Abstract
Variations in the Toll‐interacting protein (TOLLIP) gene have been identified in genome‐wide association studies to correlate with risk of disease, mortality, and response to N‐acetylcysteine therapy in idiopathic pulmonary fibrosis. Although TOLLIP is known to modulate innate immune responses, its relevance in organ fibrogenesis remains unknown. Prior work in the literature suggests TOLLIP dampens transforming growth factor beta (TGFβ) signaling in human cell lines. In this study, we examined the role of TOLLIP in mouse lung fibroblast (MLF) responses to TGFβ and in the bleomycin model of experimental lung fibrosis using Tollip−/− mice. We hypothesize that if TOLLIP negatively regulates TGFβ signaling, then Tollip−/− mouse lung fibroblasts (MLFs) would have enhanced response to TGFβ treatment, and Tollip−/− mice would develop increased fibrosis following bleomycin challenge. Primary MLFs were stimulated with TGFβ (1 ng/mL) for 24 h. RNA was obtained to assess global transcriptional responses by RNA‐seq and markers of myofibroblast transition by qPCR. Functional assessment of TGFβ‐stimulated MLFs included cell migration by scratch assay, cell proliferation, and matrix invasion through Matrigel. In the in vivo model of lung fibrosis, Tollip−/− mice and wild‐type (WT) littermates were administered bleomycin intratracheally and assessed for fibrosis. We further examined TGFβ signaling in vivo after bleomycin injury by SMAD2, ERK1/2, and TGFβR1 Western blot. In response to TGFβ treatment, both WT and Tollip−/− MLFs exhibited global transcriptional changes consistent with myofibroblast differentiation. However, Tollip−/− MLFs showed greater number of differentially expressed genes compared to WT MLFs and greater upregulation of Acta2 by qPCR. Functionally, Tollip−/− MLFs also exhibited increased migration and Matrigel invasiveness compared to WT. We found evidence of enhanced TGFβ signaling in Tollip−/− through SMAD2 in vitro and in vivo. Tollip−/− mice experienced lower survival using a standard weight‐adjusted dosing without evidence of differences in fibrosis at Day 21. With adjustment of dosing for sex, no differences were observed in fibrosis at Day 21. However, Tollip−/− mice had greater weight loss and increased bronchoalveolar lavage fluid total protein during early resolution at Day 14 compared to WT without evidence of differences in acute lung injury at Day 7, suggesting impaired resolution of lung injury.
Keywords: bleomycin, fibroblasts, lung injury, myofibroblasts, TGFβ, Toll‐interacting protein, TOLLIP, transforming growth factor beta
Toll‐interacting protein (TOLLIP) deficiency enhances TGFβ signaling in mouse lung fibroblasts and lungs of bleomycin‐injured lung. Despite enhanced fibroblast activation and impaired negative regulation of TGFβ signaling in mouse lungs, Tollip−/− mice did not demonstrate worse lung fibrosis in vivo. However, increased weight loss and total protein in the bronchoalveolar lavage fluid were observed in Tollip−/− following bleomycin‐induced lung injury, suggesting other factors in lung repair and injury resolution may be affected by TOLLIP deficiency.
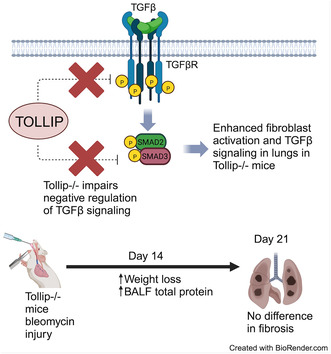
1. INTRODUCTION
Toll‐interacting protein (TOLLIP) is an intracellular adaptor protein consisting of three functional domains: an N‐terminal Tom1‐binding domain (TBD), a conserved 2 (C2) domain, and a C‐terminal ubiquitin‐to‐endoplasmic reticulum (CUE) domain. 1 TOLLIP has been studied extensively in the context of innate immunity, where it has been shown to play a role in the modulation of immune responses through interactions with adaptors of IL‐1R, TLR2, and TLR4 signaling pathways. 1 , 2 Recent studies have also investigated its role in adaptive immunity and noninfectious lung diseases. 3 , 4 , 5 , 6
To date, there are few studies examining the role of TOLLIP in fibrogenesis. Transforming growth factor beta (TGFβ) is a pro‐fibrotic cytokine that is central to fibrosis. In one study, overexpression of TOLLIP in human cell lines (HepG2 and HEK293T) impaired TGFβ signaling through modulation of SMAD7, an inhibitory SMAD in the canonical TGFβ signaling pathway, and trafficking of TGFβ type I receptor. 7 Thus, TOLLIP appears to dampen TGFβ signaling in vitro. How TOLLIP affects this signaling cascade in primary mouse lung fibroblasts (MLFs), the cells responsible for matrix deposition and architectural distortion in lung fibrosis, and whether TOLLIP deficiency affects outcomes in experimental fibrosis remain unknown.
In this study, we utilized transgenic Tollip knockout mice (Tollip−/−) to examine the effect of TOLLIP deficiency in an experimental model of lung fibrosis. We hypothesized that if TOLLIP negatively regulates TGFβ signaling, then mouse lung fibroblasts (MLFs) from Tollip−/− mice would exhibit enhanced transcriptional and functional responses to TGFβ stimulation in vitro. Furthermore, Tollip−/− mice would develop worse fibrosis in the experimental murine model of bleomycin‐induced pulmonary fibrosis.
2. METHODS
2.1. Animals and reagents
The University of Washington (UW) Institutional Animal Care and Use Committees approved all experiments involving the use of mice described in this report. Tollip−/− mice (B6.Cg‐Tolliptm1Kbns/Cnrm) were obtained from the European Mutant Mouse Archive (https://www.infrafrontier.eu) 8 and generously shared by Dr. Javeed Shah (University of Washington). 4 , 5 Male and female transgenic mice between ages of 8–16 weeks and at least 16 g in body weight were included in animal experiments. Investigators were blinded to genotypes of animals until after harvesting and processing of samples for analysis. Bleomycin (SICOR Pharmaceuticals, Inc. or TEVA) in lyophilized form was obtained from the UW Medical Center.
2.2. In vitro experiments
2.2.1. Mouse lung fibroblasts
Primary mouse lung fibroblasts (MLFs) were isolated from mouse lung digests as previously described. 9 Fibroblasts were passaged in complete growth media consisting of DMEM supplemented with 1% penicillin/streptomycin and 10% FBS until treatment. For TGFβ stimulation in vitro, MLFs were used between passages 1–3.
2.2.2. Bulk RNA‐seq analysis
For RNA‐seq and RT‐PCR experiments, primary mouse lung fibroblasts (MLFs) from WT and Tollip−/− were seeded at a density of 2.5 × 105 cells per well in a 6‐well tissue culture plate. Cells were cultured until 70% confluence at P1, then serum‐starved in serum‐free media (DMEM supplemented with 1% penicillin/streptomycin without FBS) overnight, and then exposed to either serum‐free media or TGFβ (1 ng/mL) for 24 h. RNA was collected using NucleoSpin® RNA kit (Macherey‐Nagel). RNA‐seq was performed at Northwest Genomics Center at the University of Washington, Seattle. Briefly, after RNA quality check (RNA integrity number ranged from 9.5 to 10), libraries were prepared using TruSeq Stranded mRNA kit (Illumina), sequenced using NovaSeq6000 (~50 million reads/sample), demultiplexed, and aligned to GRCm39 with reference transcriptome GENCODE release m30 using STAR (v2.6.1d). Transcript‐level counts were then generated using RNA‐SeQC (v2.3.3). Multidimensional scaling using principal component analysis (PCA) was performed using Partek Flow software (St. Louis, MO). Differentially expressed genes (DEGs) were identified using DESeq2 package in R. 10 We used an Benjamini–Hochberg (BH) adjusted p‐value < 0.05 and absolute Log2[fold change] >1 to define differentially expressed genes. Gene lists ranked by the DESeq2 statistic value were used for gene set enrichment analysis (GSEA) 11 using the clusterProfiler R package 12 based on gene ontology's (GO) biological processes. Significantly enriched pathway nodes with BH adjusted p‐value < 0.01 that shared connections (edges) with more than three other gene sets were selected for the enriched map visualization of the GSEA results using the enrichplot R package.
2.2.3. Real‐time PCR
Total RNA was isolated from homogenized lung tissue using NucleoSpin® RNA kit (Macherey‐Nagel) in conjunction with DNase treatment as per manufacturers' specifications. Total RNA was reverse‐transcribed to cDNA using Applied Biosystems High‐Capacity cDNA Archive Kit or iScript Reverse Transcription SuperMix (Bio‐Rad). Real‐time PCR was done using an ABI 7900HT instrument with the use of predesigned primers and probes (ABI TaqMan Gene Expression Assays and IDT PrimeTime® Std qPCR Assays). Quantification of gene expression was normalized to Hprt (endogenous controls). Predesigned primers and probes include: Acta2 (Mm01546133_m1, ABI), Col1a1 (Mm00801666_g1, ABI), Fn1 (Mm01256744_m1, ABI), Hprt (Mm03024075_m1, ABI). Analysis was performed in MS Excel calculating RQ by 2−ΔΔCT method. Gene expression level was normalized to averaged value for WT control group.
2.2.4. Western blot
Equal amounts of protein (5 μg) were separated by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE), and electrophoretically transferred to PVDF membrane. Membranes were blocked with 5% nonfat dry milk/0.05% Tween‐20/TBS (TBST) for 1 h at room temperature, incubated with one of the following primary antibodies over night at 4°C: rabbit anti‐phospho‐SMAD2 antibody (1:1000) (Cell Signaling, Cat. #3108), rabbit anti‐SMAD2/3 antibody (1:1000) (Cell Signaling, Cat. #3102), mouse anti‐GAPDH antibody (1:10,000) (Calbiochem, Cat. #CB1001), rabbit anti‐Erk1/2 antibody (1:5000) (Cell Signaling, Cat. #9102), rabbit anti‐phospho‐Erk1/2 antibody (1:5000) (Cell Signaling, Cat. #4376), or rabbit anti‐TGFRβ1 (1:2000) (Invitrogen, Cat. #PA5‐86551). Following primary antibody incubation, membranes were washed with 0.1% TBST, incubated with horseradish peroxidase‐conjugated goat anti‐rabbit IgG (1:10,000) (Invitrogen, Cat. 65–6120) or goat anti‐mouse IgG (1:10,000) (Invitrogen, p/n: 626520) for 1 h at RT, washed with 0.1% TBST and then developed with enhanced chemiluminescence (ECL) technique (Pierce). Phospho‐protein intensities were normalized to total proteins (presented in Figures 1, 2, 3, 4, 5, 6) and to GAPDH (presented in Figures [Link], [Link]). TGFRβ1 intensities were normalized to GAPDH.
FIGURE 1.
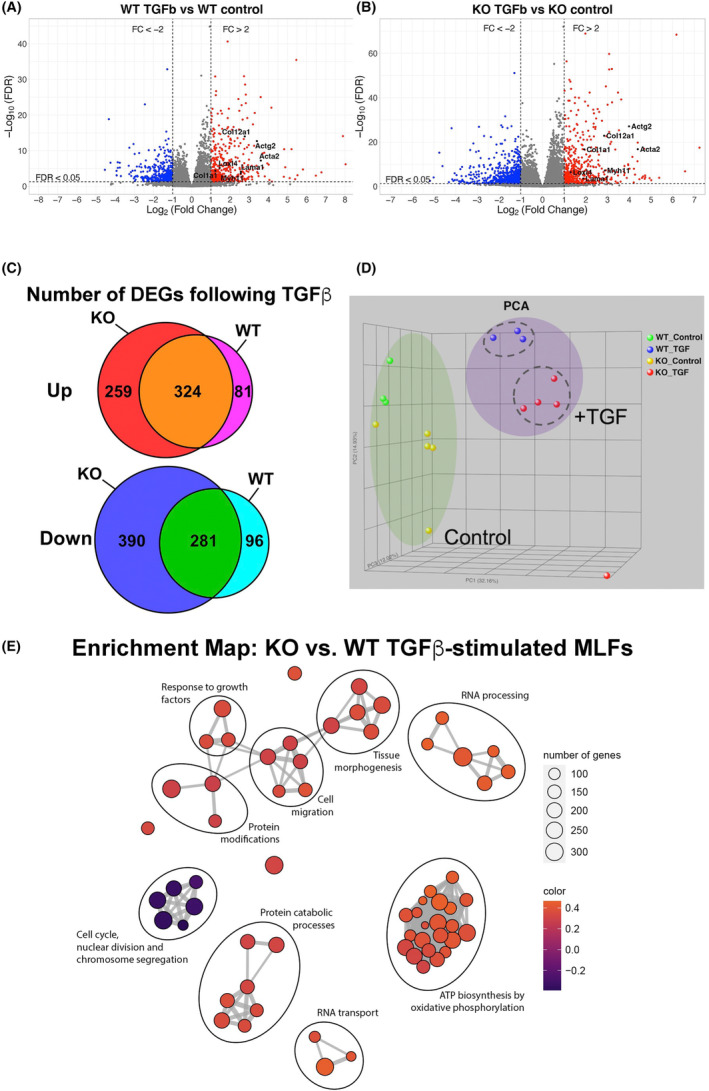
(A, B) Volcano plots of differentially expressed genes (DEGs) identified by RNA‐seq in WT (A) and Tollip−/− (B) MLFs following 24 h treatment with TGFβ. x‐axis represents fold‐change (FC) in gene expression over unstimulated control MLFs (log2 scale), and y‐axis presents false discovery rate (FDR in −Log10 scale). Dotted lines represent threshold for significance (Log2[FC] >1 or <−1, FDR < 0.05). Example of genes upregulated with myofibroblastic transition (Acta2, Actg2, Col12a1, Col1a1, Myh11, Lama1, Loxl4) are annotated in the figure. (C) Diagram illustrating the number of DEGs that were significantly upregulated and downregulated in each genotype. (D) PCA showing untreated WT MLFs (green dots), untreated Tollip−/− MLFs (yellow dots), TGFβ‐treated WT MLFs (blue dots), and TGFβ‐treated Tollip−/− MLFs (red dots). As expected, molecular signatures of MLFs treated with TGFβ differ vastly from those of untreated MLFs. Tollip−/− MLFs can be distinguished from the WT within the TGFβ‐treated group although the differences in their transcriptional signatures are less pronounced than TGFβ treatment versus no treatment. One outlier sample (TGFβ KO) was identified and excluded from all downstream analyses. (E) Network visualization of differentially enriched biological processes between WT and Tollip−/− groups after TGFβ treatment. Programs related to cell migration, tissue morphogenesis, response to growth factors, protein modifications, protein catabolic processes, RNA transport, RNA processing, and ATP biosynthesis by oxidative phosphorylation are enriched in TGFβ‐stimulated Tollip−/− MLFs.
FIGURE 2.
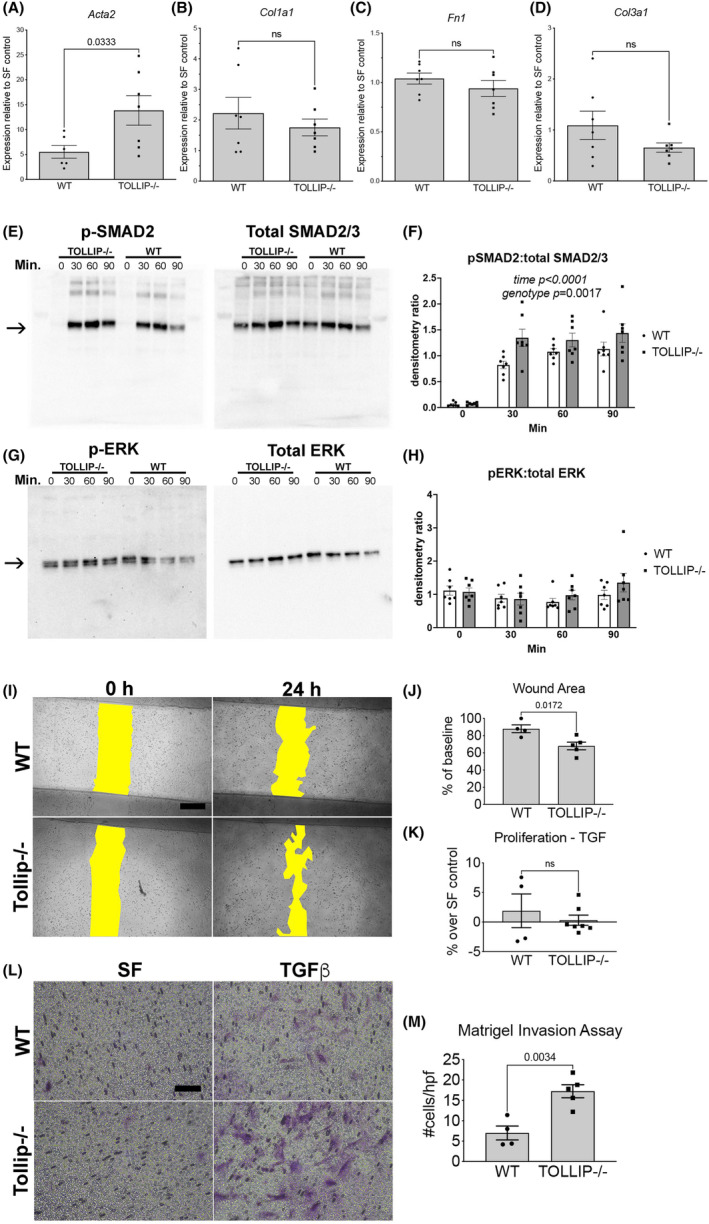
(A–D) QPCR of select genes (Acta2, Col1a1, Fn1, Col3a1) in MLFs following 24 h stimulation with TGFβ. Graphs represent fold change in expression over untreated MLF controls segregated by genotype. (Mean ± SEM, n ≥ 6/group, ns = not significant) (E, F) Western blot images of MLF protein lysates probed for phospho‐SMAD2 (left) and total SMAD2 (right) at 0, 30, 60, and 90 min after TGFβ stimulation. Graph shows densitometry results of pSMAD2‐to‐total SMAD2/3 ratio. (Mean ± SEM, n = 7/group, time factor p < 0.0001, genotype factor p = 0.0017 by two‐way ANOVA). Comparison at individual timepoints with Bonferroni's correction for multiple comparisons showed significant difference between WT and Tollip−/− at 30 min (p = 0.008). (G, H) Western blot images of MLF protein lysates probed for phospho‐ERK1/2 (left) and total ERK1/2 (right) at 0, 30, 60, and 90 min after TGFβ stimulation. Graph shows densitometry results of pERK1/2‐to‐total ERK1/2 ratio. (Mean ± SEM, n = 7/group, time and genotype factors were not significant by 2‐way ANOVA) (I) Representative 4× images of scratch assay. Wound areas at 0 and 24 h are shown in yellow. (Scale bar = 500 μm) (J) Quantification of wound area at 24 h expressed as % of baseline area at 0 h. (Mean ± SEM, n ≥ 4/group) (K) Proliferation assay at 24 h following TGFβ treatment, expressed as % over serum‐free (SF) control. (ns = not significant) (L, M) Representative 20× images of Matrigel invasion assay (scale bar = 100 μm). Images shown are underside of Matrigel‐coated transwell membranes with stained (purple) invasive MLFs following 24 h treatment in SF or TGFβ. Graph represents average number of invasive cells per high power field (hpf) from five random images per biological sample. (Mean ± SEM, n ≥ 4/group).
FIGURE 3.
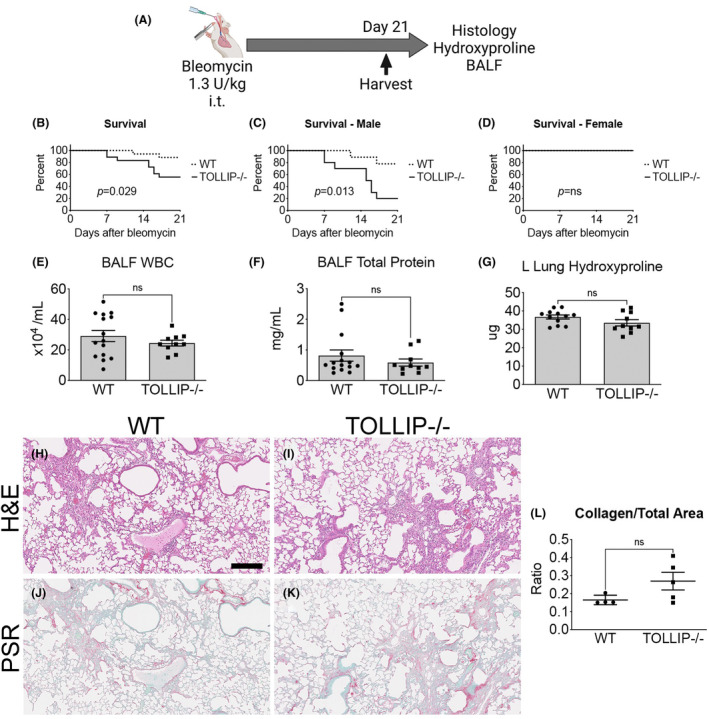
(A) Schematic of standard bleomycin dose model by intratracheal (i.t.) instillation. (B–D) Survival curves of subjects from Day 0–21 (n ≥ 17/grp). Male (C) and female (D) animal survival curves were further analyzed separately. (E, F) BALF analysis. (Mean =/− SEM, n ≥ 10/group, ns = not significant) (G) Left lung hydroxyproline content. (Mean =/− SEM, n ≥ 10/group, ns = not significant) (H–L) Histological analysis of fibrosis at Day 21. Representative H&E images (H–I) and picrosirius red (PSR) images (J, K) are shown (scale bar = 100 μm). Quantification of collagen content by histology is presented as PSR+ area‐to‐total lung area on whole slide scanning (Visiopharm®). (Mean ± SEM, n = 5/group, ns = not significant).
FIGURE 4.
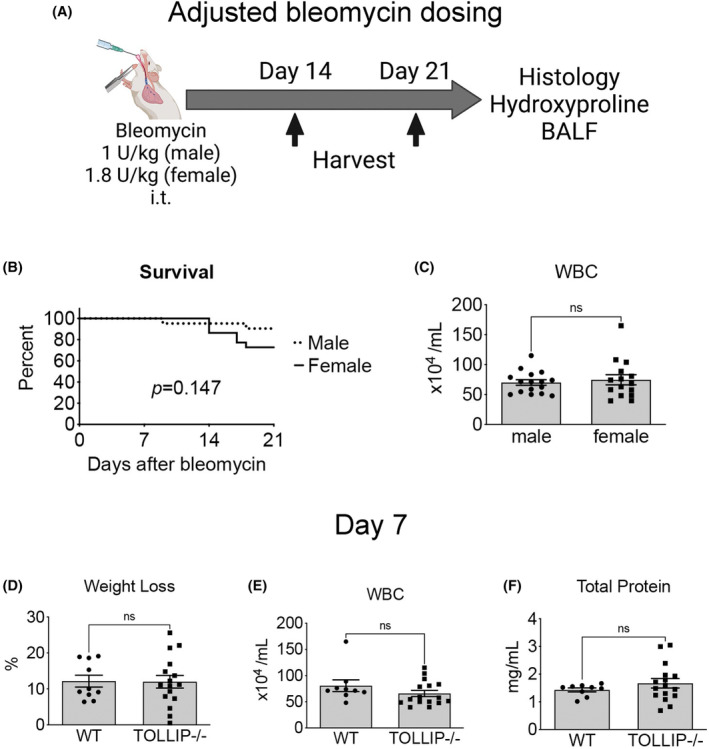
(A) Schematic of modified bleomycin dosing to analyze early repair (Day 14) and established fibrosis (Day 21). (B) Survival curve stratified by male and female subjects showing no difference between sexes in the modified regimen (n ≥ 21/group). (C) BALF WBC at Day 7 segregated by sex. (Mean ± SEM, n ≥ 21/group, ns = not significant) (D–F) Weight loss, BALF WBC, and BALF total protein at Day 7. (Mean ± SEM, n ≥ 10/group, ns = not significant).
FIGURE 5.
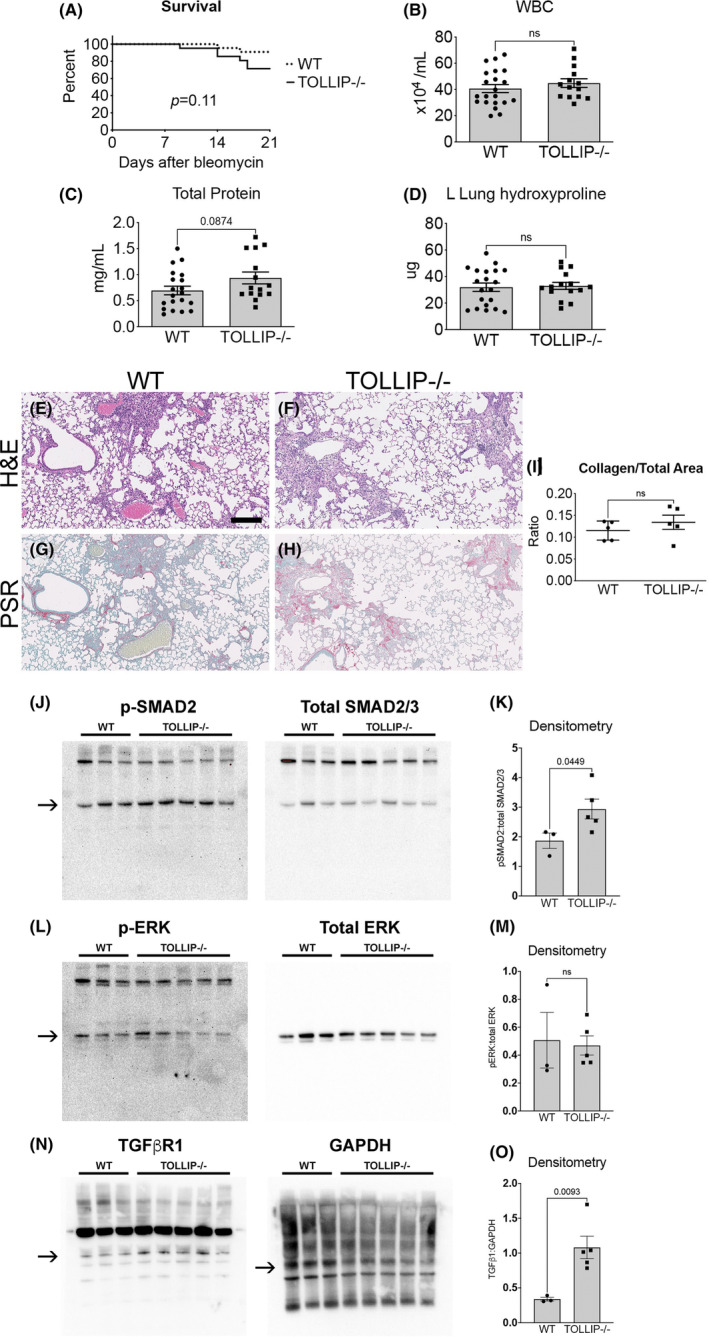
(A) Survival is similar between WT and Tollip−/− animals in the modified regimen model (n ≥ 21/group). (B–D) BALF WBC, total protein, left lung hydroxyproline at Day 21. (Mean ± SEM, n ≥ 15/group, ns = not significant) (E–I) Histological analysis of fibrosis at Day 21. Representative H&E images (E, F) and picrosirius red (PSR) images (G, H) are shown (scale bar = 100 μm). (I) Quantification of collagen content by histology is presented as PSR+ area‐to‐total lung area on whole slide scanning (Visiopharm®). (Mean ± SEM, n = 5/group, ns = not significant). (J–O) Lung homogenates from WT and Tollip−/− mice at Day 21 following the modified bleomycin regimen were probed for p‐SMAD2 and SMAD2/3, p‐ERK1/2 and ERK1/2, and TGFβR1 and GAPDH. Representative Western blots are depicted (J, L, N, respectively). Densitometries of p‐SMAD2:SMAD2/3 (K), p‐ERK1/2:ERK1/2 (M), and TGFβR1:GAPDH (O) are shown. (Mean ± SEM, n ≥ 3/group, ns = not significant).
FIGURE 6.
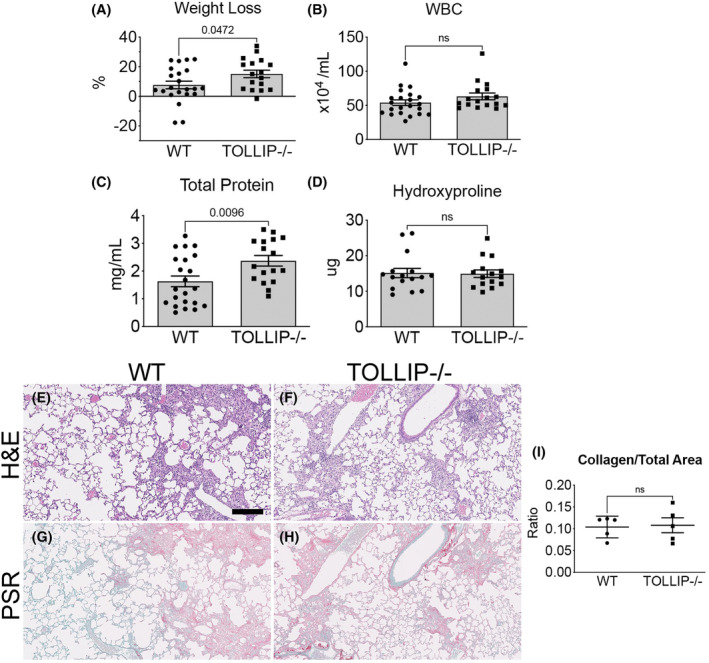
(A) Weight loss at Day 14. (Mean ± SEM, n ≥ 17/group, p = 0.0472) (B, C) BALF WBC, total protein, and left lung hydroxyproline at Day 14. (Mean ± SEM, n ≥ 17/group, ns = not significant) (E–I) Histological analysis of fibrosis at Day 21. Representative H&E images (E, F) and picrosirius red (PSR) images (G, H) are shown (scale bar = 100 μm). Quantification of collagen content by histology is presented as PSR+ area‐to‐total lung area on whole slide scanning (Visiopharm®). (Mean ± SEM, n = 5/group, ns = not significant).
Densitometric analysis of relative band intensities was performed by analyzing scanned blots with NIH Image J.
2.2.5. Scratch assay
Mouse lung fibroblast migration was assessed by in vitro scratch assay as previously described with minor modifications. 13 In brief, MLFs were seeded in 24‐well tissue culture plates at a density of 1.25 × 105 cells per well and allowed to adhere overnight in complete growth media. At or near confluence, cells were placed in serum‐free media overnight. A linear scratch was applied to the confluence cells using a 200‐μL pipette tip perpendicular to parallel lines drawn at the bottom of the well as landmark. Cells were then placed in serum‐free DMEM (control media) or treatment (TGFβ 1 ng/mL in SF DMEM). Micrographs of linear scratches were taken under 4× objective on a phase‐contrast microscope at 0 h and 24 h. Wound areas at 0 h and 24 h were analyzed using ImageJ software, and wound closure (migration) was expressed as the ratio of wound area at 24 h to area at 0 h.
2.2.6. Matrix invasion assay
CytoSelect™ 24‐well Cell Invasion Assay (Cell Biolabs, Cat # CBA‐110) was used to assess MLFmatrix invasion. We conducted the experiment per manufacturer protocol with the following specifications: Insert membranes were pre‐wetted with serum‐free DMEM for 10 min at room temperature. Following the step, serum‐free DMEM was added to the wells (lower chamber). We seeded the inserts (upper chamber) with 1.5 × 105 cells in complete growth media. The cells were allowed to attach to the membrane at 37°C, after which the media were removed from the lower and upper chambers. Cell culture media corresponding to the experimental conditions were then placed in the upper and lower chambers (serum‐free or TGFb 1 ng/mL) and the cells were allowed to incubate for 24 h until fixation and cell counting per manufacturer protocol.
2.2.7. Proliferation assay
Cell proliferation was evaluated using CyQuant Cell Proliferation Kit (ThermoFisher, #C7026). Briefly, MLFs were seeded at a density of 1 × 104 cells per well in 96‐well tissue culture‐treated black plates with clear bottom for fluorescence applications (Corning Cat. #3904). Cells were allowed to attach for 3 h in complete growth medium, after which the medium was replaced with treatment media (serum‐free or TGFβ 1 ng/mL). Cells were incubated with treatment media for 24 h, gently washed in sterile PBS once, and then frozen in −80°C. After thawing, proliferation assay was performed per manufacturer's protocol. Fluorescence was detected using Synergy 4 microplate reader (Biotek). Proliferation is reported as relative fluorescence units compared to treatment control (media only).
2.3. Bleomycin lung injury model
2.3.1. Bleomycin‐induced lung injury
Lyophilized bleomycin was reconstituted in sterile saline. Bleomcyin was delivered to anesthetized mice by intratracheal instillation (at 1 U/kg, 1.3 U/kg, or 1.8 U/kg) under isoflurane anesthesia as previously described. 9 Mice were monitored per protocol and harvested at Days 7, 14, or 21.
2.3.2. Protein assay
Protein measurements of bronchoalveolar lavage fluid (BALF) collected from animals were performed using Bradford Protein Assay (Bio‐Rad) per manufacturer's instructions. Lung homogenates of left lungs were prepared in Bead Ruptor (Omni International) in lysis buffer with phosphatase/protease inhibitors (Pierce, Cat. 1860932). Homogenates were clarified by ultracentrifugation at 14,000 g × 15 min at 4°C, and the supernatants were collected and protein concentration assayed by commercial kit (Pierce BCA Protein Assay, Cat 23225).
2.3.3. Hydroxyproline assay
Left lungs from mice were collected at selected timepoints following bleomycin administration. Left lungs were weighed and homogenized in 1‐mL deionized water. 100 μL of lung homogenate was mixed in 100 μL of 37% hydrochloric acid (Sigma #258148) and heated at 120°C for 3 h. 50 μL of the acid‐hydrolyzed samples were added to 96‐well flat‐bottom microplates in duplicates. Hydroxyproline content was then measured using the Hydroxyproline Assay Kit (Sigma #MAK0008) per manufacturer's protocol. Hydroxyproline measurements were expressed as μg hydroxyproline/left lung.
2.3.4. Histology
Right lungs were inflated with 10% neutral buffered formalin (VWR) at 25 cm H2O pressure and fixed for 24 h at room temperature. 4 μm lung sections prepared from formalin‐fixed paraffin‐embedded (FFPE) tissue blocks were stained with hematoxylin and eosin (H&E) for histology and with picrosirius red/fast green for histological evaluation of collagen deposition (Histology and Imaging Core at the University of Washington, Seattle). Stained whole slides were digitized (Hamamatsu NanoZoomer), and analysis of collagen‐to‐total lung area was performed on picrosirius‐stained sections following masking of collagen staining around central airways and vasculature (Viziopharm®).
2.3.5. Statistics
Means of data with two independent variables (time and genotype) were compared using two‐way analysis of variance (ANOVA) with Bonferroni's correction for multiple comparisons. We used Student t‐test for comparison of paired parametric data and Mann–Whitney's U test for nonparametric data. All tests were two‐tailed, and p values ≤ 0.05 were considered significant. Statistical analysis was performed using GraphPad Prism for Macintosh version 10. All experiments were repeated with a minimum of three biological replicates, and summary statistics presented as mean ± SEM.
3. RESULTS
3.1. Tollip−/− mouse lung fibroblasts exhibit enhanced response to TGFβ
Overexpression of TOLLIP in HEK293T and HepG2 cell lines reduced TGFβ responsiveness by promoting degradation of TGFβ Type I receptor, suggesting TOLLIP may be a novel modulator of TGFβ signaling. 7 Whether TOLLIP deficiency alters TGFβ responsiveness in primary lung fibroblasts remains unknown. We therefore interrogated global transcriptional response of WT and Tollip−/− MLFs to TGFβ (1 ng/mL) treatment by RNA‐seq. We identified one Tollip−/− MLF sample treated with TGFβ (KO_TGF) that was an outlier with high Tollip expression. This data point is shown in the principal component analysis (Figure 1D), but it was excluded from downstream analyses of differential gene expression. Both WT and KO mouse lung fibroblasts upregulated genes associated with myofibroblast differentiation such as Acta2, Col1a1, and Loxl1 in response to TGFβ stimulation (Figure 1A,B). However, the number of differentially upregulated (red dots) and downregulated (blue dots) genes was significantly higher in KO fibroblasts (Figure 1A,B). WT MLFs exposed to TGFβ for 24 h had 405 differentially upregulated and 377 downregulated genes compared to serum‐free control, whereas Tollip−/− MLFs exhibited 583 differentially upregulated and 671 downregulated genes compared to serum‐free control (Figure 1C). Principal component analysis revealed that TGFβ treatment exerted a strong effect on the transcriptional signature of MLFs regardless of genotype, as would be expected (Figure 1D). However, the number of differentially expressed genes in Tollip−/− MLFs after 24 h of TGFβ stimulation compared to WT MLFs suggests Tollip−/− MLFs have greater responsiveness to TGFβ stimulation.
Gene set enrichment analysis revealed downregulation of biological processes related to nuclear division and chromosome segregation and upregulation of pathways linked to morphogenesis and cell migration in Tollip−/− versus WT MLFs under serum‐free conditions (control) (Figure S1A). With TGFβ treatment, Tollip−/− MLFs upregulated biological processes involved in cell migration, tissue morphogenesis, response to growth factors, ATP biosynthesis, protein catabolic processes, RNA metabolism, and downregulated cell cycle‐associated pathways compared to WT MLFs (Figure 1E).
In independent in vitro stimulation experiments, we evaluated the upregulation of common profibrotic genes in MLFs following TGFβ stimulation (1 ng/mL) for 24 h by qPCR. Tollip−/− MLFs showed significantly greater upregulation of Acta2 compared to WT MLFs, but no significant differences were observed with Col1a1, Fn1, or Col3a1 (Figure 2A–D). We further examined SMAD2 phosphorylation in Tollip−/− and WT MLFs at 0, 30, 60, and 90 min following TGFβ (1 ng/mL) exposure in vitro. Tollip−/− MLFs showed increased phospho‐SMAD2‐to‐total SMAD2/3 ratio at 30 min, 60 min, and 90 min following TGFβ stimulation (Figure 2E,F). Investigation of alternative TGFβ signaling through ERK1/2 pathway, however, did not reveal significant differences between WT and Tollip−/− MLFs following TGFβ stimulation (Figure 2G,H).
To examine the functional effects of TGFβ stimulation on Tollip−/− MLFs, we studied other aspects of myofibroblast activation including migration by scratch assay, cell proliferation, and matrix invasion through Matrigel. These functional outcomes relate to biological processes such as cell migration, response to growth factors, and cell division, which were altered in stimulated Tollip−/− MLFs based on RNA‐seq data (Figure 1E). TGFβ‐treated Tollip−/− MLFs exhibited greater migration (smaller wound area) at 24 h compared to WT MLFs (Figure 2I,J). This difference could not be explained by differences in proliferation at 24 h which was similar between the two groups (Figure 2K). Furthermore, TGFβ‐treated Tollip−/− MLFs exhibited greater Matrigel invasion compared to TGFβ‐treated WT MLFs (Figure 2L,M). These data suggest TOLLIP deficiency in MLFs enhanced their responsiveness to TGFβ signaling and augmented aspects of myofibroblast activation including cell migration and matrix invasion.
3.2. Bleomycin exposure leads to decreased survival in Tollip−/− mice but no difference in fibrosis
Based on the in vitro data showing enhanced TGFβ responsiveness in Tollip−/− MLFs, we speculated that Tollip−/− mice would demonstrate increased fibrosis in the bleomycin‐induced lung injury model. Bleomycin (1.3 U/kg) was administered to Tollip−/− and wild‐type littermates (WT) by intratracheal instillation (i.t.) and lungs were harvested at D21 (Figure 3A). Bleomycin‐induced lung injury led to decreased survival in Tollip−/− mice compared to WT (p = 0.029, n ≥ 17/grp) (Figure 3B). When separated by sex, the survival difference was driven by male mice (78% WT vs. 20% Tollip−/−, p = 0.013, n ≥ 9/grp), whereas this dose did not affect survival in either Tollip−/− or WT female mice (Figure 3C,D). At Day 21, BALF WBC, total protein, and L lung hydroxyproline were not significantly different between Tollip−/− and WT. Histological analysis of collagen staining by picrosirius red did not reveal significant differences between the two groups (n ≥ 4/grp) (Figure 3H–L). Sub‐analysis of BALF total protein and hydroxyproline by sex also did not reveal a significant difference (Figure S3A,B). Together, these data suggest male Tollip−/− mice exhibited increased susceptibility to lung injury from bleomycin without histological or biochemical difference in lung fibrosis compared to WT. The lack of difference at Day 21, however, may be confounded by survivor bias in surviving Tollip−/− males (20%). Furthermore, the low 21‐day survival rate of Tollip−/− males limited the feasibility of assessing fibrosis outcomes at Day 21.
3.3. Adjusted bleomycin dose leads to similar acute injury between sexes and no difference in fibrosis
To circumvent the sex differences in bleomycin susceptibility and high mortality in Tollip−/− males, we next adjusted the bleomycin administered to male and female mice (Figure 4A). Using the modified dose (1 U/kg for male mice, 1.8 U/kg for female mice), we observed comparable survival at Day 21 (n ≥ 21/group) (Figure 4B) and BALF WBC at Day 7 between male and female mice (Figure 4C), suggesting the modified strategy induced comparable acute lung injury in male and female mice without significant survival differences.
Using this dosing strategy, we first evaluated acute lung injury in WT and Tollip−/− mice at Day 7. There was no significant difference in weight loss, BALF WBC, or BALF total protein between WT and Tollip−/− (Figure 4D–F). Sub‐analysis of weight loss and BALF total protein by sex at Day 7 also did not reveal a significant difference (Figure S3C,D).
At Day 21, there was no significant difference in survival, BALF WBC, and BALF total protein although there was a nonsignificant trend towards increased BALF total protein in Tollip−/− mice (Figure 5A–C). Measures of fibrosis including L lung hydroxyproline (Figure 5D) and collagen staining by picrosirius red (Figure 5G,H, quantitative analysis Figure 5I) did not reveal significant difference between WT and Tollip−/−. No significant difference in survival was observed in male or female cohorts, and BALF total protein as well as left lung hydroxyproline were also not significantly different when analyzed by sex (Figure S3E,F). Examination of TGFβ signaling in lung homogenates revealed similar findings to in vitro studies. P‐SMAD2 signaling was enhanced in Tollip−/− mouse lungs but not p‐ERK1/2 (Figure 5J–M). Furthermore, WT lung homogenates exhibited lower expression levels of TGFβR1 compared to Tollip−/−, suggesting the impaired negative regulation of TGFβ signaling through SMAD may be occurring at the level of TGFβ receptor in Tollip−/− mice (Figure 5N,O).
When we examined at an earlier timepoint at Day 14, we observed increased weight loss in the Tollip−/− group compared to WT (15 ± 2.5% vs. 7.6 ± 2.6%, mean ± SEM, n ≥ 17/grp, p = 0.0472) (Figure 6A). We also observed increased total protein in BALF in the Tollip−/− mice (2.4 ± 0.2 mg/mL vs. 1.6 ± 0.2 mg/mL, mean ± SEM, n ≥ 17/grp, p = 0.0096) but BALF WBC was not significantly different between the two groups (Figure 6B,C). L lung hydroxyproline and collagen staining were also not significantly different between the two groups (Figure 6D, Figure 6G–I). When segregated by sex, no significant difference in weight loss was observed (although there was a trend towards more weight loss in female Tollip−/− mice vs. WT mice) and female Tollip−/− mice showed greater BALF total protein compared to WT mice. However, no difference was observed in left lung hydroxproline content (Figure S3G–I).
4. DISCUSSION
The human TOLLIP gene is located on chromosome 11 at the p15.5 region. 1 In a genome‐wide association study (GWAS), SNPs in the TOLLIP gene, which conferred decreased TOLLIP mRNA expression, were associated with increased risk of developing idiopathic pulmonary fibrosis (IPF) and mortality. 14 Furthermore, genotype within TOLLIP gene (rs3750920) modified treatment response to N‐acetylcysteine (NAC) in patients with IPF. 15 Collectively, these data from human genomic studies suggest TOLLIP expression may have a role in disease pathogenesis and response to therapy in IPF. Expression of TOLLIP has been shown to be lower in lung tissues from IPF subjects compared to controls. 6 A recent observational study on chronic hypersensitivity pneumonitis found that subjects with the GG genotype at the rs5743899 locus in the TOLLIP gene experienced more rapid clinical decline as defined by change in forced vital capacity over time. 16 The GG genotype is associated with lower transcription and translation of TOLLIP, as well as higher phospho‐SMAD2 in lung tissues, suggesting enhanced TGFβ signaling in these subjects. Despite the association between reduced TOLLIP expression and lung fibrosis in population studies, mechanistic insight remains sparse in the literature.
In this study, we hypothesized that TOLLIP deficiency leads to enhanced TGFβ signaling in mouse lung fibroblasts and increased fibrosis in the bleomycin injury model. We confirmed enhanced response to TGFβ signaling in Tollip−/− MLFs, demonstrating increased phosphorylation of canonical SMAD pathway and functional differences including migration and matrix invasiveness after activation with TGFβ. Acta2 expression was significantly increased in TGFβ‐treated Tollip−/− MLFs compared to WT by qPCR, though the upregulation of Col1a1 was not different between the genotypes. Expression of Fn and Col3a1 was not increased in stimulated MLFs of either genotype, although transcriptional response in these genes may not be apparent at 24 h. These results are consistent with the transcriptional profiling of MLFs by RNA‐seq following TGFβ treatment for 24 h. In the differentially expressed genes analysis, Acta2 and Col1a1 were both significantly upregulated compared to unstimulated controls, whereas Col3a1 and Fn1 were not (significance defined by a cutoff of fold change >2, false discovery rate (FDR) < 0.05).
Given the strong profibrotic effect of TGFβ on cultured fibroblasts, we anticipated that stimulated WT and Tollip−/− MLFs would undergo myofibroblastic transcriptional changes as reflected in the RNA‐seq analysis. Both WT and Tollip−/− MLFs upregulated matrix‐associated and contractile genes including Col12a1, Col11a1, Actg2, Timp1, Myh11, Acta2, Serpinh1, Col1a1, and Lox in response to TGFβ stimulation. While both genotypes shared many myofibroblast features with stimulation, Tollip−/− MLFs maintained higher numbers of differentially regulated genes compared to WT MLFs at 24 h. Together with the functional responses to TGFβ stimulation and increased pSMAD2 signals in Tollip−/− fibroblasts, our study provides supportive evidence for a regulatory role of TOLLIP in TGFβ signaling in lung fibroblasts.
Limited evidence in human cell lines suggests TOLLIP downregulates SMAD2/3 signaling, thereby acting as a “brake” against unchecked and persistent TGFβ stimulation. TGFβ receptors reside on cell membranes and are constantly recycled. 17 Receptor binding to TGFβ leads to heterodimerization of TGFβ receptors I and II and canonical SMAD and noncanonical pathway signaling downstream to regulate the expression of TGFβ responsive genes. 17 , 18 Signaling through TGFβ receptors is modulated by receptor availability. 19 Ubiquination of TGFβ receptor I upon binding to its ligand targets the receptor for intracellular degradation. 20 , 21 In a study using human epithelial tumor cell lines (HepG2 and HEK293T), Zhu and colleagues showed that human TOLLIP, in conjunction with SMAD7, interacts with ubiquinated TGFβ receptor I through the ubiquitin binding domain on TOLLIP. 7 Overexpression of human TOLLIP accelerated degradation of TGFβ receptor I, thereby dampening the effect of TGFβ. Modulation of TGFβ signaling in primary MLFs may occur through similar receptor‐level controls where Tollip−/− mice lack the “braking” effect by TOLLIP. Tollip−/− lung homogenates from injured lungs collected at Day 21 exhibited greater p‐SMAD2 levels similar to MLFs. Furthermore, protein expression of TGFβR1 was higher in Tollip−/−, suggesting impaired negative regulation of TGFβ signaling has mechanistic relevance in this in vivo model.
While not extensively reported in the literature, weight‐based bleomycin dosing leads to greater lung injury in male mice than in female mice. In our study, male mice experienced lower survival with standard dosing (1.3 U/kg) compared to female mice. These results are consistent with previously reported sex dimorphism in bleomycin‐induced lung injury in which male mice exhibit greater lung injury compared to female mice when administered a weight‐based bleomycin regimen. 22 To ensure adequate survival of male mice to Day 21 and to ensure similar lung injury between the sexes, we repeated the study with a sex‐ and weight‐based (modified) regimen.
Neither the standard nor the modified bleomycin regimen led to a significant difference in lung fibrosis, despite the enhanced response of Tollip−/− MLFs to TGFβ. One potential confounder is the well‐recognized modulatory role of TOLLIP in acute inflammation through IL‐1R and TLRs, 2 which may bias outcomes in fibrosis. In our study, we examined acute inflammatory responses (Day 7) and did not find significant differences in markers of acute injury including BALF WBC and total protein. Thus, our findings in fibrosis were not likely influenced by early differences in acute lung injury.
Interestingly, TOLLIP deficiency affected survival and resolution in this model. The survival difference was apparent in the standard dosing (1.3 U/kg). Tollip−/− male mice exhibited markedly lower survival than WT male mice at this dose. While the modified dose of bleomycin (1 U/kg for male, 1.8 U/kg for female) did not lead to a survival difference between genotypes, Tollip−/− mice nevertheless showed persistent and significant increase in weight loss and BALF total protein compared to WT at Day 14, suggesting impaired early resolution.
The mechanism for the survival difference (at the standard bleomycin dose) and the impaired early resolution (at the adjusted dose) is incompletely understood. Possible explanations include greater susceptibility to oxidative stress and altered repair functions in Tollip−/− alveolar epithelial cells. Although we did not test for these possibilities in this study, there is evidence in the literature to suggest these processes may have contributed to increased bleomycin susceptibility in Tollip−/− animals. 6 Furthermore, how TOLLIP deficiency alters the composition and function of monocyte and macrophage subpopulations in the bleomycin model remains unknown. Our study further highlights the complex mechanistic roles TOLLIP may play in lung repair through modulation of the TGFβ pathway, in MLFs and likely in other cellular populations critical to normal lung repair including epithelial cells, monocytes, and macrophages. Investigation of these mechanistic roles will require further studies employing lineage‐specific deletion of TOLLIP in relevant injury models.
In summary, TOLLIP deficiency enhances TGFβ signaling in MLFs. Tollip−/− mice experience lower survival and impaired resolution of lung injury in the bleomycin model. Despite these differences, organ fibrosis is not altered. Future directions will focus on the role of TOLLIP in regulating resistance to oxidative stress in lung epithelial cells and how TOLLIP deficiency affects repair by different cellular populations of the lung including alveolar epithelial cells (type 1 and 2), monocytes, and macrophages.
AUTHOR CONTRIBUTIONS
CFH, WCL, and WAA were involved in research design; CFH and YHC were involved in acquiring data; CFH, CLM, and SAG were involved in analyzing data; CFH provided reagents; CFH, CLM, and SAG were involved in RNA‐seq analysis; CFH, CLM, WCL, WAA, and SAG were involved in writing of this manuscript.
FUNDING INFORMATION
Funding for this work is provided by Boehringer Ingelheim Pharmaceuticals, Inc. (BIPI) to CFH.
CONFLICT OF INTEREST STATEMENT
The author(s) meet criteria for authorship as recommended by the International Committee of Medical Journal Editors (ICMJE). This was an independent, investigator‐initiated study supported by Boehringer Ingelheim Pharmaceuticals, Inc. (BIPI). BIPI had no role in the design, analysis, or interpretation of the results in this study; BIPI was given the opportunity to review the manuscript for medical and scientific accuracy as it relates to BIPI substances, as well as intellectual property considerations.
Supporting information
Figure S1.
Figure S2.
Figure S3.
Data S1.
ACKNOWLEDGMENTS
CFH is supported by NIH grants R03HL155075 and R01HL166273‐01. CLM is supported by FPU ayudas de movilidad para estancias breves (EST22/00404), from Ministerio de Universidades and by CIBERES ayudas de perfeccionamiento y movilidad, from Instituto de Salud Carlos III.
Chow Y‐H, López‐Martínez C, Liles WC, Altemeier WA, Gharib SA, Hung CF. Toll‐interacting protein inhibits transforming growth factor beta signaling in mouse lung fibroblasts. FASEB BioAdvances. 2024;6:12‐25. doi: 10.1096/fba.2023-00054
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in the methods and/or supplementary material of this article. Detailed RNA‐seq information and raw data meeting MINSEQE (Minimum Information About a Next‐generation Sequencing Experiment) are deposited at Gene Expression Omnibus (https://www.ncbi.mlm.nih.gov/geo/).
REFERENCES
- 1. Capelluto DG. Tollip: a multitasking protein in innate immunity and protein trafficking. Microbes Infect. 2012;14:140‐147. [DOI] [PubMed] [Google Scholar]
- 2. Kowalski EJA, Li L. Toll‐interacting protein in resolving and non‐resolving inflammation. Front Immunol. 2017;8:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dakhama A, al Mubarak R, Pavelka N, et al. Tollip inhibits ST2 signaling in airway epithelial cells exposed to type 2 cytokines and rhinovirus. J Innate Immun. 2020;12:103‐115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shah JA, Emery R, Lee B, et al. TOLLIP deficiency is associated with increased resistance to Legionella pneumophila pneumonia. Mucosal Immunol. 2019;12:1382‐1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Venkatasubramanian S, Pryor R, Plumlee C, et al. TOLLIP optimizes dendritic cell maturation to lipopolysaccharide and mycobacterium tuberculosis. J Immunol. 2022;209:435‐445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li X, Kim SE, Chen TY, et al. Toll interacting protein protects bronchial epithelial cells from bleomycin‐induced apoptosis. FASEB J. 2020;34:9884‐9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu L, Wang L, Luo X, et al. Tollip, an intracellular trafficking protein, is a novel modulator of the transforming growth factor‐beta signaling pathway. J Biol Chem. 2012;287:39653‐39663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burns K, Clatworthy J, Martin L, et al. Tollip, a new component of the IL‐1RI pathway, links IRAK to the IL‐1 receptor. Nat Cell Biol. 2000;2:346‐351. [DOI] [PubMed] [Google Scholar]
- 9. Hung CF, Rohani MG, Lee SS, Chen P, Schnapp LM. Role of IGF‐1 pathway in lung fibroblast activation. Respir Res. 2013;14:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545‐15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilson CL, Stephenson SE, Higuero JP, Feghali‐Bostwick C, Hung CF, Schnapp LM. Characterization of human PDGFR‐beta‐positive pericytes from IPF and non‐IPF lungs. Am J Physiol Lung Cell Mol Physiol. 2018;315:L991‐L1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Noth I, Zhang Y, Ma SF, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome‐wide association study. Lancet Respir Med. 2013;1:309‐317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oldham JM, Ma SF, Martinez FJ, et al. TOLLIP, MUC5B, and the response to N‐acetylcysteine among individuals with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2015;192:1475‐1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Katayanagi S, Setoguchi Y, Kitagawa S, Okamoto T, Miyazaki Y. Alternative gene expression by TOLLIP variant is associated with lung function in chronic hypersensitivity pneumonitis. Chest. 2022;161:458‐469. [DOI] [PubMed] [Google Scholar]
- 17. Vander Ark A, Cao J, Li X. TGF‐beta receptors: in and beyond TGF‐beta signaling. Cell Signal. 2018;52:112‐120. [DOI] [PubMed] [Google Scholar]
- 18. di Guglielmo GM, le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF‐beta receptor signalling and turnover. Nat Cell Biol. 2003;5:410‐421. [DOI] [PubMed] [Google Scholar]
- 19. Derynck R, Budi EH. Specificity, versatility, and control of TGF‐beta family signaling. Sci Signal. 2019;12:eaav5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ebisawa T, Fukuchi M, Murakami G, et al. Smurf1 interacts with transforming growth factor‐beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477‐12480. [DOI] [PubMed] [Google Scholar]
- 21. Kavsak P, Rasmussen RK, Causing CG, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365‐1375. [DOI] [PubMed] [Google Scholar]
- 22. Redente EF, Jacobsen KM, Solomon JJ, et al. Age and sex dimorphisms contribute to the severity of bleomycin‐induced lung injury and fibrosis. Am J Physiol Lung Cell Mol Physiol. 2011;301:L510‐L518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Figure S2.
Figure S3.
Data S1.
Data Availability Statement
The data that support the findings of this study are available in the methods and/or supplementary material of this article. Detailed RNA‐seq information and raw data meeting MINSEQE (Minimum Information About a Next‐generation Sequencing Experiment) are deposited at Gene Expression Omnibus (https://www.ncbi.mlm.nih.gov/geo/).


