Summary
Recent work has identified dozens of non-coding loci for Alzheimer’s disease (AD) risk, but their mechanisms and AD transcriptional regulatory circuitry are poorly understood. Here, we profile epigenomic and transcriptomic landscapes of 850k nuclei from prefrontal cortexes of 92 individuals with and without AD to build a map of the brain regulome, including epigenomic profiles, transcriptional regulators, co-accessibility modules, and peak-to-gene links in a cell-type-specific manner. We develop methods for multimodal integration and detecting regulatory modules using peak-to-gene linking. We show AD risk loci are enriched in microglial enhancers and for specific TFs including SPI1, ELF2, and RUNX1. We detect 9,628 cell-type-specific ATAC-QTL loci, which we integrate alongside peak-to-gene links to prioritize AD variant regulatory circuits. We report differential accessibility of regulatory modules in late-AD in glia and in early-AD in neurons. Strikingly, late-stage AD brains show global epigenome dysregulation indicative of epigenome erosion and cell identity loss.
Introduction
Alzheimer’s disease (AD) is a highly heritable neurodegenerative disorder that progresses from early-stage mild memory impairment to late-stage dementia and affects tens of millions of individuals worldwide1–5. Despite high AD heritability and decades of research revealing dozens of robust and reproducible genetic loci underlying AD risk, mechanistic basis of these loci and of AD regulatory circuitry remains largely unknown at the transcriptional, epigenomic, and cellular level, thus hindering the search for new therapeutics6,7.
In the last decade, genome-wide association studies (GWAS) have tied dozens of genomic loci to AD risk 8,9, broadly implicating amyloid-beta plaque formation and clearance, inflammation, and diverse microglial functions10,11. The vast majority of these AD risk loci are hypothesized to regulate genes by disrupting the non-coding genome, but complete mechanistic explanations require their: (1) regulatory elements, (2) cell types of action, (3) target genes, (4) causal variants in these loci, and (5) upstream regulators whose binding they disrupt. In addition, measuring variants’ quantitative effects on local chromatin accessibility or expression, e.g. finding quantitative trait loci (QTLs), is a crucial missing piece for elucidating disease-associated variant function. While mapping disease risk loci with QTLs from bulk, healthy tissue, has met with limited success12,13, possibly due to masked cell-type-specific effects, approaches calling QTLs in the blood at the single-cell resolution have successfully illuminated genetic variation linked to blood and immune traits14,15. In AD, previous work has mapped epigenome QTLs in bulk brain and sorted microglia16,17, but to date no studies have been powered to do this in the brain at the single-cell level.
Systematic maps of epigenomic and transcriptional function at single-cell resolution can help overcome some of these challenges, and enable studying how molecular circuits are altered during disease progression across different cell types and biological processes. Both GWAS-targeted and single-cell studies of the aging brain have shed light into the transcriptomic involvement of many brain cell types and roles of AD GWAS risk loci in the disease’s stages18–22. Other studies have dissected the bulk or cell-type-specific epigenomic signatures of the healthy or AD PFC, in part to build epigenomic maps for locus dissection23–25. Evaluating the AD regulome across disease progression as well as across cell types and their gene regulatory networks, would shed light on both disease-associated variation and dysregulation in AD. In addition, despite recent evidence for increased somatic mutational burden, transcriptional dysregulation, and epigenomic erosion in aging and AD26,27, no study has examined cell-type-specific genome-wide epigenomic changes across chromatin states in AD.
Here, we present a single-cell-resolution population-scale map of transcriptomic and epigenomic changes in the aging brain and in AD in the context of genetic and phenotypic differences. We profile transcriptional and epigenomic landscapes of 414k (snRNA-seq) and 437k individual nuclei (snATAC-seq) from prefrontal cortex (PFC) samples of each of 92 human post mortem brains with matched whole genome sequencing (WGS) data, across 48 non-AD, 29 early-stage AD, and 15 late-stage AD individuals. We develop an iterative machine learning framework to integrate multi-modality single-nucleus data and create a method to annotate high-resolution modules of co-accessible sites to build a map of the regulome across 61 cell types in 7 major groups including cell-type-specific profiles, putative regulators, co-accessibility modules, and enhancer-gene linking, which we make available for visualization, download and exploration at http://compbio.mit.edu/ad_epigenome. This resource enables us to map AD GWAS loci at the cell type and regulator levels and to jointly investigate genetic drivers of expression and chromatin accessibility in the brain. We dissect regulome changes by AD stage, reporting compositional changes and differential accessibility at the module level in AD. We find a global shift in the epigenome reflecting epigenomic erosion and loss of cell type identity in late AD, which is accompanied by a disruption of 3D genome structure and compartmentalization and up-regulation of cohesin in neurons along AD progression that we concurrently report (Dileep et al28, Mathys et al.29, in this issue).
Results
Single-nuclei RNA and ATAC profiling of post-mortem prefrontal cortices
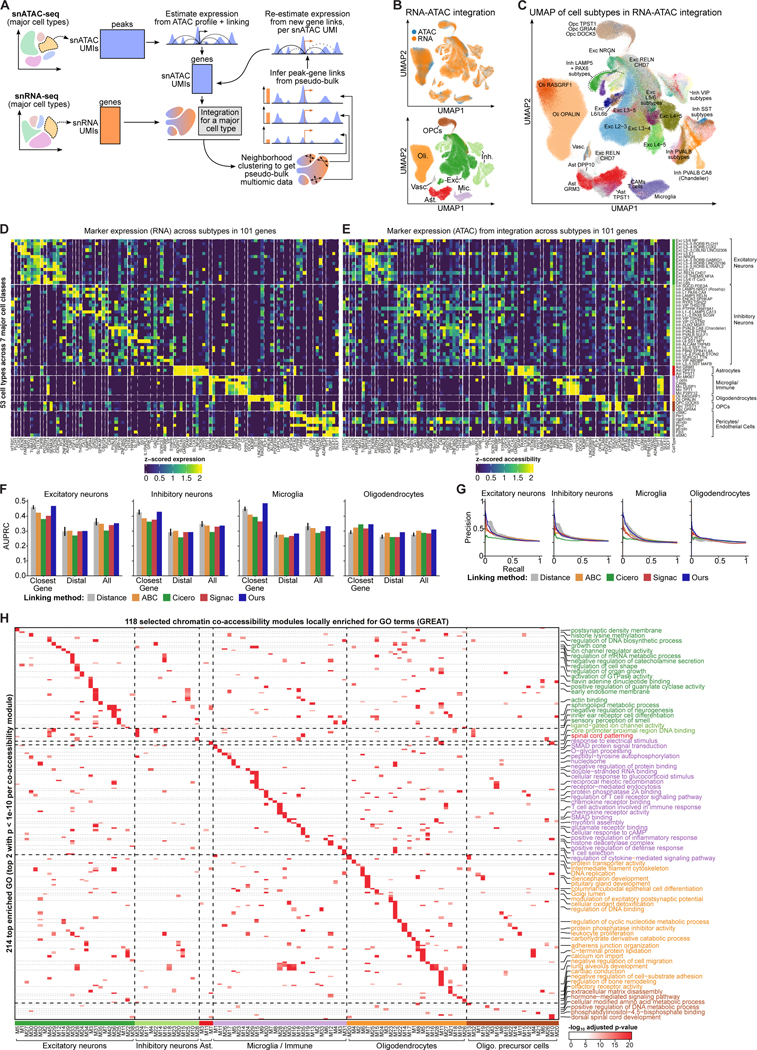
We profiled 92 post-mortem human brain prefrontal cortex (PFC) samples from the Religious Order Study (ROS) and the Rush Memory and Aging Project (MAP) including AD and age-matched controls30. We grouped individuals into non-AD (i.e. control; n=48), early-stage AD (n=29), and late-stage AD (n=15) using 7 clinico-pathological measurements, as previously defined19 (Figure 1A; Table S1). We generated single-nucleus transcriptomic (snRNA) and single-nucleus chromatin accessibility based on the Assay for Transposase-Accessible Chromatin (snATAC) profiling using the 10X Genomics platform, resulting in 414,964 high-quality transcriptomes and 171,000 high-quality epigenomes (Figure 1A) after stringent quality control (Methods).
Figure 1. Study design and epigenomic landscape across human brain cell types.
A. Overview of study design, sample collection, single-cell profiling and analyses.
B. UMAP for snRNA-seq across major brain cell types.
C. UMAP for snATAC-seq across major brain cell types based on the 500 bp tile matrix.
D. ATAC-inferred gene expression across the marker gene sets from major cell types curated by PsychENCODE (mean, column-scaled).
E. Number of peaks per cell type (union: peaks in 1+ cell types). TSS proximal defined as regions within 2 kb of TSS , distal as intergenic regions >2kb from TSSes.
F. GREAT enrichment annotation of cell-type-specific peaks.
G. ChromVar motif enrichment of the 44 candidate TF regulators identified across cell types. Left panel shows Pearson correlation between chromVar and ATAC-inferred TF expression.
H. Inferred gene expression of the regulators (ATAC gene-score using ArchR).
I. Regulator expression (snRNA-seq) across cell types. Right panel shows Pearson correlation between expression and ATAC-inferred TF expression.
J. Motif enrichment (left) and inferred gene expression (right) of candidate TF regulators (NeuroD6 in excitatory neurons, SOX10 in oligodendrocytes, and SPI1 in microglia).
We annotated the 414k snRNA-seq cells from matched individuals with seven major cell type groups and identified high-resolution cellular subtypes, including 14 excitatory, 25 inhibitory subtypes, and 22 glial and vascular subtypes (Figure 1B, S1A; Mathys et al.28, in this issue). For snATAC-seq cells, we performed stringent QC based on transcription start site (TSS) enrichment and number of unique fragments (Figure S1B, Methods). Chromatin accessibility information alone unambiguously distinguished the 7 major cell types in the brain and resulted in highly-compact clusters for each of the cell types (Figure 1C–D, Figure S1C–D). These included 22k excitatory neurons, 21k inhibitory neurons, 101k oligodendrocytes, 10k astrocytes, 7.2k oligodendrocyte progenitor cells (OPCs), 8.6k microglia, and 1.2k vascular cells (Figure 1C), and showed negligible technical or batch effects (Figure S1B).
We identified 367,242 unique ATAC peaks overall, with 80k to 200k in each cell type (except for vascular cells, likely due to their limited cell number) (Figure 1E; Figure S1E). We found 130,193 peaks that showed cell type specificity (Figure S1F), defined based on the fold change of peak intensity over other cell types (Methods). Cell-type-specific peaks were consistent with cell type gene expression specificity (Figure S1G), and were enriched for cell-type-specific functional pathways, including synaptic signaling for neurons, axon ensheathment for oligodendrocytes, extracellular matrix organization and vasculature development in pericytes and endothelial cells, and chondroitin sulfate biosynthesis in OPCs (Figure 1F).
To predict candidate upstream regulators that may be shaping the chromatin accessibility landscape of brain cells, and contributing to cell type identity and cell fate specification, we used chromVar to calculate the enrichment of transcription factors (TF) as measured by their motif accessibility (Methods; Table S2; Figure 1G). To better identify bona-fide regulators, we also measured regulator expression and its correlation with motif enrichment across cells (Figure 1H). We discovered 44 high-confidence candidate regulators that show strong correlations between TF enrichment and ATAC-inferred expression, and between ATAC-inferred expression and RNA-measured expression (Pearson>0.5, FDR<0.01) (Methods; Figure 1G–I), including NEUROD2/D6, MEF2C, and EGR family TFs in excitatory neurons, STAT4 for inhibitory neurons, MEF2A sharing between the two neuron types and microglia, EMX2 and NFIA in astrocytes and OPCs, SOX2/10 and FOXN2 in oligodendrocytes, and SPI1/B, and IRF5/8 in microglia. NEUROD6, SOX10 and SPI1 are displayed as examples of candidate regulators functioning in excitatory, oligodendrocyte, and microglia cells, respectively (Figure 1J). The cell-type-specificity of these regulators was further supported by the presence of TF footprints in the appropriate cell types (Figure S1H).
A cell-type-specific RNA-ATAC integration framework
In order to jointly resolve intra-cell heterogeneity within each major cell type and build a high-quality map of regulatory circuitry, we developed a cell-type-specific snRNA-snATAC integration framework that draws on distal regulatory element linking to enable high-resolution snATAC annotation. Our method iteratively refines a cell-cell graph across snRNA and snATAC, alternating between improving peak-to-gene links and using links to re-estimate gene expression from ATAC profiles in each snATAC-seq cell (Figure 2A). We integrated each major cell type separately in order to model intra-cell type heterogeneity instead of inter-cell type heterogeneity. We merged the cell-type-specific integrations to build a joint cell-cell graph between modalities and transferred high resolution subtype annotations from the snRNA cells to the snATAC cells (Figure 2B–C).
Figure 2. snATAC and snRNA integration enables peak-to-gene link calling.
A. Schematic for the cell-type-specific integration framework.
B. UMAP of joint ATAC and RNA cell embedding (top) with major cell type assignments (bottom).
C. UMAP of joint embedding colored with the high-resolution sub-cell-type annotation.
D. Gene expression of marker genes across sub-cell-types in snRNA.
E. Estimated gene-score of marker genes across sub-cell-types in snATAC.
F. Recovery (AUPRC) of cell-type-specific PLAC-seq by inferred links (ABC: activity-by-contact method).
G. Precision-recall curves for cell-type-specific PLAC-seq.
H. GREAT enrichment of cell-type-specific ATAC modules.
A joint UMAP from this graph shows how our ATAC-inferred gene-score matches the RNA gene expression distribution (Figure 2B). The integrated and transferred RNA and ATAC subtypes show well-aligned marker gene expression and gene-score (Figure 2D–E), with much improved alignment between RNA and ATAC profiles of subtype marker genes in our integration over scGLUE and Seurat, two state-of-the-art single-cell modality integration methods based on deep learning and canonical correlation analysis, respectively (Figure S2A)31,32. We profiled an additional 19 medial frontal cortex (MFC) samples using single-nucleus multiome sequencing (joint ATAC and RNA) for a subset of individuals in our cohort, which we used as a ground truth dataset in which to evaluate our integration method. Our method independently assigned both modalities of a multiome cell to the same high-resolution cell type 85.4% of the time, whereas Seurat had a lower matching performance (79.5% of the time).
We computed a final set of peak-to-gene links for each cell type using the snATAC-snRNA integration, first building a set of z-scored peak-gene co-accessibility and accessibility-expression correlations and then training regression models on these features to classify high-confidence peak-to-gene links (Figure S2B, Table S3). Resulting peak-to-gene links have on average 21 linked peaks per gene (Figure S2C) and decay in frequency as genomic distance increases (Figure S2D). These cell-type-specific peak-to-gene links showed superior recovery of Plac-seq 11 interactions (Proximity ligation-assisted ChIP-seq) in four cell types compared to other existing approaches, especially in the cases of distal peak-gene interactions where a peak is not linked to the nearest gene (Figure 2F–G, Figure S2E–F).
Epigenomic landscapes and regulatory circuitry
Notably, we did not observe large effect size interactions between any single pair of peaks and genes, suggesting that expression is coordinated by multiple distal accessible sites. We developed a framework to group peaks into co-accessibility modules for each major cell type (Table S4, Methods). Briefly, our method calculates a joint decomposition of snATAC profiles and estimated gene scores per-cell to calculate clusters of peaks per cell type (modules). We identified 219 co-accessibility modules across cell types, each with an average of 954 peaks which split into distal (17% on average), TSS-proximal (21%), exonic (17%), and intronic peaks (45%). We found that 66.2% of modules had over 50% of their peaks within a single chromosome, possibly capturing local cis-regulatory structure, but the majority of modules spanned multiple chromosomes (on average 15 per module), which may reflect trans-regulation. Additionally, on average each module had 524 unique nearest genes to its peaks, and each gene was linked to 8 distinct modules, indicating that these modules cooperate to influence gene expression.
Many peak modules were significantly enriched for upstream TF regulators (Figure S2G, Table S4), connecting upstream regulators with downstream targets and suggesting specific sub-programs in each cell type. Astrocyte modules showed highly stratified TF enrichments, including for SOX9/13 (M3), ETV6/7 (M6), RORA/B/C (M7), and STAT1/STAT3 for M2 (Figure S2H)33. Other notable enrichments included ZBED1 (OPC M1), which is linked to cellular proliferation34, NFKB1/2 (microglia M7), and NFY family TFs (oligodendrocyte M10), which regulate metabolic pathways including lipid and cholesterol biosynthesis35. Multiple modules were enriched for AP-1 family TFs, and a large set of modules showed broad enrichments for a number of enhancer-associated factors, including KLF and ETV family motifs, suggesting that modules stratify by promoter and enhancer drivers.
These peak modules act on distinct downstream pathways even within the same cell types (Figure 2H). Across 219 modules we found 2,364 unique highly enriched Gene Ontology (GO) terms (via GREAT)36, with an average of 54 GO terms enriched per module (Table S4), suggesting that these modules capture functional units, not just local chromatin compartments or broad cell identity programs. Cell-type-specific module enrichments included synapse and ion channel-related terms in neurons, immune activation and inflammatory terms in microglia, carbohydrate metabolism and myelination in oligodendrocytes, and ECM remodeling in OPCs (Figure 2H). These highly specific TF and functional enrichments suggest that peak co-accessibility modules often represent specific regulatory programs describing the AD epigenomic landscape and that they also would form a suitable basis set for ATAC profile aggregation in differential peak analysis.
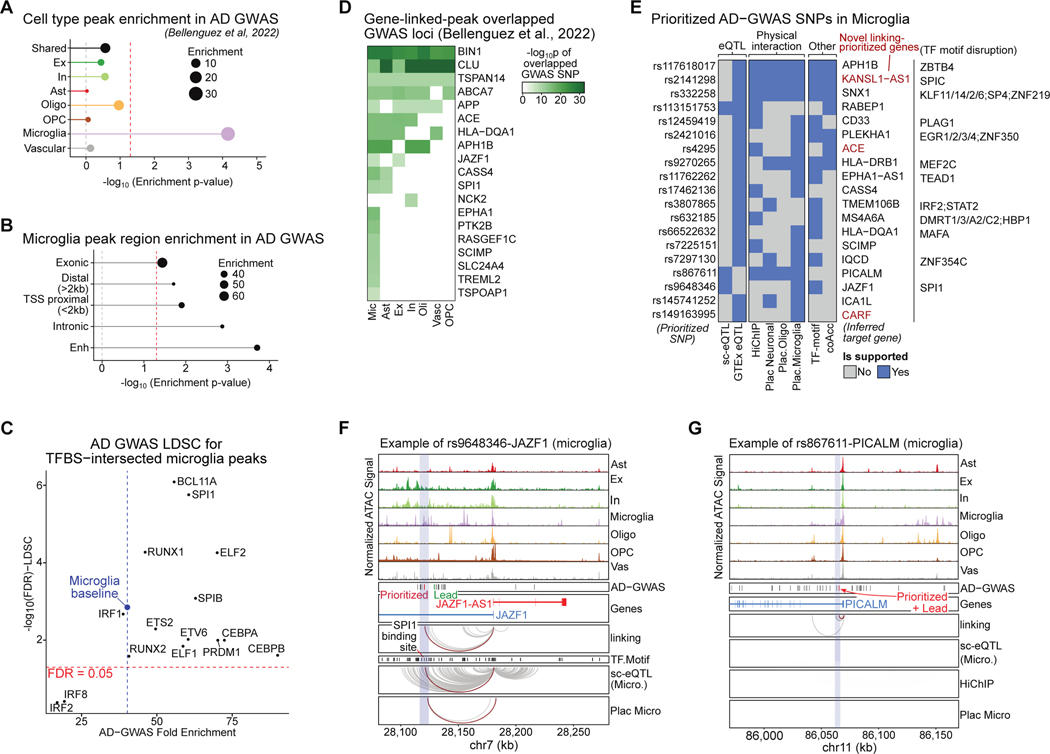
Cell-type-specific epigenomic enrichment of AD-GWAS
We next performed LD score regression (LDSC)37 to estimate the heritability enrichment of risk loci identified in the most recent AD GWAS study8 in each of the major cell types’ ATAC peak sets. As expected, we found that microglia peaks are strongly and specifically enriched for AD heritability, and peaks from other cell types and constitutive peaks (in 5+ cell types) show no enrichment (Figure 3A)10. Analysis of peaks independently identified in AD and control samples showed a consistent microglia-specific enrichment signal that was modestly but not significantly higher in AD (Figure S3A–B). Further partitioning microglia peaks based on their genomic annotations showed that enhancers are most enriched for heritability (Figure 3B). We extended this GWAS enrichment analysis into 24 other traits, and observed that other brain-related traits including schizophrenia, bipolar disorder, neuroticism were enriched for neuronal epigenomes and that the microglial epigenome was more relevant to immune-related disorders (Figure S3C; Table S5). LDSC analysis at the sub-cell-type level further revealed the cell types of action at a higher resolution (Figure S3D; Table S5).
Figure 3. AD-GWAS enrichment and variant prioritization in microglia.
A. Heritability enrichment of peaks in each cell type (using stratified-LDSC; shared peaks: 5+ cell types).
B. Heritability enrichment of microglia peaks partitioned by genomic location.
C. Scatterplot of heritability enrichment of TFBS-intersected microglia peaks (blue dotted line is overall microglia enrichment).
D. AD-GWAS loci located in gene-linked peaks for each cell type.
E. Multiple lines of evidence supporting prioritized SNP-gene pairs in microglia. TFBS containing the prioritized variants are shown on the right.
F. Example of prioritized AD-GWAS variant rs9648346 predicted to target JAZF1 in microglia. rs9648346 is in microglia-specific peak, interferes with SPI1 binding sites, and the link is supported by microglia eQTL and microglia PLAC-seq.
G. Example of prioritized variant rs867611 that is predicted to target PICALM in microglia. rs867611 is a AD-GWAS lead variant at the locus and is prioritized by the multi-evidence framework.
To identify upstream regulators of AD risk in microglia, we intersected microglia peaks with the transcription factor binding sites (TFBS) of the candidate TF regulators in microglia (Figure 1G–I), estimated heritability enrichment of the TFBS-containing peaks (Table S5), and discovered that 6 of 12 TFs showed higher fold enrichment of AD-GWAS signal compared to microglia baseline (with weaker enrichment p-values, likely due to the smaller peak sets) (Figure 3C). The binding sites of two important microglial transcriptional regulators, RUNX1, a TF regulating microglial maturation and proliferation, and SPI1, an AD risk locus itself, showed higher heritability enrichment both by fold enrichment and significance level, suggesting active regulatory roles for these TFs in AD risk loci38. By contrast, the binding sites of interferon-regulatory factor family TFs IRF1/2/5/8, which had clear microglia epigenome-specific enrichments and TF footprinting, were relatively depleted for AD-GWAS loci (Figure S3E; Figure 3C). This is consistent with the previous observation that IRF family TFs regulate microglia function through an indirect effect downstream of BIN139. Microglia TFBS-containing peaks display preferences in their genomic distribution, which may correspond to different modes of regulation (Figure S3F). For instance, SP4/KLF13-intersected peaks are preferentially located in TSS proximal regions, whereas SPI1 and RUNX1-intersected peaks are biased towards enhancer regions, suggesting a role for AD risk loci in modifying enhancer function at binding sites for enhancer-specific TFs.
Cell-type-specific prioritization of AD-GWAS variants
We next sought to prioritize AD-GWAS variants based on our epigenomic annotations and peak-to-gene links. We first overlapped genome-wide significant (p<5×10−8) AD GWAS variants with peaks that had linked genes, and found cell-type-specific overlaps for 19 loci (Figure 3D). Almost all loci overlapped microglial peaks (18 loci), consistent with the heritability enrichments, and astrocytes and excitatory neurons overlapped half of the loci (9 and 8 loci, respectively). Three loci were solely found in microglia peaks, including peaks near microglial TF SPI1 and myeloid receptors TREM2/TREML2, and 8 loci appeared in multiple cell types, including BIN1, CLU, and APP (Figure 3D).
We then prioritized functional AD-GWAS variants for each overlapped locus based on peak-to-gene links supported by multiple lines of evidence, including genetic links (expression quantitative trait loci; eQTLs) and physical interactions from HiChIP and PLAC-seq11,24 (Figure S3G; Table S5). We discovered eight genes that were specifically prioritized in microglia, including PICALM, SCIMP, JAZF1, HLA−DRB1, and several loci that were prioritized in multiple cell types (Figure S3H). While 16 of the 19 genes prioritized in microglia were previously finemapped8,11,16,40, we prioritized three genes that are potentially downstream targets of AD-GWAS loci, including KANSL1-AS1, ACE and CARF. Among the 19 AD GWAS variants prioritized in microglia, 12 of them were located within a TF motif (Figure 3E). These TFs included putatively microglial and immune regulators such as SPI1/C, SP4, and IRF2, which suggested that the prioritized variants may disrupt microglial-specific regulatory circuitry. For example, our analysis prioritized the rs9648346 variant in the JAZF1 locus (lead variant rs1160871; AD-GWAS p=9.8e-9), which is located in a microglia-specific peak, disrupts an SPI1 binding site, targets the JAZF1 gene, and is supported by microglia-specific eQTLs and PLAC-seq (Figure 3F). JAZF1 is a glucose production-promoting transcriptional repressor that has been tied to PI3K/Akt, which in turn are implicated in tau phosphorylation in AD through GSK3B inactivation41,42. Similarly, the lead variant for the PICALM locus, rs867611, is prioritized to regulate the PICALM gene in microglia based on both single-cell eQTL and interaction evidence (Figure 3G). Notably, in both the JAZF1 and PICALM loci, the genes’ promoters are constitutively open across all major cell types but distal peak accessibility and regulatory circuitry are cell-type-specific for microglia.
Single-cell ATAC-QTLs colocalize with AD-GWAS
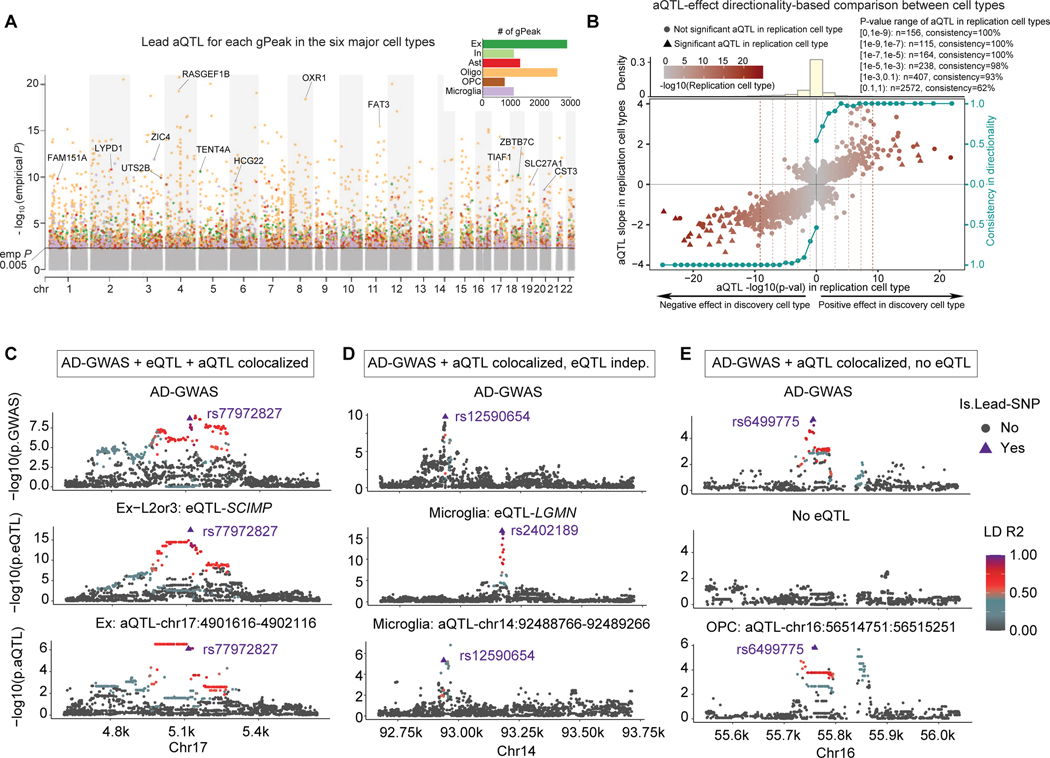
To identify cell-type-specific genetic drivers of chromatin accessibility, we performed ATAC-QTL (aQTL) calling at the major cell type level. Before calling aQTL, we first evaluated potential allelic bias from read mapping and found no difference in peak activity between minor and major alleles (Figure S4A). We identified 9,628 genetically-associated peaks (gPeaks), across 2,895 in excitatory neurons, 1,096 in inhibitory neurons, 1,285 in astrocytes, 2,561 in oligodendrocytes, 754 in OPCs, and 1,064 in microglia (Figure 4A; Table S5). While the microglia epigenome is strongly enriched for AD-GWAS signal, including the AD status of individuals as a covariate when calling aQTLs had a negligible effect on microglia aQTL identification (Figure S4B). The aQTLs tended to be located near their associated gPeaks, at a median of distance of 23kb (aQTLs were called within 100kb of peaks), with 18.2% of within 10kb and 7.0% within 1kb of their gPeak (Figure S4C). To investigate the sharing and specificity of aQTLs between cell types, we studied consistency of aQTLs directionality across cell types, mitigating false cell-type-specificity calls caused by p-value threshold selection (Methods)43. For the aQTLs that are significant in a discovery cell type but do not pass the significance threshold in the replication cell type, aQTL effect directions are over 93% consistent once the nominal p-value in the replication cell type is below p=0.1, and 100% consistent when the nominal p-value reaches 1×10−5 (Figure 4B, Figure S4D). We estimated the aQTL sharing for each pair of major cell types using this method and found that aQTL sharing is consistent with the cell type similarity (Figure S4E, Methods).
Figure 4. ATAC-QTL analysis and colocalization with AD-GWAS.
A. ATAC-QTL (aQTL) Manhattan plot (lead SNP shown), colored by cell types. Nearest genes of the top aQTL loci from each cell type are indicated. Barplot inset shows the number of genetically-associated peaks (gPeak) discovered in each cell type.
B. Directionality consistency (shown by the right axis) analysis of aQTL effect size (left y-axis) between cell types. For the significant aQTLs in the discovery cell type, the consistency increases as the p-value significance (x-axis) of the replication cell type increases for aQTLs with both positive-effect (right half-plane) and negative-effect (left half-plane). The top panel represents the distribution of aQTL loci within each p-value bin. Separated plots that show the pairwise cell-type comparison are shown in Figure S4C.
C. Example of consistent genetic colocalization across AD-GWAS, cell-type-specific eQTL, and aQTL for the SCIMP locus in excitatory neurons (lead variant shown as triangle).
D. Example of aQTL-AD-GWAS colocalization near the LGMN locus in microglia, with a non-colocalized eQTL in the locus.
E. Example of aQTL-AD-GWAS colocalized locus without any eQTL signal observed in the locus.
We next searched for AD-GWAS risk variants with aQTL effects in order to find candidate regulatory mechanisms for AD-GWAS loci. We found 631 variants that showed shared genetic effects at a sub-threshold level (5×10−8<p-value<1×10−5) in microglia as both aQTLs and AD risk loci (Figure S4F). AD GWAS and aQTL loci were highly orthogonal, with the vast majority (>99%) being either aQTL or AD-GWAS only (considering both genome-wide significant and sub-threshold loci), even for microglia aQTLs (Figure S4F). Heritability enrichment analysis confirmed that AD-GWAS risk loci were not enriched for aQTL gPeaks in any cell type (Figure S4G). To better understand the regulatory effects of shared loci, we jointly co-localized aQTLs with AD-GWAS loci and overlaid these loci with cell-type-matched single-cell eQTLs (Methods). We discovered a total of 69 aQTL-GWAS colocalization events (Figure S4H, Table S5). As an example, rs77972827, near the SCIMP locus, is a lead SNP for the AD-GWAS, a sub-threshold aQTL for excitatory neurons, and an eQTL for SCIMP in L2–3 excitatory neurons (Figure 4C). However, outside of 20% (n=14) of aQTL-GWAS colocalized loci with a concordant eQTL effect, most either show a discordant eQTL effect (70%, 48 loci, Figure 4D) or are missing an eQTL (10% n=7, Figure 4E).
AD-relevant transcriptomic and epigenomic changes
To identify the genes and regulatory regions that are potentially involved in AD etiology, we performed AD-differential gene and ATAC peak calling in each major cell type, and found that gene expression and ATAC peak changes were broadly consistent in each cell type (Figure S5A; Table S6). Peaks linked to differential genes in microglia were enriched for AD risk loci, driven by AD up-regulated microglial genes, which may play an important role in AD heritability (Figure S5B). To map altered regulatory programs in AD, we also asked which co-accessibility modules had differential ATAC accessibility across AD stages (Figure S5C; Table S6). We found 15 differential modules between non-AD and early-AD, dominated by excitatory neuron changes, including increased accessibility of modules related to RNA metabolism, apoptosis-linked mitochondrial function, and phospholipase D activity, which has been implicated in AD lipid regulation44. By contrast, 74 differential modules between early-AD and late-AD showed mostly glial differences, with almost two thirds (45) in OPCs and oligodendrocytes, including accessibility changes for modules related to protein processing and in metabolism associated modules related to protein lipidation and glycosylation. Microglia showed increased accessibility in regulatory modules tied to ECM remodeling and interleukin-11 signaling (Figure S5C, Figure 5A). Module-level differential analysis also mitigated power issues driven by ATAC sparsity, as a more typical single-peak approach did not yield any individually-significantly differential peaks after multi-testing correction, even when only testing peaks linked to differentially expressed genes.
Figure 5. AD-differential cell composition and ATAC changes.
A. GREAT functional enrichments of AD-differentially accessible (*p<0.05, **p<0.01, ***p<0.001).
B. Major cell type compositional changes in snRNA-seq comparing non-AD, early-AD and late-AD participants (Anova using propeller). Right panel is the total number of cells.
C. Major cell type compositional changes in snATAC-seq.
D. Sub-cell-type compositional changes in snATAC-seq between non-AD, early-AD and late-AD participants (fractions within each major cell type).
We next asked if cell type composition is affected by AD progression, based on both our epigenomic and transcriptomic single-nucleus datasets. Despite AD neuronal loss reported in the hippocampus and entorhinal cortex45, we found that our prefrontal cortex snRNA-seq samples did not exhibit significant changes in the fraction of neurons (Figure 5B). By contrast, our snATAC-seq data showed a substantial decrease of neurons, and especially excitatory neurons, perhaps indicating an epigenome-specific loss of neuronal identity (Figure 5C). Among glial cells, astrocytes showed a significant compositional decrease during AD progression in the snRNA-seq but not the snATAC-seq (Figure 5B–C). We also asked whether specific subtypes of neurons and glia showed compositional changes during AD progression (Figure 5D, Figure S5D). High-resolution subtypes within each major cell type showed consistent directionality of compositional change during AD progression between ATAC and RNA cells, including a significant decrease in an inhibitory subtype (Inh ENOX2 SPHKAP) (Figure 5D, Figure S5D).
Cell identity loss in late-stage AD
The discrepancy in cell composition changes during AD progression between ATAC and RNA cells prompted us to look into both snATAC-specific quality control filtering steps and cell identity loss in the AD samples. We first observed that the late-stage AD samples showed a significant decrease in TSS-enrichment compared to the non-AD and early-stage AD individuals (Figure S6A), which was not accounted for by either samples’ post-mortem interval (PMI) or individuals’ age of death (Figure S6B). Furthermore, the number of fragments per cell, another commonly-used quality metric, showed no noticeable difference between the AD groups (Figure S6A), suggesting that this TSS enrichment depletion was linked to AD pathology and not sample quality.
Given the observation that the TSS enrichment difference is related to AD pathology, we reclaimed the snATAC UMIs that were initially removed by requiring TSS enrichment>6 in order to properly study cell changes in late-stage AD (Methods). We confirmed that the late-stage AD samples were significantly enriched for the reclaimed cells (Figure S6C). We then re-performed dimensionality reduction and cell clustering on the resulting 437k nuclei, which represents a 2.5-fold increase in the number of kept snATAC UMIs. Interestingly, in addition to the seven major cell types that were clearly separated, we further identified populations that showed weaker cell identity scores that we broadly termed “de-identified” cells. These included de-identified cells originating from oligodendrocytes (Oligo-deid; n=68k), excitatory neurons (Ex-deid; n=58k), inhibitory neurons (In-deid; n=22k), astrocytes (Ast-deid; n=3.4k), OPCs (OPC-deid; n=4.3k), and microglia (Mic-deid; n=5.1k), as well as a cell group that did not show any cell type signature (de-identified; n=34k) (Figure 6A).
Figure 6. Cell identity loss and epigenome erosion in late-stage AD.
A. UMAP for snATAC-seq (TSS-enrichment>1), colored by major cell types.
B. Major and de-identified cell type compositional changes in snATAC-seq comparing late-stage AD to control and early-stage AD (Anova using propeller, inset for magnification).
C. Scatterplot of the fraction of de-identified oligodendrocytes in this study versus the fraction of undetermined/dying snRNA cells from Kousi et al.46 for the 9 individuals shared between the two studies. Boxplots show cell-fraction comparisons between AD groups within each study (Wilcoxon).
D. Log fold change of the fraction of reads in chromatin states (ChromHMM) between de-identified cells vs. the corresponding normal cells for four major cell types.
E. Example pseudo-bulk ATAC signal for normal and de-identified cells in cell-type-specific peaks (blue box) and constitutive peaks (yellow). Right panels show the signal distribution in intergenic regions.
F. UMAP of cell-level erosion scores, quantified based on the distribution of reads in ChromHMM states (higher score represents increased erosion).
G. Erosion score comparison between different AD groups in each cell type (Wilcoxon).
H. Average oligodendrocyte ATAC profiles at compartment A/B boundaries (left/right) for non-AD, early-AD and late-AD participants. Barplots show difference of means within compartments after/before A/B boundaries (insets) and the difference between the aggregate signals (bottom panels).
I. Difference between boundary A and B for each AD group in each cell type.
J. Joint immunostaining of Lamin-B1 and NeuN on a low-erosion MFC sample. The inset shows a single z-stack with LaminB1/NeuN/Hoechst together (top left), NeuN (bottom left), Lamin B1 (top right) and Hoechst (bottom right) in the bottom panel.
K. Examples of low and high erosion samples at two magnifications (left and center, Lamin B1 and Hoechst). Intensity of Lamin B1 expression was measured on masks created around NeuN-positive cell surfaces. Quantification of LaminB1 intensity (right) across 5 low (n= 91 to 164 NeuN-positive cell surfaces) and 5 high erosion samples (n= 85 to 173) (t-test).
L. Differential TF motif enrichments between de-identified cells versus normal cells for excitatory neurons (green), inhibitory neurons (light green) and oligodendrocytes (orange) based on chromVar (from TF regulators in Figure 1G).
We found that late-AD, rather than early-AD samples, were significantly enriched for the de-identified cells and depleted for the corresponding normal cells across multiple major cell types (Figure 6B). This was consistent with the decreased TSS enrichment seen in late-AD samples and suggested that these de-identified cells were driven by disease progression, rather than technical artifacts. We have previously reported transcriptomic cell identity loss during AD progression in well-based single-cell RNA-seq profiling, where QC cutoffs for read counts do not affect the recovery of individual nuclei46. For the 9 donors shared by our current and previous studies, (2 non-AD, 4 early-AD and 3 late-AD, Table S7), we found a very strong correspondence (R2=0.87, p=2.4e-4) in the fraction of de-identified cells, despite the different modalities and independent studies (Figure 6C). Moreover, for the samples that were not shared between the two studies, the late-AD samples consistently showed a significantly higher proportion of de-identified cells in each study separately (side panels, Figure 6C), indicating that the relative increase of de-identified cells in late-AD is robustly observed across modalities.
Global epigenomic alteration during cell identity loss
We further studied the epigenomic landscapes of de-identified cells during AD progression to understand why their epigenomes cluster separately from those of normal cells. Cell identity loss in bulk tissue has been proposed to be driven by epigenomic erosion, a progressive loss of clear epigenomic delineations between active, inactive, and repressed regions47–49. However, since erosion has not been studied or reported at individual-cell resolution, it is unclear whether previously-observed erosion signatures might in part result from changes in cell composition. We thus sought scATAC-based evidence of epigenomic erosion at single-cell resolution, and evaluated whether these helped explain epigenomic changes of de-identified cells in AD patients.
Indeed, we found that de-identified oligodendrocyte, excitatory, and inhibitory neuron cells were less open in active epigenomic regions as defined by ChromHMM states in prefrontal cortex50,51, including enhancers and promoters, and more open in repressive regions, including heterochromatic, quiescent, and zinc fingers gene and repeat-associated states (ZNF/Rpts) (Figure 6D), consistent with signatures of epigenome erosion previously revealed at the bulk level. To gain a better intuition of changes in accessibility during epigenome erosion, we evaluated the broader epigenomic context surrounding the GAD2 locus, a known marker gene for inhibitory neurons (Figure 6E). Although the de-identified cells retain cell-type-specific ATAC signals at the TSSes in the locus, they have a dramatically increased non-peak intergenic background signal, suggesting that these cells have a less tightly controlled chromatin accessibility landscape.
To quantify erosion at the single-cell level, we defined and calculated an integrated “erosion score” based on the relative distribution of reads in repressive versus active chromatin states genome-wide for each cell (Figure 6F, Methods). Despite the fact that these scores were calculated independently from cluster and marker-driven de-identified cell calls, cell-level erosion scores were highly predictive for de-identified status (p<2.2e-16, R=0.56) (Figure 6A), and were unrelated to other quality metrics, such as the number of fragments per cell (R=0.1, Figure S6D–E). We compared the stage-specific distributions of erosion scores and confirmed that late-AD samples had significantly higher epigenomic erosion than non-AD and early-AD samples while non-AD and early-AD samples had similar distributions of erosion scores (Figure 6G, Figure S6F), consistent with late-stage cell identity loss (Figure 6B). As additional evidence for the erosion of active chromatin states in AD, we compared AD up-regulated and down-regulated regions from a published AD ChIP-seq study profiling H3K27ac, a marker of active epigenomic regions52 (Figure S6G). Indeed, we found that AD down-regulated H3K27ac regions were preferentially located in active enhancer and promoter regions, whereas AD up-regulated regions were enriched for repressive chromatin states, conclusion that our AD-associated changes observed based on chromatin accessibility are also reflected in histone modifications (Figure 6D).
Since one of the hallmarks of epigenome erosion is decreased 3D genome compartmentalization49, we next examined the distribution of the ATAC signals surrounding active (A) and repressive (B) compartments (Figure 6H–I). In oligodendrocytes, as an example, late-AD samples showed a more diffuse increase and decrease of chromatin accessibility at A and B compartment boundaries, respectively, compared to non-AD and early-AD samples (Figure 6H). Moreover, late-AD samples showed dramatically reduced differences between ATAC signal at compartment A versus signal at compartment B (bottom panel of Figure 6H). We quantified the difference in signal between compartments across all of the major cell types, and found that late-AD samples consistently exhibited a reduced change in accessibility between compartments A and B (Figure 6I), indicating weaker compartmentalization and reduced 3D genome organization in AD, which we report concurrently (Dileep et. al28).
We generated single-nucleus multiome profiles (mentioned above for integration benchmarking) using 12 and 7 MFC samples from individuals with low and high epigenomic erosion in the prefrontal cortex, respectively, in order to validate and further explore the epigenomic erosion observed in the snATAC-seq of late-AD individuals (Table S1). We repeated the analysis of the original cohort in the snATAC data modality of the multiome for cells with TSS enrichment >1 (147,754 cells in total). Just as in the original snATAC-seq dataset, the multiome dataset contained multiple clusters of deidentified cells with eroded cellular identity (Figure S6H), which were highly enriched in the high erosion samples (Figure S6I–J). The eroded cells from MFC samples recapitulated the chromatin state changes observed in the PFC samples (Figure S6K), and matched the erosion score distributions from the PFC data (Figure S6L).
We then performed joint immunostaining of Lamin-B1 and NeuN (as a neuronal marker) on MFC samples from 6 and 6 individuals with and without erosion signatures in their PFC epigenome (Figure 6J). We found that Lamin-B1 signal in neurons was significantly lower in the high erosion samples than in the samples from individuals without epigenome erosion (Figure 6K). This is consistent with the reduction of nuclear lamina components observed in the epigenome of aging mice53 and is an orthogonal validation of the epigenomic erosion and disruption of the 3D genome in late-stage AD, showing a striking difference in the nuclear lamina of AD and control individuals of similar ages.
Finally, we asked which regions were differentially accessible during erosion, both between normal and de-identified cells and in cells binned by erosion score. We found that up-regulated regions in eroded cells were enriched for developmental processes and cell-cell adhesion pathways, whereas down-regulated regions were enriched for DNA damage and repair processes (Figure S6M; Table S7). These epigenomic and transcriptomic results parallel the changes observed during the de-differentiation process of neuronal cells in AD etiology54,55. We also calculated which genes were differentially expressed with increasing erosion in the transcriptomes of deidentified multiome cells. Enrichments for erosion-associated DEGs (434 up, 387 down, Figure S6N; Table S7) paralleled those of the epigenomic analysis, with up-regulated genes enriched for cell-cell adhesion and GTPase signaling and down-regulated genes enriched for neuronal components and synapse-associated terms (Figure S6O). Down-regulated enrichments were driven by important neuronal-function related genes such as SYNPR, TENM1, and NRXN3. Top up-regulated genes in eroded cells included MAML2, a NOTCH-related transcriptional coactivator, and neuronal development-implicated TF ZFHX3, suggesting a reset of the transcriptional landscape for neurons (Figure S6N)56,57. Erosion DEGs significantly overlapped AD DEGs (Mathys et al.29, in this issue) and showed the strongest overlaps with up-regulated DEGs for global cognition and tangle density (Figure S6P). In addition, we asked which TF binding sites showed differential accessibility in erosion to identify regulators potentially implicated in epigenome erosion (Figure 6L). Intriguingly, while most TFs showed reduced TF motif enrichment in eroded cells, zinc finger E-box binding homeobox transcription factor 1 (ZEB1), involved in both epithelial-to-mesenchymal transition (EMT) and cellular senescence, instead had significantly more accessible binding sites in excitatory neurons with high erosion scores58.
Discussion
Our study reports the transcriptional and epigenomic landscapes of over 800k individual nuclei from matched post-mortem prefrontal cortex samples of 92 ROSMAP individuals in the context of AD. We develop an iterative computational framework to integrate ATAC and RNA nuclei and concurrently build peak-to-gene regulatory circuits. With this resource, we interrogate the regulome dynamics of AD progression, identify ATAC-QTLs, prioritize AD GWAS variants and reveal their target genes, and uncover cell identity loss driven by epigenomic erosion at the late-stage of AD progression.
Inspecting both epigenome and transcriptome allows us to observe coordinated changes that would otherwise not be detected in isolation. We demonstrate that similar pathways are dysregulated in both the epigenome and transcriptome in the same cell types and see similar changes in cell type proportions when we evaluate epigenomic and transcriptomic landscapes. Despite sparse measurements of genes or peaks in single-cell data, our approaches, aggregating similarly regulated portions of the epigenome, enable us to connect non-coding enhancers to target genes, uncover upstream regulators, their downstream pathways, and identify AD differential changes.
Despite AD’s high heritability and tens of AD-GWAS-associated genetic loci, functional interpretation and finemapping of variants remains challenging as many lie in the non-coding genome8,12,13. A common hypothesis for non-coding variants is that they disrupt cell-type-specific regulatory elements to affect gene expression and thus downstream endophenotypes, but high-resolution and cell-type-specific QTLs and regulome maps are required to prioritize candidate variants and their functions. Our integrated resource across AD patients at different stages of the disease allowed us to prioritize functional AD-GWAS variants and could be used in the future to understand other brain-related and neuron-centric diseases. Interestingly, heritability enrichment analysis suggested that disease-specific maps may modestly outperform disease-agnostic ones.
AD-GWAS studies have been very successful in identifying non-coding common variants. However, rare and often coding variants, including known mutations in APP and PSEN1/2, are largely neglected by GWAS studies and are studied separately from non-coding variants. The integrated epigenomic and transcriptomic regulatory circuitry from our study represents a rich resource and significant foundation to integrate common and rare variants contributing to AD and look for convergence in functional effects.
In addition to the local epigenomic changes that mostly take place during the early-stage of AD progression, we see global dysregulation of the epigenome in cells from late-AD individuals and wide-spread cell identity loss at the late-stage of the disease. This type of dysregulation, termed “epigenomic erosion” in aging cells, is marked by repressive chromatin regions becoming more and open-chromatin regions becoming less accessible47–49. Interestingly, we observe a gradient of erosion across cells, suggesting that there is a spectrum of epigenome dysregulation and cell-identity loss that reflects disease progression. Functional studies are warranted to elucidate whether epigenome erosion is the cause of dysregulation, erosion is a downstream effect of cell identity loss, or if these two processes occur jointly.
Overall, our datasets, analyses, and the resulting resources provide a cell-type-specific foundation to understand AD regulatory circuitry, reveal epigenomic and transcriptomic dynamics with regards to AD, and prioritize and map genetic variants to their targets at the cell type resolution in the human brain. Additionally, we report significant epigenomic dysregulation at the genome scale in individual cells in the late-stage of AD progression, opening up new directions for mechanistic investigation.
Limitations of the Study
We envision multiple future directions and improvements based on our work. First, although ATAC-seq generates a comprehensive epigenomic landscape, the resulting chromatin accessibility alone lacks indication of fine-grained chromatin states (e.g. enhancers vs. promoters; active vs. poised elements). Therefore, complementing ATAC-seq with histone ChIP-seq will allow us to gain a more comprehensive view of the epigenomic dynamics at different chromatin states during AD progression. The majority of the ATAC and RNA nuclei in the study were measured separately. Even though we computationally align the two modalities, the time-displaced dynamics of the transcriptome relative to the epigenome, especially in the disease context, means that concurrent profiling will enable a more accurate map of the regulatory circuitry. Finally, while we identify and characterize epigenomic erosion as a hallmark of late AD, we lack a clear picture of the progression of epigenomic erosion in AD. Putting together a time-series picture of AD progression that includes epigenomic erosion must be addressed at multiple scales: across individuals, brain regions, and at the cellular level; and from multiple angles, especially in the context of regional and local AD pathology, molecular changes in these cells before and after erosion, and in the spatial context of eroded cells.
STAR Methods
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Manolis Kellis (manoli@mit.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Single-nucleus ATAC-seq, RNA-seq, and multiome data will be available through the AD Knowledge Portal on Synapse upon publication of this manuscript. The data will be available here: https://www.synapse.org/#!Synapse:syn52293426. The data are available under controlled use conditions set by human privacy regulations. To access the data, a data use agreement is needed. This registration is in place solely to ensure the anonymity of the ROSMAP study participants. A data use agreement can be agreed with either Rush University Medical Center (RUMC) or with SAGE, which maintains Synapse, and can be downloaded from their websites (https://adknowledgeportal.synapse.org/). Additional processed data, matrices, metadata, and integration are available https://personal.broadinstitute.org/bjames/AD_snATAC/, with the overview and detailed file explanation available at http://compbio.mit.edu/ad_epigenome/. Integrative visualization and exploration of the single-cell data are available with UCSC Cell Browser interface https://rosmap-ad-aging-brain.cells.ucsc.edu59.
Code for analysis, including snATAC and snRNA data processing, GWAS analysis, QTL calling are available at https://github.com/KellisLab/AD_regulome_analysis and Zenodo https://doi.org/10.5281/zenodo.8287248. The code for single-cell module detection for ATAC-seq, multiomics, and peak-gene linking are available at https://github.com/KellisLab/epiclust and Zenodo https://zenodo.org/record/8292785.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental Model and Study participant details
Data reporting
We did not use statistical methods to predetermine the sample size.
Human subjects
We selected 427 individuals from ROSMAP from the Religious Orders Study and Rush Memory and Aging Project (ROSMAP)30, both ongoing longitudinal clinical-pathologic cohort studies of aging and dementia, in which all participants are brain donors. The studies include clinical data collected annually, detailed post-mortem pathological evaluations, and extensive genetic, epigenomic, transcriptomic, proteomic, and metabolomic bulk-tissue profiling and performed snRNA-seq on the prefrontal cortex region of these samples (Mathys et al.29). For 92 of the samples, we carried out snATAC profiling as well. The 92 samples are with a variety of AD-related pathological and cognitive measurements, and are grouped into 48 controls (22 female; 26 male), 29 early-stage AD (15 female; 14 male) and 15 late-stage AD (9 female; 6 male) samples. The AD grouping was based on a k-means clustering using seven pathological phenotypes, including a global AD pathology, molecularly specific amyloid-β and tangles, and global cognition proximate to death. The ages between AD groups are matched, without significant difference (Table S1). Informed consent was obtained from each subject, and the Religious Orders Study and Rush Memory and Aging Project were each approved by an Institutional Review Board (IRB) of Rush University Medical Center. Participants also signed an Anatomic Gift Act, and a repository consent to allow their data to be repurposed.
Methods details
Nuclei isolation from frozen post-mortem prefrontal cortex
Frozen postmortem brain tissue was used to isolate nuclei as previously described (Mathys et al., Nature 2019) with some modifications. Briefly the brain tissue was homogenized in Homogenization buffer (700μL), the homogenate was filtered through a 40μm cell strainer, Working solution (450μL) was added and loaded as a 25% Iodixanol solution on top of a 30%−40% Iodixanol density gradient (750μL 30% Iodixanol solution, 300μL 40% Iodixanol solution). The nuclei were separated by centrifugation on a fixed rotor, tabletop centrifuge (5 minutes, 10000g, 4˚C). Nuclei pellet was collected and transferred on a new tube, washed twice with 1mL ice-cold PBS/0.04% BSA (centrifuged 3 minutes, 300g, 4˚C) and resuspended in 100μL PBS/0.04% BSA.
Droplet-based snRNA-seq
For the snRNA profiling, nuclei were counted using a manual hemocytometer and concentration was adjusted to 1000 nuclei per μL. The droplet-based 10x scRNA-seq assay was used to target 5000 nuclei per brain region and individual and libraries were prepared with the 10x Genomics Chromium Single-Cell 3′ Reagent Kit v3 according to manufacturer’s protocol (CG000183 Rev.A). Pooled libraries were sequenced using the NovaSeq 6000 S2 sequencing kits (100 cycles, Illumina).
Droplet-based snATAC-seq
For the snATAC profiling the remaining nuclei were spun down, resuspended in 1x Diluted Nuclei buffer and their concentration was adjusted to 2500 nuclei per μL. For the library construction, 5000 nuclei per brain region and individual were targeted using the 10x Genomics Chromium Single-Cell ATAC Reagent Kit v1 following the manufacturer’s protocol (CG000168 Rev.D). Pooled libraries were sequenced using the NovaSeq 6000 S2 sequencing kits (100 cycles, Illumina).
Droplet-based single cell multiome-seq (Gene Expression- and ATAC-seq)
The single cell ATAC and gene expression (GEX) profiles were generated using the 10x Genomics Chromium Next GEM Single Cell Multiome ATAC + GEX assay kit, which allows preparation of paired ATAC and GEX libraries from the same nuclei sample, following manufacturer’s instructions (CG000338, Rev. A). Nuclei were isolated as described above and resuspended in 1x Diluted Nuclei buffer containing RNase inhibitors and 1mM DTT. Nuclei were counted with a manual hemocytometer and used at a final concentration of 2500–5000 nuclei per μL to achieve 7000 targeted nuclei recovery per sample. Generated libraries were pooled and sequenced on NovaSeq 6000 using S2 sequencing kit (100 cycles, Illumina).
Experimental validation of epigenome erosion using Lamin-B1 staining
Fixed human medial frontal cortex human tissue was sectioned at 40μm using a vibratome. Blocking buffer (PBS containing 5% bovine serum albumin, 1% normal donkey serum and 0.3% Triton X-100) was used to block the sections for one hour at room temperature (RT). The sections were then incubated for 24 hours at 4°C with the primary antibodies in blocking buffer. Tissue sections were probed with NeuN (Synaptic Systems, #266004 1:1000) and Lamin B1 (Abcam, #ab229025 1:500). Following primary antibody incubation, the sections were washed three times for 5 minutes each at RT in PBS and then incubated with secondary antibodies (dilution 1:1000) and Hoechst 33342 (Sigma-Aldrich; Cat#94403) for two hours at RT. After three more 5-minute washes in PBS, the slices were mounted on Superfrost Plus Slides (Fisherbrand; 870 Cat#12–550-15) and incubated in TrueBlack (Biotium; Cat#23007) for 90 seconds. After a final round of washes, the coverslips were applied to Fluromount-G (SouthernBiotech; Cat#0100–01). Primary antibodies were visualized with Alexa-Fluor 488 and Alexa-Fluor 555, antibodies (Molecular Probes), and cell nuclei were visualized with Hoechst 33342 using a confocal microscope (LSM 900; Zeiss) with a 20× or 63× objective. For this analysis, a subset of 10 subjects from the larger cohort in our study were selected; 5 subjects were assigned as controls and 5 subjects were assigned as high erosion based on scores computed by our snATAC-seq analysis. For quantification, 3D reconstructions were rendered through IMARIS (Oxford Instruments) from a series of 12 Z-steps at 20x for all samples. The intensity of Lamin B1 expression was measured on masks created around NeuN-positive cell surfaces after maximal projection. On average, 129 NeuN-positive cell surfaces were quantified from control subjects (n= 91 to 164 NeuN-positive cell surfaces per 5 subjects in this group) and 122 NeuN-positive cell surfaces from high erosion subjects (n= 85 to 173 NeuN-positive cell surfaces per 5 subjects in this group). The images were manually assessed in IMARIS for quality control to avoid the potential inclusion of autofluorescence lipofuscin granules if present. The data was plotted with Graphpad Prism (Version 10.0.2) and statistical significance determined with a student t-test.
Quantification and Statistical Analysis
snATAC data processing and major cell type annotation
We used “cellranger-atac mkfastq” (V1.1.0) to demultiplex BCL files into Fastq raw sequencing data, and then ran “cellranger-atac count” to map the reads onto human reference genome (GRCh38) and obtain the fragment file of each sample. We then utilized ArchR (V1.0.1) to process the snATAC-seq data, using fragment files as input60. We performed the first round of doublet removal within each sample using the “filterDoublets” function.
For the “high-quality cell” analysis, we selected the cells with TSS enrichment > 6 and number of fragments between 1000 to 100,000. We performed Iterative LSI dimension reduction and clustering using a 500 bp tile matrix, with parameters “iterations=4, resolution=0.2, varFeat=50000”. Cell embedding visualization was performed using UMAP. We estimated gene expression score using ArchR, and then quantified the cell type signature of each cluster based on PsychENCODE marker genes61. We calculated the mean of gene scores of all the cell marker genes in each cell type for each cluster, which were then used to assign cell types. We performed a second round of doublet removal at cluster level by discarding the cell clusters that show ambiguous cell type marker signal and meanwhile locate between two clusters that show clear cell type signature.
For the “erosion” analysis, we selected the cells with TSS enrichment > 1 and number of fragments between 1000 to 100,000. We performed Iterative LSI dimension reduction and clustering using a 500 bp tile matrix, with parameters “iterations=3, resolution=0.2, varFeat=50000”. Cell embedding visualization was performed using UMAP. We performed a second round of doublet removal at cluster level using the same strategy as “TSS enrichment > 6” analysis described above.
After cell annotation, we performed peak calling at pseudobulk level using MACS2, with a q-value threshold 0.01. Peak functional annotation was carried out using GREAT, and the functional terms from molecular functions (MF), biological process (BP) and cellular component (CC) are selected and shown. Cell-type-specific peaks are identified using the getMarkers function from ArchR, with “FDR <= 0.01 & Log2FC >= 1” as thresholds, and adjusted for TSS enrichment and library size.
Candidate TF regulator identification
We first used chromVar implemented in ArchR to perform motif enrichment analysis and calculate z-score across all the cells. The genome-wide motif scanning map from CIS-BP database was used for the chromVar analysis. To better identify functional TF in each cell type, we calculated the Pearson correlation between the motif enrichment z-score against the ATAC-inferred gene expression, across the low-overlapping cell aggregates using the “correlateMatrices” function (FDR<0.01 & correlation>0.5). Moreover, we performed Pearson correlation analysis between ATAC-inferred gene expression and RNA-measured gene expression across the 7 major cell types, and applied a second filtering (FDR<0.1 & correlation>0.5), and the TFs that passed both correlation thresholds are defined as the candidate TF regulators (Table S2).
snRNA clustering and cell assignments
snRNA counts were gathered using the CellRanger 10x pipeline. Analysis using the Scanpy framework consisted of count normalization by (1) normalizing per-cell depth to 10000 counts per cell, then (2) log(1+x) normalization, followed by (3) highly variable gene selection across batches to 5000 highly variable genes. Next, batch correction was applied on these variable genes using the ComBat framework within Scanpy. Finally, a low level UMAP embedding was generated using the standard Scanpy analysis framework of PCA, approximate nearest neighbors, and UMAP calling. Clusters were identified using the Leiden algorithm, and cell types were checked against marker genes.
Integration overview
We developed an approach for snATAC-snRNA integration that iteratively updates the snATAC gene score matrix, the snATAC-snRNA integration, and the peak-gene links. In order to perform this integration, we start from an initial estimate of the gene score matrix for snATAC cells and have no prior knowledge for the distal regulatory links or the snRNA-snATAC integration. At a high-level, the following steps are taken:
Integrate snRNA (gene expression matrix) and snATAC (estimated gene score matrix)
Assign sub-cell types to the snATAC cells from the snATAC-snRNA joint space
Update peak-gene links based on the snRNA-snATAC integration
Re-estimate the gene score matrix for snATAC data using the updated peak-gene links
Go back to step (1) and re-perform snRNA and snATAC integration.
These steps are each described in the sections below (integration, cell type assignment, peak-gene linking). Algorithm convergence is defined as when the Pearson correlation between snRNA gene expression and the refined snATAC gene score reaches a plateau. In the case of the PFC data, we found that after 2 iterations of updating links and re-estimating the gene score matrix there is little to no improvement. Alternatively, since each iteration is performed manually, the user can determine when an integration is sufficient. Since major cell types can be identified in both snATAC and snRNA, we integrate each major cell type separately to study the variability within cell types more accurately, especially for neurons.
Integration
Our initial estimate of the snATAC gene score matrix is done by inferring gene-level expression from chromatin accessibility in a manner similar to ArchR60. Briefly, from a set of 200 bp tiles across the genome, we count the number of reads residing within 5kb of each gene body, weighted by a score that exponentially decays by distance (e-|distance_to_genebody/5000| +e−1), thus up-weighing proximal peaks and down-weighing distal peaks. Additionally, we set each gene’s possible contributing distal peaks to stop at the next gene’s gene body at first. Finally, we add an additional gene score weight to scale gene length inversely. This weight corresponds to the inverse of the gene length such that the smallest genes get a score of 5 and the largest genes get a score of 1. Finally, the UMI counts for each snATAC cell get scaled such that the total count across all genes is 10,000, similar to snRNA-based normalization strategies.
Once we have peak-to-gene linking information in subsequent rounds of integration, we incorporate these links in the snATAC gene score matrix estimation. Specifically, we reweight the contribution of distal peaks using the computed peak-to-gene linking probability scores output by the logistic regression model. When we use peak-gene links to calculate the gene score we do not stop at nearby gene boundaries and instead use all gene-linked peaks (peak-gene links computed for peaks within 1Mb of the gene TSS).
Once gene scores are obtained for both the ATAC and RNA modalities, each cell type is integrated separately. For each cell type, we scale the ATAC and RNA gene matrices to 10,000 total counts per cell, then log(1+x) scaled. Since we must severely correct between ATAC and RNA, simple batch correction cannot correct full effects. We run ComBat62, Harmony63, and BB-kNN64 sequentially through ScanPy to adjust ATAC-RNA differences. Next, we apply a diffusion map as implemented in Scanpy over the nearest neighbor matrix to generate a latent space from which we calculate a UMAP embedding.
Cell type assignment
Using an ATAC-RNA integration, cell types are assigned by counting the number of sub cell types that are nearest neighbors from the nearest neighbor matrix used to build the integrated UMAP. Leiden clustering65 is also applied on this neighbor matrix, with two graphs from resolution parameters of 1 and 2 to build two levels of meta-cells from which peak-gene linking is estimated. However, for the final integration UMAP, we instead use predicted cell type labels to create meta-cells to predict links.
Peak-gene linking
We develop a method for computing peak-gene links from either snATAC alone (first iteration of the integration) or from integrated snATAC-snRNA (subsequent iterations). At a high level, we take the following steps for the first iteration.
Estimate and rescale the gene accessibility (gene score) matrix
Perform TF-IDF on and rescale the peak matrix
Concatenate the peak and gene accessibility matrices
Perform PCA on this concatenated matrix
Re-scale the PCA embeddings with different weights and concatenate these rescaled embeddings
Take dot product between peak and gene across embeddings to estimate their correlation
Compute the partial correlation adjusted for quality covariates, and transform by arctanh
Bin each peak-gene pair into a bin based on the peak sparsity and the gene sparsity
Compute per-bin statistics (mean, sd) of the correlation (over all (peak,gene) pairs in a bin)
Adjust the scores for individual pairs by z-scoring with the (mean, sd) statistics from their bin
Predict link probability from these scores using a logistic regression model
Multiply link probability by peak accessibility and normalize this as in ABC (accounting for gene density and distance) to obtain the final linking scores
We describe these steps and the modifications for subsequent iterations in more detail below:
Peak-gene correlation is based on combined co-accessibility (i.e. within ATAC-seq) and peak-gene correlation (i.e. across assays). We calculated snATAC gene accessibility matrix (or gene score matrix) as part of the integration as outlined above, either using the ArchR method alone (in initialization) or peak-gene links reweighting the relative contribution of distal peaks (where the links used are from the previous round of integration). We first calculate peak-gene accessibility PC embeddings per UMI for co-accessibility and peak-gene accessibility-gene expression PC embeddings per metacell for peak-gene expression linking. Only peaks within 1Mb of the gene TSS are considered for peak-gene linking.
To accurately scale peaks and gene accessibility, we ensure that each of these modalities has a per-UMI sum of 10,000. To do so, gene accessibility scores are scaled to 10,000 per UMI. However, due to the sparsity inherent in an snATAC peak matrix, we utilize term frequency inverse document frequency (TF-IDF) to initially scale the peak matrix. Next, we scale the TF-IDF matrix to 10,000 per-UMI counts to ensure compatibility with gene accessibility. Finally, we concatenate the 10,000 per-UMI counts and log(1+x) scale the concatenated matrix before we call truncated PCA with 100 principal components.
After we perform an initial integration with snRNA, we include that data in order to more accurately capture correlations with transcripts. Similar to co-accessibility embeddings, we calculate peak-gene embeddings in a similar manner. However, instead of calculating the embeddings per-UMI, we utilize a metacell like framework to combine ATAC and RNA modalities in a high enough resolution that preserves accuracy. Here we use sub cell types per individual as meta cells. Similar to co-accessibility estimation, we use TF-IDF for the peak matrix followed by scaling to 10000 counts per meta cell as well as 10,000 counts per UMI for both gene accessibility and gene expression. In total, when combined, each meta-cell has 30,000 counts. Then, we utilize log(1+x) scaling followed by truncated PCA with 100 components to form a similar embedding.
This framework utilizes varying powers of the eigenvalues of the PC embeddings to generate five distinct embeddings scaling from zero-phase component analysis (ZCA)66 to principal component analysis (PCA). Reweighting is performed following the equation below, with eigenvalues s, eigenvectors V from PCA and power p as one of (0, 0.25, 0.5, 0.75, 1), where the resulting embeddings are adjusted by the eigenvalues, centered, and normalized:
When done for both co-accessibility and peak-gene embeddings, we use 10 total embeddings; when gene expression meta-cells are included, we use 15 total embeddings. In these embeddings, we subtract their per-UMI means and Euclidean-norm the resulting matrix so that the dot product in that space approximates Pearson correlation.
From these embeddings, we calculate initial scores between peaks and genes by taking the dot product between two indices in the embeddings. In order to remove the effect of TSS enrichment and number of fragments on these correlations, we compute partial correlations over the peak-gene embeddings instead of raw correlations. Partial correlations allow for adjustment due to these factors, but the classical partial correlation algorithm requires a matrix inverse of the autocorrelation matrix, which is infeasible for peak-peak or peak-gene correlations. Instead, we compute partial correlations as:
Given a correlation , covariate correlations as , and covariate auto-correlation matrix as . We reduce this to the following equation, which we use to compute the partial correlation:
Since and can be computed beforehand, the pairwise partial correlations can be easily calculated from this formula. The sign can be calculated using intermediate products. We transform these correlations using the hyperbolic arctangent (Fisher’s transform), to stabilize the variance of these correlations, which is especially important when quantifying high correlations.
Next, we stratified all peaks by their sparsity into 50 bins and all gene accessibility scores into another 50 bins by sparsity. We group peaks and genes into these bins by sparsity in order to find sparsity bin-specific baselines from which we can remove contributions to correlations inflated by technical issues. For each pair of bins (peak sparsity and gene sparsity), we calculate the mean and standard deviations of all the scores for peak and gene tuples that fall into this pair of bins. However, due to the computational intractability of calculating all peak-gene pairs to estimate the mean and standard deviation of peak-gene correlations is infeasible computationally, we estimate these metrics by sampling. Since Fisher transformed Pearson correlations have a known standard error, we perform a power calculation (Z=2 and margin of error of 0.05) to compute how many peak-gene pairs to sample for our estimates for each sparsity bin (gene, peak) pair. Finally, we smooth these means and standard deviations across sparsity bins using a Gaussian smoothing filter.
Once we set the mean and standard deviation for each peak-gene pair, we calculate z-scores for each peak-gene pair, such that the transformed and z-scored partial correlation between gene i and peak j is:
Where the mean and standard deviation are the bin-wise estimates described above. We then normalize the Fisher transformed partial correlations by the mean and standard deviation derived from the bins mentioned above to produce z-scores for each of the 10 embeddings. In order to find baselines for each gene for peak-gene linking, we calculate corresponding random-peak to gene z-scores by taking 5 random peaks and correlating them to the target gene for each peak.
Finally, we train a logistic regression model to distinguish peak-to-gene z-scores against 5 random peak-to-gene z-scores per candidate peak-gene link as mentioned above. We sample the random peaks from a different chromosome to guarantee that they are not cis-acting regulators of the gene. We separately compute the peak accessibility, multiply it by the logistic regression probability, and normalize it as in ABC, accounting for distance and gene density67. Specifically, we use , using the Hi-C power of −0.87 and scale parameter of 5.41 (parameters to the ABC model) to estimate contact, with activity defined by pseudo-bulk accessibility, and the probability as mentioned above. We call links as peak-gene pairs with scores above a cutoff of 0.001, which we determined based on testing against PLAC-seq 11.
Peak modules
The peak-gene accessibility embeddings and adjustments generated for the peak to gene linking provide methodology to cluster peaks together. Moreover, we utilize a custom distance function given to the high-performance UMAP library to find 10 approximate nearest neighbors according to the z-scores. Using methodology inspired by BB-kNN64, both top peak-peak correlations, peak-gene accessibility correlations, and gene accessibility-gene accessibility correlations are concatenated to provide a balanced nearest neighbor graph across peaks and gene accessibility.
Since clustering peaks only requires high confidence correlation scores, nodes (peaks or genes) are filtered such that each embedding contains at least one correlation z-score in one of its embeddings in the 95th percentile. Thus, peaks with low connectivity do not hurt the clusters.
Because these nearest neighbor graphs are calculated for each of the five co-accessibility embeddings, we utilize the multiplex Leiden community detection algorithm over these five graphs with a resolution parameter of 1 and a maximum community size of 2500 under a Reichardt and Bornholdts Potts model with a configuration null model. The community size restriction ensures that large modules do not dominate; however, this means that modules with similar profiles may coexist.
Differentially accessible peak modules
Differentially accessible modules are calculated by summing all counts in a module per cell, then using a combination of RUVseq and Nebula to calculate differential modules at the single cell level68,69. We utilize 10 RUV pseudobulk terms and the covariates: age, sex, post-mortem interval, and number of fragments. We demonstrate (Supplementary Data) that this approach has similar results to using edgeR70; however, this approach leverages the single cell nature of snATAC-seq.
Module enrichments
Module enrichments for Gene Ontology terms were calculated via GREAT and the rGREAT package, using a background set of all module peaks36. Adjusted p-values were found via the HyperFdrQ column from returned tables and a cutoff of HyperFdrQ < 0.01 was used. Additional cutoffs to ensure relevance included: GO terms below 500 genes per term, at least 50 foreground regions per GO enrichment, and at least five foreground genes. TF binding sites were calculated from the JASPAR 2022 core motif set using the motifmatchr and TFBStools packages71. Motif enrichments were then calculated via a hypergeometric test per module by comparing the number of peaks intersecting motif binding sites to all peaks in the module. A background set of peaks consisted of all peaks across that cell type’s peak modules.
TF enrichments for AD DEG-linked peaks
AD-DEG linked peaks were enriched for motifs in a similar manner to module TF enrichments using a peak set of AD-DEG linked peaks with a score above 0.001; however, background peaks used were all linked peaks with a linking score above 0.001.
GWAS heritability enrichment
GWAS heritability enrichment analysis was carried out using S-LDSC (v1.0.1) based on the tutorial37. The enrichment was quantified as the proportion of heritability normalized by the proportion of SNPs covered; the standard error was then calculated and used for p-value calculation. AD-GWAS studies from Bellenguez et al., 2022, Jansen et al., 2019 and Kunkle et al., 2019 were used. Summary statistics files of other GWAS studies that are curated by LDSC were downloaded and used for the analysis (https://data.broadinstitute.org/alkesgroup/LDSCORE/independent_sumstats/). For the microglia peaks partitioned by genomic regions, the enhancers are from a published study11, defined as peaks with H3K27ac but not H3K4me3. For the LDSC analysis on aQTL loci, we took the aQTL lead SNPs in each major cell-type, extended 5k from each end, and then performed LDSC for the resulting 10k aQTL regions.
sc-eQTLs analysis
For each major cell type, we used a poisson-gamma model to calculate the average gene expression, with a sequencing depth parameter estimated at the cell level, and a mean parameter estimated at the individual level. The individual-level mean parameter was estimated using an integrative variation inference framework72. The confounding factors underlying the individual-level gene expression matrix were then estimated in an adaptive way. We defined a control gene set for each gene by using the top correlated genes located across different chromosomes. We identified the principal components (PCs) and took the first 50 PCs as covariates. Next, we built knock-off filters73 for a genotype dosage matrix sampled from the 1MB cis-window surrounding the TSS for each gene. We used a singular value decomposition (SVD) to estimate the PCs across the sampled genotype matrix. The top PCs that account for over 80% of variation was used to estimate the confounding factors that correspond to the genetic structures. We estimated a knock-off design matrix using scalable techniques reported previously74–76. Lastly, we carried out a mulit-variate regression for each gene between the pseudo-bulk profiles, versus the observed and knock-off genotype matrix, respectively. A Beyesian spike-and-slab regression was applied before measuring the posterior inclusion probability (PIP) for each SNP. The difference between the PIP on the observed variable vs the knock-off counterpart was used to calculate the knock-off statistics. The optimal threshold of false discovery rate (FDR) was determined using the R knock-off package77.
ATAC-QTL (aQTL) calling and colocalization with AD-GWAS
We generated a peak by individual row count pseudo-bulk matrices for each major cell type by aggregating the cells from each individual in each cell type, and then calculated a normalization factor to scale the raw library size for each matrix. We then used the voom function from Limma78 to transform the scaled data to Log2(counts per million) and estimated the mean-variance relationship, which was then used to compute appropriate observation-level weight. The resulting matrix was sent for PEER factor estimation to uncover potential unwanted variation. We included the first 35, 40, 35, 12, 30, 35 peer factors for the excitatory neuron, inhibitory neuron, astrocyte, oligodendrocyte, OPC and microglia for aQTL calling, which maximize the sensitivity of QTL identification. We used FastQTL permute to identify gPeaks and meanwhile perform multi-testing correction of SNPs being tested for each peak79. The adjusted empirical p-value for each SNP was extrapolated based on the beta distribution of each peak. We utilized empirical p-value < 0.005 on the lead variant to define significant gPeak. Within each gPeak, we calculated the nominal p-value that corresponds to the adjusted empirical p-value of 0.005 and used it to define aQTLs for each locus.
We then performed colocalization between aQTL and AD-GWAS (Jansen et al., 2019) using coloc80 for the loci with p-value<1×10−5 to identify the sub-optimal GWAS loci that can be explained by aQTLs. Colocalization analysis was performed with the hg19 coordinates. The aQTL-AD-GWAS colocalization was defined by PP4>0.1. We then include single-cell eQTLs to perform multiple-trait-colocalization for the aQTL-AD-GWAS colocalized loci using moloc81. The aQTL-eQTL-AD-GWAS colocalized loci were defined by PPA.abc>0.1 (PPA.abc denotes the posterior probability that all three genetics components colocalize) and PPA.abc>PPA.ac.b+PPA.ac (where PPA.ac.b and PPA.ac represent the posterior probability AD-GWAS and aQTL colocalize, but eQTL does not and that no eQTL effect is observed at the locus, respectively). LD expansion for AD GWAS loci (Figure S3G) was based on the Phase 3 of European ancestry genome structure curated by 1000 Genomes82.
Cell-type sharing of aQTL
For each pair of cell-types across all the cell types, we estimated aQTL sharing for the aQTLs whose targeting peaks are shared between the two cell-types using a previously-reported directionality-dependent method 43. For all the lead aQTLs that are significant in at least one cell-type (discovery cell type), we tested whether their aQTL-effect directionalities are consistent in each of the rest of the cell types (replication cell type). The directionality consistency was then calculated as the percentage of gPeak-aQTL pairs that show consistent directionality (either positive or negative) between the discovery cell type and the replication cell type. We further group the aQTLs into six bins depending on their nominal p-values in the replication cell type: (i) P<10−9, (ii) 10−9<P<10−7, (iii) 10−7<P<10−5, (iv) 10−5<P<10−3, (v) 10−3<P<0.1, and (vi) no effect (P>0.1), and calculated the aQTL directionality consistency within each of the P-value bins. Lastly, the proportion of cell-type-sharing (Figure S4C) aQTLs was quantified as:
where the represents the percentage of aQTLs of bin i, and the represents the aQTL directionality consistency of bin i.
Cell fraction change during AD progression
The cell composition change between non-AD, early-AD and late-AD was first carried out at the major cell type level using the snATAC and snRNA cells, respectively. The fraction of each cell type in each individual was calculated and compared across different AD groups. All of the composition analysis was done using propeller (Figure 5B–D, Figure S5D and Figure 6B), a published method designed to test for compositional changes in single-cell data83. The statistical tests were based on the Anova testing implemented in propeller.
We included age of death, sex, and post-mortem interval (pmi) as covariates when testing the significance of composition change against AD status. For Figures 5B–D and S5D, we tested across all three AD groups (non-AD, early-AD, and late-AD). For Figure 6B, we test late-AD versus non/early-AD, as we are specifically looking at the late-AD change in cell fraction associated with epigenomic erosion. For the subtypes with more than 500 cells, we further performed composition change analysis within each major cell type.
Identification of de-identified ATAC cells
We used a very loose TSS enrichment threshold (>1) for ATAC cell filtration to make sure that the biologically-induced de-identified cells are included in the analysis. After dimension reduction and clustering as described in the “snATAC data processing and major cell type annotation” section, we examined the marker expression signature of each major cell type for each cluster. The cluster that does not show clear marker expression signatures for any of the major cell types was defined as the “de-identified” cell population. For the clusters that show relatively weak cell type marker signature compared to the corresponding normal major cell types that they are contiguous to, and meanwhile stretched to the “de-identified” cell population, they are assigned as the de-identified cell groups for the corresponding major cell types. We then interrogated the distribution of normal vs. de-identified cells during AD progression, and confirmed that the late-stage AD samples are enriched for the de-identified cells, and meanwhile depleted for the corresponding normal cell populations.
Per-cell level epigenomic erosion score estimation
We quantified epigenome erosion at per-cell level based on the fragment distribution in ChromHMM states for the dorsolateral prefrontal cortex (E073)51 with the following steps: 1) for each cell, we calculate the fraction of reads in each of the 18 ChromHMM states, resulting in a 414k x 18 cell by state matrix; 2) we perform center log ratio (CLR) normalization across cells; 3) for each cell in each chromatin state, we calculate its deviation to the mean of that ChromHMM state; 4) for each cell, the deviations from 18 ChromHMM states were summed up, with the active states (Enhancers; TSS; Tx) assigned negative sign, and the repressive regions (Quies, Repr, Het, TxWk) assigned positive sign, such that a higher positive score represents that the repressive regions are more opened, and therefore the corresponding cell is more eroded in the epigenome.
Additional Resources
Overview and detailed file explanation of dataset files available at http://compbio.mit.edu/ad_epigenome/. Integrative visualization and exploration of the single-cell data are available with UCSC Cell Browser interface https://rosmap-ad-aging-brain.cells.ucsc.edu.
Supplementary Material
Acknowledgements
We thank the study participants and staff of the Rush Alzheimer’s Disease Center. We thank Y. Tanigawa, J. Yang, Z. Liu, S. Mohammadi, J. D-Velderrain, H. Meharena and all members from the Kellis Lab and Tsai Lab for their help and suggestions on the project. We thank A. Grayson and P. Purcell for their help on structuring the manuscript. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. 1745302 awarded to Benjamin T. James. This work was supported in part by NIH grants AG054012, AG058002, AG062377, NS110453, NS115064, AG062335, NS127187 (M.K., L.-H.T.); AG067151, MH109978, MH119509, HG008155, DA053631 (M.K.); P30AG10161, P30AG72975, R01AG15819, R01AG17917, U01AG46152, U01AG61356, and R01AG57473 (D.A.B.); and the Cure Alzheimer’s Foundation CIRCUITS consortium (M.K., L.-H.T).
Footnotes
Declaration of Interests
L.-H.T. is a member of the Scientific Advisory Board of Cognito Therapeutics, 4M Therapeutics, Cell Signaling Technology, and Souvien Therapeutics, which has no association to the work described in this manuscript.
References
- 1.Wilson RS, Boyle PA, Yu L, Barnes LL, Sytsma J, Buchman AS, Bennett DA, and Schneider JA (2015). Temporal course and pathologic basis of unawareness of memory loss in dementia. Neurology 85, 984–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett DA, Wilson RS, Schneider JA, Evans DA, Beckett LA, Aggarwal NT, Barnes LL, Fox JH, and Bach J. (2002). Natural history of mild cognitive impairment in older persons. Neurology 59, 198–205. [DOI] [PubMed] [Google Scholar]
- 3.Bennett DA, Schneider JA, Wilson RS, Bienias JL, and Arnold SE (2004). Neurofibrillary tangles mediate the association of amyloid load with clinical Alzheimer disease and level of cognitive function. Arch. Neurol 61, 378–384. [DOI] [PubMed] [Google Scholar]
- 4.Braak H, and Braak E. (1995). Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol. Aging 16, 271–278; discussion 278–284. [DOI] [PubMed] [Google Scholar]
- 5.Querfurth HW, and LaFerla FM (2010). Alzheimer’s disease. N. Engl. J. Med 362, 329–344. [DOI] [PubMed] [Google Scholar]
- 6.Ridge PG, Mukherjee S, Crane PK, Kauwe JSK, and Alzheimer’s Disease Genetics Consortium (2013). Alzheimer’s disease: analyzing the missing heritability. PLoS One 8, e79771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson RS, Barral S, Lee JH, Leurgans SE, Foroud TM, Sweet RA, Graff-Radford N, Bird TD, Mayeux R, and Bennett DA (2011). Heritability of different forms of memory in the Late Onset Alzheimer’s Disease Family Study. J. Alzheimers. Dis 23, 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellenguez C, Küçükali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N, Naj AC, Campos-Martin R, Grenier-Boley B, Andrade V, et al. (2022). New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet 54, 412–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartzentruber J, Cooper S, Liu JZ, Barrio-Hernandez I, Bello E, Kumasaka N, Young AMH, Franklin RJM, Johnson T, Estrada K, et al. (2021). Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new Alzheimer’s disease risk genes. Nat. Genet 53, 392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gjoneska E, Pfenning AR, Mathys H, Quon G, Kundaje A, Tsai L-H, and Kellis M. (2015). Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 518, 365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nott A, Holtman IR, Coufal NG, Schlachetzki JCM, Yu M, Hu R, Han CZ, Pena M, Xiao J, Wu Y, et al. (2019). Brain cell type-specific enhancer-promoter interactome maps and diseaserisk association. Science 366, 1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mostafavi H, Spence JP, Naqvi S, and Pritchard JK (2022). Limited overlap of eQTLs and GWAS hits due to systematic differences in discovery. bioRxiv, 2022.05.07.491045. 10.1101/2022.05.07.491045. [DOI] [Google Scholar]
- 13.Connally NJ, Nazeen S, Lee D, Shi H, Stamatoyannopoulos J, Chun S, Cotsapas C, Cassa CA, and Sunyaev SR (2022). The missing link between genetic association and regulatory function. Elife 11. 10.7554/eLife.74970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez RK, Gordon MG, Subramaniam M, Kim MC, Hartoularos GC, Targ S, Sun Y, Ogorodnikov A, Bueno R, Lu A, et al. (2022). Single-cell RNA-seq reveals cell type–specific molecular and genetic associations to lupus. Science 376, eabf1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gate RE, Cheng CS, Aiden AP, Siba A, Tabaka M, Lituiev D, Machol I, Gordon MG, Subramaniam M, Shamim M, et al. (2018). Genetic determinants of co-accessible chromatin regions in activated T cells across humans. Nat. Genet 50, 1140–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosoy R, Fullard JF, Zeng B, Bendl J, Dong P, Rahman S, Kleopoulos SP, Shao Z, Girdhar K, Humphrey J, et al. (2022). Genetics of the human microglia regulome refines Alzheimer’s disease risk loci. Nat. Genet 54, 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamazon ER, Segrè AV, van de Bunt M, Wen X, Xi HS, Hormozdiari F, Ongen H, Konkashbaev A, Derks EM, Aguet F, et al. (2018). Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat. Genet 50, 956–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang AC, Vest RT, Kern F, Lee DP, Agam M, Maat CA, Losada PM, Chen MB, Schaum N, Khoury N, et al. (2022). A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature. 10.1038/s41586-021-04369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, Menon M, He L, Abdurrob F, Jiang X, et al. (2019). Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grubman A, Chew G, Ouyang JF, Sun G, Choo XY, McLean C, Simmons RK, Buckberry S, Vargas-Landin DB, Poppe D, et al. (2019). A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci 22, 2087–2097. [DOI] [PubMed] [Google Scholar]
- 21.Yeh FL, Wang Y, Tom I, Gonzalez LC, and Sheng M. (2016). TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia. Neuron 91, 328–340. [DOI] [PubMed] [Google Scholar]
- 22.Ulland TK, Song WM, Huang SC-C, Ulrich JD, Sergushichev A, Beatty WL, Loboda AA, Zhou Y, Cairns NJ, Kambal A, et al. (2017). TREM2 Maintains Microglial Metabolic Fitness in Alzheimer’s Disease. Cell 170, 649–663.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morabito S, Miyoshi E, Michael N, Shahin S, Martini AC, Head E, Silva J, Leavy K, Perez-Rosendahl M, and Swarup V. (2021). Single-nucleus chromatin accessibility and transcriptomic characterization of Alzheimer’s disease. Nat. Genet 53, 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corces MR, Shcherbina A, Kundu S, Gloudemans MJ, Frésard L, Granja JM, Louie BH, Eulalio T, Shams S, Bagdatli ST, et al. (2020). Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat. Genet 52, 1158–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bendl J, Hauberg ME, Girdhar K, Im E, Vicari JM, Rahman S, Fernando MB, Townsley KG, Dong P, Misir R, et al. (2022). The three-dimensional landscape of cortical chromatin accessibility in Alzheimer’s disease. Nat. Neurosci 25, 1366–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller MB, Huang AY, Kim J, Zhou Z, Kirkham SL, Maury EA, Ziegenfuss JS, Reed HC, Neil JE, Rento L, et al. (2022). Somatic genomic changes in single Alzheimer’s disease neurons. Nature 604, 714–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J-H, Griffin PT, Vera DL, Hayano M, Meer MV, Salfati EL, Su Q, Munding EM, Blanchette M, Bhakta M, et al. (2019). Erosion of the Epigenetic Landscape and Loss of Cellular Identity as a Cause of Aging in Mammals. bioRxiv, 808642. 10.1101/808642. [DOI] [Google Scholar]
- 28.Neuronal DNA double-strand breaks lead to chromosomal structural variations and 3D genome disruption in neurodegeneration Paperpile. https://paperpile.com/app/p/f3162b05-bfa3-0021-86f6-eb419a9a65d3. [DOI] [PMC free article] [PubMed]
- 29.Placeholder Paperpile (Co-submitted paper in this issue). https://paperpile.com/app/p/f72bc3f2-8466-08e7-bc55-4a1344246486.
- 30.Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, and Schneider JA (2018). Religious Orders Study and Rush Memory and Aging Project. J. Alzheimers. Dis 64, S161–S189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao Z-J, and Gao G. (2022). Multi-omics single-cell data integration and regulatory inference with graph-linked embedding. Nat. Biotechnol 10.1038/s41587-022-01284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Priego N, Zhu L, Monteiro C, Mulders M, Wasilewski D, Bindeman W, Doglio L, Martínez L, Martínez-Saez E, Ramón Y Cajal S, et al. (2018). STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis. Nat. Med 24, 1024–1035. [DOI] [PubMed] [Google Scholar]
- 34.Jin Y, Li R, Zhang Z, Ren J, Song X, and Zhang G. (2020). ZBED1/DREF: A transcription factor that regulates cell proliferation. Oncol. Lett 20, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reed BD, Charos AE, Szekely AM, and Weissman SM (2008). Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, and Bejerano G. (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium, Patterson N, Daly MJ, Price AL, and Neale BM (2015). LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kierdorf K, and Prinz M. (2013). Factors regulating microglia activation. Front. Cell. Neurosci 7, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sudwarts A, Ramesha S, Gao T, Ponnusamy M, Wang S, Hansen M, Kozlova A, Bitarafan S, Kumar P, Beaulieu-Abdelahad D, et al. (2022). BIN1 is a key regulator of proinflammatory and neurodegeneration-related activation in microglia. Mol. Neurodegener 17, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novikova G, Kapoor M, Tcw J, Abud EM, Efthymiou AG, Chen SX, Cheng H, Fullard JF, Bendl J, Liu Y, et al. (2021). Integration of Alzheimer’s disease genetics and myeloid genomics identifies disease risk regulatory elements and genes. Nat. Commun 12, 1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou M, Xu X, Wang H, Yang G, Yang M, Zhao X, Guo H, Song J, Zheng H, Zhu Z, et al. (2020). Effect of central JAZF1 on glucose production is regulated by the PI3K-Akt-AMPK pathway. FASEB J. 34, 7058–7074. [DOI] [PubMed] [Google Scholar]
- 42.Manning BD, and Toker A. (2017). AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong X, Hou L, Park YP, Molinie B, GTEx Consortium, Gregory RI, and Kellis M. (2021). Genetic drivers of mA methylation in human brain, lung, heart and muscle. Nat. Genet 53, 1156–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliveira TG, and Di Paolo G. (2010). Phospholipase D in brain function and Alzheimer’s disease. Biochim. Biophys. Acta 1801, 799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leng K, Li E, Eser R, Piergies A, Sit R, Tan M, Neff N, Li SH, Rodriguez RD, Suemoto CK, et al. (2021). Molecular characterization of selectively vulnerable neurons in Alzheimer’s disease. Nat. Neurosci 24, 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kousi M, Boix C, Park YP, Mathys H, Sledzieski S, Peng Z, Bennett DA, Tsai L-H, and Kellis M. (2022). Single-cell mosaicism analysis reveals cell-type-specific somatic mutational burden in Alzheimer’s Dementia. bioRxiv, 2022.04.21.489103. 10.1101/2022.04.21.489103. [DOI] [Google Scholar]
- 47.Bertucci EM, and Parrott BB (2020). Is CpG Density the Link between Epigenetic Aging and Lifespan? Trends Genet. 36, 725–727. [DOI] [PubMed] [Google Scholar]
- 48.Kosan C, Heidel FH, Godmann M, and Bierhoff H. (2018). Epigenetic Erosion in Adult Stem Cells: Drivers and Passengers of Aging. Cells 7. 10.3390/cells7120237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Z, Ji Q, Ren J, Yan P, Wu Z, Wang S, Sun L, Wang Z, Li J, Sun G, et al. (2022). Large-scale chromatin reorganization reactivates placenta-specific genes that drive cellular aging. Dev. Cell 57, 1347–1368.e12. [DOI] [PubMed] [Google Scholar]
- 50.Ernst J, and Kellis M. (2012). ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marzi SJ, Leung SK, Ribarska T, Hannon E, Smith AR, Pishva E, Poschmann J, Moore K, Troakes C, Al-Sarraj S, et al. (2018). A histone acetylome-wide association study of Alzheimer’s disease identifies disease-associated H3K27ac differences in the entorhinal cortex. Nat. Neurosci 21, 1618–1627. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Amaral ML, Zhu C, Grieco SF, Hou X, Lin L, Buchanan J, Tong L, Preissl S, Xu X, et al. (2022). Single-cell epigenome analysis reveals age-associated decay of heterochromatin domains in excitatory neurons in the mouse brain. Cell Res. 32, 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caldwell AB, Liu Q, Schroth GP, Galasko DR, Yuan SH, Wagner SL, and Subramaniam S. (2020). Dedifferentiation and neuronal repression define familial Alzheimer’s disease. Sci Adv 6. 10.1126/sciadv.aba5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mertens J, Herdy JR, Traxler L, Schafer ST, Schlachetzki JCM, Böhnke L, Reid DA, Lee H, Zangwill D, Fernandes DP, et al. (2021). Age-dependent instability of mature neuronal fate in induced neurons from Alzheimer’s patients. Cell Stem Cell 28, 1533–1548.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu L, Sun T, Kobayashi K, Gao P, and Griffin JD (2002). Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol. Cell. Biol 22, 7688–7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miura Y, Tam T, Ido A, Morinaga T, Miki T, Hashimoto T, and Tamaoki T. (1995). Cloning and characterization of an ATBF1 isoform that expresses in a neuronal differentiation-dependent manner. J. Biol. Chem 270, 26840–26848. [DOI] [PubMed] [Google Scholar]
- 58.Liu Y, El-Naggar S, Darling DS, Higashi Y, and Dean DC (2008). Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development 135, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Speir ML, Bhaduri A, Markov NS, Moreno P, Nowakowski TJ, Papatheodorou I, Pollen AA, Raney BJ, Seninge L, Kent WJ, et al. (2021). UCSC Cell Browser: Visualize Your Single-Cell Data. Bioinformatics 37, 4578–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Granja JM, Corces MR, Pierce SE, Bagdatli ST, Choudhry H, Chang HY, and Greenleaf WJ (2021). ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet 53, 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.PsychENCODE Consortium, Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, Jaffe AE, Pinto D, Dracheva S, et al. (2015). The PsychENCODE project. Nat. Neurosci 18, 1707–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson EN, Burns TC, Hayda RA, Hospenthal DR, and Murray CK (2007). Infectious complications of open type III tibial fractures among combat casualties. Clin. Infect. Dis 45, 409–415. [DOI] [PubMed] [Google Scholar]
- 63.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P-R, and Raychaudhuri S. (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Polański K, Young MD, Miao Z, Meyer KB, Teichmann SA, and Park J-E (2019). BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19. 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bell AJ, and Sejnowski TJ (1997). The “independent components” of natural scenes are edge filters. Vision Res. 37, 3327–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fulco CP, Nasser J, Jones TR, Munson G, Bergman DT, Subramanian V, Grossman SR, Anyoha R, Doughty BR, Patwardhan TA, et al. (2019). Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat. Genet 51, 1664–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Risso D, Ngai J, Speed TP, and Dudoit S. (2014). Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol 32, 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He L, Davila-Velderrain J, Sumida TS, Hafler DA, Kellis M, and Kulminski AM (2021). NEBULA is a fast negative binomial mixed model for differential or co-expression analysis of large-scale multi-subject single-cell data. Commun Biol 4, 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Berhanu Lemma R, Turchi L, Blanc-Mathieu R, Lucas J, Boddie P, Khan A, Manosalva Pérez N, et al. (2021). JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 50, D165–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park YP, and Kellis M. (2021). CoCoA-diff: counterfactual inference for single-cell gene expression analysis. Genome Biol. 22, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barber RF, and Candès EJ (2015). Controlling the false discovery rate via knockoffs. aos 43, 2055–2085. [Google Scholar]
- 74.Wang Y, and Blei DM (2019). The Blessings of Multiple Causes. J. Am. Stat. Assoc 114, 1574–1596. [Google Scholar]
- 75.Zhu Z, Fan Y, Kong Y, Lv J, and Sun F. (2021). DeepLINK: Deep learning inference using knockoffs with applications to genomics. Proc. Natl. Acad. Sci. U. S. A 118. 10.1073/pnas.2104683118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jiang T, Li Y, and Motsinger-Reif AA (2021). Knockoff boosted tree for model-free variable selection. Bioinformatics 37, 976–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Candes E, Fan Y, Janson L, and Lv J. (2018). Panning for gold:’model‐X’knockoffs for high dimensional controlled variable selection. Journal of the Royal. [Google Scholar]
- 78.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ongen H, Buil A, Brown AA, Dermitzakis ET, and Delaneau O. (2016). Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics 32, 1479–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, and Plagnol V. (2014). Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giambartolomei C, Zhenli Liu J, Zhang W, Hauberg M, Shi H, Boocock J, Pickrell J, Jaffe AE, CommonMind Consortium, Pasaniuc B, et al. (2018). A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics 34, 2538–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH-Y, et al. (2015). An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Phipson B, Sim CB, Porrello ER, Hewitt AW, Powell J, and Oshlack A. (2022). propeller: testing for differences in cell type proportions in single cell data. Bioinformatics 38, 4720–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Johnson WE, Li C, and Rabinovic A. (2007). Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127. 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 86.Tan G, and Lenhard B. (2016). TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics 32, 1555–1556. 10.1093/bioinformatics/btw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-nucleus ATAC-seq, RNA-seq, and multiome data will be available through the AD Knowledge Portal on Synapse upon publication of this manuscript. The data will be available here: https://www.synapse.org/#!Synapse:syn52293426. The data are available under controlled use conditions set by human privacy regulations. To access the data, a data use agreement is needed. This registration is in place solely to ensure the anonymity of the ROSMAP study participants. A data use agreement can be agreed with either Rush University Medical Center (RUMC) or with SAGE, which maintains Synapse, and can be downloaded from their websites (https://adknowledgeportal.synapse.org/). Additional processed data, matrices, metadata, and integration are available https://personal.broadinstitute.org/bjames/AD_snATAC/, with the overview and detailed file explanation available at http://compbio.mit.edu/ad_epigenome/. Integrative visualization and exploration of the single-cell data are available with UCSC Cell Browser interface https://rosmap-ad-aging-brain.cells.ucsc.edu59.
Code for analysis, including snATAC and snRNA data processing, GWAS analysis, QTL calling are available at https://github.com/KellisLab/AD_regulome_analysis and Zenodo https://doi.org/10.5281/zenodo.8287248. The code for single-cell module detection for ATAC-seq, multiomics, and peak-gene linking are available at https://github.com/KellisLab/epiclust and Zenodo https://zenodo.org/record/8292785.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.