Abstract
Stable isotope tracers are applied in vivo and in vitro studies to reveal the activity of enzymes and intracellular metabolic pathways. Most often, such tracers are used with gas chromatography coupled to mass spectrometry (GC-MS) due to its ease of operation and reproducible mass spectral databases. Differences in isotope tracer performance of classic GC-quadrupole MS instrument and newer time-of-flight instruments are not well-studied. Here, we used three commercially available instruments for the analysis of identical samples from a stable isotope labeling study that used [U-13C6] d-glucose to investigate the metabolism of bacterium Rothia mucilaginosa with respect to 29 amino acids and hydroxyl acids involved in primary metabolism. The prokaryote Rothia mucilaginosa belongs to the family of Micrococcaceae and is present and metabolically active in the airways and sputum of cystic fibrosis patients. Overall, all three GC-MS instruments (low-resolution GC-SQ MS, low-resolution GC-TOF MS, and high-resolution GC-QTOF MS) can be used to perform stable isotope tracing studies for glycolytic intermediates, TCA metabolites and amino acids, yielding similar biological results, with high-resolution GC-QTOF MS offering additional capabilities to identify chemical structures of unknown compounds that might show significant isotope enrichments in biological studies.
Graphical Abstract

Introduction
Metabolomics has been widely used to study human diseases in the past 20 years.1–7 While snapshot (steady-state) of metabolite profiles provides important information on cellular metabolism and physiology, such data do not necessarily reflect enzyme or pathway activity directly.8 Instead, stable isotope tracing can be used to track the fate of labeled atoms in metabolite substrates, quantify the rates of changes of these metabolites and elaborate the direction of pathway activities.9–11 When isotope tracers are used along with absolute (molar) quantifications, fluxes across pathways can be discerned.12–13 Isotope tracers can delineate the alternative use of substrates in uncontrolled cell growth in malignant tumors14–16, determine the target pathways of novel drugs for disease therapy development,17–19 investigate the metabolic cross-feeding between bacteria,20–22 and assign/quantify the global plant metabolome that is riddled with heavy matrix interference.23–24
Mass spectrometry (MS) has played a central role in metabolomics and stable isotope tracing studies.9 Gas chromatography with mass spectrometry (GC-MS) has been used for a long time for stable isotope labeling studies because many target compounds are directly amenable to GC-MS, including metabolites in the oxidative part of glycolysis, the TCA cycle and adjacent amino acids.25–27 For many compounds including sugars and hydroxyl acids,28–31 GC-MS offers an easier route to chromatographic separation than liquid chromatography (LC)-MS unless specific procedures like ion-pairing LC are used.32 Small molecules analyzed by GC-MS require derivatization to decrease their polarity and to increase volatility, selectivity, and sensitivity, especially for metabolites bearing acidic protons.30, 33 A typical silylation reagent used for derivatization in stable isotope tracing is N-Methyl-N-tert-butyldimethylsilyltrifluoroacetamide (MTBSTFA), which replaces non-sterically hindered active hydrogens (e.g., on −COOH, −OH, NH2, −HN, −SH groups) with tert-butyldimethylsilyl (TBDMS). MTBSTFA-derivatives produce characteristic fragmentation patterns with three major ions [M]+, [M–57]+, and [M–131]+. The [M–57]+ fragment is generally the dominant ion and harbors the entire original metabolite7, 34 and is therefore best suited for isotope tracer studies. In comparison, trimethylsilylation (TMS) reagents do not yield abundant molecular fragment ions and are less stable and more sensitive to moisture.35 On one hand, due to steric hindrance, TBDMS derivatives only replace one active hydrogen per amino group, leading to better precision in quantitative analysis of amino-containing compounds than smaller TMS reagents36–37 that generate several derivatives. On the other hand, because of this effect, TBDMS-derivatization cannot derivatize sugars or sugar alcohols, limiting the use of TBDMS derivatization for general untargeted metabolomics compared to TMS reagents.
A wide range of GC-MS instruments are available varying in resolution and mass separation. However, comparison of the performance of different GC-MS instruments using stable isotope labeling technique is not well-studied. Isotope tracing studies investigate either isotopomer or isotopologue information of metabolites.38–41 Isotopologue refers to molecules that differ only in their isotopic compositions. Isotopomers also consider the constitutional differences of isotopes within molecule structures.42 Isotope enrichment studies are not often conducted together with absolute quantifications43–44 which were used for the determination of absolute fluxes.45 To date, GC-MS are widely used for isotopologue tracer studies for which we here compared on three types of GC-MS instruments.
We have applied [U-13C6] d-glucose to investigate the metabolism of Rothia mucilaginosa and reported the incorporation of glucose derived 13C into glycolysis metabolites and some amino acid biosynthesis pathways using a low-resolution GC-single quadrupole-MS (GC-SQ MS).20 Here, we investigate the same samples but used three different GC-MS instruments (Figure 1, Figure S1), with either low-resolution single quadrupole MS (GC-SQ MS), low-resolution time-of-flight MS (GC-TOF MS) or high-resolution GC-Quadrupole Time-of-Flight MS (GC-QTOF MS).
Figure 1. Evaluating the performance of isotope measurements for three GC-MS instruments using chemical standards and isotope tracer biological experiments.

Isotope enrichments are calculated by the increases of the fully labeled carbons in molecules, such as increases in M+3/M for 3-carbon molecules, indicated by dotted lines versus solid lines in the mass spectra.
Methods
Samples
A quality control mixture of 29 unlabeled metabolite standards (Table S1) was prepared to reach a final concentration of 1 mg/mL as stock solution. 50 μL aliquot from each stock solution was combined into a new tube and dried down. Then, the mixture was diluted to reach a final concentration of 50 μg/mL working solution.
Rothia mucilaginosa strain RmFLR01 was isolated from a cystic fibrosis (CF) patient at the UC San Diego Adult CF Clinic.20, 46 R. mucilaginosa cultures were grown in triplicates in artificial-sputum medium47 spiked with 100 mM [U-13C6] d-glucose (Cambridge Isotope Laboratory, Tewksbury, MA, USA) under anaerobic and aerobic conditions (5% CO2) at 37°C and harvested at 4, 8, 12, and 24 h for isotope tracer analyses.
Sample preparation
For the 29 unlabeled standards mixture, 10 μL of the final solution was dried down for GC-MS measurement. Derivatization and data acquisition of mixture aliquots by GC-MS were reproduced on three days for inter-day precision. Dried mixtures were derivatized by adding 10 μL of 40 mg/mL methoxyamine hydrochloride (Sigma-Aldrich, St. Louis, MO, USA) in pyridine (Sigma-Aldrich, St. Louis, MO, USA) and shaking at 30°C for 1.5 h. Subsequently, 90 μL MTBSTFA (Sigma-Aldrich, St. Louis, MO, USA) was added with 13 fatty acid methyl esters (FAMEs) as retention index markers and shaken at 80°C for 30 min. Samples were immediately transferred to crimp top vials and injected onto each GC-MS instrument.
Same samples of R. mucilaginosa cultures were extracted using published methods.20 The extraction solvent was prepared by mixing 3 volume of acetonitrile (Fisher Scientific), 3 volume of isopropanol (Fisher Scientific), and 3 volume of water (Fisher Scientific) together and was then degassed by directing a gentle stream of nitrogen through the solvent for 5 min. The solvent was cooled to −20°C prior to extraction. 30, 36 Samples were added 1 mL pre-chilled, degassed acetonitrile: isopropanol: water (v/v/v 3:3:2, Fisher Scientific) followed by vortexing 30 s and shaking at 4°C for 5 min. Samples were centrifuged for 2 min at 12,210 × g to precipitate debris from extracts. Supernatants were collected and split into two equal portions. One aliquot was dried to completeness in a Labconco cold trap centrifuge evaporator and then resuspended in 0.5 mL degassed acetonitrile: water (v/v 1:1, Fisher Scientific) to remove triacylglycerides. Resuspension solutions were vortexed for 30s and centrifuged for 2 min. Supernatants were transferred into clean Eppendorf tubes and dried down completely. Dried extracts were derivatized as given above.
Gas chromatographic conditions
Each mass spectrometer was coupled to an Agilent 7890 GC system (Santa Clara, CA) installed with a Restek (Bellefonte, PA) RTX-5Sil MS column (30m length, 0.25 mm i.d, 0.25 μM df, 95% dimethyl/5%diphenyl polysiloxane film) with an additional 10m guard column. For the quadrupole-MS and QTOF MS analyses, identical GC parameters were used by injecting 1 μL of derivatized sample into the GC in splitless mode at an injection temperature of 250°C and a constant flow of 1 mL/min. The initial oven temperature was held at 60°C for 0.5 min, and ramped at a rate of 10°C/min to 325°C that was maintained for 10 min for a total run time of 37 min.
For the low-resolution LECO TOF MS (St. Joseph, MI, USA) instrument, 1 μL of derivatized sample was injected in splitless mode with an injection temperature of 275°C. The oven temperature started at 50°C for 1 min, increased at 20°C/min to 330°C, and kept isothermal for 5 min.
Each standards mixture sample was injected 6 times to obtain intra-day precision data for each instrument. The comparison of these samples measured on three separate days gave data for inter-day precisions.
Each of the R. mucilaginosa biological triplicate samples was injected 10 times on each of the three GC-MS instruments to discriminate true biological isotope enrichment effects from the impact of technical imprecision.
MS analysis
We compared the high-resolution Agilent 7200 QTOF (Santa Clara, CA, USA), the low-resolution Agilent 5977 MSD (Santa Clara, CA, USA), and the low resolution LECO Pegasus IV TOF MS (St. Joseph, MI, USA) in this study. All three instruments used electron ionization (EI) mode at +70 eV electron voltage. Mass spectra were acquired from 85–700 m/z at 17 Hz and 250°C source temperature on the Pegasus IV TOF MS, 50–1050 m/z at 1.5 Hz and 240°C source temperature on the quadrupole-MS and from 50–1050 m/z at 5 Hz and 230°C source temperature on the QTOF MS instrument.
Data processing
Because of the chromatography parameter differences on three instruments, retention times (RT) were initially determined by running each metabolite as chemical standard on the high-resolution GC-QTOF MS. During derivatization, FAMEs (fatty-acid methyl esters) with 8 to 30 fatty acyl carbon lengths were added into MTBSTFA. These FAMEs were used as internal standards for retention index markers. This FAME marker set yields a stable set system with fixed differences in retention times.48 Despite other differences between the instruments, all three GC-MS instruments used the same electron ionization (EI) mode at +70 eV electron voltage and the spectra were very comparable among three instruments to be used for annotation with the NIST library. An example is given in Figure S1.
LECO TOF MS raw data files (.CDF format) were converted to MassHunter formats (.D format) using Agilent GCMS translator software. MassHunter Quantitative Analysis B.07.00 version was used to process the data that acquired from all three GC-MS instruments to avoid potential differences caused by different data processing software parameters. We used identical signal-to-noise ratio (S/N) and RT tolerance settings for quantifying isotope tracers on the GC-QTOF and the GC-TOF MS instruments. Threshold of S/N was set to peak height ≥ 1% of the largest peak. RT tolerance was set to ± 0.08 min. For the GC-quadrupole MS, the S/N was set to peak height ≥ 0.2%. All peak integrations were manually validated within the vendor software to exclude potential artifacts based on possible differences in software settings. Theoretical isotope ratios were calculated for [M–57]+ ion of each derivatized structure of the 29 standards using an online isotope calculator.49 As this study is not for absolute quantification and absolute flux calculations, we normalized over-enrichment of isotopes to the unlabeled M0 monoisotope ion, i.e., the [M-57]+ fragment. Hence, normalization was performed as ratio M+1/M (or M+2/M or M+3/M) from peak heights of extracted ion chromatograms (data available in Table S5–6). Averages and standard deviations as well as technical errors and isotope enrichments were calculated in Microsoft Excel by correcting for natural isotope abundances. Statistical analysis and corresponding plots were obtained using GraphPad Prism Version 8.4.3. The data sets are available in the Metabolomics Workbench data repository with accession numbers 2231(ST001613) for low-resolution GC-TOF MS and 2278 (ST001608) for the other two GC-MS instruments.
Results and Discussion
Precision of isotope abundance measurements for three GC-MS instruments using 29 unlabeled metabolite standards
MTBSTFA derivatization replaces active hydrogens on polar functional groups and forms tert-butyldimethylsilyl (TBDMS) derivatives which are fragmented during electron impact ionization to generate the most abundant ion [M–57]+ with tert-butyl cleavage (Figure 2A). TBDMS derivatization therefore has the advantage to yield a very abundant molecular fragment ion [M–57]+ which contains the entire underivatized molecular skeleton with only one derivative product per metabolite. In addition, it had been previously shown that TBDMS derivatization yields better reproducibility than trimethylsilyl derivatization for amino acids which are important target compounds to be analyzed in metabolic isotope tracer studies. Using silylation reactions has a big advantage for isotope abundance analyses because the Si-atom has a high natural abundance of 5.1% for the M+1 isotope and 3.4% for the M+2 isotope, leading to easily detectable signals in GC-MS analyses even for low abundant metabolites in biological studies.
Figure 2. Overall performance of isotope detection using three GC-MS instruments for 29 unlabeled standards.
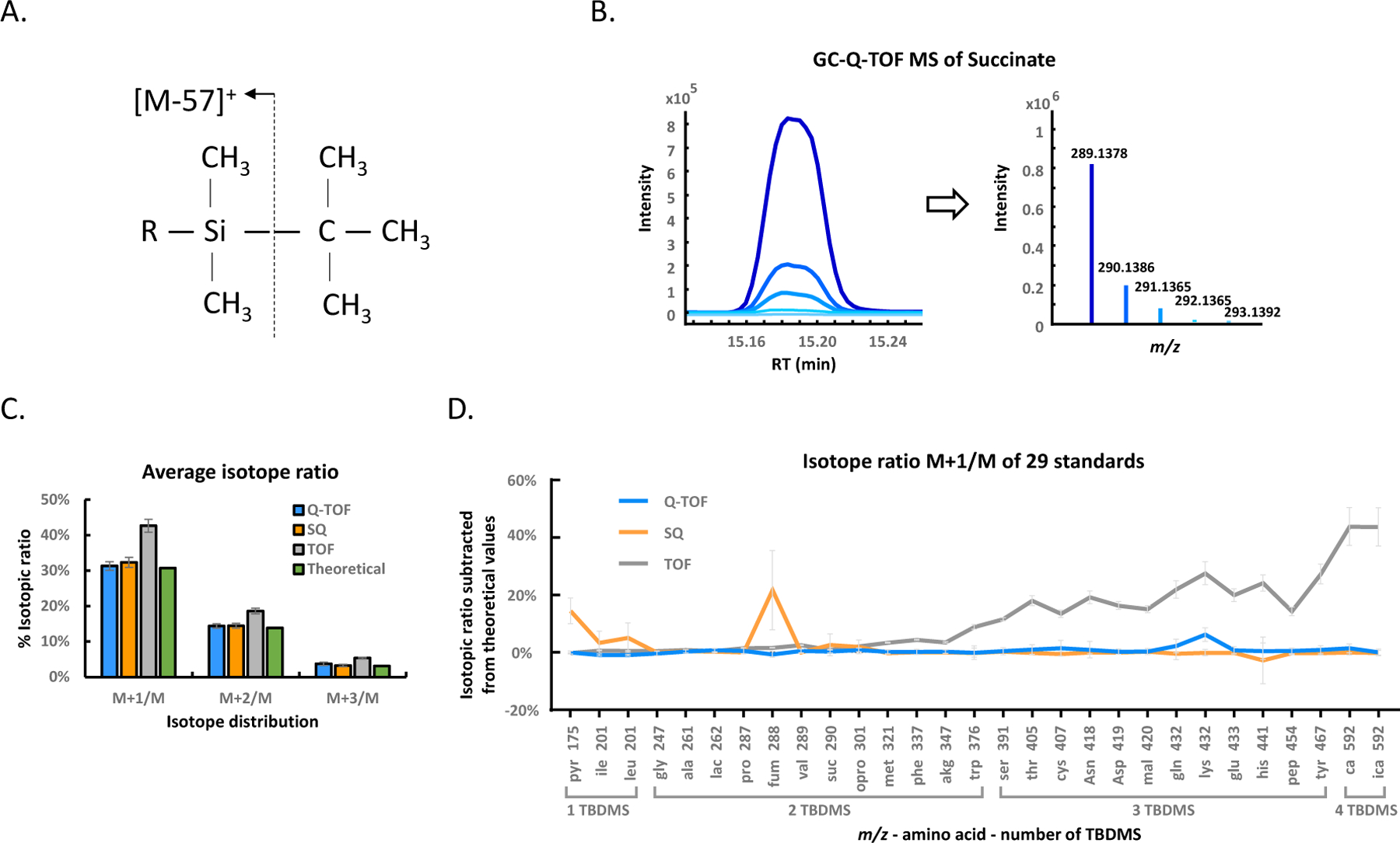
(A) MTBSTFA derivation leads to [M–(tert-butyl)]+ fragments that are used as M+ surrogate ions. (B) Example of chromatographic ion traces and corresponding averaged mass spectrum for [M–57]+ fragment of succinate.2TBDMS derivative at m/z 289 and its isotopologue intensities. (C) Average isotope ratios across all 29 metabolite standards and three GC-MS instruments. QTOF = high-resolution quadrupole time-of-flight MS, SQ = low-resolution single quadrupole MS, TOF = low-resolution time-of-flight MS. Isotope ratios are represented as arithmetic means ±1 σ. (D) Measured M+1/M ratios for 29 metabolites corrected by their theoretical abundances, sorted by number of TBDMS derivatizations and m/z values.
We measured 29 unlabeled metabolite standards that are typically investigated in metabolic flux studies including glycolysis intermediates, TCA metabolites, and amino acids on three GC-MS instruments. We first evaluated the overall performance of three instruments by calculating relative and absolute errors for the three most important isotope ratios (M+1/M, M+2/M and M+3/M). Our results showed that the low-resolution GC-TOF MS instrument yielded a significant systematic overrepresentation of the average isotope ratios for M+1/M, M+2/M, and M+3/M (Figure 2C, Table S5) compared to GC-QTOF MS and GC-SQ MS instruments. The GC-TOF MS instrument also showed greater absolute errors (Table 1). These errors in the low-resolution GC-TOF MS instrument were clearly dependent on the molecular size of the chemicals (Figure 2D) as molecule [M–57] + fragments at < m/z 350 had isotopic ratios that were close to the theoretical values whereas at > m/z 350, systematic errors increased dramatically. These systematic errors are likely due to the way of this specific instrument accounting for ions by optimizing ion detection for speed, not for resolution, leading to non-optimal accounting for mass defects at higher m/z values. For the single quadrupole, we found three outlier compounds, leucine and fumarate that showed detector saturation at [M–57]+ monoisotopic ion peak heights, and mass spectral undersampling for pyruvate as very low abundant compound (Table S5). The GC-QTOF MS showed the most accurate isotope ratios with only one compound (lysine) that had more than 5% absolute error for the M+1/M isotope ratio (Figure 2D, Table S5). This excellent accuracy of the GC-QTOF MS (Table 1) might be due to the high mass resolving power that at least partly separates co-elution isobaric noise ions and therefore leads to better peak integration of low abundant isotope ions.
Table 1. Comparison of M+1/M and M+2/M isotope ratios of non-labeled standards from three instruments.
QTOF = high-resolution quadrupole time-of-flight MS, SQ = low-resolution single quadrupole MS, TOF = low-resolution time-of-flight MS
| intra-day precision (%CV) | inter-day precision (%CV) | absolute error (ratio %) | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| QTOF | SQ | TOF | QTOF | SQ | TOF | QTOF | SQ | TOF | |
| M+1/M | 2.7% | 3.3% | 1.1% | 3.7% | 5.8% | 2.3% | 0.8% | 3.1% | 12.0% |
| M+2/M | 3.4% | 3.7% | 1.5% | 4.2% | 6.1% | 2.7% | 0.6% | 1.5% | 4.7% |
The mixture of 29 metabolite standards was injected six times each day to determine intra-day precision and reproduced for three days to test inter-day precision that were calculated as coefficient of variation (%CV). The low-resolution GC-TOF MS gave significantly better intra- and inter-day precisions than the other two instruments. For M+1/M (Table 1), GC-TOF MS instrument yielded only 1.1% CV on average for intra-day precision, compared to more than 2% CV for GC-quadrupole and GC-QTOF MS instruments. Similarly, for inter-day precision of M+1/M ratios, the low-resolution GC-TOF MS had best precision with 2.3% CV compared to 4–6% CV for the other two instruments. The same trends were observed for M+2/M with significantly better intra-day and inter-day precisions for the low-resolution GC-TOF MS instrument compared to the other two mass spectrometers (Table S5). For M+3/M ratios, overall signal intensities were too low to obtain reliable data with 4–10% CV for intra-day and 5–13% CV for inter-day precisions (Table S5). We verified the excellent precision but also the high absolute error for the low-resolution GC-TOF MS on four separate instruments (Table S3, Table S9) for TMS-derivatized standards, analyzed on different days and by different personnel. The superior precision of the low-resolution GC-TOF MS instruments compared to the other two instruments was also verified by additional analysis of underivatized fatty acid methyl esters (Table S4, Table S10). In combination, these findings indicate that isotope ratio measurements correspond to the physics of the ion optics and ion detection system of each instrument type, rather than to small differences between individual instruments of the same type or the type of chemical derivatizations.
Interestingly, despite its excellent isotope accuracy, we did not find improved precision for the high-resolution QTOF MS instrument compared to classic unit-resolution quadrupole instrument. Instead, the better precision observed for the GC-TOF MS might be explained by its faster spectral acquisition rate of the. Using this instrument, spectra are acquired at 500 spectra/s and then accumulated to 17 spectra/s during initial raw data processing. In comparison, high-resolution GC-QTOF MS and classic quadrupole MS instruments are typically scanning at 2–5 spectra/s. At this spectral acquisition rate, data might be under-sampled for peaks that have band widths of 3 s (i.e. a total of 6–12 data points per peak) such as pyruvate measured on GC-SQ MS (Figure 2D), especially for lower-abundant M+1 and M+2 ions. Such undersampling of data points may certainly lead to lower precision.
Performance of three GC-MS instruments on 13C labeled R. mucilaginosa samples
Isotope tracing in biochemistry can highlight active enzymes and pathways that may differ between biological conditions. We therefore tested the performance of three GC-MS instruments to determine whether each of the instruments would yield the same biological conclusions. We here used a [U-13C6] d-glucose feeding experiment for which we previously published the overall biological insights using the GC-quadrupole-MS system,20 and reproduced the study three times to compare GC-SQ MS to the GC-TOF MS and the GC-QTOF MS systems while also increasing the number of metabolite targets to low abundant molecules such as phosphoenolypyruvate, tryptophan, cysteine, methionine and others (Table S6).
Rothia mucilaginosa (formerly Stomatococcus mucilaginosus) is a gram-positive coccus of the family Micrococcaceae and was found as a cause of oral, cutaneous, and central nervous system infections.50 All 29 metabolites were detected in R. mucilaginosa cultures grown under anaerobic and aerobic conditions from 4–24 hours. Three independent cultures per time point were harvested and measured repeatedly for ten times. Isotope enrichment of three primary metabolites, pyruvate, serine and citrate all showed clear incorporation of labeled carbon into the metabolite structures at 24h (Figure 3A–C). Isotope enrichment analysis showed that the 3-carbon molecule pyruvate significantly enriched its M+3 isotope (Figure 3A) with 8–9 folds higher abundance than the unlabeled M+ monoisotopic parent molecule. Incomplete incorporation by one or two labeled carbons was much less pronounced for pyruvate. In comparison, the 3-carbon molecule serine (Figure 3B) showed only modest, but notable incorporation of labeled carbon into the M+1, M+2 and M+3 isotopologues. For the 6-carbon molecule citrate (under aerobic condition, Figure 3C) that is formed by addition of acetyl-groups using citrate synthase, we investigated the M+2, M+4 and M+6 isotopologues and also found small but detectable labeling. Citrate under aerobic conditions were still detected at low abundance, thus the isotope enrichment among three instruments were slightly different but gave overall similar 13C incorporation patterns (Table S6). These comparisons showed that all three GC-MS instruments correctly detected a large metabolic flux into the endpoint of glycolysis, pyruvate, with only small flux into off-stream biosynthesis of serine under anaerobic conditions. The instruments also all found an overall limited metabolic flux from pyruvate into the TCA cycle.
Figure 3. Isotope enrichment measured on three GC-MS instruments for selected metabolites in R. mucilaginosa.

QTOF = high-resolution quadrupole time-of-flight MS, SQ = low-resolution single quadrupole MS, TOF = low-resolution time-of-flight MS. (3A–3C) Isotope enrichment of selected metabolites pyruvate, serine, and citrate in R. mucilaginosa grown for 24h under anaerobic or aerobic conditions depending on different [M–57]+ fragment size. Isotope enrichments were corrected for natural abundance and are represented as arithmetic means ±1 σ. Standard deviation σ was calculated from 30 injections using 3 biological sample replicates, each injected 10 times.
Importantly, pyruvate did not show a large difference between the GC-SQ and the other two instruments (Figure 3A), unlike for the unlabeled mixture experiment (Figure 2D). Moreover, despite the differences in reported isotope enrichment for each of the metabolites on the three GC-MS instruments, the biological conclusions would be the same, with high degree of 13C incorporation in pyruvate but low 13C enrichment for citrate and serine (Figure 3A–C). This biological conclusion would be even clearer if raw measurements were to be corrected for the systematic errors observed in the GC-TOF MS instrument by a post hoc mass-dependent regression, which can be further explored in future studies.
The growth and metabolism of bacteria are sensitive to oxygen conditions. Thus, we examined the impact of oxygen condition on the metabolism of R. mucilaginosa and studied whether biological conclusions would differ between the three GC-MS instruments with respect to isotope enrichment over the course of one day under both aerobic and anaerobic conditions, correcting for natural isotope abundances (Figure 4A–I). The increase pattern in fully labeled pyruvate M+3 was similar for all three GC-MS instruments (Figure 4A–C) as well as for amino acids, citrate and other TCA cycle metabolites (Table S7, Table S8). All three instruments showed a decrease of fully labeled pyruvate M+3 under aerobic conditions at 24h compared to 12h, but not for anaerobic conditions (Figure 4A–C). Interestingly, all three instruments found much higher incorporation of labeled carbon into lactate under aerobic conditions than under anaerobic media between 8–24h growth (Figure 4D–F), reaching a plateau at 12h. Under anaerobic conditions, all three instruments showed only enrichment for lactate only at 24h (Figure 4D–F). For amino acid metabolite valine, greater isotope enrichment was observed under anaerobic conditions compared to aerobic growth conditions (Figure 4G–I). This indicated the incorporation rates of glucose derived 13C into valine were faster under anaerobic condition than aerobic environment. Similar biological results were observed by all three types of GC-MS instruments for tryptophan that showed clearly higher enrichment for tryptophan M+3 under aerobic conditions than anaerobic conditions (Figure S2A–C). Only when absolute ion intensities were too low, for example for phosphoenolpyruvate (Figure S2D–F), differences between the instruments might interfere with detection of biological differences. Specifically, temporal differences in isotope enrichment in PEP from 4–24h growth were found only when using the quadrupole and QTOF instruments, but not the TOF instrument (Figure S2D–F). While we have not tested for biological 15N incorporation,51 similar performance of three GC-MS instruments as in 13C labeling isotope enrichment analyses can be expected because such isotopologue enrichment analyses only consider the differences between full mass units, not the isotopic fine structures.38, 52
Figure 4. Increase in isotope enrichments measured on three GC-MS instruments for pyruvate, lactate and valine in R. mucilaginosa.
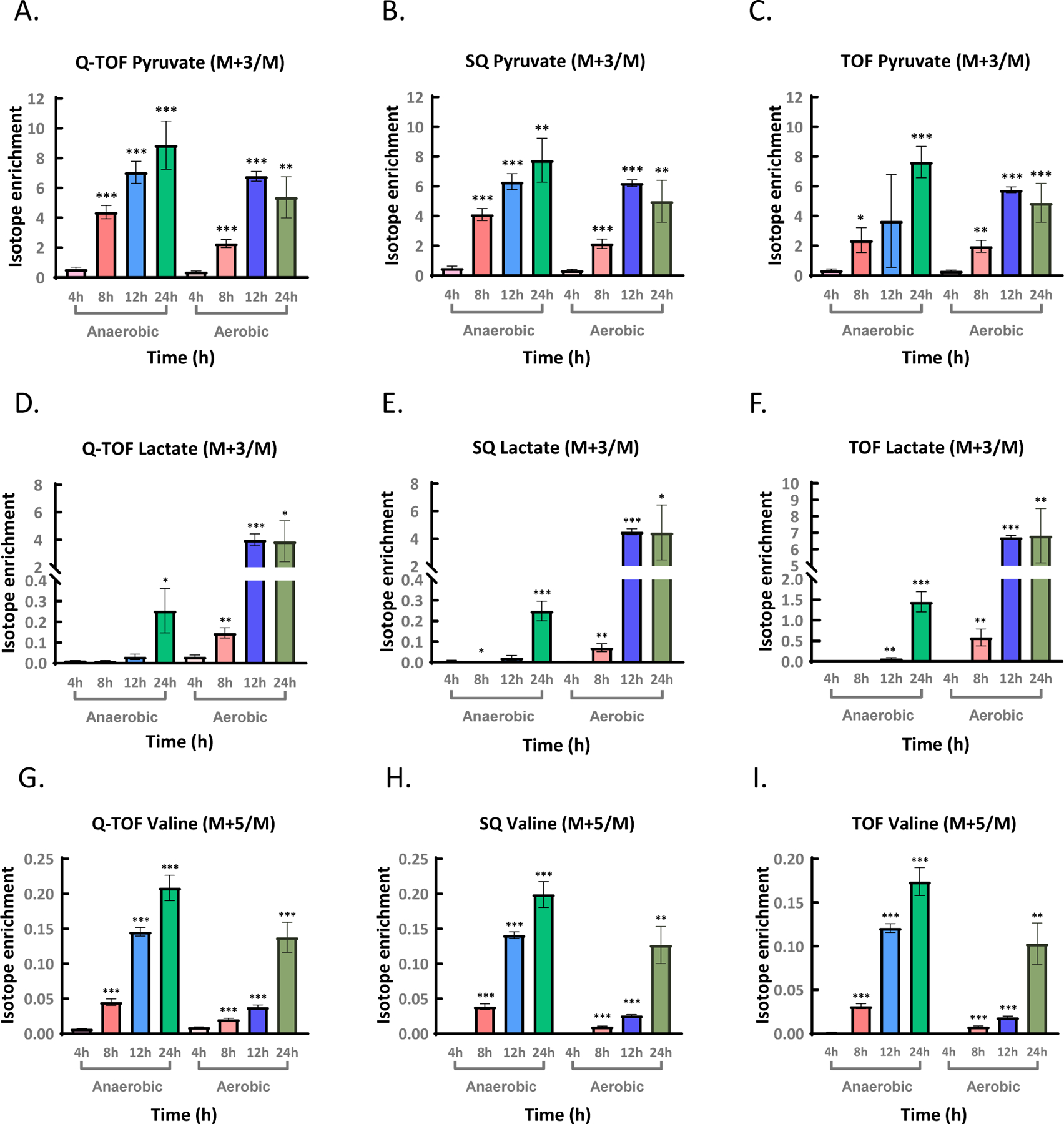
QTOF = high-resolution quadrupole time-of-flight MS, SQ = low-resolution single quadrupole MS, TOF = low-resolution time-of-flight MS. Student t-tests were used to estimate significance levels from three biological replicates for enrichment at each time point compared to 4h time point in R. mucilaginosa grown for 4h–24h under anaerobic or aerobic conditions (Table S8). *p < 0.05, **p < 0.01, ***p < 0.001 with values represented as arithmetic means ±1 σ. (4A–4C) Increase in isotope enrichments for pyruvate M+3/M. (4D–4F) Increase in isotope enrichments for lactate M+3/M. (4G–4I) Increase in isotope enrichments for valine M+5/M.
Conclusion
Our results show that the three tested types of GC-MS instruments were all capable of performing 13C-based stable isotope ratio enrichments studies for primary metabolites. While the GC-QTOF MS showed better accuracy than the other two instruments, the low-resolution GC-TOF MS was superior in precision of 13C labeled isotopologue analysis. It is important to note that none of these instruments enables isotopomer (13C positional) analyses or studies combining 13C, 18O, and 15N labeled substrates. Instruments to enable such studies for discerning the fine isotope structure of the labeled isotopes must have resolving power with R > 250,000.38, 52 The nominal-mass GC-TOF MS showed systematic overrepresentation of isotope ratios for high m/z metabolites while the nominal-mass GC-single quadrupole yielded saturation effects at high metabolite concentrations. Nevertheless, when used in an example biological study on 13C-fed Rothia bacteria, all three GC-MS instruments yielded similar biological interpretations. While the low-resolution instruments are cheaper and easier to operate and maintain than high-resolution GC-MS instruments, accurate mass GC-QTOF MS instruments offer additional capabilities to identify chemical structures of unknown compounds.53–54 This study exceeded the number of metabolites analyzed in the prior biology-focused study that only used GC-quadrupole MS instrumentation.20 Interestingly, the inclusion of such a wider range of metabolic targets discovered differences in metabolic 13C-enrichment for low abundant compounds that were not reported in the prior study.20 For example, stark differences in isotope enrichment for low abundant tryptophan were observed with all three instruments between aerobic and anaerobic cultures. In addition, temporal 13C-enrichment differences in some low abundant metabolites such as phosphoenolpyruvate were only detected by GC-QTOF MS and GC-SQ MS instruments, but not by low-resolution GC-TOF mass spectrometry. Hence, this study shows how the number of metabolites in 13C-enrichment analysis can be easily enlarged, offering the potential to extend the use of GC-MS to untargeted metabolome enrichment studies. If the aim of using 13C-enrichment is to report absolute fluxes in biological studies, multiple time points would need to be analyzed in combination with absolute quantifications. Overall, we conclude that different GC-MS instruments showed to be highly useful for isotopic enrichment analysis in metabolomic studies.
Supplementary Material
ACKNOWLEDGMENT
This work was supported as a pilot project by NIH U24 DK097154 and NIH U2C ES030158. K.L.W. is supported by a Gilead CF Research Scholars Award (app_00b072) and NIH NHLBI (grant 5R01 HL136647–04). T.G. is supported through the National Science Foundation’s Integrative Graduate Education and Research Traineeship (IGERT) program (grant DGE-1144901).
REFERENCES
- 1.McCartney A; Vignoli A; Biganzoli L; Love R; Tenori L; Luchinat C; Di Leo A, Metabolomics in breast cancer: A decade in review. Cancer Treat Rev 2018, 67, 88–96. [DOI] [PubMed] [Google Scholar]
- 2.Yu L; Li K; Zhang X, Next-generation metabolomics in lung cancer diagnosis, treatment and precision medicine: mini review. Oncotarget 2017, 8 (70), 115774–115786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guasch-Ferre M; Hruby A; Toledo E; Clish CB; Martinez-Gonzalez MA; Salas-Salvado J; Hu FB, Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes Care 2016, 39 (5), 833–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rauschert S; Uhl O; Koletzko B; Hellmuth C, Metabolomic biomarkers for obesity in humans: a short review. Ann Nutr Metab 2014, 64 (3–4), 314–24. [DOI] [PubMed] [Google Scholar]
- 5.Jacob M; Lopata AL; Dasouki M; Abdel Rahman AM, Metabolomics toward personalized medicine. Mass Spectrom Rev 2019, 38 (3), 221–238. [DOI] [PubMed] [Google Scholar]
- 6.Misra BB; Olivier M, High Resolution GC-Orbitrap-MS Metabolomics Using Both Electron Ionization and Chemical Ionization for Analysis of Human Plasma. J Proteome Res 2020, 19 (7), 2717–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higashi RM; Fan TW; Lorkiewicz PK; Moseley HN; Lane AN, Stable isotope-labeled tracers for metabolic pathway elucidation by GC-MS and FT-MS. Methods Mol Biol 2014, 1198, 147–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zamboni N; Saghatelian A; Patti GJ, Defining the metabolome: size, flux, and regulation. Mol Cell 2015, 58 (4), 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang C; Chen L; Rabinowitz JD, Metabolomics and Isotope Tracing. Cell 2018, 173 (4), 822–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkinson DJ, Historical and contemporary stable isotope tracer approaches to studying mammalian protein metabolism. Mass Spectrom Rev 2018, 37 (1), 57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heuillet M; Bellvert F; Cahoreau E; Letisse F; Millard P; Portais JC, Methodology for the Validation of Isotopic Analyses by Mass Spectrometry in Stable-Isotope Labeling Experiments. Anal Chem 2018, 90 (3), 1852–1860. [DOI] [PubMed] [Google Scholar]
- 12.Liu L; Shah S; Fan J; Park JO; Wellen KE; Rabinowitz JD, Malic enzyme tracers reveal hypoxia-induced switch in adipocyte NADPH pathway usage. Nat Chem Biol 2016, 12 (5), 345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang P; Guo L; Jaini R; Klempien A; McCoy RM; Morgan JA; Dudareva N; Chapple C, A (13)C isotope labeling method for the measurement of lignin metabolic flux in Arabidopsis stems. Plant Methods 2018, 14, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Courtney KD; Bezwada D; Mashimo T; Pichumani K; Vemireddy V; Funk AM; Wimberly J; McNeil SS; Kapur P; Lotan Y; Margulis V; Cadeddu JA; Pedrosa I; DeBerardinis RJ; Malloy CR; Bachoo RM; Maher EA, Isotope Tracing of Human Clear Cell Renal Cell Carcinomas Demonstrates Suppressed Glucose Oxidation In Vivo. Cell Metab 2018, 28 (5), 793–800 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roci I; Gallart-Ayala H; Schmidt A; Watrous J; Jain M; Wheelock CE; Nilsson R, Metabolite Profiling and Stable Isotope Tracing in Sorted Subpopulations of Mammalian Cells. Anal Chem 2016, 88 (5), 2707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan TW; Lane AN; Higashi RM; Farag MA; Gao H; Bousamra M; Miller DM, Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM). Mol Cancer 2009, 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan TW; Lorkiewicz PK; Sellers K; Moseley HN; Higashi RM; Lane AN, Stable isotope-resolved metabolomics and applications for drug development. Pharmacol Ther 2012, 133 (3), 366–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane AN; Higashi RM; Fan TW, Preclinical models for interrogating drug action in human cancers using Stable Isotope Resolved Metabolomics (SIRM). Metabolomics 2016, 12 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan M; Kremer DM; Huang H; Breitkopf SB; Ben-Sahra I; Manning BD; Lyssiotis CA; Asara JM, Ex vivo and in vivo stable isotope labelling of central carbon metabolism and related pathways with analysis by LC-MS/MS. Nat Protoc 2019, 14 (2), 313–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao B; Gallagher T; Zhang Y; Elbadawi-Sidhu M; Lai Z; Fiehn O; Whiteson KL, Tracking Polymicrobial Metabolism in Cystic Fibrosis Airways: Pseudomonas aeruginosa Metabolism and Physiology Are Influenced by Rothia mucilaginosa-Derived Metabolites. mSphere 2018, 3 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan BF; Zhu QF; Guo N; Zheng SJ; Wang YL; Wang J; Xu J; Liu SJ; He K; Hu T; Zheng YW; Xu FQ; Feng YQ, Comprehensive Profiling of Fecal Metabolome of Mice by Integrated Chemical Isotope Labeling-Mass Spectrometry Analysis. Anal Chem 2018, 90 (5), 3512–3520. [DOI] [PubMed] [Google Scholar]
- 22.Hartl J; Kiefer P; Kaczmarczyk A; Mittelviefhaus M; Meyer F; Vonderach T; Hattendorf B; Jenal U; Vorholt JA, Untargeted metabolomics links glutathione to bacterial cell cycle progression. Nat Metab 2020, 2 (2), 153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freund DM; Hegeman AD, Recent advances in stable isotope-enabled mass spectrometry-based plant metabolomics. Curr Opin Biotechnol 2017, 43, 41–48. [DOI] [PubMed] [Google Scholar]
- 24.Doppler M; Bueschl C; Kluger B; Koutnik A; Lemmens M; Buerstmayr H; Rechthaler J; Krska R; Adam G; Schuhmacher R, Stable Isotope-Assisted Plant Metabolomics: Combination of Global and Tracer-Based Labeling for Enhanced Untargeted Profiling and Compound Annotation. Front Plant Sci 2019, 10, 1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wittmann C; Heinzle E, Modeling and experimental design for metabolic flux analysis of lysine-producing Corynebacteria by mass spectrometry. Metab Eng 2001, 3 (2), 173–91. [DOI] [PubMed] [Google Scholar]
- 26.Fischer E; Zamboni N; Sauer U, High-throughput metabolic flux analysis based on gas chromatography-mass spectrometry derived 13C constraints. Anal Biochem 2004, 325 (2), 308–16. [DOI] [PubMed] [Google Scholar]
- 27.Beale DJ; Pinu FR; Kouremenos KA; Poojary MM; Narayana VK; Boughton BA; Kanojia K; Dayalan S; Jones OAH; Dias DA, Review of recent developments in GC-MS approaches to metabolomics-based research. Metabolomics 2018, 14 (11), 152. [DOI] [PubMed] [Google Scholar]
- 28.Boets E; Gomand SV; Deroover L; Preston T; Vermeulen K; De Preter V; Hamer HM; Van den Mooter G; De Vuyst L; Courtin CM; Annaert P; Delcour JA; Verbeke KA, Systemic availability and metabolism of colonic-derived short-chain fatty acids in healthy subjects: a stable isotope study. J Physiol 2017, 595 (2), 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lima VF; de Souza LP; Williams TCR; Fernie AR; Daloso DM, Gas Chromatography-Mass Spectrometry-Based (13)C-Labeling Studies in Plant Metabolomics. Methods Mol Biol 2018, 1778, 47–58. [DOI] [PubMed] [Google Scholar]
- 30.Fiehn O, Metabolomics by Gas Chromatography-Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr Protoc Mol Biol 2016, 114, 30 4 1–30 4 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McConnell BO; Antoniewicz MR, Measuring the Composition and Stable-Isotope Labeling of Algal Biomass Carbohydrates via Gas Chromatography/Mass Spectrometry. Anal Chem 2016, 88 (9), 4624–8. [DOI] [PubMed] [Google Scholar]
- 32.Meissen JK; Pirman DA; Wan M; Miller E; Jatkar A; Miller R; Steenwyk RC; Blatnik M, Phenotyping hepatocellular metabolism using uniformly labeled carbon-13 molecular probes and LC-HRMS stable isotope tracing. Anal Biochem 2016, 508, 129–37. [DOI] [PubMed] [Google Scholar]
- 33.Lai Z; Fiehn O, Mass spectral fragmentation of trimethylsilylated small molecules. Mass Spectrom Rev 2018, 37 (3), 245–257. [DOI] [PubMed] [Google Scholar]
- 34.Schummer C; Delhomme O; Appenzeller BM; Wennig R; Millet M, Comparison of MTBSTFA and BSTFA in derivatization reactions of polar compounds prior to GC/MS analysis. Talanta 2009, 77 (4), 1473–82. [DOI] [PubMed] [Google Scholar]
- 35.Pietrogrande MC; Bacco D; Mercuriali M, GC-MS analysis of low-molecular-weight dicarboxylic acids in atmospheric aerosol: comparison between silylation and esterification derivatization procedures. Anal Bioanal Chem 2010, 396 (2), 877–85. [DOI] [PubMed] [Google Scholar]
- 36.Fiehn O; Wohlgemuth G; Scholz M; Kind T; Lee DY; Lu Y; Moon S; Nikolau B, Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J 2008, 53 (4), 691–704. [DOI] [PubMed] [Google Scholar]
- 37.Kind T; Wohlgemuth G; Lee DY; Lu Y; Palazoglu M; Shahbaz S; Fiehn O, FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem 2009, 81 (24), 10038–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhoads TW; Rose CM; Bailey DJ; Riley NM; Molden RC; Nestler AJ; Merrill AE; Smith LM; Hebert AS; Westphall MS; Pagliarini DJ; Garcia BA; Coon JJ, Neutron-encoded mass signatures for quantitative top-down proteomics. Anal Chem 2014, 86 (5), 2314–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bequette BJ; Sunny NE; El-Kadi SW; Owens SL, Application of stable isotopes and mass isotopomer distribution analysis to the study of intermediary metabolism of nutrients. J Anim Sci 2006, 84 Suppl, E50–9. [DOI] [PubMed] [Google Scholar]
- 40.Cline GW; Lepine RL; Papas KK; Kibbey RG; Shulman GI, 13C NMR isotopomer analysis of anaplerotic pathways in INS-1 cells. J Biol Chem 2004, 279 (43), 44370–5. [DOI] [PubMed] [Google Scholar]
- 41.Moseley HN; Lane AN; Belshoff AC; Higashi RM; Fan TW, A novel deconvolution method for modeling UDP-N-acetyl-D-glucosamine biosynthetic pathways based on (13)C mass isotopologue profiles under non-steady-state conditions. BMC Biol 2011, 9, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buescher JM; Antoniewicz MR; Boros LG; Burgess SC; Brunengraber H; Clish CB; DeBerardinis RJ; Feron O; Frezza C; Ghesquiere B; Gottlieb E; Hiller K; Jones RG; Kamphorst JJ; Kibbey RG; Kimmelman AC; Locasale JW; Lunt SY; Maddocks OD; Malloy C; Metallo CM; Meuillet EJ; Munger J; Noh K; Rabinowitz JD; Ralser M; Sauer U; Stephanopoulos G; St-Pierre J; Tennant DA; Wittmann C; Vander Heiden MG; Vazquez A; Vousden K; Young JD; Zamboni N; Fendt SM, A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr Opin Biotechnol 2015, 34, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metallo CM; Gameiro PA; Bell EL; Mattaini KR; Yang J; Hiller K; Jewell CM; Johnson ZR; Irvine DJ; Guarente L; Kelleher JK; Vander Heiden MG; Iliopoulos O; Stephanopoulos G, Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2011, 481 (7381), 380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan J; Kamphorst JJ; Rabinowitz JD; Shlomi T, Fatty acid labeling from glutamine in hypoxia can be explained by isotope exchange without net reductive isocitrate dehydrogenase (IDH) flux. J Biol Chem 2013, 288 (43), 31363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zamboni N; Fendt SM; Ruhl M; Sauer U, (13)C-based metabolic flux analysis. Nat Protoc 2009, 4 (6), 878–92. [DOI] [PubMed] [Google Scholar]
- 46.Phan J; Meinardi S; Barletta B; Blake DR; Whiteson K, Stable isotope profiles reveal active production of VOCs from human-associated microbes. J Breath Res 2017, 11 (1), 017101. [DOI] [PubMed] [Google Scholar]
- 47.Quinn RA; Whiteson K; Lim YW; Salamon P; Bailey B; Mienardi S; Sanchez SE; Blake D; Conrad D; Rohwer F, A Winogradsky-based culture system shows an association between microbial fermentation and cystic fibrosis exacerbation. ISME J 2015, 9 (4), 1024–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skogerson K; Wohlgemuth G; Barupal DK; Fiehn O, The volatile compound BinBase mass spectral database. BMC Bioinformatics 2011, 12, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scientific Instrument Services. (2005, May 22). [Online]. Available: http://www.sisweb.com/referenc/tools/massdes.htm. [Google Scholar]
- 50.Collins MD; Hutson RA; Baverud V; Falsen E, Characterization of a Rothia-like organism from a mouse: description of Rothia nasimurium sp. nov. and reclassification of Stomatococcus mucilaginosus as Rothia mucilaginosa comb. nov. Int J Syst Evol Microbiol 2000, 50 Pt 3, 1247–1251. [DOI] [PubMed] [Google Scholar]
- 51.Lane AN; Tan J; Wang Y; Yan J; Higashi RM; Fan TW, Probing the metabolic phenotype of breast cancer cells by multiple tracer stable isotope resolved metabolomics. Metab Eng 2017, 43 (Pt B), 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frey AJ; Feldman DR; Trefely S; Worth AJ; Basu SS; Snyder NW, LC-quadrupole/Orbitrap high-resolution mass spectrometry enables stable isotope-resolved simultaneous quantification and (1)(3)C-isotopic labeling of acyl-coenzyme A thioesters. Anal Bioanal Chem 2016, 408 (13), 3651–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lai Z; Tsugawa H; Wohlgemuth G; Mehta S; Mueller M; Zheng Y; Ogiwara A; Meissen J; Showalter M; Takeuchi K; Kind T; Beal P; Arita M; Fiehn O, Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat Methods 2018, 15 (1), 53–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiu Y; Moir R; Willis I; Beecher C; Tsai YH; Garrett TJ; Yost RA; Kurland IJ, Isotopic Ratio Outlier Analysis of the S. cerevisiae Metabolome Using Accurate Mass Gas Chromatography/Time-of-Flight Mass Spectrometry: A New Method for Discovery. Anal Chem 2016, 88 (5), 2747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


