-shapes). Each contains labels, all included in the loss . We train another network on the Buckner40 anatomical labels instead of shapes (sm-brains) and optimize the largest brain labels in We emphasize that no acquired intensity images are used.
Analyses
We conduct extensive hyperparameter analyses (not shown due to space limitations) using skull-stripped HCP-A T1w pairs that do not overlap with the test set and select the values shown in Table 1. Accuracy is mildly sensitive to regularization, and we choose . Using higher numbers of convolutional features per layer boosts accuracy; we choose a network width of .
Table 1.
Hyperparameters. Spatial measures are given in voxels. Our data are 160×160×192 volumes. For fields sampled at a lower resolution , we obtain the volume size by multiplying each dimension by and rounding up (e.g. 4×4×5 for ).
| Hyperparameter | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Value | 1 | 1:32 | 100 | 25 | 225 | 5 | 25 | 1:40 | 0.3 | 1 | 0.25 | 1:16 | 3 |
Results
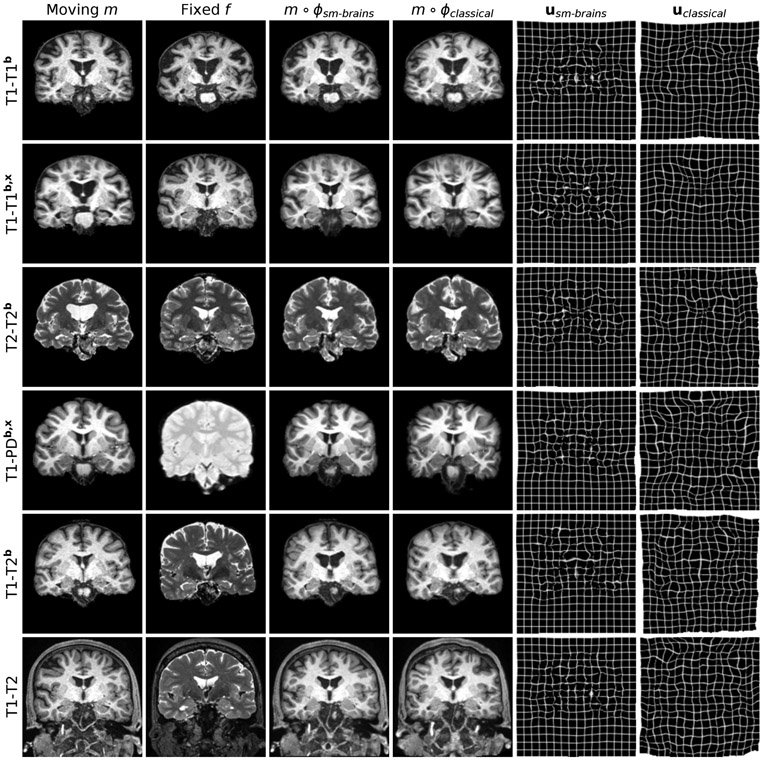
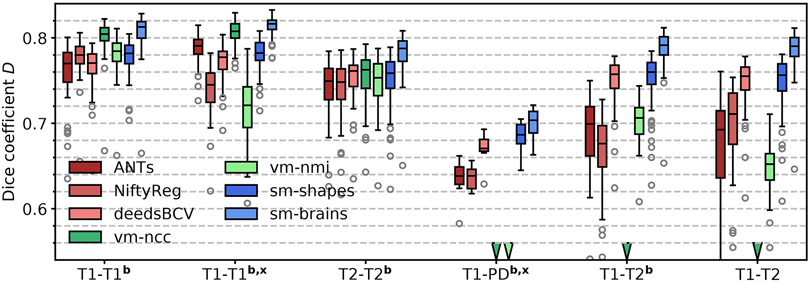
We show typical examples in Figure 4. Figure 5 compares Dice scores across structures. Exploiting the information in a set of brain labels, sm-brains achieves the highest accuracy throughout, even though no real images are used for training: for T1w-T2w/PDw pairs, sm-brains leads by at least 3 Dice points. The shape and contrast-agnostic network sm-shapes matches the best classical methods at same-contrast and exceeds all methods except sm-brains at cross-contrast registration, despite never having seen real MR images nor brain anatomy. The learning baselines perform well but break down for unseen contrast pairs, while SynthMorph remains robust. The learning methods take less than 1 second per 3D registration on an Nvidia Tesla V100 GPU, whereas NiftyReg, ANTs and deedsBCV take ~0.5 h, ~1.2 h and ~3 min on a 3.3-GHz Intel Xeon CPU, respectively (single-threaded).
Fig. 4.
Typical results for sm-brains and classical methods. Each row shows a pair from the datasets indicated on the left, where the letters b and x mark skull-stripping and registration across datasets (e.g. OASIS-HCP), respectively. We show the best-performing classical baseline: NiftyReg on the 1st, ANTs on the 2nd, and deedsBCV on all other rows.
Fig. 5.
Registration accuracy: overlap of brain structures for 30 subject pairs (8 for PD). The letters b and x indicate skull-stripping and registration across datasets (e.g. OASIS-HCP).
4. CONCLUSION
We introduce a general framework for learning registration that does not require any real imaging data during training. Instead, we synthesize random label maps and images with widely varying shape and contrast from noise distributions. This strategy produces powerful contrast-agnostic networks that surpass classical accuracy at all contrasts tested, while being substantially faster. Learning methods like VoxelMorph yield results similar to SynthMorph for contrasts observed at training but break down for unseen (new) image types. In contrast, networks trained in the SynthMorph framework remain robust, obviating the need for retraining given a new sequence. In the case where (few) brain segmentations are available, training on images synthesized from these produces networks that substantially outperform the state of the art.
5. COMPLIANCE WITH ETHICAL STANDARDS
This retrospective research study uses human subject data. Analysis of locally acquired HCP-A data was last approved on 2020-12-22 by IRBs at Washington University in St. Louis (201603117) and Mass General Brigham (2016p001689). No ethical approval was required for retrospective analysis of open access data from OASIS, BIRN and fMRIDC.
6. ACKNOWLEDGMENTS
This research is supported by ERC StG 677697 and NIH grants NICHD K99 HD101553, NIA R56 AG064027, R01 AG064027, AG008122, AG016495, NIBIB P41 EB015896, R01 EB023281, EB006758, EB019956, R21 EB018907, NIDDK R21 DK10827701, as well as grants NINDS R01 NS0525851, NS070963, NS105820, NS083534, and R21 NS072652, U01 NS086625, U24 NS10059103, SIG S10 RR023401, RR019307, RR023043, BICCN U01 MH117023, Blueprint for Neuroscience Research U01 MH093765.
Data are provided in part by OASIS Cross-Sectional (PIs D. Marcus, R. Buckner, J. Csernansky, J. Morris; grants P50 AG05681, P01 AG03991, AG026276, R01 AG021910, P20 MH071616, U24 R021382). HCP-A: Research reported in this publication is supported by grants U01 AG052564 and AG052564-S1 and by the 14 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research, by the McDonnell Center for Systems Neuroscience at Washington University, by the Office of the Provost at Washington University, and by the University of Minnesota Medical School.
Footnotes
BF has a financial interest in CorticoMetrics, a company focusing on brain imaging and measurement technologies. BF’s interests are reviewed and managed by Massachusetts General Hospital and Mass General Brigham in accordance with their conflict of interest policies. The authors have no other relevant financial or non-financial interests to disclose.
7. REFERENCES
- [1].Fischl Bruce, “Freesurfer,” Neuroimage, vol. 62, no. 2, pp. 774–81, 2012, 20 YEARS OF fMRI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Frackowiak RSJ et al. , Human Brain Function, Aca-demic Press, 2nd edition, 2003. [Google Scholar]
- [3].Puonti Oula, Iglesias Juan Eugenio, and Van Leemput Koen, “Fast and sequence-adaptive whole-brain segmentation using parametric bayesian modeling,” NeuroImage, vol. 143, pp. 235–49, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].McRobbie Donald W. et al. , MRI from Picture to Proton, Cambridge university press, 2017. [Google Scholar]
- [5].Sridharan Ramesh et al. , “Quantification and analysis of large multimodal clinical image studies: Application to stroke,” in International Workshop on Multimodal Brain Image Analysis. Springer, 2013, pp. 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ashburner John, “A fast diffeomorphic image registra-tion algorithm,” Neuroimage, vol. 38, no. 1, pp. 95–113, 2007. [DOI] [PubMed] [Google Scholar]
- [7].Avants BB et al. , “Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain,” Medical Image Analysis, vol. 12, no. 1, pp. 26–41, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Modat Marc et al. , “Fast free-form deformation using graphics processing units,” Computer Methods and Programs in Biomedicine, vol. 98, no. 3, pp. 278–84, 2010, HP-MICCAI 2008. [DOI] [PubMed] [Google Scholar]
- [9].Rueckert Daniel et al. , “Nonrigid registration using free-form deformations: application to breast mr images,” IEEE TMI, vol. 18, no. 8, pp. 712–21, 1999. [DOI] [PubMed] [Google Scholar]
- [10].Heinrich Mattias P. et al. , “Mrf-based deformable registration and ventilation estimation of lung ct,” IEEE TMI, vol. 32, no. 7, pp. 1239–1248, 2013. [DOI] [PubMed] [Google Scholar]
- [11].Balakrishnan Guha et al. , “Voxelmorph: A learning framework for deformable medical image registration,” IEEE Trans. Med. Im, vol. 38, no. 8, pp. 1788–00, 2019. [DOI] [PubMed] [Google Scholar]
- [12].de Vos Bob D. et al. , “End-to-end unsupervised deformable image registration with a convolutional neural network,” in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, pp. 204–12. Springer, 2017. [Google Scholar]
- [13].Li Hongming and Fan Yong, “Non-rigid image registration using fully convolutional networks with deep self-supervision,” arXiv preprint arXiv:1709.00799, 2017. [Google Scholar]
- [14].Rohé Marc et al. , “SVF-Net: Learning deformable image registration using shape matching,” MICCAI, pp. 266–74, 2017. [Google Scholar]
- [15].Wu Guorong et al. , “Scalable high-performance image registration framework by unsupervised deep feature representations learning,” IEEE TBME, vol. 63, no. 7, pp. 1505–16, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yang Xiao et al. , “Quicksilver: Fast predictive image registration–a deep learning approach,” NeuroImage, vol. 158, pp. 378–96, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ronneberger Olaf, Fischer Philipp, and Brox Thomas, “U-net: Convolutional networks for biomedical image segmentation,” CoRR, vol. abs/1505.04597, 2015. [Google Scholar]
- [18].Dalca Adrian V. et al. , “Unsupervised learning of probabilistic diffeomorphic registration for images and surfaces,” Medical Image Analysis, vol. 57, pp. 226–36, 2019. [DOI] [PubMed] [Google Scholar]
- [19].Milletari Fausto, Navab Nassir, and Ahmadi Seyed-Ahmad, “V-net: Fully convolutional neural networks for volumetric medical image segmentation,” in 2016 3DV, 2016, pp. 565–71. [Google Scholar]
- [20].Arsigny Vincent et al. , “A log-euclidean framework for statistics on diffeomorphisms,” in MICCAI. Springer, 2006, pp. 924–31. [DOI] [PubMed] [Google Scholar]
- [21].Ashburner John and Friston Karl J., “Unified segmentation,” NeuroImage, vol. 26, pp. 839–51, 2005. [DOI] [PubMed] [Google Scholar]
- [22].Van Leemput Koen et al. , “Automated model-based tissue classification of mr images of the brain,” IEEE TMI, vol. 18, pp. 897–908, 1999. [DOI] [PubMed] [Google Scholar]
- [23].Wells William M. et al. , “Adaptive segmentation of MRI data,” IEEE TMI, vol. 15, no. 4, pp. 429–42, 1996. [DOI] [PubMed] [Google Scholar]
- [24].Billot Benjamin et al. , “A learning strategy for contrast-agnostic mri segmentation,” in PMLR, Montreal, QC, Canada, 06–08 Jul 2020, vol. 121 of Proceedings of Machine Learning Research, pp. 75–93. [Google Scholar]
- [25].Belaroussi Boubakeur et al. , “Intensity non-uniformity correction in MRI: existing methods and their validation,” Medical image analysis, vol. 10, no. 2, pp. 234–46, 2006. [DOI] [PubMed] [Google Scholar]
- [26].Hou Zujun, “A review on mr image intensity inhomogeneity correction,” International journal of biomedical imaging, vol. 2006, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Krebs Julian et al. , “Learning a probabilistic model for diffeomorphic registration,” IEEE TMI, vol. 38, no. 9, pp. 2165–176, 2019. [DOI] [PubMed] [Google Scholar]
- [28].Bookheimer Susan Y. et al. , “The Lifespan Human Connectome Project in Aging: An overview,” NeuroImage, vol. 185, pp. 335–48, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Harms Michael P. et al. , “Extending the human connectome project across ages: Imaging protocols for the lifespan development and aging projects,” NeuroImage, vol. 183, pp. 972–84, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Marcus Daniel S. et al. , “Open access series of imaging studies (oasis): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults,” Journal of cognitive neuroscience, vol. 19, no. 9, pp. 1498–507, 2007. [DOI] [PubMed] [Google Scholar]
- [31].Keator David B et al. , “A national human neuroimaging collaboratory enabled by the biomedical informatics research network (BIRN),” IEEE TITB, vol. 12, no. 2, pp. 162–172, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fischl Bruce et al. , “Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain,” Neuron, vol. 33, no. 3, pp. 341–355, 2002. [DOI] [PubMed] [Google Scholar]
- [33].Van Horn John D et al. , “The functional magnetic resonance imaging data center (fMRIDC): the challenges and rewards of large–scale databasing of neuroimaging studies,” Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences, vol. 356, no. 1412, pp. 1323–1339, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Reuter Martin et al. , “Highly accurate inverse consistent registration: A robust approach,” NeuroImage, vol. 53, no. 4, pp. 1181–96, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pustina Dorian and Cook Philip, “Anatomy of an antsRegistration call,” 2017. [Google Scholar]