SUMMARY
Neurotransmitter receptors partition into nanometer-scale subdomains within the postsynaptic membrane that are precisely aligned with presynaptic neurotransmitter release sites. While spatial coordination between pre- and postsynaptic elements is observed at both excitatory and inhibitory synapses, the functional significance of this molecular architecture has been challenging to evaluate experimentally. Here we utilized an optogenetic clustering approach to acutely alter the nanoscale organization of the postsynaptic inhibitory scaffold gephyrin while monitoring synaptic function. Gephyrin clustering rapidly enlarged postsynaptic area, laterally displacing GABAA receptors from their normally precise apposition with presynaptic active zones. Receptor displacement was accompanied by decreased synaptic GABAA receptor currents even though presynaptic release probability and the overall abundance and function of synaptic GABAA receptors remained unperturbed. Thus, acutely repositioning neurotransmitter receptors within the postsynaptic membrane profoundly influences synaptic efficacy, establishing the functional importance of precision pre-/postsynaptic molecular coordination at inhibitory synapses.
In brief
Olah et al. developed an optogenetic approach to acutely disrupt the nanoscale localization of postsynaptic neurotransmitter receptors relative to presynaptic neurotransmitter release sites. They use the approach to establish the functional importance of precision pre-/postsynaptic nano-alignment at GABAergic inhibitory synapses.
Graphical abstract
INTRODUCTION
Synaptic function is largely governed by the molecular composition of the postsynaptic membrane. Neurotransmitter receptors, adhesion proteins, signaling molecules, and cytoskeletal elements interact in transient but highly regulated ways to shape neurotransmission. Modular scaffolding proteins play a decisive role in this organization.1,2 A large body of evidence at excitatory synapses indicates that postsynaptic scaffold proteins are not homogeneously distributed across the synaptic membrane but are concentrated into one or more subsynaptic “nanodomains.”3–6 The clustered organization of synaptic scaffolds is mirrored by the neurotransmitter receptors to which they bind. For example, AMPA-type glutamate receptors (AMPARs) form overlapping nanodomains at excitatory synapses with the scaffold protein PSD-95, while GABAA receptors (GABAARs) are concentrated at nanodomains defined by the inhibitory synaptic scaffold protein gephyrin.7,8 In both cases, postsynaptic neurotransmitter receptor clusters appear to be situated directly opposite presynaptic neurotransmitter release sites, forming trans-synaptic “nanocolumns.”3,9 The potential functional importance of nanocolumnar organization is supported by theoretical modeling at excitatory synapses suggesting postsynaptic AMPARs in the immediate vicinity of presynaptic vesicle fusion sites are more likely to be activated by released glutamate than receptors at more distal sites within the post synaptic density (PSD).10–13 Thus, the precise subsynaptic localization of receptors within the postsynaptic membrane could play a central role in synaptic transmission. While there is emerging experimental evidence for this model at excitatory synapses,4,14–16 little is known about how nanoscale positioning of neurotransmitter receptors influences the function of inhibitory synapses.
Here we developed an optogenetic approach to acutely manipulate GABAergic inhibitory synapse size and nanoscale molecular organization. Paradoxically, enlarging the inhibitory postsynaptic scaffold network resulted in decreased synaptic transmission. While acute synapse growth did not perturb presynaptic properties, nor postsynaptic GABAAR number or function, GABAARs were displaced from their normal nanoscale registration with presynaptic active zones. Thus, disruption of trans-synaptic nanocolumns profoundly influences synaptic efficacy, establishing the functional importance of accurate trans-synaptic molecular coordination at inhibitory synapses.
RESULTS
A tool for manipulating inhibitory postsynaptic scaffolding with light
We set out to generate an optogenetic approach for real-time manipulation of inhibitory synaptic function. A number of different plant photoreceptor proteins have been adapted for inducible spatiotemporal control of protein-protein interactions and localization.4,17–19 For example, a variant of the Arabidopsis thaliana photoreceptor cryptochrome 2 (CRY2), CRY2olig, undergoes rapid and robust clustering by self-association within seconds of blue light exposure.17 We tested whether we could use CRY2olig for remote control of inhibitory synapse size and strength. We fused CRY2olig to a genetically encoded fibronectin intrabody generated by mRNA display (FingR) that binds to endogenous gephyrin,20 the predominant inhibitory postsynaptic scaffold. Activation of CRY2olig-GephFingR should nucleate the recruitment of additional CRY2olig-GephFingR to the inhibitory synapse, allowing us to tune synapse size and potentially function with light (Figure 1A). We first confirmed that CRY2olig retained its ability to cluster by directly imaging mScarlet (mScar)-fused or GFP-fused CRY2olig-GephFingR in dissociated hippocampal neurons before and after blue light exposure. Illumination with a brief pulse of blue light led to a rapid increase in CRY2olig-GephFingR-mScar signal at pre-existing CRY2olig-GephFingR-mScar puncta (Figures 1B and 1C). These puncta were confirmed to be synapses by post hoc fixation and immunostaining for endogenous gephyrin, surface GABAARγ2 (a prevalent synaptic GABAAR subunit),21,22 and vesicular GABA transporter (VGAT) (Figure 1B). In addition to increased total CRY2olig-GephFingR-mScar signal (Figure 1C), light treatment stabilized CRY2olig-GephFingR at synapses, measured by fluorescence recovery after photobleaching (Figure S1A).
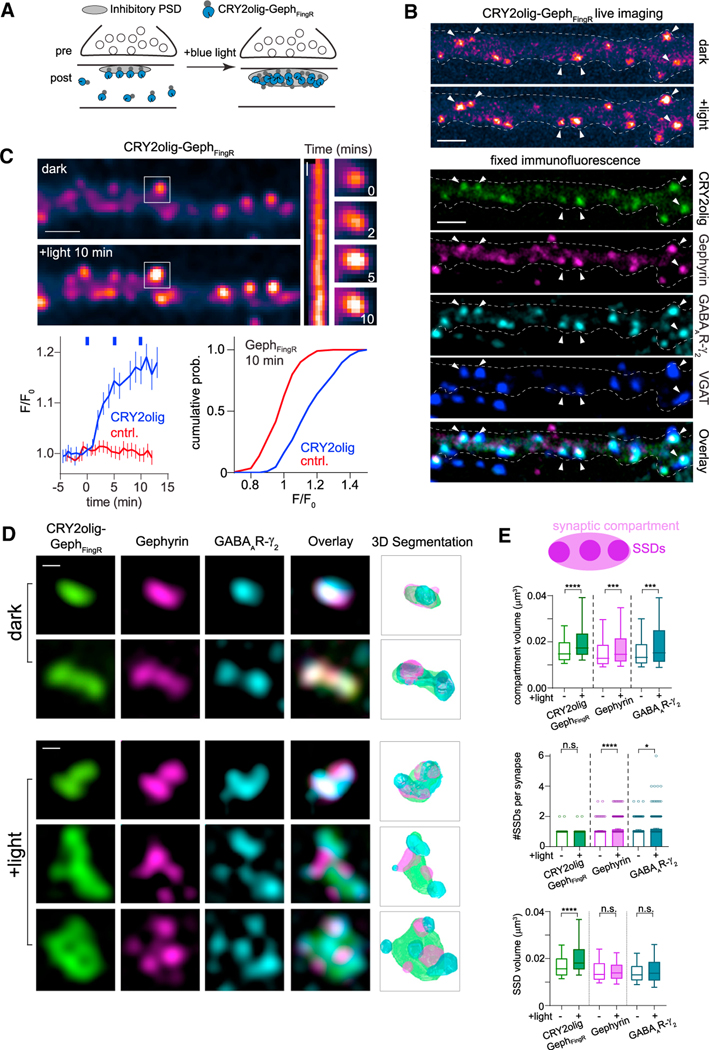
Figure 1. A tool for manipulating the inhibitory postsynaptic scaffolding network with light.
(A) Schematic of optogenetic oligomerization strategy. CRY2olig fused to GephFingR localizes to the postsynaptic membrane of inhibitory synapses. Light exposure induces oligomerization of CRY2olig-GephFingR at the synapse and recruits cytosolic CRY2olig-GephFingR.
(B) CRY2olig-GephFingR-GFP localizes to inhibitory synapses where it nucleates clustering and recruitment of additional CRY2olig-GephFingR-GFP within minutes following blue light exposure. Top panels, live-cell imaging of CRY2olig-GephFingR-GFP before and 10 min following illumination with 488-nm light. Bottom panels, the same cell as in top panels after fixation and immunofluorescent labeling of endogenous gephyrin, GABAAR, and VGAT. Arrows indicate the same synapses across images. Scale bar represents 2 μm.
(C) Confocal images of CRY2olig-GephFingR-mScarlet distribution before (top) and after (bottom) light exposure; scale bar represents 2.5 μm. Kymograph and example images of CRY2olig-GephFingR intensity at a single synapse (denoted by white box) are shown to the right. Kymograph scale bar represents 2 min. Bottom left, kinetics of synaptic CRY2olig-GephFingR clustering. Error bars represent 95% CI. The blue dashes indicate timing of 10-s pulses of 488-nm illumination (n = 80–200 synapses, from 6–7 cells, from 2 independent cultures). Bottom right, cumulative distribution of fluorescent intensity changes for synaptic CRY2olig-GephFingR (blue) and GephFingR control (red) 10 min following onset of blue light exposures.
(D) Structured illumination microscopy (SIM) demonstrates the changes in synapse size and morphology after light-induced CRY2olig-GephFingR clustering. Cells expressing CRY2olig-GephFingR-GFP (green) were stained for endogenous gephyrin (magenta) and GABAARγ2 (cyan) before (dark, top rows) or 10 min following blue light illumination (+light, bottom rows); scale bars represent 250 nm.
(E) Schematic representation of SIM measurements. Compartment volume represents total synaptic volume, encompassing all subsynaptic domains (SSDs). The boxplots below show compartment volumes (top) and mean number and volume of individual subsynaptic domains (SSDs) (middle, bottom) at inhibitory synapses before and after 10 min blue light illumination for CRY2olig-GephFingR-GFP (green), endogenous gephyrin (magenta), and GABAARγ2 (teal). Boxes represent second and third quartile, and bars represent 10th–90th percentile for each dataset. *p < 0.05; **p < 0.01; ****p < 0.0001; n.s. non-significant; Mann-Whitney test. n = 300–420 synapses from three independent cultures.
We next used three-dimensional structured illumination microscopy (3D-SIM) to better quantify the effect of CRY2olig-GephFingR-GFP clustering on inhibitory synapse size and morphology. 3D-SIM imaging of endogenous gephyrin and surface GABAARγ2 revealed the total volume of the inhibitory scaffold/receptor network was indeed larger in light-treated, CRY2olig-GephFingR-GFP-expressing neurons compared to dark controls, with some synapses adopting atypical morphology (Figure 1D). In the most extreme cases, postsynaptic structures formed a ring or annular morphology reminiscent of potentiated excitatory PSDs (Figure 1D; S1B).23,24 Following light treatment, CRY2olig-GephFingR-GFP signal often appeared to envelop smaller endogenous gephyrin puncta, suggesting free (i.e., not bound to gephyrin) CRY2olig-GephFingR-GFP is also recruited to the synapse, potentially dispersing the postsynaptic scaffold network. Indeed, CRY2olig-GephFingR-GFP clustering led to a significant increase in the number of discrete gephyrin and GABARg2 subsynaptic domains (SSDs) 10 min following light exposure compared to dark controls (Figure 1E). These perturbations appeared specific to gephyrin and GABAARs as SSD size and number of neuroligin-2 (NL2), a central inhibitory synapse adhesion protein, were not significantly altered (Figures S1B and S1C). SSD volumes for both gephyrin and GABAARs did not appear significantly different following CRY2olig-GephFingR-GFP clustering; however these structures are at the limit of SIM resolution, and therefore, these measurements would not report small-scale changes. Nevertheless, these results demonstrate CRY2olig can be used to rapidly enlarge the postsynaptic specialization of inhibitory synapses and may impact the nanoscale localization of gephyrin and GABAARs residing in the postsynaptic membrane.
CRY2olig-GephFingR clustering perturbs the nanodomain organization of postsynaptic scaffolds and receptors
To gain a more precise and quantitative measure of the nano-scale reorganization that occurs at the postsynaptic membrane following CRYolig-GephFingR activation, we used direct stochastic optical reconstruction microscopy25 (dSTORM). Because of the increased resolution of the technique, subsynaptic clusters or high-density regions (HDRs) measured by dSTORM appeared considerably smaller than SSDs identified by 3D-SIM, potentially allowing us to resolve finer-scale alterations to synaptic architecture.26 We imaged live hippocampal neurons expressing CRY2olig-GephFingR-GFP to visualize light-triggered clustering, followed by post hoc fixation and gephyrin/GABAARγ2 immunostaining for retrospective dSTORM of the same synapses visualized in live cells (Figure 2A). Consistent with our 3D-SIM data, light activation of CRY2olig-GephFingR-GFP increased the postsynaptic area occupied by both GABAARγ2 and gephyrin (Figures 2B and 2C) compared to GephFingR-GFP controls. We also compared the degree of CRY2olig-GephFingR-GFP synaptic recruitment (measured in live cells) with the total postsynaptic GABAARγ2 area measured by dSTORM (Figure 2C). Although we observed a trend in which synapses that had accumulated more CRY2olig-GephFingR-GFP appeared larger by dSTORM, there was not a statistically significant linear correlation (Figure 2C). However, in agreement with our 3D-SIM data, we observed a significant increase in the number of individual HDRs within the synapse for both GABAARγ2 and gephyrin and significantly decreased HDR area for GABAARγ2 following CRY2olig-GephFingR-GFP clustering (Figures 2D and 2E). The decreased size of GABAARγ2 HDRs offset their increased number, resulting in no significant difference in the total HDR area per synapse. Thus, the total number of receptors may not be altered as the postsynaptic structure grows (Figure 2F). Indeed, we only observed a modest increase in total synaptic GABAARγ2 levels when we performed quantitative immunolabeling/confocal imaging 10 min following CRY2olig-GephFingR-GFP clustering, compared to dark controls (Figure 2G). To account for the high degree of heterogeneity in GABAAR levels at different synapses, we also measured GABAARs before and after CRY2olig-GephFingR clustering at the same synapses in live cells using expressed, Halotag-fused GABAARα2 labeled with membrane-impermeant JF635i Halotag ligand.27 In darkness, Halo-GA-BAARα2 colocalized with CRY2olig-GephFingR, confirming its localization at synaptic sites. We observed no obvious change in synaptic GABAARα2-Halo signal following CRY2olig-GephFingR clustering, indicating little change in receptor number, at least for expressed α2-containing GABAARs (Figure S2). Together, these results show that CRY2olig-GephFingR clustering leads to increased synapse size and a dramatic reorganization of postsynaptic nanoarchitecture with little or no change in total synaptic GABAAR levels.
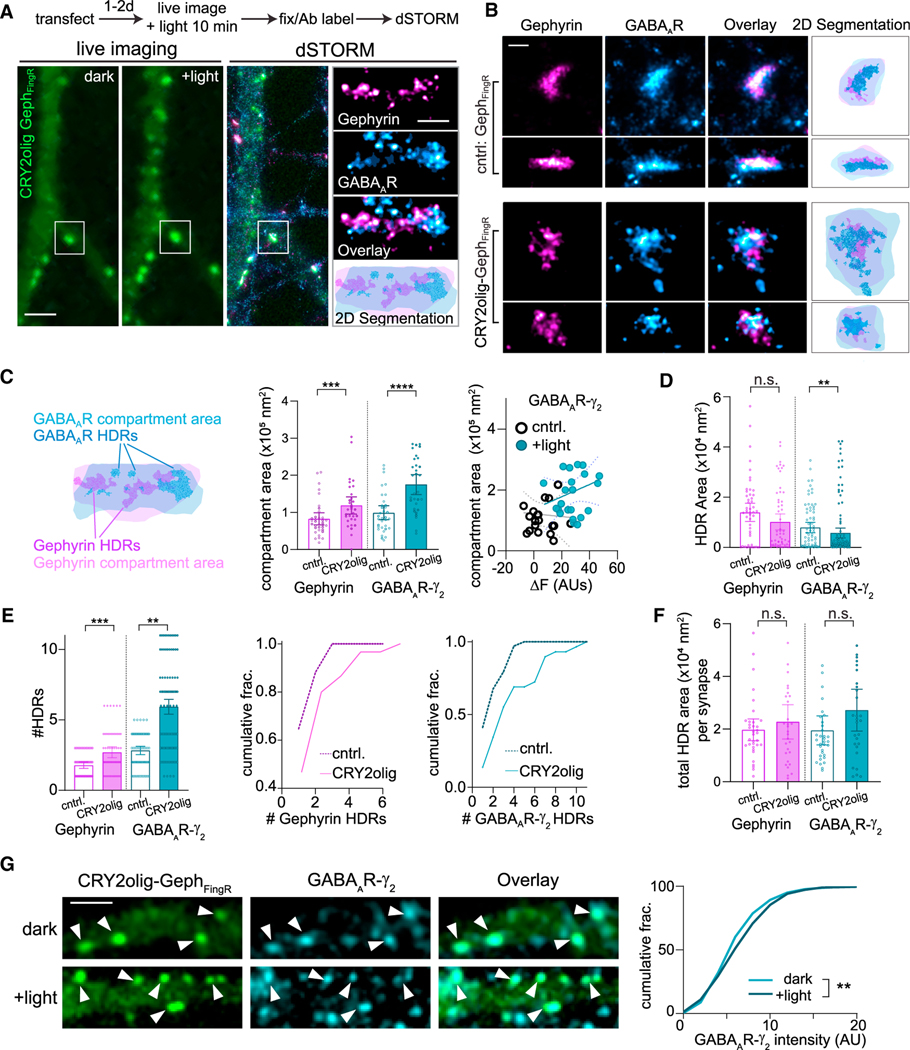
Figure 2. CRY2olig-GephFingR clustering perturbs the nanodomain organization of postsynaptic scaffolds and receptors.
(A) Experimental timeline and representative images for live-cell and retrospective dSTORM imaging protocol. Cultured cells expressing CRY2olig-GephFingR-GFP or GephFingR-GFP were imaged live before (dark) and 10 min after light exposure (+light). Cover slips were then fixed and labeled for endogenous gephyrin and GABAARγ2, and the same cells were imaged by dSTORM. dSTORM localization data for gephyrin (magenta) and GABAARγ2 (blue) are shown to the right, overlayed on the confocal image of CRY2olig-GephFingR. Scale bar represents 2.5 μm. An individual synapse (white box) is magnified to the right. The 2D segmentation image (bottom) displays boundaries defined by minimum localization densities (light shaded areas) and high-density regions (darker shaded regions) for gephyrin (magenta) and GABAARγ2 (blue). Scale bar represents 250 nm.
(B) High-resolution dSTORM example images of inhibitory synapses expressing CRY2olig-GephFingR-GFP or GephFingR-GFP after light-induced clustering. Scale bar represents 200 nm.
(C) Left, diagram showing the delineation of GABAAR and gephyrin high-density regions (HDRs) (darker shades) and postsynaptic compartment regions (lighter shades) in dSTORM reconstructions. Middle, mean compartment area of gephyrin and GABAARγ2 at synapses in cells expressing CRY2olig-GephFingR-GFP or GephFingR-GFP after light-induced clustering. Error bars represent 95% CI. ***p < 0.001; ****p < 0.0001; Mann-Whitney test. n = 63–70 synapses, from three independent cultures. Right, plotted is the the GABAAR compartment area, measured by dSTORM, for synapses from cells expressing CRY2olig-GephFingR-GFP (teal) or GephFingR-GFP (black) vs. the live-cell change in synaptic GephFingR fluorescence intensity (ΔF) following light exposure, quantified from the same synapses.
(D) Mean HDR area for gephyrin and GABAARγ2 in cells expressing CRY2olig-GephFingR-GFP or GephFingR-GFP (cntrl) 10 min after light treatment. Error bars represent 95% CI. **p < 0.01; Mann-Whitney test. n = 63–70 synapses, from three independent cultures.
(E) Mean number of HDRs per synapse for gephyrin and GABAARγ2 in cells expressing CRY2olig-GephFingR-GFP or GephFingR-GFP (cntrl) 10 min after light treatment. Error bars represent 95% CI. **p < 0.01; ****p < 0.0001; Mann-Whitney.n = 63–70 synapses, from three independent cultures. Right, same data plotted as cumulative fractions.
(F) Total summed area of all HDRs within a synapse in cells expressing CRY2olig-GephFingR-GFP (cntrl) or GephFingR-GFP 10 min after light treatment. Error bars represent 95% CI. n.s., not significant; Mann-Whitney test. n = 63–70 synapses, from three independent cultures.
(G) Representative confocal images of CRY2olig-GephFingR-GFP and GABAARs from cells maintained in darkness and 10 min following blue light exposure. Scale bar represents 2.5 μm. Right, cumulative distribution of GABAARγ2 synapse fluorescence intensity in darkness and 10 min after light exposure. **p < 0.01; Kruskal-Wallis non-parametric test. n = 200–300 synapses per condition, from 10 cells in two independent cultures.
CRY2olig-GephFingR oligomerization disrupts the subsynaptic registration of GABAARs and presynaptic GABA release zones
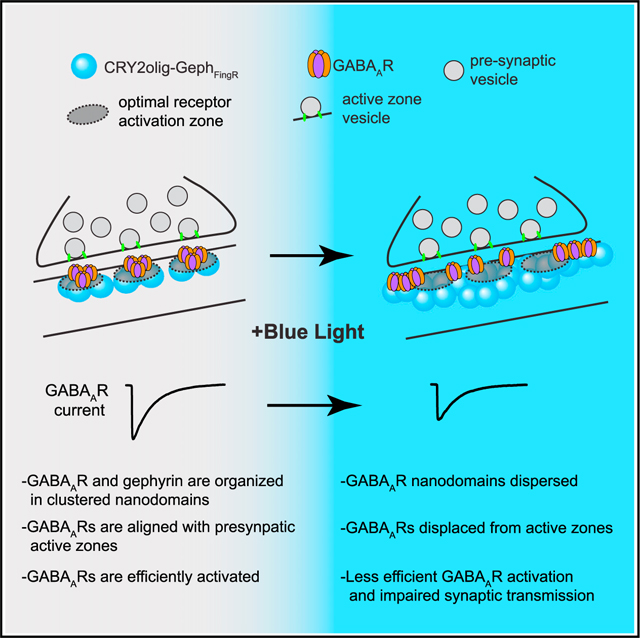
Given the rapid fragmentation of postsynaptic GABAAR nanodomains upon CRY2olig-GephFingR clustering, we wondered if this manipulation also disrupted their spatial coordination with presynaptic release sites. We used both 3D-SIM and stimulated emission depletion (STED) microscopy to visualize postsynaptic GABAARγ2 clusters relative to presynaptic Rab3-interacting molecule (RIM1), which labels presynaptic active zones at both excitatory and inhibitory synapses7,28 (Figure 3A). We first confirmed nano-alignment between RIM1 and GABAARγ2 in control conditions using STED to quantify 3D distances between the intensity center of mass of each RIM1 SSD and the nearest GABAARγ2 SSD (Figure S3A). The experimentally measured distances were significantly smaller than simulated control data, where the same number of points were randomly distributed within the same synaptic volumes (Figures S3A and S3B). We next tested whether CRY2olig-GephFingR-GFP activation disrupted RIM1/GABAAR alignment. Consistent with our dSTORM data, light treatment increased the total volume of synaptic GABAARγ2 signal (Figures 3B, S3C, and S3D). However, we did not observe a compensatory increase in the number or volume of individual presynaptic RIM1 clusters (Figures 3B–3D). Moreover, the average 3D distance between each GABAARγ2 SSD and the nearest RIM1 SSD was significantly increased following CRY2olig-GephFingR-GFP activation compared to dark controls in both our STED and SIM datasets (Figures 3E and S3E). Nearly identical results were obtained when we measured the distance from each RIM1 SSD to the nearest GABAAR SSD (Figures 3E and S3E). Consistent with increased separation between GABAAR and RIM1 nanodomains, we observed decreased overlap between their respective signals following CRY2oligGephFingR-GFP clustering in our SIM datasets (Figure S3F). We also observed a positive correlation between total postsynaptic volume (measured with CRY2olig-GephFingR-GFP signal) and GABAARγ2-to-RIM1 distances in light-treated samples (Figure 3F). This correlation was not observed across the range of naturally occurring synapse sizes in dark controls. Together, these data show that acutely enlarging the postsynaptic scaffold structure leads to a misalignment between GABAARs and GABA release sites, yielding a unique opportunity to assess the functional consequences of breaking pre-/postsynaptic nanocolumn organization in real time.
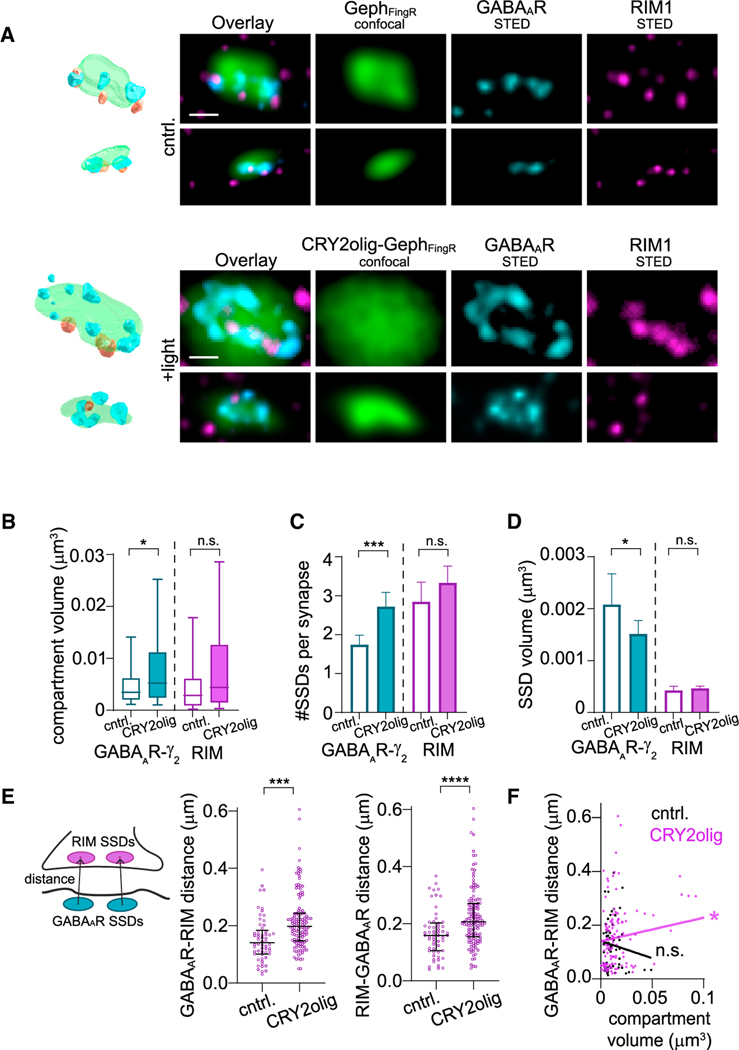
Figure 3. CRY2olig-GephFingR oligomerization disrupts the subsynaptic registration of GABAARs and presynaptic active zones.
(A) High-resolution STED images and 3D re-constructions of synaptic GABAARγ2 and RIM1 in cells expressing GephFingR-GFP (cntrl, top rows) or CRY2olig-GephFingR-GFP (bottom rows), 10 min after blue light exposure. Scale bar represents 200 nm.
(B) Compartment volume of GABAARγ2 and RIM1 at synapses in cells expressing CRY2olig-GephFingRGFP or GephFingR-GFP 10 min after light treatment. Boxes represent second and third quartile, and bars represent 10th–90th percentile for each dataset.. *p < 0.05; Mann-Whitney test. n = 58–128 synapses, from three independent cultures.
(C) Mean number of SSDs per synapse for bothGABAARγ2 and RIM1 in cells expressing CRY2olig-GephFingR-GFP or GephFingR-GFP 10 min after light treatment. Error bars represent 95% CI. ***p < 0.001; Welch’s t test. n = 58–128 synapses, from three independent cultures.
(D) Mean SSD volume for GABAARγ2 and RIM1 in cells expressing CRY2olig-GephFingR-GFP or GephFingR-GFP 10 min after light treatment. Error bars represent 95% CI. *p < 0.05; Mann-Whitney test. n = 58–128 synapses, from three independent cultures.
(E) Left, schematic diagram for GABAAR-RIM1 distance, calculated as the distance between an individual GABAAR SSD center of mass (COM) and the COM for the nearest RIM1 SSD. Middle, median GABAAR to RIM1 and, right, median RIM1 to GABAAR distance per synapse in cells expressing CRY2olig-GephFingR-GFP or GephFingR-GFP, 10 min after light treatment. Error bars represent interquartile range. ***p < 0.001; ****p < 0.0001; MannWhitney test. n = 58–128 synapses, from three independent cultures.
(F) Correlation of average GABAAR to RIM1 distances per synapse vs. synaptic compartment volume in cells expressing CRY2olig-GephFingR-GFP (magenta) or GephFingR-GFP (black) after light-induced clustering. *p < 0.05; n.s. not significant; linear regression test.
CRY2olig-GephFingR clustering impairs inhibitory synaptic function with no change in the probability of presynaptic neurotransmitter release or GABAAR number/function
To assess inhibitory synapse function, we performed whole-cell voltage-clamp recordings of GABAAR currents in the same neurons before and after CRY2olig-GephFingR clustering. For a direct comparison to our super-resolution imaging experiments, we first recorded evoked inhibitory postsynaptic currents (eIPSCs) from dissociated hippocampal neurons expressing CRY2olig-GephFingR-GFP before and after light treatment. In neurons expressing CRY2olig-GephFingR-GFP, eIPSC amplitudes decreased following blue light exposure with similar kinetics to CRY2olig-GephFingR-GFP clustering, reaching an average value of 54% ± 10% below dark baseline values (Figures 4A and 4B). eIPSCs measured from control neurons expressing GephFingR-GFP alone (without CRY2olig) were unaffected by light (Figures 4B and 4C). eIPSC kinetics were not significantly altered by light treatment (Figure S4A). Spontaneous IPSCs (sIPSCs) collected during the inter-stimulus intervals of our evoked recordings also trended toward reduced amplitude following light treatment in CRY2olig-GephFingR-GFP-expressing cells compared to controls (Figure 4D). No change in sIPSC frequency was observed (ΔsIPSC [post light – pre light]: GephFingR-GFP control = 0.015 ± 0.32 Hz, CRY2olig-GephFingR-GFP = −0.13 ± 0.32 Hz, two-way ANOVA). Since evoked and spontaneous vesicle fusion activate distinct GABAAR pools,29,30 we also recorded miniature IPSCs (mIPSCs) before and after light exposure. mIPSC amplitude also decreased following light exposure, albeit to a lesser degree than evoked IPSCs, while frequency remained unaltered (Figure 4E). Light treatment of CRY2olig-GephFingR-GFP-expressing cells did not affect tonic GABAAR current, measured as the difference in baseline holding current before and after blocking GABAARs with picrotoxin (Figure 4F). Finally, we observed no change in paired-pulse IPSC ratios, indicating decreased synaptic transmission following light treatment is unlikely to be a consequence of reduced presynaptic release probability (Figure 4G).
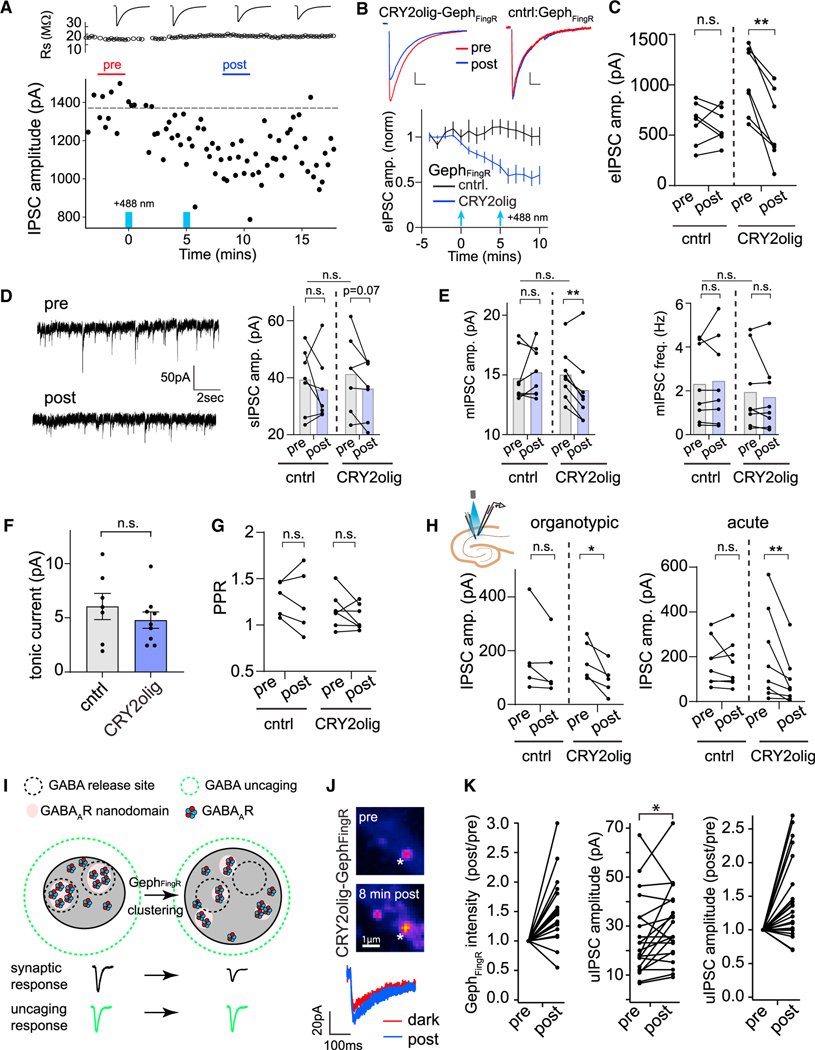
Figure 4. CRY2olig-GephFingR clustering impairs evoked and spontaneous synaptic transmission through GABAAR displacement within the postsynaptic membrane.
(A) Plotted are electrically evoked IPSC amplitudes measured from individual sweeps before and after 2-s light exposures at 0 min and 5 min (denoted by blue bars) recorded from a dissociated hippocampal neuron expressing CRY2olig-GephFingR-GFP. Series resistance measurements are plotted above.
(B) Averaged evoked IPSC amplitudes are plotted as a function of time before and after blue light illumination (2-s pulse, denoted by blue arrows) in dissociated hippocampal neurons expressing GephFingR-GFP (black) or CRY2olig-GephFingR-GFP (blue). Error bars are 95% CI. Example IPSC traces before (red) and 10 min following light exposure (blue) are shown above. Scale bar left, 250 pA, 35 ms. Scale bar right, 150 pA, 45 ms. *p < 0.05; two-way ANOVA. n = 7 cells each condition, from 5 independent cultures.
(C) Paired comparisons of IPSC peak amplitudes before (pre) and 10 min following blue light illumination (post) are shown for hippocampal neurons expressing GephFingR-GFP (cntrl) or CRY2olig-GephFingR-GFP (CRY2olig). **p < 0.01, n.s. non-significant; paired t test, n = 7 cells, from 5 independent cultures.
(D) Left, spontaneous IPSC (sIPSC) example traces within the inter-stimulus intervals from cells in (A)–(C). Right, paired comparison of sIPSC amplitude from the same cells expressing either GephFingR-GFP (cntrl) or CRY2olig-GephFingR-GFP (CRY2olig), before (pre) and 10 min following blue light exposure (post). p = 0.07, n.s. non-significant; paired t test. Comparison of population means for before light (pre) conditions on top. n.s. non-significant; Student’s t test. n = 7 cells, from 5 independent cultures.
(E) mIPSC amplitude (left) and frequency (right) are shown, paired from the same cells expressing either GephFingR-GFP (cntrl) or CRY2olig-GephFingR-GFP (CRY2olig) before (pre) and 10 min following blue light exposure (post). **p < 0.01; paired t test. n.s. non-significant. Comparison of population means for before light (pre) conditions on top. n.s. non-significant; Student’s t test. n = 8 cells.
(F) Tonic GABAAR currents were measured by the change in holding current before and after the addition of picrotoxin in light-treated cells expressing GephFingR-GFP (cntrl) or CRY2olig-GephFingR-GFP (CRY2olig). Error bars represent SEM. n.s. non-significant; Student’s t test, n = 7–9 cells.
(G) Paired-pulse ratios (PPRs) were measured (100-msec interval) from cells expressing GephFingR-GFP (cntrl) or CRY2olig-GephFingR-GFP (CRY2olig) before (pre) and 10 min following blue light exposure (post). n.s. non-significant; paired t test, n = 5–7 cells.
(H) Paired comparisons of IPSC peak amplitudes before (pre) and 10 min after blue light activation (post) are shown for CA1 pyramidal neurons expressing GephFingR-GFP (cntrl) or CRY2olig-GephFingR-GFP (CRY2olig). Data were taken from biolistically transfected organotypic slices (left) or acute slices from AAV-infected mice (right). *p < 0.05; **p < 0.01; paired t test, n = 5 cells organotypic slice, from 5 independent cultures and 8 cells acute slices, from 3 animals.
(I) Model for activation of receptors within subsynaptic domains apposed to GABA release sites (black dashed circles) before and after expansion of the postsynaptic membrane by GephFingR clustering. If electrically evoked IPSCs are reduced due to nanoscale displacement within the postsynapse, uncaging-evoked IPSCs should remain unperturbed as the diffraction-limited uncaging volume releases GABA across the entire postsynaptic membrane (green dashed circle).
(J) Top, images of a hippocampal neuron expressing CRY2olig-GephFingR-Halo (labeled with JF646) before and after light-induced clustering. White asterisk marks location of focal GABA uncaging. Bottom, representative uIPSCs (average of two sweeps) are shown before (red) and 8 min following CRY2olig-GephFingR-Halo clustering (blue).
(K) Paired comparisons of normalized CRY2-GephFingR-Halo intensity (left), raw uIPSC amplitudes (middle), and normalized uIPSC amplitudes (right) before and after CRY2olig-GephFingR-Halo clustering. n = 21 synapses from 7 neurons from 3 independent cultures. Statistical comparisons were only carried out on the raw, non-normalized uIPSC amplitude data. *p < 0.05, paired t test.
Because neurotransmitter receptor dynamics, synapse organization, and size can be significantly different in intact neural circuitry, we also tested the functional effects of CRY2olig-GephFingR clustering in both organotypic and acute hippocampal slices. CRY2olig-GephFingR-GFP was introduced biolistically (organotypic slices) or through viral transduction with adeno-associated virus (AAV) injected at p21, with acute slices prepared 2 weeks later (Figures S4B–S4D). We measured evoked IPSCs from CA1 pyramidal neurons with the stimulation electrode placed in stratum radiatum. In both cases, we observed light-triggered impairment of evoked IPSC amplitude in CA1 neurons expressing CRY2olig-GephFingR-GFP but not control cells expressing GephFingR alone (Figures 4H, S4B, and S4D). Overall, the effect of CRY2olig-GephFingR-GFP clustering on IPSCs in intact circuitry was nearly identical to that observed in primary dissociated neurons (Figures S4B–S4E).
Taken together, our imaging and functional experiments are consistent with a model where displacing GABAARs from presynaptic GABA release sites results in decreased receptor activation. Alternatively, CRY2olig-GephFingR clustering could directly impair synaptic GABAAR function through structural or biochemical alterations. To control for this possibility, we measured GABAAR function before and after light-triggered clustering using focal RuBi-GABA uncaging at individual synapses while simultaneously recording uncaging-evoked IPSCs (uIPSCs). For direct comparison to our super-resolution experiments, we performed these recordings in dissociated hippocampal neurons. We calibrated the uncaging laser pulse intensity and duration such that uIPSC amplitudes were similar in amplitude to quantal mIPSCs. The uncaging volume is limited by diffraction (~250 nm in x/y) and therefore delivers GABA over a volume that encompasses the entire postsynaptic area. Thus, uncaging-evoked GABAAR currents should be insensitive to small receptor displacements within the postsynaptic structure (Figure 4I). In striking contrast to electrically evoked responses, we observed a small but significant increase in uIPSC amplitude following CRY2olig-GephFingR-GFP clustering, consistent with the slight increase in GABAARγ2 signal we observed by immunolabeling (Figures 4J, 4K, and 2G). Thus, reduced electrically evoked and miniature GABAAR currents following CRY2olig-GephFingR clustering are not a consequence of reduced GABAAR number or function. Instead, these data support a critical role for nanocolumnar organization between postsynaptic receptors and presynaptic vesicle release sites for efficient synaptic transmission.
DISCUSSION
We initially set out to develop an optogenetic tool to control the function of inhibitory synapses. Through characterizing the approach, we gained insights into the functional role of molecular nanoarchitecture for inhibitory synaptic transmission. Notably, while CRY2olig-GephFingR clustering led to a rapid increase in the overall size of the inhibitory postsynaptic scaffold network, evoked and spontaneous miniature GABAAR currents grew smaller. Decreased synaptic transmission could be due to direct structural or biochemical perturbations to GABAARs or their dissociation from the postsynaptic membrane. However, when GABA was delivered using focal uncaging at individual synapses, we observed slightly larger GABAAR currents. Thus, GABAAR number, ligand binding, and channel function do not appear to be directly impaired by CRY2olig-GephFingR clustering. Alternatively, transmission could be impaired because CRY2olig-GephFingR clustering physically disrupts synaptic architecture, and GABAARs are no longer optimally positioned for activation by released neurotransmitter. Indeed, previous studies suggest synaptic GABAARs are not saturated during single vesicle release and therefore may be sensitive to subsynaptic positioning.31–35 Consistent with these data, our super-resolution imaging experiments show precise apposition of postsynaptic GABAAR clusters with RIM1 under control conditions, providing additional support for trans-synaptic nanocolumn structures at inhibitory synapses, as described in previous work.7,36 Our experiments acutely disrupting nanocolumnar alignment revealed that even relatively small receptor displacements can significantly impact inhibitory neurotransmission. The fact that evoked GABAAR currents were more robustly impacted by CRY2olig-GephFingR clustering than spontaneous currents suggests that GABAARs associated with evoked release sites are more sensitive to nanoscale positioning compared to receptors activated during spontaneous neurotransmission, which may be localized to more peripheral regions of the postsynaptic membrane.15,29
Surprisingly, presynaptic release zones defined by RIM1 did not laterally track with postsynaptic receptor clusters as they dispersed in the postsynaptic membrane as would be expected if inhibitory nanocolumns were maintained through stable trans-synaptic adhesion complexes. These observations suggest normal pre-/post-apposition may be regulated by indirect or weak associations of GABAARs and/or gephyrin with trans-synaptic organizing molecules. However, it is also possible that alignment is normally tightly coupled, and that CRY2olig-GephFingR clustering interferes with biochemical modifications or scaffolding interactions that otherwise localize receptors to nanocolumns. For example, gephyrin directly interacts with a number of different postsynaptic components, including the trans-synaptic adhesion protein NL2.37,38 Future experiments will determine the molecular constituents of inhibitory nanocolumns, but the fact that NL2 nanoarchitecture was resistant to CRY2olig-GephFingR clustering suggests its association with presynaptic adhesion proteins could play a key role. It will also be interesting to determine whether new presynaptic active zones form or existing active zones mobilize to reestablish contact with displaced postsynaptic receptor clusters over longer timescales than the experiments we report here.
CRY2olig-GephFingR clustering led to diverse morphological changes at individual synapses, with some synapses nearly doubling in size, and others remaining apparently unperturbed. In the most extreme cases, CRY2olig-GephFingR clustering triggered the formation of ring-like inhibitory postsynaptic structures at a subset of synapses. Similar structures are adopted by excitatory PSDs following synapse growth and enlargement associated with long-term potentiation, leading to the apparent PSD perforations commonly observed in single-plane electron micrographs.23,24 Conversion from disc to annular morphology may be a general physical principle common to growing, interconnected membrane-associated protein condensates. Importantly, we also observed this morphology at a sparse subset of inhibitory synapses in our control datasets. Thus, while rare, this structure naturally exists and has been previously documented in somato-sensory cortex and spinal cord.39,40 On the other hand, a significant fraction of synapses did not accumulate CRY2olig-GephFingR following light treatment. Furthermore, there was considerable overlap in the distribution of synapse size and gephyrin nanodomain properties (size, number) between clustered and control synapses. This could be due to a lower abundance of gephyrin at non-affected synapses and therefore less initial CRY2olig-GephFingR to nucleate growth. Additionally, some inhibitory synapses may be more dependent on different scaffold molecules such as LHFPL4/GARLH family members38,41 and therefore less susceptible to gephyrin manipulations. The significant fraction of synapses refractory to the manipulation may explain why we did not observe a larger decrease in synaptic transmission nor significant changes in IPSC kinetics. Given the natural heterogeneity in inhibitory synapse size and GABAAR composition, it will be interesting to determine whether synapses formed by distinct interneuron populations display similar nanocolumnar organization and sensitivity to perturbations to postsynaptic nanoarchitecture. Finally, whether these same principles apply to excitatory synapses remains to be seen, but the general approach we describe here could be readily applied to scaffolds present at other synapse types or, more broadly, to any cellular signaling hub where precision nanoscale localization could influence activation and downstream signaling.
Limitations of the study
In this study we observed that light-triggered CRY2olig-GephFingR clustering led to perturbations in nanoscale gephyrin/GABAAR organization at dendritic inhibitory synapses. However, it remains unclear how subsynaptic molecular organization and function are impacted at inhibitory synapses located in different subcellular domains (e.g., dendrites, soma, axon initial segment) made by distinct interneuron populations. Furthermore, due to the availability of robust labeling reagents, we focused primarily on the γ2 subunit, which may not capture the diversity of synaptic and extrasynaptic GABAARs. Due to technical limitations, we were not able to directly compare synaptic nanostructure at the same synapses before and after CRY2olig-GephFingR clustering. Therefore our measurements are based on population averages, making it difficult to extract quantitative information about GABAAR activation at different distances from GABA release sites. Finally, our experiments report the short-term (within minutes) effects of scaffold/receptor nanodomain perturbations. The long-term effects on synaptic strength and morphology, and whether these manipulations are readily reversible, remain to be seen.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Any additional information or enquiries about reagents and resources should be directed to the Lead contact, Matthew J. Kennedy (Matthew.Kennedy@cuanschutz.edu).
Materials availability
The transfer of plasmids generated for this study will be made available upon request. A Materials Transfer Agreement may be required.
Data and code availability
No standardized datatypes are reported in this paper. All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
All procedures were conducted in accordance with guidelines approved by Administrative Panel on Laboratory Animal Care at University of Colorado, Anschutz School of Medicine, accredited by Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) (00235). Rats were obtained as pregnant dams (typically embryonic day 16) from Charles River Laboratories and housed under standard conditions. Dissociated cultures were prepared from both male and female Sprague-Dawley rat pups within 48 h of birth. Mice were bred at the University of Colorado Anschutz and were from a C57BL/6 genetic background. Mice were housed in a dedicated animal care facility maintained at 35% humidity, 21°C–23°C, on a 14/10 light/dark cycle. Mice were housed in groups of 2–5 in ventilated cages with same-sex littermates with food and water ad libitum. Animals were stereotactically injected at P21, with acute hippocampal brain slices prepared from adult (P36–42) animals.
Cell culture
Dissociated hippocampal cultures were prepared from neonatal rat pups as previously described.44 Briefly, hippocampi were dissected from the brains of postnatal day 0–2 rats and dissociated by papain digestion. Cultures were plated at an approximate density of 100,000 cells per well on 18-mm poly-D-lysine-coated (Sigma) coverslips in 12-well cell culture dishes in MEM and 10% FBS (Hyclone) containing penicillin/streptomycin. After 1 day, the medium was replaced with Neurobasal-A medium (Invitrogen) supplemented with B27 (Invitrogen) and Glutamax (Thermo Fisher). Neurons were maintained at 37°C in a humidified incubator at 5% CO2. Neurons were then fed on days in vitro (DIV) 7 or 8 by replacing half the medium with Neurobasal-A, B27, and mitotic inhibitors (uridine and fluoro dexyuridine). On DIV 15–18, neurons were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendation, wrapped in aluminum foil to protect from light exposure and allowed to express for 24–48 h. All experiments were conducted between days in vitro (DIV) 16 to 21.
METHOD DETAILS
Molecular cloning and expression constructs
The gephyrin intrabody (pCAG_GPHN.FingR-eGFP-CCR5TC) was a gift from Don Arnold20 (Addgene plasmid # 46296). We modified this construct using standard cloning techniques to insert Cry2Olig17 into the multiple cloning site on the N terminus, leaving the C terminus unmodified. For experiments examining GABAA receptor localization at inhibitory synapses, neurons were transfected with plasmids encoding HaloTag-tagged α2 GABAA receptor subunits. The original GABAA receptor subunit α2SE was a gift from Tija Jacob & Stephen Moss42 (Addgene plasmid # 49169). We modified this construct by excising out the Superecliptic pHluorin (SEP) tag and replacing it with a HaloTag using Gibson Assembly and standard cloning techniques.
CRY2olig light activation
We found a similar degree and kinetics of CRY2olig-GephFingR clustering using multiple different light exposure conditions. For live cell imaging studies samples were directly imaged using full field confocal illumination with 488 excitation light, both acquiring multiple z stack series continuously every 1 min or every 5 min. For fixed immunofluorescence samples were exposed to 2 s pulses of blue light treatment (461 nm delivered from a custom-built LED array, 15.6 mW/cm2) every 5 min for a total of 10 min (three pulses total). For electrophysiology experiments samples were illuminated using a GFP filter cube (469 ± 22 nm excitation) with an LED light source for 2–3 s per light pulse once every 5 min through the objective.
Live cell imaging
Live imaging of dissociated neurons was carried out at 32°C on an Olympus IX71 equipped with a spinning-disc scan head (Yoko-gawa). Excitation illumination was delivered from an acousto-optic tunable filter (AOTF) controlled laser launch (Andor). Images were acquired using a 60X Plan-Apochromat 1.4 numerical aperture (NA) objective and collected on a 1,024× 1,024 pixel Andor iXon EMCCD camera. For imaging experiments in this study, the main apical dendritic arbor approximately 0–150 μm from the cell soma was imaged. Data acquisition and analysis were performed with Metamorph (Molecular Devices), Andor IQ, ImageJ, and MATLAB software. Quantification of synaptic fluorescence intensity was performed in ImageJ. A mask of all segmented puncta was created for each image as follows: images were smoothed using the MexicanHat filter plugin (Laplacian of Gaussian) or the “Unsharp Mask” filter (Difference of Gaussians) and then thresholded manually. The average fluorescence intensity within these masked puncta was then calculated from the raw fluorescence images. Cry2Olig activation occurred through full field illumination with 488 excitation light, both acquiring multiple z stack series continuously every 1 min or every 5 min as indicated in figure legends.
Fluorescence recovery after photo bleaching (FRAP)
For quantification of receptor mobility, primary hippocampal neuronal cultures were transfected with CRY2olig-GephFingR/GephFingR-GFP and an pCAG-mCherry cell fill construct to identify expressing cells without light exposure. Photobleaching of individual synapses was carried out using galvometric steered laser excitation (FRAPPA, Andor) at single spots for 1 s. Photobleaching pulses were calibrated so that they bleached no more than 30–85% of the original signal. Recovery was monitored following photobleaching every 20 s. FRAP recovery rates were determined for different sets of synapses with or without focal blue light exposure (488 FRAPPA, 10% laser power, 0.1 s) from the same neuron. Non-photobleached synapses were used to correct for photobleaching and recovery curves were fit to a single component exponential fit I(t) = A*(1-exp(−τ*t)), where I(t) represents the fluorescence intensity at time t, (1-A) represents the immobile fraction, and t represents the recovery rate.
Immunohistochemistry Sample preparation
24–48 h after transfection (wrapped in aluminum foil after transfection and kept in dark) cultured neurons were exposed to 2 s pulses of blue light treatment (461 nm delivered from a custom-built LED array, 15.6 mW/cm2) every 5 min for a total of 10 min (three pulses total) or kept in the dark. After light treatment, neurons were immediately fixed in 4% PFA solution [4% sucrose, 1× PBS and 50 mM HEPES (pH7.5)] for 5 min at room temperature followed by three washes with 1× PBS. Neurons were blocked for 30 min (5% BSA, 2% Normal Goat Serum). For surface GABAAR labeling, antibody was added before permeabilization (GABAAR-γ2 (1:500 Synaptic Systems Guinea Pig – 224 004). Neurons were washed three times in 1x PBS and then permeabilized for 5 min (5% BSA, 2% Normal Goat Serum, 0.2% NP-40 and 13 PBS) and blocked for 1 h (5% BSA, 2% Normal Goat Serum 13 PBS). Neurons were incubated with antibodies to Gephyrin mAb7a (1:500 Synaptic Systems 147 011), RIM1 (1:150 Synaptic Systems Rabbit – 140 003), VGAT (1:1000 Synaptic Systems 131 004) and NL2 (1:500 Synaptic Systems 129 203) diluted in block solution for 1 h. Neurons were washed three times in 1× PBS and then labeled with secondary antibodies for 1 h at room temperature (for 3D-SIM; 1:1000 ThermoFisher Alexa Fluor 488, 568 and 647), or overnight (for dSTORM; 1:1000 Alexa Fluor 647 anti-mouse; 1:1000 Sigma-Aldrich CF568 anti-rabbit; 1:1000 ThermoFisher Alexa Fluor 488 anti-guinea pig) and (for STED; 1:200 anti-guinea pig Abberior STAR ORANGE, 1:200 anti-rabbit Abberior STAR RED). Coverslips were washed four times in 1× PBS and mounted on glass microscope slides using ProLong Gold Antifade mounting media for confocal, 3D-SIM and STED. For dSTORM, coverslips were washed and post fixed with 4% PFA in 1× PBS for 5 min at room temperature followed by final washes where they remained in 1× PBS until imaging.
dSTORM Sample preparation and imaging
Samples for dSTORM and SIM imaging were prepared as previously described.26 Images were acquired on a Zeiss Elyra P.1 TIRF microscope equipped with a Zeiss alpha Plan-Apochromat TIRF 100x/1.6 NA oil objective; tube lens for an additional factor of 1.6x magnification; and quad-band dichroic (405/488/561/642). For both the CF568 and Alexa 647 dyes, an Andor iXon+ EMCCD camera captured a sequential time-series of 20,000 frames each at a gain setting of 100 with an integration time of 18ms. Image size was 256X256 pixels, with a pixel size of 100 nm xy. Alexa 647 molecules were ground-state depleted and imaged with a 100mW 642 laser at 100% AOTF transmission in ultra-high-power mode (condensed field of illumination), corresponding to approximately 1.4W/cm2. Emission light passed through an LP 655 filter. CF-568 molecules were ground-state depleted and imaged with a 200mW 561 laser at 100% AOTF transmission in ultra-high-power mode, corresponding to approximately 2.5W/cm2. Emission light was passed through a BP 570–650 + LP 750 filter. For each dye, ground-state return was elicited by continuous illumination with a 50mW 405 laser at 0.01 to 0.1% AOTF transmission.
Electrophysiology in organotypic slices and cultured neurons
Whole-cell and cell-attached voltage-clamp recordings were performed on dissociated hippocampal neurons perfused with an extracellular solution containing (in mM) 10 HEPES, 130 NaCl, 5 KCl, 30 d glucose, 2 CaCl2, and 1 MgCl2 equilibrated with 95% O2/5% CO2. Recordings were conducted at approximately 30°C. For evoked IPSC recordings, neurons were held at −70mV and excitatory transmission was blocked with DL-AP5 (100 μM) and NBQX (30 μM). For mIPSC recordings, TTX (1 μM) was included in the bath solution. eIPSC and sIPSC recordings were conducted with a holding potential of −70mV using high chloride internal. High chloride internal solution contained (in mM): 67.5 CsCl, 67.5 CsMeSO4, 0.1 CaCl2, 2 MgCl2, 10 HEPES, 0.1 EGTA, 0.5 Na3GTP, 3 Na2ATP, 10 phosphocreatine, pH was adjusted to 7.25 with CsOH. mIPSC recordings were conducted using a holding potential of −20 mV using a standard internal solution. Standard internal solution contained 130 CsMeSO4, 3 Na2ATP, 0.5 Na3GTP, 0.5 EGTA, 10 phosphocreatine, 5 MgCl2, 2.5 NaCl, 10 HEPES (285–300 mOsm). Under these conditions, the addition of picrotoxin (100 μM) eliminated all synaptic events. Picrotoxin (100 μM) also produced a shift in the holding current, consistent with the blockade of extrasynaptic GABAA receptors mediating the tonic current. For evoked IPSCs, stimulation was performed using a theta glass electrode or a concentric bipolar electrode (FHC, Bowdoin, ME) placed just above the culture, approximately 200 μm from the targeted neuron.
Data were collected using a Multiclamp 700b amplifier and digitized using a National Instruments DAQ board at 10kHz, filtered at 2 kHz (single pole Bessel filter), and collected with WinLTP software (University of Bristol). For Cry2olig activation, we illuminated the preparation using a GFP filter cube (469 ± 22 nm excitation) with an LED light source for 2–3 s per light pulse through the objective. The exact light stimulation used in each experiment can be found in the corresponding figure legends. The light intensity used to image a single frame of GFP is fully sufficient to activate Cry2olig, thus phototoxicity has not been a concern.45 Control (dark treated, or light insensitive GephFingR expressing neurons) and experimental (light-treated or CRY2olig-GephFingR expressing neurons) were interleaved. Evoked IPSC traces were analyzed using WinLTP software; mIPSC traces were analyzed using Mini Analysis (Synaptosoft). For all recordings, cells were excluded from analysis of the series resistance grew larger than 25 MU. All electrophysiology experiments were replicated using at least 3 independent preparations, with sample sizes for each experiment determined by the precision of the measurements from independent cells. We found that a minimum of 5 cells yielded an acceptable standard error (typically <15% of the measured value).
For GABA uncaging experiments, we included 50μM RuBi-GABA in the bath solution and focally stimulated the preparation using galvanometric mirrors (FRAPPA, Andor technologies) to steer a diffraction-limited 488 nm spot. An AOTF was used to gate a 1 ms pulse of 488 nm light, with the intensity adjusted to trigger an approximately quantal (15–30pA) GABAAR current. Intensities ranged from 3 to 4% of total laser power from a 100mW 488nm laser that was fiber coupled to an FRAPPA laser scanning unit. Uncaging spatial precision was measured by recording currents when the uncaging spot was moved different distances from an inhibitory synapses with currents falling off with a space constant of 2.3 μm, similar to previous reports,46 which allows us to measure responses at individual synapses, but not within subsynaptic domains.
Stereotactic viral injections
Mice were bred at the University of Colorado Anschutz and were from a C57BL/6 genetic background. Mice were housed in a dedicated animal care facility maintained at 35% humidity, 21°C–23°C, on a 14/10 light/dark cycle. Mice were housed in groups of 2–5 in ventilated cages with same-sex littermates with food and water ad libitum. Animals were stereotactically injected at P21, and all other experiments were performed during adulthood (P36–42). All procedures were conducted in accordance with guidelines approved by Administrative Panel on Laboratory Animal Care at University of Colorado, Anschutz School of Medicine, accredited by Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) (00235).
Stereotactic injections were performed on P21 mice. Animals were induced with 5% isoflurane, maintained at 1–2% isoflurane, and then head fixed to a stereotactic frame (KOPF). After drilling small holes in the skull using a handheld drill, 0.5 μL solutions of adeno associated viruses (AAVs) were injected with pulled glass micropipettes into hippocampus at a rate of 11–14 μL/h using a syringe pump (World Precision Instruments). Control virus was injected in one hemisphere and the Cry2Olig containing virus was injected into the contralateral hemisphere within the same mouse. Coordinates (in mm) were: rostrocaudal: −3.41, mediolateral: +/− 3.17 (relative to bregma), and dorsoventral: −3.45 (relative to pia). All AAVs used in this study were packaged in-house: AAV hSYN-mRuby and AAV hSyn-CRY2olig-GephFingR-GFP or hSyn-GephFingRGFP was used at 1:1 ratio.
Ex vivo whole-cell electrophysiology
Animals were deeply anesthetized with isoflurane and decapitated. Brains were rapidly dissected and 300 μm horizontal slices were sectioned with a vibratome (Leica VT1200) in ice-cold high-sucrose cutting solution containing (in mM) 85 NaCl, 75 sucrose, 25 D-glucose, 24 NaHCO3, 4 MgCl2, 2.5 KCl, 1.3 NaH2PO4, and 0.5 CaCl2. Slices were transferred to 31.5°C oxygenated ACSF containing (in mM) 126 NaCl, 26.2 NaHCO3, 11 D-Glucose, 2.5 KCl, 2.5 CaCl2, 1.3 MgSO4-7H2O, and 1 NaH2PO4 for 30 min, then recovered at room temperature for at least 1 h before recordings. Whole-cell patch clamp recordings were made at 29.5°C ACSF, with 4–6 MU patch pipettes and cells were voltage-clamped at −70mV. Room illumination was removed, and all preparations/recordings were made in the dark. All recordings were acquired using Molecular Devices Multiclamp 700B amplifier and Digidata 1440 digitizer with Axon pClamp 9.0 Clampex software, lowpass filtered at 2 kHz and digitized at 10–20 kHz.
Inhibitory postsynaptic currents
For acute slice experiments, we used a cesium-based internal solution containing (in mM) 117 cs-methanesulfonate, 15 CsCl, 10 TEA-Cl, 10 HEPES, 10 phosphocreatine, 8 NaCl, 4 Mg2-ATP, 1 MgCl2, 0.5 Na2-GTP, and 0.2 EGTA for experiments measuring inhibitory currents. Inhibitory currents were isolated pharmacologically by including 10μM NBQX and 50μm D-AP5 in the ACSF. Virus expressing cells were visually identified and targeted by mRuby expression. To optically activate Cry2Olig expressing pyramidal cells, slices were illuminated with 470 nm LED light (ThorLabs M470L2-C1) continuously or via 20s pulses through the 40x dipping objective located directly over the recorded cell. With an illumination area of 33.18mm2 the tissue was exposed to an irradiance of 0.17 mW/mm2. For electrical stimulation a homemade nichrome stimulating electrode was placed ~200 μm from the patched cell and pulsed at 0.1 Hz at 100mA (A-M Systems 2100 Isolated pulse stimulator). The stimulation electrode was placed in the stratum radiatum.
QUANTIFICATION AND STATISTICAL ANALYSIS
dSTORM Image processing
Raw data was processed through a custom written pipeline written in MATLAB (Mathworks) made up of several modular elements, described briefly. The Bio-Formats MATLAB toolbox47 was used to read Zeiss raw data files into MATLAB. Image data was transferred between MATLAB and FIJI using MIJI (http://bigwww.epfl.ch/sage/soft/mij/). If necessary, raw data was pre-processed with a temporal median filter48 to remove non-homogeneous background. The filter radius was set at 51 frames, with a key frame distance of 10 (filter is explicitly calculated only for every 10 frames and interpolated between), the quantile for the filtering was set at 20%. Localization of dye emitters was performed using the ThunderSTORM ImageJ plugin.49 The camera EM gain was set to 100, which resulted in a photon-to-ADU of 1.65. When the temporal median filter was used, the Offset was set to zero. Image filtering was done with the Wavelet filter setting, with a B-Spline order of 3 and scale of 2.0. A first pass approximate localization of molecules was achieved by finding local maximum with a peak intensity threshold of 3*std(Wave.F1) and 8-neighborhood connectivity. Weighted least squares fitting of the PSF to achieve sub-pixel localizations was achieved by use of an integrated Gaussian with a fitting radius of four pixels and an initial sigma of 1.2. Localizations were filtered based on the attributes of uncertainty (<20 nm), sigma (50–150 nm), and intensity (<10,000 for CF568 and <15,000 for Alx647). Localizations within 50 nm were merged with a framegap allowance of 1.
Before each experiment a calibration was calculated to correct for shifts and distortions between the acquired fluorescent channels. Sub-diffraction size beads, labeled with fluorophores in both channels were imaged. The bead positions were fitted and registered between the fluorescent channels. Registered localizations from multiple bead images were compiled into one dataset. Calibration matrices of the shift in x and y direction between the imaging channels across the full field of view were calculated by either applying a 2D polynomial fit or a localized weighted averaging to the registered bead localizations. In the raw data, the shift and distortion between the imaging channels was up to 100 nm. Applying the calibration to the STORM data yields an RMS error of less than 15 nm for the channel misalignment. Drift correction was performed using the redundant cross-correlation method described in.50 The segmentation parameter was set at 500 frames, the bin size used in the cross-correlation was 10 nm, and the error threshold for the recalculation of the drift was five pixels.
Localizations were rendered into images using the ThunderSTORM visualization module using the method of average shifted histograms with a magnification of 10 and lateral shift of 2 nm.
dSTORM analysis
Coordinate analysis of our dSTORM data is conceptually similar to methods previously used to classify nanoscale organization at the excitatory synapse.3 Synapses for downstream analysis were selected manually from a composite rendered image and ROI coordinates were recorded using a custom ImageJ macro. ROI details were imported into MATLAB using the ReadImageJROI function (github.com/DylanMuir/ReadImageJROI). The gephyrin scaffold and GABAAR localizations were segmented using a coordinate-by-coordinate density calculation. Briefly, because labeling density could vary greatly, the thresholding parameter was determined from the overall density range of the ROI. Localizations with a local density in the lower 10% of that range were considered to be outside of the synaptic region/clusters. Boundaries for these regions were delineated using MATLAB’s alphaShape function, with an α value of 100. Only gephyrin regions with an area of 1.5 × 103 nm2 or greater were considered for analysis. High-density regions (HDRs) were defined by a cutoff determined by randomizing the experimental localizations assuming a uniform distribution across the synaptic region. The local density threshold for an experimental coordinate to be considered as part of an HDR was set at the mean local density of the randomized dataset plus 2 standard deviations. The geometric boundaries of individual HDRs were again delineated using MATLAB’s alphaShape function, with an α value of 7.
3D structured illumination microscopy (3D-SIM) and analysis
Images were acquired with a Nikon SIM-E Structured Illumination super-resolution microscope equipped with a 100x, 1.49 NA objective; an ORCA-Flash 4.0 sCMOS camera (Hamamatsu); and Nikon Elements software. To maximize signal to noise and reduce photo-bleaching, acquisition conditions and camera integration time were set as previously described.7,26 Synapses were within the entire z stack, and each selection was based on CRY2olig-GephFingR-GFP/GephFingR-GFP positive expression. A high throughput pipeline for analysis is as follows: synapses were processed by background subtraction (ImageJ), image segmentation (split-Bregman/MOSAIC suite51 and geometric analysis (MATLAB) as previously detailed.7,26 For image segmentation, the following parameters were utilized: ‘Subpixel segmentation’, ‘Exclude Z edge’, Local intensity estimation ‘Medium’, Noise Model ‘Gauss’. All 3D-SIM imaging analysis was performed blind to experimental condition. For the RIM/GABAAR distance analysis we identified the center of mass intensity point within each SSD. The binary mask created by the object-segmentation was applied to the original intensity image, creating an object-bound intensity map. The center points of regional intensity maximum were then identified using the MATLAB function [imregionalmax]. The nearest neighbor regional intensity max point pairs in the corresponding channels (RIM and GABAAR) were assigned based on the [knnsearch] function in MATLAB.
STED imaging and analysis
STED super-resolution images were acquired using the commercial Abberior STEDYCON addition to an Olympus confocal microscope equipped with the following: an Olympus UPLXAPO100XO 100X magnification and 1.45 NA. objective, 4 excitation lasers; 405(cw), 485nm(pulsed), 561nm(pulsed), 640nm(pulsed), 4 corresponding single-photon counting APD detectors (avalanche photodiodes), and a 775nm laser for stimulated emission depletion. Samples were stained using secondary antibodies labeled with Abberior STAR RED and/or STAR ORANGE and mounted for 24+ hours in prolong gold mounting medium. A minimum of 3 z-stacks at 0.15 μm spacing were acquired for each image and deconvolved using the SVI Huygens deconvolution software with the Standard Mode Express Deconvolution.
Image segmentation was carried out on deconvolved STED images using the same SIM pipeline as described in the SIM and Analysis methods. Changes to the image segmentation Bregman/MOSAIC suite51 parameters were made to better match the data: Local intensity estimation ‘High’, Noise Model ‘Poisson’.
To determine whether RIM1 and GABAAR alignment was significantly correlated across the synapse we compared experimental data with a simulated dataset. To ensure realistic synaptic geometries and object numbers, randomized data was generated on a ROI to ROI basis as follows: For each synapse, a segmented mask of the entire GABAARγ2 containing post synaptic density (PSD) was generated using the Bregman/MOSAIC suite51: Local intensity estimation ‘Medium’. Random locations within a minimum cube encompassing the GABAARɣ2 PSD volume were generated using the MATLAB ‘rand’ function and only points falling within the GABAARγ2 PSD volume were kept. For each synapse the number of simulated points matched the number of identified GABAARɣ2 SSDs in the actual data. The average of 20 simulations was used for quantification.
Electrophysiology analysis
All IPSC kinetics were fit using ClampFit Software. Rise-time represents 10–90% of peak time. The decay constant represents the weighted decay lifetime from a two-term exponential fit. Values and weights were calculated by fitting the decay slope to a standard, two-term exponential functions (Levenberg-Marquardt method). The decay tau was calculated as TW=((T13A1)+(T23A2))/(A1+A2). Paired data values represent fits to the average of 8 sweeps at the indicated times.
Spontaneous and miniature IPSC amplitude and frequency were quantified using ClampFit ‘Event Detection’ Software. A ‘Template Search’ was used and defined with a custom generated template based on a few manually identified example miniature IPSC events. All events were manually inspected and only accepted if they changed significantly from the baseline variability.
Statistical analysis
Quantification for imaging experiments was carried out on raw fluorescent images using ImageJ to measure pixel intensities. Background values estimated by pixel intensities in regions with no detectable signal were routinely subtracted. Statistical analysis was conducted using Graphpad Prism 10.0 software. A normality test was first carried out on all populations using both the D’Agostino-Pearson test and the Shapiro-Wilk test. For all distributions that were not classified as normal under either test, we used the non-parametric Mann-Whitney test. For all distributions that were classified as normal we used a Student’s t-test. For statistical analysis of time course data or paired datasets, we used 2-way ANOVAs or paired t-tests, as denoted in figure legends. To test for linear correlation, we used the ‘simple linear regression’ analysis in Prism without constraints to determine if the slope deviated significantly from zero. For all statistical analyses, p values < 0.05 were considered significant. All data are presented as ± 95% CI unless otherwise noted. Definitions of “n” values, “N” values, statistical tests, and p values can be found in corresponding figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| GABAAR-γ2 (imaging) | Synaptic Systems | Cat. #224 004; RRID:AB_10594245 |
| Gephyrin (mAB7a; imaging) | Synaptic Systems | Cat. #147 011; RRID:AB_887717 |
| VGAT (rabbit; imaging) | Synaptic Systems | Cat. #131 003; RRID:AB_887869 |
| RIM1 (imaging) | Synaptic Systems | Cat. #140 003; RRID:AB_887774 |
| Neuroligin 2 (NL2; imaging) | Synaptic Systems | Cat. #129 511; RRID:AB_2619813 |
| Guinea pig Alexa Fluor 647 | Abcam | Cat. #150187; RRID:AB_2827756 |
| Mouse AF568 | ThermoFisher | Cat. #A-11004; RRID:AB_2534072 |
| Rabbit DyLight405 | ThermoFisher | Cat. #35550; RRID:AB_1965945 |
| Guinea Pig STAR 580 | Abberior | Cat. #ST580-1006 |
| Rabbit STAR 635p | Abberior | Cat. #ST635P-1002; RRID:AB_2893229 |
| Mouse CF568 | Biotium | Cat. #20100-1; RRID:AB_10853601 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Lipofectamine 2000 | ThermoFisher | Cat. #11668027 |
| JF635i-Halotag ligand | HHMI Janelia Materials | https://janeliamaterials.azurewebsites.net/ |
| DL-AP5 sodium salt | Tocris Bioscience | Cat. #3693 |
| Tetrodotoxin citrate | Tocris Bioscience | Cat. #1069 |
| Picrotoxin | Tocris Bioscience | Cat. #1128 |
| NBQX disodium salt | Tocris Bioscience | Cat. #0373 |
| TetraSpeck microspheres | ThermoFisher | Cat. #T7279 |
|
| ||
| Critical commercial assays | ||
|
| ||
| ZymoPURE™ II Plasmid Maxiprep Kit | Zymo Research | Cat. #D4203 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Rat, Sprague Dawley Charles River | Charles River | RRID: RGD_734476 |
| PV-IRES-Cre: B6; 129P2-Pvalb tm1(cre)Arbr/J | Gift, Dr. Diego Restrepo, CU Anschutz | JAX: 008069, RRID:IMSR_JAX:008069 |
|
| ||
| Recombinant DNA | ||
|
| ||
| Gephyrin-FingR | Gross et al.20 | RRID:Addgene_46296 |
| CRY2olig-mCherry | Taslimi et al.17 | RRID:Addgene_60032 |
| GABA(A)R-α2 | Tretter et al.42 | RRID:Addgene_49169 |
| CRY2Olig-Gephyrin-FingR-GFP | This paper | N/A |
| CRY2Olig-Gephyrin-FingR-HaloTag | This paper | N/A |
| CRY2Olig-Gephyrin-FingR-mScarlet | This paper | N/A |
| Gephyrin-FingR-mScarlet | This paper | N/A |
| AAV- hSyn-CRY2Olig-Gephyrin-FingR-GFP | This paper | N/A |
| AAV- hSyn-Gephyrin-FingR-GFP | This paper | N/A |
| AAV hSYN-mRuby | Liu et al.,43 gift from Dr. Jason Aoto, CU Anschutz | N/A |
| GABA(A)R- α2-HaloTag | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Prism 10 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| NIS Elements | Nikon | https://www.microscope.healthcare.nikon.com/products/software |
| Mosaic Suite (FIJI/ImageJ plugin) | Mosaic Group | https://sbalzarini-lab.org/ |
| MATLAB | Mathworks | https://www.mathworks.com |
| Mini analysis | Synaptosoft | http://www.synaptosoft.com; RRID:SCR_002184 |
| Andor IQ3 acquisition software package | Andor Technologies | http://www.andor.com/scientific-software/iq-live-cell-imaging-software |
| Metamorph | Molecular Devices | https://www.moleculardevices.com/systems/metamorph-research-imaging/metamorphmicroscopy-automation-and-imageanalysis-software |
| Huygens Essential: STED Deconvolution Software | Scientific Volume Imaging | https://svi.nl/Huygens-STED-Software |
| ClampFit 11.2 | Molecular Decives | http://www.moleculardevices.com/ |
Highlights.
GABAARs clusters are normally localized directly opposite GABA release sites
Acutely clustering gephyrin disperses GABAARs within the postsynaptic membrane
Gephyrin clustering impairs synaptic transmission but not GABAAR number/function
Precise GABAAR alignment with GABA release is required for optimal synapse function
ACKNOWLEDGMENTS
The authors would liketothank Mark Dell’Aqua, Carmen Hernández-Candia, and Chandra Tucker for critical discussions. We thank Dominik Stich for assisting with dSTORM/STED imaging, performed at the University of Colorado Anschutz Medical Campus Advanced Light Microscopy Core. This work was supported by National Institute of Mental Health, National Institute of Neurological Disorders and Stroke, National Institute on Drug Abuse, and National Institute of General Medical Sciences: F32MH123053 (S.S.O); F31NS130979 (D.J.K.); F30DA0507053 and 5T32GM007635 (C.N.M.); R01MH116901 (J.A.); R01MH119154 (K.R.S.); R35NS116879, UF1NS107710, and R21MH134019 (M.J.K.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113331.
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
REFERENCES
- 1.Hanus C, Ehrensperger MV, and Triller A. (2006). Activity-Dependent Movements of Postsynaptic Scaffolds at Inhibitory Synapses. J. Neurosci. 26, 4586–4595. 10.1523/JNEUROSCI.5123-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison DW, Chervin AS, Gelfand VI, and Craig AM (2000). Postsynaptic Scaffolds of Excitatory and Inhibitory Synapses in Hippocampal Neurons: Maintenance of Core Components Independent of Actin Filaments and Microtubules. J. Neurosci. 20, 4545–4554, 20/12/4545 [pii]. 10.1523/JNEUROSCI.20-12-04545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang AH, Chen H, Li TP, Metzbower SR, MacGillavry HD, and Blanpied TA (2016). A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 536, 210–214. 10.1038/nature19058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinnen BL, Bowen AB, Forte JS, Hiester BG, Crosby KC, Gibson ES, Dell’Acqua ML, and Kennedy MJ (2017). Optogenetic Control of Synaptic Composition and Function. Neuron 93, 646–660.e5. 10.1016/j.neuron.2016.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nair D, Hosy E, Petersen JD, Constals A, Giannone G, Choquet D,and Sibarita J-B (2013). Super-Resolution Imaging Reveals That AMPA Receptors Inside Synapses Are Dynamically Organized in Nanodomains Regulated by PSD95. J. Neurosci. 33, 13204–13224. 10.1523/JNEUROSCI.2381-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hruska M, Henderson N, Le Marchand SJ, Jafri H, and Dalva MB(2018). Synaptic nanomodules underlie the organization and plasticity of spine synapses. Nat. Neurosci. 21, 671–682. 10.1038/s41593-018-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crosby KC, Gookin SE, Garcia JD, Hahm KM, Dell’Acqua ML, and Smith KR (2019). Nanoscale Subsynaptic Domains Underlie the Organization of the Inhibitory Synapse. Cell Rep. 26, 3284–3297.e3. 10.1016/j.celrep.2019.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennacchietti F, Vascon S, Nieus T, Rosillo C, Das S, Tyagarajan SK, Diaspro A, Del Bue A, Petrini EM, Barberis A, and Cella Zanacchi F. (2017). Nanoscale Molecular Reorganization of the Inhibitory Post-synaptic Density Is a Determinant of GABAergic Synaptic Potentiation. J. Neurosci. 37, 1747–1756. 10.1523/JNEUROSCI.0514-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mondin M, Labrousse V, Hosy E, Heine M, Tessier B, Levet F, Poujol C, Blanchet C, Choquet D, and Thoumine O. (2011). Neurexin-Neuroligin Adhesions Capture Surface-Diffusing AMPA Receptors through PSD-95 Scaffolds. J. Neurosci. 31, 13500–13515. 10.1523/JNEUROSCI.6439-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu G, Choi S, and Tsien RW (1999). Variability of neurotransmitter concentration and nonsaturation of postsynaptic AMPA receptors at synapses in hippocampal cultures and slices. Neuron 22, 395–409. 10.1016/S0896-6273(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 11.Lisman JE, Raghavachari S, and Tsien RW (2007). The sequence of events that underlie quantal transmission at central glutamatergic synapses. Nat. Rev. Neurosci. 8, 597–609. 10.1038/nrn2191. [DOI] [PubMed] [Google Scholar]
- 12.Franks KM, Stevens CF, and Sejnowski TJ (2003). Independent sources of quantal variability at single glutamatergic synapses. J. Neurosci. 23, 3186–3195. 10.1523/jneurosci.23-08-03186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raghavachari S, and Lisman JE (2004). Properties of quantal transmission at CA1 synapses. J. Neurophysiol. 92, 2456–2467. 10.1152/jn.00258.2004. [DOI] [PubMed] [Google Scholar]
- 14.Kellermayer B, Ferreira JS, Dupuis J, Levet F, Grillo-Bosch D, Bard L, Linarè s-Loyez J, Bouchet D, Choquet D, Rusakov DA, et al. (2018). Differential Nanoscale Topography and Functional Role of GluN2-NMDA Receptor Subtypes at Glutamatergic Synapses. Neuron 100, 106–119.e7. 10.1016/j.neuron.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Ramsey AM, Tang AH, LeGates TA, Gou XZ, Carbone BE,Thompson SM, Biederer T, and Blanpied TA (2021). Subsynaptic positioning of AMPARs by LRRTM2 controls synaptic strength. Sci. Adv. 7, eabf3126. 10.1126/sciadv.abf3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haas KT, Compans B, Letellier M, Bartol TM, Grillo-Bosch D, Sejnowski TJ, Sainlos M, Choquet D, Thoumine O, and Hosy E. (2018). Pre-post synaptic alignment through neuroligin-1 tunes synaptic transmission efficiency. Elife 7, e31755. 10.7554/eLife.31755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ, and Tucker CL (2014). An optimized optogenetic clustering tool for probing protein interaction and function. Nat. Commun. 5, 4925–4929. 10.1038/ncomms5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmerman SP, Hallett RA, Bourke AM, Bear JE, Kennedy MJ, and Kuhlman B. (2016). Tuning the Binding Affinities and Reversion Kinetics of a Light Inducible Dimer Allows Control of Transmembrane Protein Localization. Biochemistry 55, 5264–5271. 10.1021/acs.biochem.6b00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H,Bear JE, and Kuhlman B. (2015). Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. USA 112, 112–117. 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross GG, Junge JA, Mora RJ, Kwon HB, Olson CA, Takahashi TT, Liman ER, Ellis-Davies GCR, McGee AW, Sabatini BL, et al. (2013). Recombinant Probes for Visualizing Endogenous Synaptic Proteins in Living Neurons. Neuron 78, 971–985. 10.1016/j.neuron.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Essrich C, Lorez M, Benson JA, Fritschy JM, and Lüscher B. (1998). Postsynaptic clustering of major GABAA receptor subtypes requires the γ2 subunit and gephyrin. Nat. Neurosci. 1, 563–571. 10.1038/2798. [DOI] [PubMed] [Google Scholar]
- 22.Crestani F, Lorez M, Baer K, Essrich C, Benke D, Laurent JP, Belzung C, Fritschy JM, Lüscher B, and Mohler H. (1999). Decreased GABA(A)-receptor clustering results in enhanced anxiety and a bias for threat cues. Nat. Neurosci. 2, 833–839. 10.1038/12207. [DOI] [PubMed] [Google Scholar]
- 23.Gore A, Yurina A, Yukevich-Mussomeli A, and Nahmani M. (2022). Synaptic spinules are reliable indicators of excitatory presynaptic bouton size and strength and are ubiquitous components of excitatory synapses in CA1 hippocampus. Front. Synaptic Neurosci. 14, 968404. 10.3389/fnsyn.2022.968404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen RS, and Siekevitz P. (1978). Form of the postsynaptic density. Aserial section study. J. Cell Biol. 78, 36–46. 10.1083/jcb.78.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heilemann M, Van De Linde S, Schüttpelz M, Kasper R, Seefeldt B, Mukherjee A, Tinnefeld P, and Sauer M. (2008). Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem., Int. Ed. Engl. 47, 6172–6176. 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 26.Gookin SE, Taylor MR, Schwartz SL, Kennedy MJ, Dell’Acqua ML, Crosby KC, and Smith KR (2022). Complementary Use of Super-Resolution Imaging Modalities to Study the Nanoscale Architecture of Inhibitory Synapses. Front. Synaptic Neurosci. 14, 14. 10.3389/FNSYN.2022.852227/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimm JB, Muthusamy AK, Liang Y, Brown TA, Lemon WC, Patel R, Lu R, Macklin JJ, Keller PJ, Ji N, and Lavis LD (2017). A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat. Methods 14, 987–994. 10.1038/nmeth.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaeser PS, Deng L, Wang Y, Dulubova I, Liu X, Rizo J, and Südhof TC (2011). RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell 144, 282–295. 10.1016/j.cell.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guzikowski NJ, and Kavalali ET (2022). Nano-organization of spontaneous GABAergic transmission directs its autonomous function in neuronal signaling. Cell Rep. 40, 111172. 10.1016/j.cel-rep.2022.111172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horvath PM, Piazza MK, Monteggia LM, and Kavalali ET (2020). Spontaneous and evoked neurotransmission are partially segregated at inhibitory synapses. Elife 9, e52852. 10.7554/eLife.52852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frerking M, Borges S, and Wilson M. (1995). Variation in GABA mini amplitude is the consequence of variation in transmitter concentration. Neuron 15, 885–895. 10.1016/0896-6273(95)90179-5. [DOI] [PubMed] [Google Scholar]
- 32.Perrais D, and Ropert N. (1999). Effect of zolpidem on miniature IPSCs and occupancy of postsynaptic GABA(A) receptors in central synapses. J. Neurosci. 19, 578–588. 10.1523/jneurosci.19-02-00578.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards FA, Konnerth A, and Sakmann B. (1990). Quantal analysis of inhibitory synaptic transmission in the dentate gyrus of rat hippocampal slices: a patch-clamp study. J. Physiol. 430, 213–249. 10.1113/jphysiol.1990.sp018289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barberis A, Petrini EM, and Cherubini E. (2004). Presynaptic source of quantal size variability at GABAergic synapses in rat hippocampal neurons in culture. Eur. J. Neurosci. 20, 1803–1810. 10.1111/j.1460-9568.2004.03624.x. [DOI] [PubMed] [Google Scholar]
- 35.Petrini EM, Nieus T, Ravasenga T, Succol F, Guazzi S, Benfenati F, and Barberis A. (2011). Influence of GABAAr monoliganded states on GABAergic responses. J. Neurosci. 10.1523/JNEUROSCI.1453-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X, Le Corronc H, Legendre P, Triller A, and Specht CG (2021). Differential regulation of glycinergic and GABAergic nanocolumns at mixed inhibitory synapses. EMBO Rep. 22, e52154. 10.15252/embr.202052154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poulopoulos A, Aramuni G, Meyer G, Soykan T, Hoon M, Papadopoulos T, Zhang M, Paarmann I, Fuchs C, Harvey K, et al. (2009). Neuroligin 2 Drives Postsynaptic Assembly at Perisomatic Inhibitory Synapses through Gephyrin and Collybistin. Neuron 63, 628–642. 10.1016/j.neuron.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 38.Yamasaki T, Hoyos-Ramirez E, Martenson JS, Morimoto-Tomita M,and Tomita S. (2017). GARLH Family Proteins Stabilize GABAA Receptors at Synapses. Neuron 93, 1138–1152.e6. 10.1016/j.neuron.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maynard SA, Rostaing P, Schaefer N, Gemin O, Candat A, Dumoulin A, Villmann C, Triller A, and Specht CG (2021). Identification of a stereotypic molecular arrangement of endogenous glycine receptors at spinal cord synapses. Elife 10, e74441. 10.7554/eLife.74441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santuy A, Rodríguez JR, DeFelipe J, and Merchán-Pérez A. (2018). Study of the size and shape of synapses in the juvenile rat somatosensory cortex with 3D electron microscopy. eNeuro 5, ENEURO.0377, 17.2017. 10.1523/ENEURO.0377-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davenport EC, Pendolino V, Kontou G, McGee TP, Sheehan DF,López-Doménech G, Farrant M, and Kittler JT (2017). An Essential Role for the Tetraspanin LHFPL4 in the Cell-Type-Specific Targeting and Clustering of Synaptic GABAA Receptors. Cell Rep. 21, 70–83. 10.1016/j.celrep.2017.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tretter V, Jacob TC, Mukherjee J, Fritschy JM, Pangalos MN, andMoss SJ (2008). The clustering of GABAA receptor subtypes at inhibitory synapses is facilitated via the direct binding of receptor a2 subunits to gephyrin. J. Neurosci. 28, 1356–1365. 10.1523/JNEUROSCI.5050-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q, Sinnen BL, Boxer EE, Schneider MW, Grybko MJ, Buchta WC, Gibson ES, Wysoczynski CL, Ford CP, Gottschalk A, et al. (2019). A Photoactivatable Botulinum Neurotoxin for Inducible Control of Neurotransmission. Neuron 101, 863–875.e6. 10.1016/j.neuron.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beaudoin GMJ, Lee SH, Singh D, Yuan Y, Ng YG, Reichardt LF, and Arikkath J. (2012). Culturing pyramidal neurons from the early post-natal mouse hippocampus and cortex. Nat. Protoc. 7, 1741–1754. 10.1038/nprot.2012.099. [DOI] [PubMed] [Google Scholar]
- 45.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD,and Tucker CL (2010). Rapid blue light induction of protein interactions in living cells. Nat. Methods 7, 973–975. 10.1038/NMETH.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rial Verde EM, Zayat L, Etchenique R, and Yuste R. (2008). Photorelease of GABA with visible light using an inorganic caging group. Front. Neural Circuits 2, 2. 10.3389/neuro.04.002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Linkert M, Rueden CT, Allan C, Burel JM, Moore W, Patterson A, Loranger B, Moore J, Neves C, MacDonald D, et al. (2010). Metadata matters: Access to image data in the real world. J. Cell Biol. 189, 777–782. 10.1083/jcb.201004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoogendoorn E, Crosby KC, Leyton-Puig D, Breedijk RMP, Jalink K, Gadella TWJ, and Postma M. (2014). The fidelity of stochastic single-molecule super-resolution reconstructions critically depends upon robust background estimation. Sci. Rep. 4, 3854. 10.1038/SREP03854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ovesný M, Křížek P, Borkovec J, Švindrych Z, and Hagen GM (2014). ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Schnitzbauer J, Hu Z, Li X, Cheng Y, Huang Z-L, and Huang B. (2014). Localization events-based sample drift correction for localization microscopy with redundant cross-correlation algorithm. Opt Express 22, 15982–15991. 10.1364/oe.22.015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rizk A, Paul G, Incardona P, Bugarski M, Mansouri M, Niemann A,Ziegler U, Berger P, and Sbalzarini IF (2014). Segmentation and quantification of subcellular structures in fluorescence microscopy images using Squassh. Nat. Protoc. 9, 586–596. 10.1038/nprot.2014.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No standardized datatypes are reported in this paper. All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.