Abstract
Pseudomonas aeruginosa a-type strains produce flagellin proteins which vary in molecular weight between strains. To compare the properties of a-type flagellins, the flagellin genes of several Pseudomonas aeruginosa a-type strains, as determined by interaction with specific anti-a monoclonal antibody, were cloned and sequenced. PCR amplification of the a-type flagellin gene fragments from five strains each yielded a 1.02-kb product, indicating that the gene size is not likely to be responsible for the observed molecular weight differences among the a-type strains. The flagellin amino acid sequences of several a-type strains (170018, 5933, 5939, and PAK) were compared, and that of 170018 was compared with that of PAO1, a b-type strain. The former comparisons revealed that a-type strains are similar in amino acid sequence, while the latter comparison revealed differences between 170018 and PAO1. Posttranslational modification was explored for its contribution to the observed differences in molecular weight among the a-type strains. A biotin-hydrazide glycosylation assay was performed on the flagellins of three a-type strains (170018, 5933, and 5939) and one b-type strain (M2), revealing a positive glycosylation reaction for strains 5933 and 5939 and a negative reaction for 170018 and M2. Deglycosylation of the flagellin proteins with trifluoromethanesulfonic acid (TFMS) confirmed the glycosylation results. A molecular weight shift was observed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis for the TFMS-treated flagellins of 5933 and 5939. These results indicate that the molecular weight discrepancies observed for the a-type flagellins can be attributed, at least in part, to glycosylation of the protein. Anti-a flagellin monoclonal antibody reacted with the TFMS-treated flagellins, suggesting that the glycosyl groups are not a necessary component of the epitope for the human anti-a monoclonal antibody. Comparisons between a-type sequences and a b-type sequence (PAO1) will aid in delineation of the epitope for this monoclonal antibody.
Pseudomonas aeruginosa is motile via a single polar flagellum. The flagellar filament of P. aeruginosa is made up of a single protein called flagellin. The flagellin protein can be classified into one of two major types: a-type or b-type flagellin. These classifications are made based on reactions with specific polyclonal antibodies (4, 31) and molecular weight (1). a-type flagellins are a heterologous group of proteins, with molecular masses ranging from 45 to 52 kDa, whereas b-type flagellins are a homologous group of proteins which all have a molecular mass of 53 kDa. a-type flagellins can be classified into subtypes based on H-antigenic components. All a-type flagellins were reported to have an a0 common cross-reacting antigenic component and usually in addition have one or more additional antigenic components (1, 4) which play at most a minor role in inheriting antigenicity. The a-type H-antigen flagellin subtypes (in parentheses) and molecular masses for several representative strains are as follows: for strain 5933 (a0, a1, and a2), 51 kDa; for strain 5939 (a0 and a3), 52 kDa; and for strain 170018 (a0, a3, and a4), 45 kDa (1).
Comparisons of the nucleotide and amino acid sequences of flagellins from different species have shown the N and C termini to be highly conserved and the central hypervariable region to be less conserved (7, 28, 44). For example, in Aquifex pyrophilus, the deduced primary structure of the 501-amino-acid flagellin protein revealed conserved N- and C-terminal regions and a variable central domain of 246 residues compared to other bacterial flagellins (9). A comparison of the amino acid sequence of the flagellin N terminus of Pseudomonas pseudomallei with those of Proteus mirabilis, Bordetella bronchiseptica, and P. aeruginosa PAK revealed significant homology (10). The deduced amino acid sequence of the P. aeruginosa PAK flagellin also revealed conservation in the N- and C-terminal domains compared with the amino acid sequences of other flagellins (41). Similar results were observed with Pseudomonas putida (45). Comparison of 12 P. aeruginosa a-type fliC sequences revealed that the N- and C-terminal residues were nearly identical (39).
Evidence suggests that the N- and C-terminal domains may be responsible for similar functions in all flagellate bacteria. These functions include export of flagellin and assembly of flagella (21, 28, 41). Most of the structural and functional features of flagella are determined by the N- and C-terminal conserved regions, whereas the antigenic or serological variation is found in the central portion of the flagellin (32, 44). In Salmonella spp. much of the central hypervariable region can be replaced by a nonrelated sequence without interfering with the function of the flagella. The replacement of the dominant flagellar epitope by a hepatitis B virus surface antigen into the flagellin of Salmonella did not impair the assembly of functional flagella, although removal of this epitope did impair motility (18). In Salmonella the dominant B-cell epitope in the hypervariable region IV was found to be at the surface of the flagella when assembled (18). In the flagellin of Campylobacter coli, it was concluded that when the flagellin proteins assemble, the carboxy- and amino-terminal domains become inaccessible or nonepitopic (34).
Winstanley et al. (46) have used PCR to amplify part of the flagellin gene of several P. aeruginosa strains. This was accomplished by comparing the flagellin gene sequences of two P. putida strains with the flagellin gene sequence of P. aeruginosa PAK (41). From this comparison, oligonucleotide primers specific for N-terminal (CW46) and C-terminal (CW45) conserved regions were designed. Thirty-seven presumed a-type isolates of P. aeruginosa yielded PCR amplification products of 1.02 kb, whereas 26 b-type isolates each yielded a 1.25-kb product. Due to the positioning of the oligonucleotide primers on the flagellin gene, the entire flagellin coding sequence is not amplified by this procedure. These missing N- and C-terminal sequences are in regions where there is no significant difference in length and sequence between different bacterial species (46).
Size discrepancies in the apparent and the deduced molecular mass of flagellin proteins have been observed in several bacterial species. Totten and Lory (41) reported that the predicted molecular mass of the a-type flagellin protein of P. aeruginosa PAK was 40 kDa while the apparent molecular mass as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was 45 kDa. Discrepancies have been seen in Campylobacter flagellin proteins as well (14). Posttranslational modification may be responsible for the molecular mass differences observed.
Posttranslational modification has been reported for a number of bacterial species. P. aeruginosa a-type and b-type strains were reported to contain phosphotyrosine residues (22, 23) but are also phosphorylated on threonine and serine residues in vitro (12), while Campylobacter has been shown to contain phosphoserine residues (26). Some Salmonella strains have been shown to contain N-methyl lysine on their flagellins (3, 21) as have the flagellins of Proteus morganii (8) and Spirillum serpens (15). The first report of glycosylation of eubacterial flagellin protein was reported for C. coli (14, 17). It was determined that the terminal group was a sialic acid residue (14). Glycosylation of the flagellin of several species of archaebacteria has been reported, including glycosylation in Methanospirillum hungatei (38), Sulfolobus (16), and Halobacterium halobium in the form of an N-linked sulfated glycosyl moiety (43).
The approach in this research was to employ a PCR method so that the major portion of the fliC gene could be rapidly obtained from selected a-type strains with flagellins of different molecular masses. Comparisons of PCR fragment size would indicate if the fliC gene variability could account for flagellin protein differences. Cloning and sequencing could further ascertain whether differences in amino acid composition could account for the molecular mass differences but could also ascertain what the variations compared with b-types were. It was found that gene size and amino acid content could account for differences between a- and b-types, particularly in the variable regions, but not among the a-types compared. The detection of glycosyl groups in a-type flagellins provided a reasonable explanation for a portion of the apparent molecular mass differences in the a-type flagellin proteins.
MATERIALS AND METHODS
Bacterial strains and media.
Table 1 shows the P. aeruginosa strains used in this study and the source for each. Bacteria were cultured in Luria-Bertani (LB) broth (Difco Laboratories, Detroit, Mich.). For colony blots, cells were grown on LB plates containing 1.7% agar. For ammonium sulfate precipitation of flagellin, cells were grown on mineral salts medium with 0.2% glucose instead of sodium succinate (1). For the selection of ampicillin-resistant cells, either LB broth or LB agar plates containing ampicillin (100 μg/ml) were used.
TABLE 1.
Molecular weights, PCR product sizes, and antibody reactions of P. aeruginosa strains
| Straina | Mr (103) of flagellin | PCR product size (kb) | MAb reactivityb
|
|
|---|---|---|---|---|
| a type | b type | |||
| 170018 | 45 | 1 | ++ | − |
| 5940 | 47 | 1 | + | − |
| GNB-1 | 49 | 1 | +++ | − |
| 5933 | 52 | 1 | ++ | − |
| 7191 | 51 | 1 | ++++ | − |
| 5939 | 51 | 1 | +++ | − |
| CF414iic | 1 | − | − | |
| M2 | 53 | 1.25 | − | ++ |
Strains and sources (given parenthetically) are as follows: 170018 (L. Lanyi, National Institutes of Hygiene, Budapest, Hungary); 5933, 5939, and 5940 (R. Ansorg, Collection d’Institut Pasteur, Paris, France); 7191, M2 PAO1, and GNB-1 (I. A. Holder, Shriners Burn Institute, Cincinnati, Ohio); and CF414ii (M. J. Thomassen, Cleveland Cystic Fibrosis Center, Cleveland, Ohio).
a-type and b-type MAbs are human antiflagellin MAbs.
CF414 is a nonflagellated (Fla−) cystic fibrosis sputum isolate.
PCR amplification.
The PCR method that was used to amplify the flagellin gene fragments (11, 46) utilized primers CW45 and CW46 (46), which were synthesized by Life Technologies, Inc. (Gaithersburg, Md.). The genomic DNA was isolated by resuspending approximately six large colonies from an LB agar plate in a solution containing 45 μl of distilled water and 5 μl of 50 mM EDTA. The suspension was boiled for 5 min and centrifuged at 12,000 × g in a microcentrifuge for 1 min. The supernatant was removed and used in the PCR amplification. Amplifications were carried out in 50-μl volumes containing 9 μM concentrations of each primer (CW45 and CW46), 1× PCR buffer (Promega, Madison, Wis.), 2.5 U of Taq polymerase (Promega), 4 mM magnesium chloride, and 100 mM nucleotide (dATP, dCTP, dGTP, and dTTP). Amplifications were carried out in a GeneAmp PCR System 2400 Thermocycler (Perkin-Elmer, Norwalk, Conn.). PCR amplification conditions were as follows: 94°C for 4 min, 30 cycles consisting of 94°C (40 s), 50°C (1 min), and 72°C (2 min), and an additional extension time at 72°C (10 min) after the 30 cycles were completed. After amplification, 5 μl of each sample was removed and separated by electrophoresis on a 1.0% agarose (Sigma, St. Louis, Mo.) gel to confirm the presence of an amplified product.
Colony blots.
Colonies of various strains were transferred to LB gelatin-blocked, nitrocellulose filters (Bio-Rad Laboratories, Hercules, Calif.). Primary antibody, either human anti-a flagellin monoclonal antibody (MAb) or human anti-b flagellin MAb solution (24) at a 1:1,000 dilution in 1% gelatin–phosphate-buffered saline (PBS), was added and incubated either overnight at 4°C or for 2 h at 37°C. Filters were washed, and the secondary antibody, goat anti-human immunoglobulin G (Bio-Rad Laboratories), was added at 1:5,000 dilution in 1% gelatin–PBS. Filters were again washed and developed with 4-chloro-1-naphthol and hydrogen peroxide.
Cloning of the flagellin gene fragment.
Cloning of the a-type flagellin gene fragments was accomplished by using the PCR amplification mixture containing the 1.02-kb flagellin gene fragment and the pGEM-T Vector System I (Promega). Ligation was performed by using 3 Weiss units of T4 DNA ligase, 50 ng of pGEM-T vector, 1 μl of T4 DNA Ligase 10× buffer, and 7 μl of unquantitated PCR amplification mix. The resulting plasmids were transformed into high-efficiency Escherichia coli JM109 cells. The transformants were plated onto LB–ampicillin–5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)–isopropyl-β-d-thiogalactopyranoside (IPTG) (ampicillin [100 μg/ml], 1 mg of X-Gal [Sigma], 10 mM IPTG [Sigma]) and incubated at 37°C overnight.
Lithium chloride plasmid isolation.
This procedure was obtained from Sambrook et al. (36) with one modification: 5 M lithium chloride (Mallinckrodt Specialty Chemicals Co., Paris, Ky.) was added after solution III.
Plasmid isolation and PEG precipitation.
Plasmid isolation followed by polyethylene glycol (PEG) (PEG 8000; Sigma) precipitation was used in cases where the plasmid preparation was to be used in digests or sequencing (5).
Screening transformants.
White colonies were picked; plasmids were isolated by PEG precipitation and screened by cleaving the insert from the plasmid with either SacI/SacII or SacI/ApaI (Promega). DNA samples were separated by electrophoresis in a 1% agarose gel (Sigma) containing Tris-acetate buffer (TAE) (36). The transformants that were shown to have a 1.02-kb insert were used for sequencing. The plasmid containing the 1.02-kb fragment from P. aeruginosa 170018 was designated pCB1, while the plasmid containing the 1.02-kb fragment from strain 5939 was designated pCB2 and that from strain 5933 was designated pCB3.
Southern hybridization.
The 1.7-kb fragment from plasmid pPT218 (41) was cleaved as an EcoRI/HindIII fragment and isolated according to the method of Steck (40) and then used in a PCR amplification procedure using primers CW45 and CW46 to amplify a 1.02-kb fragment of PAK fliC. This 1.02-kb fragment was biotinylated by using the BluGene Nonradioactive Nucleic Acid Detection System (Life Technologies, Inc.) and used as a probe to hybridize with the 1.02-kb fragment from pCB1 and pCB2. The 1.7-kb fragment from plasmid pPT218 was cleaved as an EcoRI/HindIII fragment to be used as a positive control, while linearized pUC18 and pGEM-T vector were used as negative controls. The Southern hybridization procedure followed the BluGene Nonradioactive Nucleic Acid Detection Systems manufacturer’s instructions.
Sequencing.
Sequencing was performed by the dideoxy chain termination method using SP6 and T7 primers at the University of Tennessee Molecular Biology Resources Facility (5). The P. aeruginosa sequences used in this study are as follows (GenBank accession numbers are shown in parentheses): 170018 (U76543), 5933 (AF003906), 5939 (AF003905), PAO1 (U55775), and PAK (M57501).
Amino acid comparisons.
The amino acid sequence of the flagellin gene fragment for the various P. aeruginosa strains was deduced from the DNA sequence by using the DNA* computer program (13). The amino acid sequences were aligned for DNA* comparisons using the CLUSTAL method according to multiple-alignment parameters (gap penalty, 10; gap length penalty, 10) and pairwise-alignment parameters (ktuple, 1; gap penalty, 3; window, 5; diagonals saved, 5). This method groups sequences into clusters by examining the distances between all pairs. Clusters are then aligned into groups with gaps introduced by the computer at points of divergence (13).
Flagellin isolation and purification.
The flagellin protein of several strains was isolated and partially purified by using ammonium sulfate precipitation (10). Luria agar (Life Technologies, Inc.) plates containing 0.2% glucose were inoculated with the designated P. aeruginosa strain and incubated for 48 h at 37°C. Six flasks containing 500 ml of glucose mineral salts medium were inoculated with the bacterial colonies from the 48-h plates and agitated at 175 rpm overnight at 37°C. The cells were removed from the culture medium by centrifugation at 7,480 × g at 4°C for 20 min. Cells were resuspended in 180 ml of 50 mM sodium phosphate buffer (pH 7.0). The cell suspension was then blended at the low setting in a Waring commercial blender for 1.5 min. The homogenate was centrifuged at 12,000 × g at 4°C for 20 min to remove the cells from the supernatant. The supernatant was saturated in 5% increments with a 4.1 M ammonium sulfate solution. After each addition of ammonium sulfate, the solutions were allowed to stir at room temperature for 4 to 5 h. The precipitated protein was pelleted by centrifugation at 12,000 × g at 4°C for 30 min. Each of the protein fractions was redissolved in 1 ml of 50 mM sodium phosphate buffer (pH 7.0) prior to being dialyzed against 50 mM sodium phosphate buffer (pH 7.0) at 4°C overnight.
PAGE.
Ammonium sulfate precipitated samples and molecular weight markers were suspended in 12.5% 0.5 M Tris-Cl–0.4% SDS buffer (pH 6.8) with 10% glycerol, 2% SDS, and 0.00005% bromophenol blue. All samples were heated to 100°C for 5 min just prior to electrophoresis. Electrophoresis was accomplished by using a 12% acrylamide separating gel (National Diagnostics, Atlanta, Ga.), with other standard ingredients in a Minigel 2-D unit (Bio-Rad Laboratories, Hercules, Calif.) using a constant current of 14 mA per gel. Proteins were stained with Coomassie brilliant blue solution (36) and stained in a 10% methanol–10% acetic acid solution.
Western blot.
After separation of proteins by SDS-PAGE, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Micron Separations Incorporated, Westborough, Mass.) by using a semidry blotter unit (Labconco, Kansas City, Mo.).
The membrane was removed and washed according to standard procedures. Primary antibody, either human anti-a flagellin MAb or human anti-b flagellin MAb (24), was added at a 1:1,000 dilution in Tris-buffered saline (TBS)–Tween and incubated overnight at 4°C with shaking. Washes were performed as outlined above with TBS-Tween. Affinity-purified goat anti-human immunoglobulin G (heavy and light chain) horseradish peroxidase conjugate antibody (Bio-Rad Laboratories) was added at a 1:5,000 dilution in TBS-Tween and incubated at 37°C, followed by development with Bio-Rad horseradish peroxidase reagents A and B.
Glycosylation assay.
Glycosylation of flagellin proteins was determined by the biotin-hydrazide glycosylation assay (14) with a few modifications. First, proteins were separated by electrophoresis (for a description of this procedure, see the section “PAGE” [above]). The proteins were then transferred to PVDF membrane using the procedure discussed previously. The membrane was washed in PBS (36) for 10 min at room temperature. The membrane was oxidized with 15 mM meta-periodate (Boehringer Mannheim, Indianapolis, Ind.) in 50 mM sodium acetate buffer (pH 5.45) for 30 min at room temperature in the dark. Aluminum foil was used to exclude light during this reaction. Three 10-min washes with PBS were performed at room temperature to remove periodate. Five millimolar biotin-hydrazide in 50 mM sodium acetate (pH 5.45) was added to the membrane and incubated at room temperature for 1 h. The membrane was then washed three times with TBS (25 mM Tris, 0.14 M NaCl, 2.7 mM KCl [pH 7.4] [36]) for 10 min each at room temperature to remove the biotin-hydrazide. Blocking was performed by incubating the membrane with casein in TBS (Pierce, Rockford, Ill.) for 30 min at room temperature followed by three washes in TBS for 10 min at room temperature. Streptavidin-alkaline phosphatase (Southern Biotechnology Associates, Inc., Birmingham, Ala.) diluted to a concentration of 1:2,000 in TBS was added to the membrane and incubated for 1 h at room temperature. Again, three washes were performed with TBS as described above. Each of the incubations involved gentle agitation. The membrane was developed with a solution of nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) (Oxford Glycosystems Inc., Bedford, Mass.) in TBS for 2 h.
Deglycosylation assay.
Removal of carbohydrate groups was accomplished with a GlycoFree kit (Oxford Glycosystems). Glycosyl groups were removed with anhydrous trifluoromethanesulfonic acid (TFMS), which has been shown to remove N- and O-linked glycosyl moieties. This procedure was performed according to manufacturer’s instructions (33).
RESULTS
Colony blots.
Reaction of human anti-a flagellin MAb with the flagellin of several P. aeruginosa strains was observed by using colony blots. Strains 170018, 5933, 5939, 5940, 7191, and GNB-1 reacted with the human anti-a flagellin MAb, while no reactivity was seen with the human anti-b flagellin MAb (Table 1). CF414ii, an a-type Fla− cystic fibrosis strain, did not react with the a-type MAb due to the lack of a flagellum (27). M2, a b-type strain, reacted with the b-type MAb as expected. These results confirm the typing of the a-type strains used in PCR amplifications of fliC.
PCR amplification of the fliC gene of several P. aeruginosa a-type strains.
PCR amplification was performed as described above for strains 170018, 5933, 5939, 5940, 7191, GNB-1, and CF414ii. In confirmation of the findings of Winstanley et al. (46), a 1.02-kb fragment of the fliC gene resulted from amplification of each a-type strain (Table 2). Although this procedure does not result in the amplification of the entire fliC gene, the majority of the N and C termini and the central hypervariable region are amplified by this method.
TABLE 2.
Molecular weight analysis of a-type flagellin proteins in P. aeruginosa
| Strain | Deduced molecular weight (103) | Apparent Mrb (103) | Adjusted molecular weightc (103) | Differenced |
|---|---|---|---|---|
| 170018 | 34.33a | 45 | 39.93 | 5.07 |
| 5933 | 33.90a | 52 | 39.50 | 12.50 |
| 5939 | 34.32a | 51 | 39.92 | 11.08 |
| PAK | 39.91 | 45 | 39.91 | 5.09 |
Molecular weight of the amino acid sequence translated from the DNA sequence of the 1-kb flagellin gene fragment.
Molecular weight as observed on SDS-polyacrylamide gel.
The adjusted molecular weight was obtained by adding the molecular weight of the missing N- and C-terminal residues of PAK (5,600). No adjustment was made for the Mr of PAK since its deduced molecular weight represents the entire gene.
Apparent Mr − adjusted molecular weight.
The PCR-amplified fliC fragment from P. aeruginosa strains 170018, 5939, and 5933 were cloned into E. coli JM109 by using the pGEM-T vector system (Promega). The clones were designated pCB1, pCB2, and pCB3, respectively. In order to confirm the presence of the 1.02-kb fliC fragment in clones pCB1 and pCB2, Southern hybridization experiments using part of the P. aeruginosa PAK fliC gene (41) as a biotinylated probe were performed (data not shown). This experiment showed that pCB1 and pCB2 contained inserts which were homologous to the PAK probe. The flagellin inserts of pCB1, pCB2, and pCB3 were sequenced.
Amino acid comparison of an a-type (170018) and a b-type (PAO1) flagellin sequence.
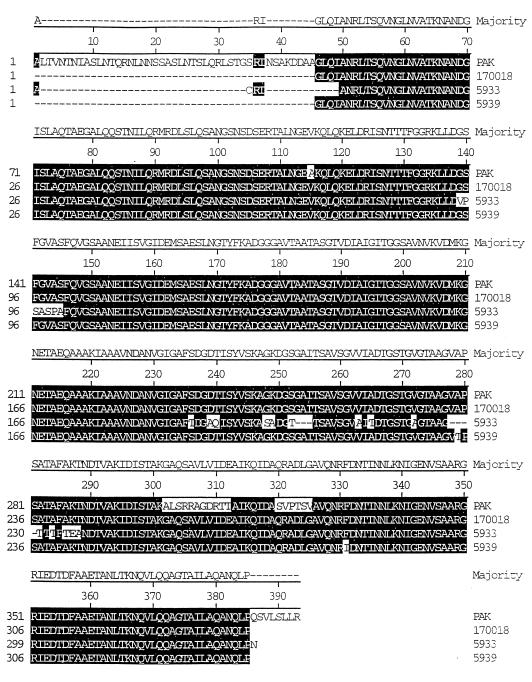
Sequencing and subsequent analysis of the deduced amino acid sequence of 170018 and comparison with a b-type (PAO1) amino acid sequence revealed significant divergence (Fig. 1). The most obvious difference between the a- and b-type flagellin sequences is that the 170018 sequence was missing large segments of sequence in the central region. Specifically, these segments were residues 231 to 281, 294 to 304, 329 to 339, 351 to 364, and 384 to 387 (numbers are according to the consensus sequence). Interestingly, many single amino acid changes were from alanine to threonine or serine or vice versa. When the sequence of the four a-types and PAO1 were compared, the N-terminal 106 amino acids and the C-terminal 68 amino acids were found to be highly conserved.
FIG. 1.
Comparison of a-type, 170018 (GenBank accession no. U76543), and b-type, PAO1 (GenBank accession no. U54775), flagellin amino acid sequences. Alignment was performed by the CLUSTAL method by using the DNA* computer program.
Amino acid comparison of the flagellin sequence of several a-type strains.
Sequencing and subsequent analysis of the deduced amino acid sequence revealed a high degree of similarity between the three a-type strains sequenced and the a-type PAK sequence from Totten and Lory (41) (Fig. 2). Residue numbers were taken from the majority or consensus sequence. There were only two amino acid differences between the sequence of 170018 and that of 5939. One of these was a change from an alanine in 170018 to a threonine in 5939 at position 279. Interestingly, the same type of change was seen between the sequences of PAK and 5939. The greatest variation was seen between 5933 and the other a-type strains. There were 31 amino acid differences between 170018 and 5933. A region containing seven amino acid differences was seen from residue 139 to 145 when comparing 5933 to the other a-type sequences (numbers are according to the majority or consensus sequence). Additionally, only 5933 showed variation at residues 239 to 240, 248 to 250, 252 to 255, 263, 265, 272, 278 to 282, 284, and 286 to 288 compared to the other a-type sequences. Another important feature of the 5933 sequence was the prevalence of single amino acid changes which involved changes from alanine to threonine at residues 265, 282, 284, and 286. Of the a-type strains compared, strain 5933 flagellin is the most divergent in amino acid sequence. Another consistent variation between the PAK sequence and the other a-type sequences was seen between residues 302 and 312 and 320 and 325.
FIG. 2.
Comparison of a-type flagellin amino acid sequences from P. aeruginosa PAK (41), 170018 (GenBank accession no. U76543), 5933 (GenBank accession no. AF003906), and 5939 (GenBank accession no. AF003905). Alignment was performed by the CLUSTAL method by using the DNA* computer program.
When comparing the sequences of 170018, 5933, and 5939 with the sequence of PAK, one can ascertain the number of N- and C-terminal residues that are missing extra amplified product, because these residues are highly conserved (Fig. 1 and 2). Approximately 45 amino acids from the N terminus and 8 amino acids from the C terminus are absent. The molecular mass of these missing amino acids is 5.6 kDa. The 5.6 kDa was added to the deduced molecular mass to give the adjusted molecular mass, which was approximately 40 kDa for each of these a-type strains and should correspond to the apparent Mr. However, the adjusted Mr was 5,000 lower for 170018 and PAK, 12,500 lower for 5933, and 11,000 lower for 5939. This discrepancy could be explained by posttranslational modification resulting in the higher apparent Mr.
Glycosylation assay.
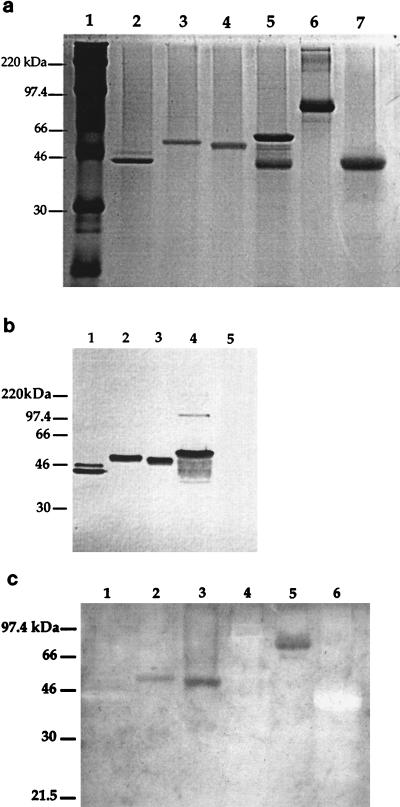
In an effort to explain the discrepancy among the a-type apparent Mr and the deduced molecular mass and to explain the variation in Mr values among various a-types, assays for glycosylation of the flagellin proteins were performed (Fig. 3). Flagellins resolved by SDS-PAGE were either stained with Coomassie brilliant blue, assayed for reactivity with human anti-a and anti-b MAbs, or assayed for glycosylation. Anti-a and anti-b MAbs were used to reveal the presence of a- and b-type flagellins on the same membrane (Fig. 3b). Colony blots (Table 1) were used to show specific reactivity of each strain with either the a- or b-type MAb. The biotin-hydrazide glycosylation assay gave a positive reaction with the flagellin of strain 5933 and 5939 and no reaction with the flagellin of strain 170018 or M2 (b-type strain) under the assay conditions used (Fig. 3c). Transferrin, a glycoprotein, gave a positive reaction, and pepsin, a nonglycosylated protein, gave a negative reaction when tested for glycosylation. Since 170018 is not glycosylated and shows a difference of 5 kDa between the apparent and adjusted molecular mass, it is possible that phosphorylation accounts for a portion of the mass.
FIG. 3.
(a) SDS-PAGE of P. aeruginosa flagellin proteins stained with Coomassie brilliant blue. Lane 1, protein standards; lane 2, 170018 flagellin (Mr, 45,000); lane 3, 5933 flagellin (Mr, 52,000); lane 4, 5939 flagellin (Mr, 41,000); lane 5, M2 (b-type) flagellin (Mr, 53,000); lane 6, transferrin; and lane 7, pepsin. (b) Western blot of flagellin proteins with human anti-a and anti-b MAbs. Anti-a MAb at a 1:1,000 dilution was added to anti-b MAb at a 1:1,000 dilution, and the mixture was added to the membrane. Lane 1, 170018 flagellin; lane 2, 5933 flagellin; lane 3, 5939 flagellin; lane 4, M2 flagellin; and lane 5, transferrin as a negative control. (c) Biotin-hydrazide glycosylation assay of the flagellin proteins of P. aeruginosa strains. Approximately 3 μg of each protein was separated by electrophoresis and transferred to a PVDF membrane. Meta-periodate (15 mM) in 50 mM sodium acetate buffer (pH 5.45) oxidized the sugars. Biotin-hydrazide (5 mM) was then added, followed by streptavidin-alkaline phosphatase. Lane 1, 170018 flagellin; lane 2, 5933 flagellin; lane 3, 5939 flagellin; lane 4, M2 flagellin; lane 5, glycoprotein transferrin as a positive control; and lane 6, pepsin as a negative control.
The M2 fraction (Fig. 3 and 4) revealed in gels a common characteristic of these flagellin preparations, the appearance of a degraded lower-molecular-mass polypeptide, which is apparently due to the presence of minute amounts of proteases which act on this flagellin during purification and storage. The M2 flagellin commonly exhibits this property, reflecting the presence of several potent proteases associated with this highly virulent strain. The loss of the lower band after TFMS treatment probably indicated further denaturation which occurred during the TFMS-acid treatment. The fragmentary flagellin but not native protein would be much more susceptible to denaturation. The presence of a doublet in the 170018 fraction was puzzling, and several interpretations are possible (see Discussion).
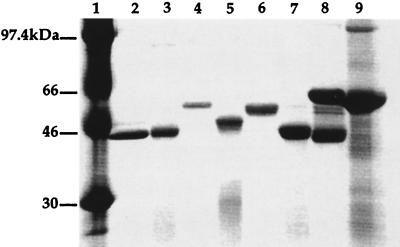
FIG. 4.
SDS-PAGE of TFMS-treated and untreated flagellins stained with Coomassie brilliant blue. The deglycosylation assay revealed a molecular mass shift for the flagellins of strains 5933, 5939, and M2 after chemical removal of the glycosyl group. Lane 1, protein standards; lane 2, 170018 flagellin; lane 3, deglycosylated 170018 flagellin; lane 4, 5933 flagellin; lane 5, deglycosylated 5933 flagellin; lane 6, 5939 flagellin; lane 7, deglycosylated 5939 flagellin; lane 8, M2 flagellin; and lane 9, deglycosylated M2 flagellin.
Deglycosylation assay.
In order to determine how much of the molecular mass discrepancy for each flagellin could be accounted for by the addition of glycosyl groups, the flagellin of each of the strains was chemically deglycosylated by the use of TFMS, which removes O- and N-linked oligosaccharides from the protein, and then run on gels. Figure 4 shows the results of deglycosylation of flagellins of 170018, 5933, 5939, and M2. These results were consistent with the glycosylation results for 170018, 5933, and 5939. The molecular masses of the flagellins of strains 170018 and M2 did not change significantly after removal of glycosyl groups from the protein. There was a molecular mass shift in the flagellin of strains 5933 and 5939 after deglycosylation. The deglycosylated 5933 flagellin protein migrated approximately 7 kDa lower than untreated 5933 flagellin. The deglycosylated 5939 flagellin protein migrated approximately 8 kDa lower than the untreated 5939 protein.
In order to confirm that the deglycosylated proteins had all glycosyl groups removed, a biotin-hydrazide glycosylation assay was performed on the TFMS-treated flagellin proteins (data not shown). Results revealed that the flagellin proteins treated with TFMS were deglycosylated since the deglycosylated proteins of 5933 and 5939 gave negative glycosylation reactions in this assay.
MAb binding to deglycosylated flagellin.
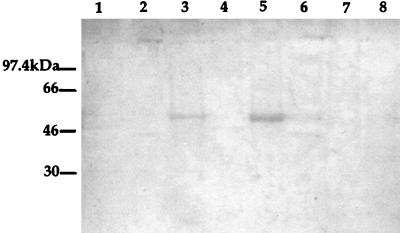
To investigate the role the glycosyl groups play in antigenicity, the deglycosylated (TFMS-treated) flagellin proteins were tested for the ability to react with the human anti-a flagellin MAb (Fig. 5). If a glycosyl group constitutes the epitope for this antibody, then the removal of glycosyl moieties by TFMS should eliminate MAb binding. One could argue that TFMS could destroy the conformation of the protein such that the monoclonal epitope is destroyed. However, the MAb binds to a-type flagellin proteins under denaturing conditions. The proteins are denatured by boiling in SDS prior to electrophoresis so that the majority of secondary and tertiary structure was eliminated. Therefore, the MAb epitope is not likely to be a conformational epitope. If the MAb were bound to the TFMS-treated proteins, then one would conclude that the glycosyl moiety is not part of the epitope. Results showed that the anti-a MAb reacted with the deglycosylated a-type flagellin proteins. In Figure 5, lane 2, which contains the 170018 flagellin protein which had been treated with TFMS, showed a decreased binding of the MAb, whereas lanes 4 and 6, which contain the deglycosylated 5933 and 5939 flagellins, respectively, showed no decrease in the binding of the antibody.
FIG. 5.
Western blot of TFMS-treated and untreated flagellin proteins with human anti-a flagellin MAb. Lane 1, 170018 flagellin; lane 2, TFMS-treated 170018 flagellin; lane 3, 5933 flagellin; lane 4, TFMS-treated 5933 flagellin; lane 5, 5939 flagellin; and lane 6, TFMS-treated 5939 flagellin.
DISCUSSION
It is clear from the PCR results reported above that both a-types and b-types of P. aeruginosa gave PCR amplification products of characteristic sizes, 1.02 and 1.25 kb respectively (11, 46). This has been demonstrated unequivocally for the a-types in these experiments using selected, immunologically characterized strains. The amino acid sequence-derived molecular mass for PAO1 (b-type) is approximately 49.3 kDa, while the derived molecular mass for the a-type is approximately 40 kDa. The major differences between the a- and b-type strains is the absence of large stretches of amino acid residues in the central region of the a-type sequences, which would account for the differences in molecular masses of the flagellin between a- and b-types. This was previously seen in the apparent differences in amino acid content by direct protein analysis (30, 35) and by SDS-PAGE (1). These sequence differences are consistent with the findings of Spangenberg et al. (39) in their comparisons of a- and b-type flagellins. It is also apparent from Spangenberg’s data comparing a-type strains that the N-terminal region is highly conserved and that only four amino acid differences occur in residues 1 through 48. Thus, the absence of amino acids from the N-terminal sequence as a result of the internal primer annealing is not an important factor in our analysis. The C terminus has been shown to be highly conserved as well and thus is not a site of comparative variation in the flagellin sequence. An interesting common feature in all a-type sequences is the absence of comparable amino acids at residues 229 to 339 and 351 through 364 (numbering is according to the consensus sequence in Fig. 1) as compared to b-types. This deletion seems to be common to all a-types reported so far (39, 41).
When a-type sequences are compared (Fig. 2), no major differences in amino acid sequences that could account for molecular mass differences are found. One important change may be the switch from alanine to serine or alanine to threonine. This change would introduce additional sites for phosphorylation or glycosylation through O linkages. It will be of interest to determine the type of linkage involved in the glycan moiety. In Campylobacter, the lack of inhibition of glycosylation by tunicamycin and the presence of seven serine residues in the designated modified site suggests an O linkage (14). In this regard, the replacement of alanine in the low-molecular-mass flagellins by serine or threonine in the high-molecular-mass flagellin sequence may involve new potential glycosylation sites. One possibility is that these sites involve phosphate bridges to the glycosyl moiety (42). It is apparent from the derived sequence data that the molecular mass falls below the values obtained by SDS-PAGE. With the b-types, this represents only a slight differential (approximately 4 kDa) and can be ascribed to phosphorylation at hydroxy amino acids. We have previously reported posttranslational modifications at Thr or Ser and Tyr and possibly tyrosine in b-type and a-type flagellins (12, 22, 23, 37).
The differences between the derived molecular masses and Mr values are more striking for the a-types than b-types (Table 2). Strain 170018 (a-type) showed a difference of approximately 5,000 between its Mr (45,000) and its derived molecular weight, while two strains with higher Mr values, 5933 (52,000) and 5939 (51,000), each gave an Mr differential of 11,000 to 12,500. This prompted us to investigate whether in addition to phosphorylation, noted previously, other posttranslational modifications might account for the difference. The presence of sialic acid residues in the flagellin of C. coli (14) suggested the possibility of glycosylation of the flagellin in P. aeruginosa. In order to assay for glycosylation, an approach similar to that of Doig et al. (14) was adopted for P. aeruginosa flagellin. A definitive positive glycosylation reaction was seen for the two high-molecular-mass flagellins, 5933 and 5939, but not the low-molecular-mass flagellin of 170018. Strain M2, having a b-type flagellin, gave a negative glycosylation reaction. These data indicate that the large discrepancy between predicted and apparent molecular masses of a-type flagellins is due in part to glycosylation. However, we cannot rule out the possibility that 170018 (Mr, 45,000) is glycosylated at a low level not detectable by this methodology. A very preliminary result obtained with PAK, a low-molecular-mass, a-type flagellin, indicated the presence of glycosyl groups (data not shown). In this regard the presence of a doublet in the 170018 fraction is difficult to interpret (Fig. 3 and 5). In the 170018 flagellin, two bands were reactive with anti-a MAb, but the upper band was only weakly stained with Coomassie blue (Fig. 3a). It is possible that the upper band (approximately 46 kDa) is weakly glycosylated while the 45-kDa band is not. Since the 46-kDa band was present at a low concentration, it would not have been revealed in the glycosylation assay. A likely alternative explanation is indicated by the strong MAb reaction of the upper band, which would suggest that it contained primarily native protein compared to the lower band (45 kDa). This observation suggests that the 45-kDa band was somewhat more denatured or resulted from a single protease clip. Additional experiments are needed to distinguish these possibilities.
The 11- to 12.5-kDa molecular mass discrepancies of 5933 and 5939 flagellins is consistent with the glycosylation results and our interpretation that the addition of glycosyl moieties to these flagellin proteins adds additional mass. Effects of glycosylation on overall charge and conformation could also contribute alterations in protein migration. Nondetectable levels of glycosylation in b-type strains could explain the lack of glycosylation previously reported for P. aeruginosa flagellin if only b-type or selected a-type strains were analyzed (46). Conversely, consistent with our results was a preliminary report that a-types are glycosylated and b-types are not (8a). The deglycosylation of the flagellin proteins revealed that the flagellin proteins of 5933 and 5939 are glycosylated, as shown by the large molecular mass shift after treatment with TFMS to remove N- and O-linked carbohydrates. Given the 7- and 8-kDa molecular mass shifts for 5933 and 5939, respectively, the percent carbohydrate for each of the flagellin proteins was calculated. The flagellin of strain 5933 contains 13% carbohydrate, while the flagellin of 5939 contains 15% carbohydrate. After treating the 5933 and 5939 flagellin proteins with TFMS in order to remove the glycosyl moieties, these proteins were assayed for glycosyl groups by the biotin-hydrazide glycosylation assay. This experiment revealed that the glycosyl groups were removed from the proteins to levels undetectable by this assay method.
The flagellin proteins that had been subjected to the deglycosylation procedure were reacted with the anti-a MAb to ascertain whether the glycosyl groups could be contributing to the epitope of the MAb. Although 170018 gave a negative glycosylation result, it still reacted with the MAb. Upon removal of the glycosyl groups from the flagellins of 5933 and 5939 with TFMS, reactivity was still observed with the anti-a MAb. These results support the prediction that the glycosyl groups would not be involved in the epitope for the MAb.
It is likely that the heterogeneity seen in the central regions of the a-type flagellins contributes to the variation in antigenic cross-reactivity among a types with several antiflagellin MAbs (24, 30). With at least one mouse MAb, i.e., that raised against the flagella of 170018, there was an association between Mr and cross-reactivity (30). These data together with the glycosylation results suggest the possibility that glycosylation has a cryptic effect on antigenicity, rendering the richly glycosylated flagellins less detectable as foreign during infection. In Neisseria gonorrhoeae infections, the strains associated with disseminated infections are serum resistant due to the presence of sialic residues on the lipopolysaccharide core carbohydrate chain. This phenomenon is due to the fact that sialic acid molecules are a surface component of erythrocytes and are thus seen as self antigen. Therefore, sialic acid residues camouflage the organism from the immune system (19). Alternatively, although less likely, the glycosyl groups could play a role in the antigenicity of the flagellin protein, constituting an immunodominant epitope. In Campylobacter, sialic acid residues are responsible for antigenicity (17). In C. coli VC167, it has been suggested that posttranslational modifications are responsible for antigenic variation seen in the two types of flagellin produced by this organism (2, 34). It was later shown by Doig et al. (14) that the posttranslational modification present in C. coli was glycosylation involving terminal sialic acid residues. Therefore, reports suggest both the involvement of sialic acid residues in protection from the immune response and their involvement in antigenicity. Such hypotheses would have to be tested more extensively to elucidate the role of the glycosyl moieties in Pseudomonas. Since glycosyl moieties are found in pathogenic and nonpathogenic bacteria, a common protective feature for both might include some protection of this exposed organelle from proteases and other enzymes (42).
It has been recently shown that at least 50% of the genera of the domain Archaea are glycosylated (20). As observed with the a-type flagellins, the archaeal flagellins have higher Mr values than would be predicted by their primary sequences. In M. hungatei and H. halobium, a reduction of 10 kDa occurred following chemical deglycosylation (20, 25, 38). Of importance is the finding that at least some of the archaeal flagellins contain a number of classical carbohydrate attachment consensus sequences (Asn-X-Ser/Thr) which represent N-linkage sites in Methanococcus spp. These are quite common, and one such linkage has been identified to be associated with a glycosylated hexapeptide (25). A number of these sequences occur in the a-type flagellins; however, we believe an O linkage is more likely for P. aeruginosa and is probably found in the case of Campylobacter (14). The archaea seem to require glycosylation for proper flagellar filament assembly (20). Evidence indicates that this posttranslational modification occurs at the cell surface, even outside the cell membrane. What is puzzling is that glycosylation may occur in one archaeal species but not be found in a closely related species. This situation parallels our finding of glycosylation in some a-types but not in b-types in P. aeruginosa.
It is clear that the posttranslational modification motif is widespread in external organelles such as flagellins, where not only glycosylation but also additions of lysine (3, 21) and phosphates (12, 22, 23, 26, 37) are observed. In the last few years, the identification of eukaryotic protein phosphokinases in the classes Myxobacteria (Myxocococcus xanthus), Cyanobacteria, and Streptomyces (47) have emphasized that the enzymatic machinery necessary has preceded evolution into eukaryotic cells. Very recently, tyrosine-phosphorylated proteins were verified in Anabaena spp. (34) and in three unrelated members of the domain Archaea. The data from some of the M. xanthus studies suggest that at least some of the Ser/Thr kinase activity is associated with metabolic signaling. In the case of Pseudomonas flagella, modifications affecting the function(s) remain to be determined.
Until recently, glycosylation was thought to be a characteristic of eukaryotic proteins and not prokaryotic proteins. However, this study has shown that some a-type flagellins of P. aeruginosa are glycosylated. This study reveals the importance and need for further investigations to characterize the glycosyl moieties on the flagellin protein, the biological role of the glycosyl moieties, and the mechanism of addition of these modifications.
ACKNOWLEDGMENTS
We thank Cynthia Peterson for her suggestions and discussions on glycosylation and Neil Quigley and Jeff Becker’s laboratory group for their aid in sequencing and computer analysis.
REFERENCES
- 1.Allison J S, Dawson M, Drake D, Montie T C. Electrophoretic separation and molecular weight characterization of Pseudomonas aeruginosa H-antigen flagellins. Infect Immun. 1985;49:770–774. doi: 10.1128/iai.49.3.770-774.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm R A, Guerry P, Power M E, Trust T J. Variation in antigenicity and molecular weight of Campylobacter coli VC167 flagellin in different genetic backgrounds. J Bacteriol. 1992;174:4230–4238. doi: 10.1128/jb.174.13.4230-4238.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambler R P, Rees M W. N-methyl-lysine in bacterial flagellar protein. Nature. 1959;184:56–57. doi: 10.1038/184056b0. [DOI] [PubMed] [Google Scholar]
- 4.Ansorg R. Flagella specific H antigenic schema of Pseudomonas aeruginosa. Zentbl Bakteriol Parasitenkd Infektkrankh Hyg Abt 1. 1978;224:228–238. [PubMed] [Google Scholar]
- 5.Applied Biosystems, Inc. High-quality template DNA for Taq cycle sequencing using dideoxy terminators: an improved preparation procedure. User bulletin 18. Foster City, Calif: Applied Biosystems, Inc.; 1991. [Google Scholar]
- 6.Applied Biosystems, Inc. ABI PRISM dye terminator cycle sequencing ready reaction kit protocol (ABI P/N 402078). Foster City, Calif: Applied Biosystems, Inc.; 1995. [Google Scholar]
- 7.Armitage J P. Behavioral responses in bacteria. Annu Rev Physiol. 1992;54:683–714. doi: 10.1146/annurev.ph.54.030192.003343. [DOI] [PubMed] [Google Scholar]
- 8.Baker B S, Smith S E, McDonough M W. The presence of several residues of N-methyl lysine in Proteus morganii flagellin. Microbios Lett. 1983;23:7–12. [Google Scholar]
- 8a.Baker, N. Personal communication.
- 9.Behammer W, Shao Z, Mages W, Rachel R, Stetter K, Schmitt R. Flagellar structure and hyperthermophily: analysis of a single flagellin gene and its product in Aquifex pyrophilus. J Bacteriol. 1995;177:6630–6637. doi: 10.1128/jb.177.22.6630-6637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brett P J, Mah D C W, Woods D E. Isolation and characterization of Pseudomonas pseudomallei flagellin proteins. Infect Immun. 1994;62:1914–1919. doi: 10.1128/iai.62.5.1914-1919.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brickman C S, Kelly-Wintenberg K, Montie T C. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Comparative analysis of flagellin genes from wild-type Fla+ and non-motile Fla− P. aeruginosa strains, abstr. D-42; p. 215. [Google Scholar]
- 12.Cha H, Nichols R K, Montie T C. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Post-translational phosphorylation of flagellin of Pseudomonas aeruginosa by an envelope kinase in vitro, abstr. D-73; p. 254. [Google Scholar]
- 13.DNA* Inc. Lasergene: biocomputing software for the Macintosh. User’s guide. Madison, Wis: DNA* Inc.; 1994. [Google Scholar]
- 14.Doig P, Kinsella N, Guerry P, Trust T J. Characterization of a post-translational modification of Campylobacter flagellin: identification of a sero-specific glycosylation moiety. Mol Microbiol. 1996;19:379–387. doi: 10.1046/j.1365-2958.1996.370890.x. [DOI] [PubMed] [Google Scholar]
- 15.Glazer A N, DeLange R J, Martinez R J. Identification of N-methyl-lysine in Spirillum serpens flagella and N-dimethyl-lysine in Salmonella typhimurium flagella. Biochim Biophys Acta. 1969;189:164–165. doi: 10.1016/0005-2795(69)90059-2. [DOI] [PubMed] [Google Scholar]
- 16.Grogan D W. Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J Bacteriol. 1989;171:6710–6719. doi: 10.1128/jb.171.12.6710-6719.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerry P, Doig, Alm R A, Burr D H, Kinsella N, Trust T J. Identification and characterization of genes required for post-translational modification of Campylobacter coli. Mol Microbiol. 1996;19:369–378. doi: 10.1046/j.1365-2958.1996.369895.x. [DOI] [PubMed] [Google Scholar]
- 18.He X-S, Rivkina M, Stocker B A D, Robinson W S. Hypervariable region IV of Salmonella gene fliCd encodes a dominant surface epitope and a stabilizing factor for functional flagella. J Bacteriol. 1994;176:2406–2414. doi: 10.1128/jb.176.8.2406-2414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hitchcock P J, Robinson E N, Jr, McGee Z A. Neisseriae: gonococcus and meningococcus. In: Satterfield T S, Napora L, Potler S, editors. Mechanisms of microbial Disease. 2nd ed. Baltimore, Md: Williams and Wilkins; 1993. p. 229. [Google Scholar]
- 20.Jarrell K F, Bayley D P, Kostyukova A S. The archeal flagellum: a unique motility structure. J Bacteriol. 1996;178:5057–5064. doi: 10.1128/jb.178.17.5057-5064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joys T M, Kim H. Identification of N-methyl-lysine residues in the phase-1 flagellar protein of Salmonella typhimurium. Microbios Lett. 1979;7:65–68. [Google Scholar]
- 22.Kelly-Wintenberg K, Anderson T, Montie T C. Phosphorylated tyrosine in the flagellum filament protein of Pseudomonas aeruginosa. J Bacteriol. 1990;172:5135–5139. doi: 10.1128/jb.172.9.5135-5139.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly-Wintenberg K D, South S L, Montie T C. Tyrosine phosphate in a- and b-type flagellins of Pseudomonas aeruginosa. J Bacteriol. 1993;175:2458–2461. doi: 10.1128/jb.175.8.2458-2461.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landsperger W J, Kelly-Wintenberg K D, Montie T C, Knight L S, Hansen M B, Huntenburg C C, Schneidkraut M J. Inhibition of bacterial motility with human antiflagellar monoclonal antibodies attenuates Pseudomonas aeruginosa-induced pneumonia in the immunocompetent rat. Infect Immun. 1994;62:4825–4830. doi: 10.1128/iai.62.11.4825-4830.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lechner J, Wieland F. Structure and biosynthesis of prokaryotic glycoproteins. Annu Rev Biochem. 1989;58:173–194. doi: 10.1146/annurev.bi.58.070189.001133. [DOI] [PubMed] [Google Scholar]
- 26.Logan S M, Trust T J, Guerry P. Evidence for posttranslational modification and gene duplication of Campylobacter flagellin. J Bacteriol. 1989;171:3031–3038. doi: 10.1128/jb.171.6.3031-3038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luzar M A, Thomassen M J, Montie T C. Flagella and motility alterations in Pseudomonas aeruginosa strains from patients with cystic fibrosis in relationship to patient clinical condition. Infect Immun. 1985;50:577–582. doi: 10.1128/iai.50.2.577-582.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macnab R M. Flagella. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 1. Washington, D.C: American Society for Microbiology; 1987. pp. 70–83. [Google Scholar]
- 29.McCartney B, Howell L D, Kennelly P J, Potts M. Protein tyrosine phosphorylation in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:2314–2318. doi: 10.1128/jb.179.7.2314-2318.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montie T C. The flagellum. In: Montie T C, editor. Pseudomonas. Biotechnology handbooks. London, United Kingdom: Plenum; 1988. [Google Scholar]
- 31.Montie T C, Anderson T R. Enzyme-linked immunosorbent assay for detection of Pseudomonas aeruginosa H (flagellar) antigen. Eur J Clin Microbiol. 1988;7:256–260. doi: 10.1007/BF01963097. [DOI] [PubMed] [Google Scholar]
- 32.Namba K, Yamashita I, Vonderviszt F. Structure of the core and central channel of bacterial flagella. Nature. 1989;342:648–654. doi: 10.1038/342648a0. [DOI] [PubMed] [Google Scholar]
- 33.Oxford Glycosystems Inc. GlycoTrack carbohydrate detection kit K-050 user’s manual. Bedford, Mass: Oxford Glycosystems Inc.; 1995. [Google Scholar]
- 34.Power M E, Guerry P, McCubbin W D, Kay C M, Trust T J. Structural and antigenic characteristics of Campylobacter coli FlaA flagellin. J Bacteriol. 1994;176:3303–3313. doi: 10.1128/jb.176.11.3303-3313.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rotering H, Dorner F. Flagella vaccine. In: Hoiby H, Pedersen S, Shand G H, Doring G, Holder I A, editors. Pseudomonas aeruginosa infection. 42. S. Basel, Switzerland: Karger AG; 1989. pp. 218–228. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.South S L, Nichols R, Montie T C. Tyrosine kinase activity in Pseudomonas aeruginosa. Mol Microbiol. 1994;12:903–910. doi: 10.1111/j.1365-2958.1994.tb01078.x. [DOI] [PubMed] [Google Scholar]
- 38.Southam G, Kalmokoff M L, Jarrell K F, Koval S F, Beveridge T J. Isolation, characterization, and cellular insertion of the flagella from two strains of the archaebacterium Methanospirillum hungatei. J Bacteriol. 1990;172:3221–3228. doi: 10.1128/jb.172.6.3221-3228.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spangenberg C, Heuer T, Burger C, Tummler B. Genetic diversity of flagellins of Pseudomonas aeruginosa. FEBS Lett. 1996;396(2-3):213–217. doi: 10.1016/0014-5793(96)01099-x. [DOI] [PubMed] [Google Scholar]
- 40.Steck T R. Use of low-melt agarose for the efficient isolation of large DNA fragments. BioTechniques. 1994;17:676–678. [PubMed] [Google Scholar]
- 41.Totten P A, Lory S. Characterization of the a-type flagellin gene from Pseudomonas aeruginosa PAK. J Bacteriol. 1990;172:7188–7199. doi: 10.1128/jb.172.12.7188-7199.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varki A. Biological roles of oligosaccharides. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wieland F, Paul G, Sumper M. Halobacterial flagellins are sulphated glycoproteins. J Biol Chem. 1985;260:15180–15185. [PubMed] [Google Scholar]
- 44.Wilson D R, Beveridge T J. Bacterial flagellar filaments and their component flagellins. Can J Microbiol. 1993;39:451–472. doi: 10.1139/m93-066. [DOI] [PubMed] [Google Scholar]
- 45.Winstanley C, Morgan J A W, Pickup R W, Saunders J R. Molecular cloning of two Pseudomonas flagellin genes and basal body structural genes. Microbiology. 1994;140:2019–2031. doi: 10.1099/13500872-140-8-2019. [DOI] [PubMed] [Google Scholar]
- 46.Winstanley C, Coulson M A, Wepner B, Morgan J A W, Hart C A. Flagellin gene and protein variation amongst clinical isolates of Pseudomonas aeruginosa. Microbiology. 1996;142:2145–2151. doi: 10.1099/13500872-142-8-2145. [DOI] [PubMed] [Google Scholar]
- 47.Zhang C. Bacterial signaling involving eukaryotic-type protein kinases. Mol Microbiol. 1996;20:9–15. doi: 10.1111/j.1365-2958.1996.tb02483.x. [DOI] [PubMed] [Google Scholar]