Abstract
Discoveries in human genetic studies have revolutionized our understanding of complex rheumatic and autoimmune diseases, including the identification of hundreds of genetic loci and single nucleotide polymorphisms that potentially predispose individuals to disease. However, in most cases, the exact disease-causing variants and their mechanisms of action remain unresolved. Functional follow-up of these findings is most challenging for genomic variants that are in non-coding genomic regions, where the large majority of common disease-associated variants are located, and/or that probably affect disease progression via cell type-specific gene regulation. To deliver on the therapeutic promise of human genetic studies, defining the mechanisms of action of these alleles is essential. Genome editing technology, such as CRISPR–Cas, has created a vast toolbox for targeted genetic and epigenetic modifications that presents unprecedented opportunities to decipher disease-causing loci, genes and variants in autoimmunity. In this Review, we discuss the past 5–10 years of progress in resolving the mechanisms underlying rheumatic disease-associated alleles, with an emphasis on how genomic editing techniques can enable targeted dissection and mechanistic studies of causal autoimmune risk variants.
Rheumatic diseases are highly complex and affect up to 7–10% of the general global population1. Characterized by chronic inflammation, intermittent flares and progressive tissue damage, these diseases can result in morbidity and mortality if left untreated. Some of the most common rheumatic diseases are caused by issues within the immune system, leading to autoimmunity and deterioration of the body. Although the aetiology differs between disorders, the risk of developing a rheumatic disorder is increased among those individuals with affected relatives; indeed, studies of monozygotic and dizygotic twins show that rheumatic diseases share a strong genetic component2.
Defining the genetic variants that increase the risk of rheumatic disease and the genes that the disease-associated variants influence will improve our ability to both diagnose and treat these complex conditions. For example, characterizing drug targets on the basis of human genetic data can increase the likelihood of developing a successful therapeutic agent3-6. In this regard, large-scale collaborative efforts over the past decade have conducted genome-wide association studies (GWAS) to identify hundreds of loci associated with autoimmune disorders7. These studies use genotyping information from millions of single nucleotide polymorphisms (SNPs) in thousands of affected individuals and thousands of unaffected individuals to pinpoint regions associated with disease. The findings of a particular SNP allele that is more (or less) frequent in individuals with a particular disease than in healthy individuals indicates that the SNP, or a nearby variant in tight linkage with that genomic location, contributes to disease risk (FIG. 1).
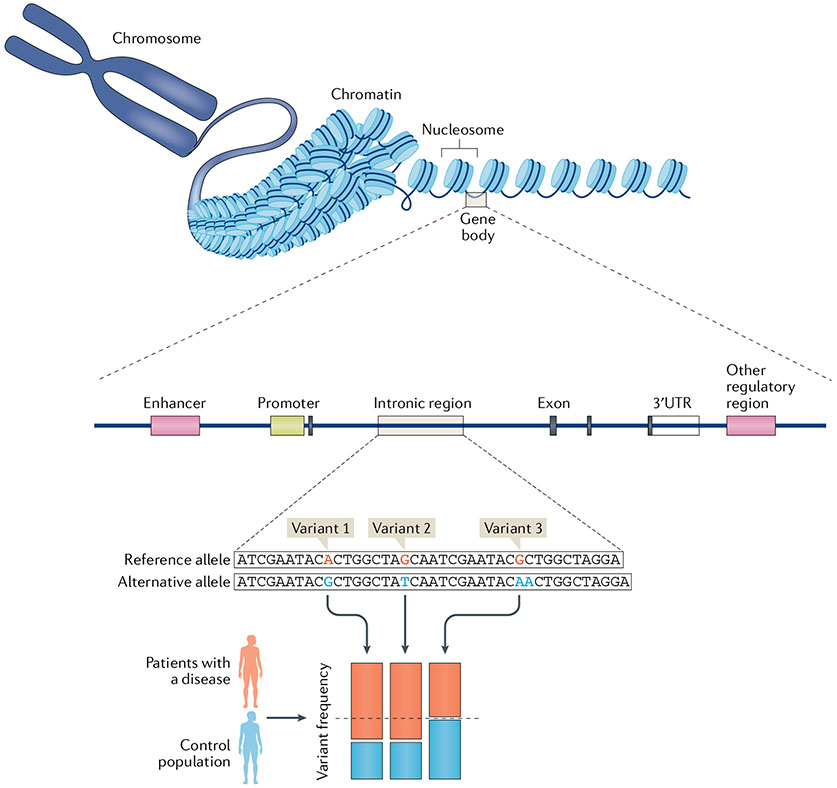
Fig. 1 ∣. Studying the genetics of rheumatic diseases.
The human genome is made up of billions of pairs of DNA and harbours thousands of protein-coding genes and well as other regulatory non-coding regions. DNA sequences are wrapped around histones, forming nucleosomes, for easy storage; an array of nucleosome form chromatin, which chain together to form chromosomes. Chromatin accessibility varies across the genome (depending on whether the chromatin has an open or closed conformation), governing the transcription and expression of the contained genes. Genes also have promoter, regulator and enhancer elements that control their expression. Particular variations or single nucleotide polymorphisms in DNA, located in both coding and non-coding regions, are linked to the development of rheumatic diseases. Genome-wide association studies identify potential causal variants and loci by comparing the frequency of particular variants in patients with a disease with that in a control population (such as in healthy individuals). In the example shown, variants 1 and 2, but not variant 3, are enriched in patients with the disease (implicating these variants in disease); here, two variants are found in linkage disequilibrium, meaning that they are generally inherited together as a block.
Despite the plethora of information gained from GWAS, the overall goal of assigning genetic mechanisms remains unexpectedly elusive. To achieve this goal, we need to ascribe causality for SNPs by identifying the affected genes and the downstream effects of the gene modulation on immune cell function. Experimental efforts to define allelic function over the past decade have been restricted largely to molecular biology assays and mouse models of disease. Both strategies are useful but have various limitations8-13. In vitro cellular assays might not retain the relevant chromatin context (that is, the chromatin organization present in the local tissue environment) and therefore cannot fully recapitulate disease, leading to technical noise. Mouse models are unable to recapitulate the full pathophysiology of human disease, including species-specific gene regulatory mechanisms. Fortunately, advances in genome editing and CRISPR–Cas technologies — the discovery of which led to Jennifer Doudna and Emmanuelle Charpentier being awarded the 2020 Nobel Prize in Chemistry — have opened up new opportunities and techniques for investigating and validating the genetics of rheumatic diseases.
At its root, CRISPR–Cas technology exploits the programmable specificity of RNA-guided nucleases14,15. Unlike previous iterations of programmable protein nucleases, CRISPR–Cas systems are guided to their target with easily tunable guide RNAs (gRNAs). Different versions of Cas proteins exist, with different functions and specifications. The most commonly used versions include Cas9, an RNA-guided DNA endonuclease, Cas12a, another RNA-guided DNA endonuclease that is more sensitive to A-T rich regions than Cas9, and Cas13, an RNA-guided RNA endonuclease. Countless other Cas proteins exist in nature and have been studied. The ease of specifying target sites practically anywhere in the genome along with the rapid democratization of these tools by the scientific community has made CRISPR–Cas easily accessible and highly versatile. Some applications of this technology include the deletion of particular genes, targeted homology-directed repair (HDR), precise nucleotide conversions and recruitment of transcriptional repressors and activators16,17. The basic principles of CRISPR–Cas editing have been reviewed in depth elsewhere16.
In this Review, we provide a detailed look at using genome editing to study the genetics of rheumatic diseases, with a focus on autoimmune rheumatic diseases. We discuss how computational and genomic editing approaches can be used to discover loci and genes associated with autoimmune conditions, identify regulatory regions, define causal variants and characterize critical cell types and cell states.
Dissecting risk loci
Traditional approaches.
Prior to the widespread utilization of GWAS, traditional approaches to understanding the genetics of rheumatic diseases focused on the investigation of candidate genes thought likely to alter immune function. Most of these candidate genes emerged from model systems, such as painstakingly created gene knockout mice. These models implicated a critical role for various protein-coding genes such as Stat4, Bach2, Ptpn22, Zap70, Foxp3 and Tnfaip3 (encoding A20), to name but a few, in the development of, or protection against, autoimmunity18-23. Some examples include Tfnaip3 knockout animals, which die prematurely because of uncontrolled and widespread inflammation, Ptpn22 knockout mice, which have increased levels of T cell activation, and Stat4 knockout mice, which are protected from arthritis induction18,20. Other subtler mouse models used cross-breeding of inbred strains to create congenic animals. In these studies, animal strains that were prone to the spontaneous development of autoimmunity were carefully bred with other strains. The resulting progeny were then genotyped to identify regions associated with the autoimmune phenotype, as has been done with New Zealand Black and White (NZB/W) mice in the investigation of systemic lupus erythematosus and non-obese diabetic (NOD) mice in the investigation of type 1 diabetes24,25. Although these early mouse studies were indispensable in discovering regions and genes that might contribute to disease, investigators recognized that not all findings translate to human disease.
To gain a comprehensive understanding of the human genetics of rheumatic disease, researchers have turned to large-scale GWAS to pinpoint variants and loci associated with autoimmunity. For example, an analysis of patients with ankylosing spondylitis, Crohn’s disease, psoriasis, primary sclerosing cholangitis or ulcerative colitis revealed extensive pleiotropy (shared genetic risk) among these diseases and identified 244 independent disease-associated loci26. Another meta-analysis of ten paediatric autoimmune disorders identified 27 disease-associated loci of genome-wide statistical significance that were enriched for loci implicated in T helper cell signalling27. Finally, in a study of 42 disorders that used data from the Biobank Japan project, researchers discovered 276 risk loci across 27 diseases, including some loci unique to the East Asian population7. Taken together, GWAS in rheumatic diseases have identified hundreds of loci that might be linked to disease and provide important genetic clues as to the mechanism of disease progression. However, despite the plethora of identified signals, associations from GWAS rarely identify the genes that are causal28. Additionally, rare (minor allele frequency <1–5%) variants that affect gene expression or function might also be missed by GWAS, as GWAS mainly analyse common variants29. Genomic editing can complement our discovery of genes involved in autoimmunity using both unbiased genome-wide knockout screens as well as targeted editing to elucidate function.
Genome editing of protein-coding regions.
To test the function of every known protein-coding gene and its possible contribution to autoimmune pathways in an unbiased fashion, researchers have employed CRISPR–Cas genome-wide screening assays. In these types of experiments, a pooled library of thousands of gRNAs is used to induce gene knockouts via the generation of indels (insertions and deletions), gene silencing with CRISPR interference (CRISPRi), or gene activation with CRISPR activation (CRISPRa) (FIG. 2). Libraries are virally packaged and transduced into cells followed by perturbation such as T cell receptor (TCR) stimulation, immune cell differentiation or drug treatment. After cell selection on the basis of cell surface expression, survival or another carefully chosen response condition, genomic DNA is extracted from the selected and non-selected (or the input) cell populations to sequence the integrated gRNAs and identify the targets that affect the cellular phenotype. This unbiased screening approach has the potential to discover and investigate the function of genes related to important immune processes such as T cell activation and proliferation, potentially identifying new links and discovering unknown functions. In the study of rheumatic diseases, this approach can be used to better understand the roles of genes implicated in disease by GWAS and discover previously unidentified targets.
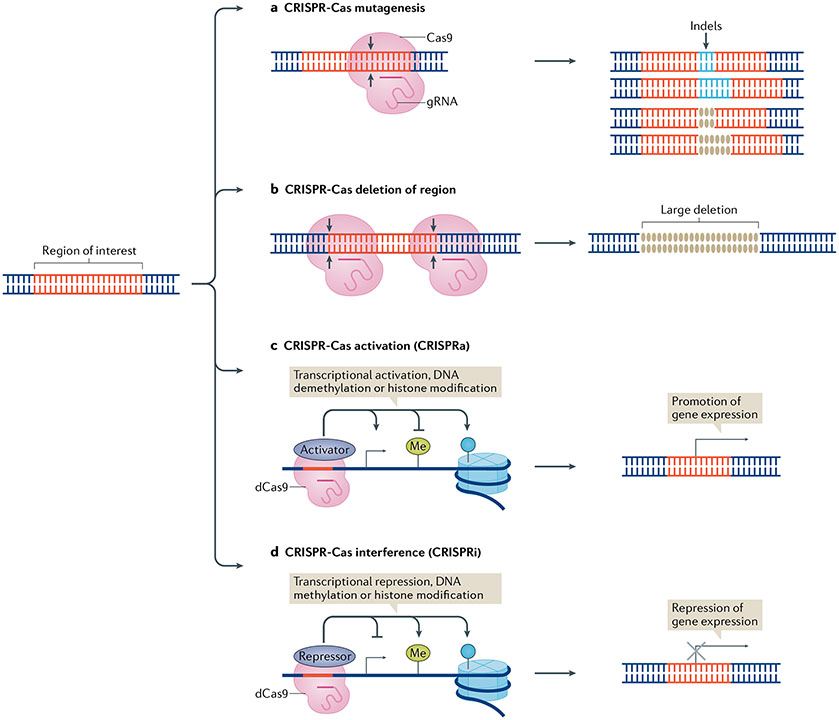
Fig. 2 ∣. Genomic editing to investigate regions of interest in rheumatic diseases.
A region of interest (for example, a gene or a regulatory non-coding region of interest) can be investigated using a number of genomic editing techniques. a ∣ CRISPR–Cas mutagenesis involves the generation of an array of small insertions or deletions (indels) in the region of interest following cleavage by the Cas nuclease. b ∣ Cas nuclease-mediated deletion can also be used to delete large sections of the region of interest. c ∣ CRISPR–Cas activation (CRISPRa) involves the fusion of proteins that activate gene expression, such as transcriptional activators, enzymes involved in DNA demethylation or histone modifiers that promote DNA accessibility, to a nuclease-deactivated Cas9 protein (dCas9) for targeted promotion of gene expression. d ∣ CRISPR–Cas interference (CRISPRi) involves the fusion of proteins that repress gene expression, such as transcriptional repressors, enzymes involved in DNA methylation (Me) or histone modifiers that epigenetically silence the area, to dCas9 for targeted repression of gene expression.
Various investigators have used this approach to screen for genes critical in immune cell activation and differentiation in cell lines and mice. For example, using a library of 250,000 gRNAs, one group of researchers investigated which genes can enhance or reduce CD69 expression following TCR stimulation in a cell line, the efforts of which led to the discovery of a previously uncharacterized regulator of TCR activation known as family with sequence similarity 49 member B (FAM49B)30. A separate study used a similar approach to study the TNF response following lipopolysaccharide stimulation in primary bone marrow-derived mouse dendritic cells, providing important insight into the genes involved in the regulation of this known autoimmune-related pathway31. CRISPR-based genome-wide screening approaches have also been used to detect genes involved in T cell differentiation. For example, in one study, a genome-wide library of gRNAs was virally transfected into naive T cells from Cas9-expressing transgenic mice that were then stimulated with a TCR agonist and IL-4 (conditions that induced T helper 2 (TH2) cell differentiation); the cells were then selected on the basis of the expression of GATA3 and other TH2 markers. This approach led to identification of various genes involved in TH2 differentiation, such as Pparg and Bhlhe40 (REF.32).
More recently, investigators have used Foxp3 reporter mice and targeted CRISPR screening to uncover the nuclear factors involved in tuning the expression of FOXP3, a critical master regulator of regulatory T (Treg) cells33. From this work, two novel modulators, Usp22 and Rnf20, were found to have opposing effects on Foxp3 expression. Taking this work a step further, the investigators also validated the role of USP22 in primary human Treg cells by knocking out the gene using CRISPR–Cas technology. Notably, Usp22 ablation in murine Treg cells exacerbated disease in experimental models of colitis and multiple sclerosis33. Together, this work highlights the utility of using CRISPR screening to identify new and relevant targets in autoimmunity.
Excitingly, these genomic screening studies have been extended to primary human immune cells (BOX 1). For example, sequential delivery of a gRNA-encoding library by lentiviral transduction and Cas9 by protein electroporation has been used to conduct a genome-wide screen in human CD8 T cells. By measuring the proliferation of the cells following restimulation with CD3 and CD28, the researchers could identify various genes that affect TCR signalling. Notably, CRISPR–Cas mediated disruption of UBASH3A and TNFAIP3 genes, two genes implicated in autoimmune disease development, resulted in enhanced proliferation of the cells, suggesting that these genes are involved in the regulation of T cell activation in humans and providing potential insight into their role in disease34.
Box 1 ∣. Genomic editing and cell lines.
The majority of research cited in this Review has been conducted in immortalized cell lines. This choice of cell lines is one of practicality. Cell lines are easier to use, capable of being propagated at the single-cell level to create clones and are amenable to complex and sequential genetic manipulations; CRISR–Cas editing efficiency is also generally higher in cell lines than in primary cells. However, cell lines also have the disadvantage of often being highly mutated and genomically unstable, reflecting, in many cases, their origin from human cancers. Cell lines are often constitutively activated, and therefore cannot fully recapitulate physiological conditions118,119. Fortunately, CRISPR–Cas technologies are largely applicable to primary human immune cells as well as haematopoietic stem cells, and studies in primary immune cells are emerging, as highlighted whenever possible in this Review. We expect that in the near future, CRISPR–Cas-based genomic editing in primary immune cells will become a standard practice.
A similar approach, though limited in scope, can be used to investigate the function of individual genes implicated in autoimmunity. For example, a group of investigators used CRISPR–Cas editing to study the function of PTPN22, PTPN2 and ZAP70 (all implicated in the development of autoimmunity) in primary human CD4 T cells. The results of this study highlighted the role of these molecules in IL-2 signalling and TCR activation, similar to findings in mouse models35. A single-gene knockout approach to autoimmunity-associated genes can also be applied to other primary human lineages, including other T cell subsets, dendritic cells, B cells or natural killer cells36-39. For example, one study that used this approach found that knockout of IRF4, a gene with a well-established link to autoimmune susceptibility, in primary dendritic cells can enhance cell activation while attenuating inflammatory nuclear factor κB (NF-κB) signalling38.
Hence, the investigation of gene function in animal models and primary human cells has been revolutionized by genomic editing tools. As the field continues to move forwards, directly studying the function of genes in primary human cells will become easier and more reliable, paving the way for direct and detailed investigations of cells from patients. These types of studies enable us to take an unbiased examination of all genes in diverse human cell types and uncover as yet unknown networks and potential therapeutic targets.
Identifying regulatory regions
Traditional approaches.
The studies described so far have broadly helped to find disease-associated genes and identify their roles in critical immune responses such as T cell proliferation and differentiation. Yet, variation between individuals is rarely the result of complete gene deletion but instead reflects SNPs and other genetic variants, such as indels and nucleotide substitutions. Although GWAS have identified more than 10,000 trait-associated variants in hundreds of loci, most loci lie in non-coding regions and are of unknown function40-42. Pruning and identifying potentially causal variants can be more effectively achieved by determining which regions have a regulatory function. One strategy is to look to epigenomic data and other functional genomic annotations, such as those being produced by the Encyclopaedia of DNA Elements (ENCODE) project43. Data on chromatin configuration, histone modifications and transcription factor-binding can be used to pinpoint functionally active spots in the genome and co-localize variants to these regions.
Examples of techniques that annotate the genome include traditional chromatin immunoprecipitation sequencing, DNase I hypersensitive sites sequencing and chromatin capture assays, which can be used to find regions of transcription factor binding and chromatin configuration44. Other more recently developed techniques, such as cleavage under targets and tagmentation (CUT&Tag) sequencing, cleavage under targets and release using nuclease (CUT&RUN) sequencing and assay for transposase-accessible chromatin using sequencing (ATAC-seq), have augmented our ability to annotate the genome with a higher accuracy and lower cell input45-48. Computational tools take advantage of this wealth of information to interrogate GWAS signals. Epigenomic annotations have been used to predict causal SNPs and have implicated shared CD4 T cell-related pathways across many autoimmune disorders49-53. Other tools, such as the inference and modelling of the phenotype-related active transcription (iMPACT) annotation tool have attempted to integrate and interrogate all known annotations to better identify cell type-specific causal autoimmune risk variants40. However, all of these approaches rely on making inferences and cannot directly confirm functional regions. A better strategy would be to discover the segments within the genome with a regulatory function using direct experimental approaches, such as genome editing.
Genome editing of regulatory regions.
Genome-editing techniques now enable us to identify functional regions in non-coding elements experimentally (FIG. 2). Such research can be performed using CRISPR–Cas screening or targeted deletion with Cas nucleases, in a similar fashion to that described in the previous section on “Dissecting risk loci” One of the first uses of pooled CRISPR–Cas screening in a non-coding region was a mutagenic screen that was applied to a BCL11a enhancer. The transcription factor BC11A has been reported to control the expression of fetal hemoglobulin and is therefore a potential therapeutic avenue for the treatment of sickle cell disease54. In this seminal work, a tiled lentiviral library of gRNA was targeted to this enhancer to systematically create deletions along the entire length of the region in a human cell line. In this way, the investigators could broadly identify nucleotide positions in the enhancer regions that affected gene expression, thereby resolving the region55. Similar CRISPR–Cas screening approaches that systematically interrogate a genomic region have been used to uncover regulatory elements in the four major pluripotency genes (Tdgf1, Zfp42, Nanog and Rpp25) in mouse embryonic stem cells56. In another study, rather than starting with a large, unknown genomic region, the researchers used an alternative method of first computationally identifying all the genome-wide binding sites of p53 and ESR1, and then using a screen to validate these regulatory regions and the downstream genes of interest regulated by these regions57.
Similar to editing individual genes, CRISPR–Cas9 editing can also be applied in a more targeted fashion to interrogate predicted enhancer regions on a case-by-case basis as opposed to a screening format. For instance, this targeted approach was used to investigate a putative enhancer region around the SNP rs13239597 in close proximity to IRF5; the investigators used CRISPR–Cas9 nucleases to delete a 1,000-base pair region around the variant in three different cell lines, which caused cell type-specific changes in IRF5 and TNPO3 expression58.
Although CRISPR–Cas systems were initially used to generate double-stranded DNA breaks to induce indels, researchers quickly adapted the technology by fusing Cas proteins to other modifiers, such as transcriptional activators, repressors and deaminases, to name a few59. These methods have greatly expanded the CRISPR–Cas toolkit to enable targeted genome investigation of regulatory regions. For example, in one study, a CRISPRa screen, which fused deactivated Cas9 with transcriptional activators such as VP64, was used to interrogate two autoimmune risk loci, CD69 and IL2RA. In this setup, the binding of the CRISPRa complex results in the recruitment of transcriptional activation machinery, enabling the identification of promoter and enhancer regions. By using a library of gRNA that saturates the genomic region around CD69 and IL2RA in Jurkat T cells, the researchers identified various enhancer areas, including one for IL2RA that overlapped with a known autoimmune disease-associated variant. Follow-up research was conducted on the newly recognized IL2RA enhancer by investigating mice that had been modified to carry the mutated variant; notably, this modification had a clear effect on T cell stimulation60. Hence, this study is a good example of CRISPR screening being applied to an autoimmunity-associated locus to pinpoint causal variants and investigate the functional outcome of these changes on T cells.
A complementary approach to CRISPRa is CRISPRi, which fuses deactivated Cas9 with repressors of gene expression such as the histone methylator KRAB. When the CRISPRi complex is targeted to a region of interest with programmable gRNAs, the associated KRAB proteins effectively silence the region by methylating nearby histones. Similar to CRISPRa, CRISPRi has been used experimentally to investigate autoimmune risk loci. An early example of this approach applied a targeted CRISPRi screen to investigate the functionality of all DNase I sites around the β-globin locus control region and HER2 (REF.61). DNase I sites mark areas of chromatin accessibility, indicating that a region is similarly available to enhancer or repressor proteins and thus potentially important for gene regulation. Some studies have used a combination of both CRISPRa and CRISPRi to explore autoimmune risk loci. For example, in one study exploring the locus TNFAIP3, researchers designed a gRNA library that targeted all areas of open chromatin around the gene and performed a CRISPRi screen62. A CRISPRa screen of the computationally identified variants was then performed in the region to find regulatory areas of overlap, that is, those areas that resulted in a loss or gain of expression when targeted by both CRISPRa and CRISPRi machinery. In comparison to other techniques, including the in vitro massively parallel reporter assay, CRISPRi had the highest likelihood of detecting true hits62.
Many of the approaches mentioned so far have relied on reporter cell lines or cell surface markers to measure changes in gene expression and facilitate screening. To move beyond cell lines and target any gene, as opposed to just genes with validated antibodies, investigators have paired CRISPRi screening with fluorescent in situ hybridization. This novel approach enabled the rapid testing of over 3,500 enhancer regions in 30 different genes63.
CRISPR–Cas screening of thousands of gRNA is a powerful tool for resolving large Mb regions of the genome and generating new hypotheses. However, such scale can be prohibitively expensive and technically challenging. As an alternative, these techniques can also be applied in a more targeted, hypothesis-testing manner to investigate precise regions of interest (<1 kb) of the genome on a case-by-case basis. As an example, to study a potentially causal distal enhancer in a risk locus associated with CD4 Treg cell-mediated suppression of colitis, researchers applied targeted CRISPRa in primary human CD4 T cells only to a handful of pre-selected variants in the region of interest64. Using this focused approach, they discovered that only gRNAs near rs11236797 could induce changes in the expression of the nearby gene GARP. In this way, the researchers were able to decipher a known colitis-associated region, identify the downstream gene and provide a direct link to Treg cell function.
In the past 5 years, researchers have begun to couple single-cell RNA sequencing (scRNA-seq) with CRISPR screening to rapidly test multiple regions and measure corresponding gene expression in a highly parallelized system. For example, a recent publication used these new methods to test approximately 6,000 putative enhancers with a single cell CRISPRi screen. Using this highly multiplexed technology, the researchers profiled approximately 300,000 single cells and were able to experimentally confirm functional versus non-functional enhancer regions in the genome. This approach allowed the investigators to then discern which genome annotations are most associated with functional enhancer regions, identifying p300, histone H3 acetylation at lysine 27 (H3K27ac) and cell-line-specific transcription factor binding as the most important signatures65.
Defining non-coding variation is a major challenge in the investigation of rheumatic diseases. In the near future, as these large-scale genomic studies continue to rapidly increase in size and scope, our ability to accurately annotate the human genome will considerably improve. In turn, such advances will enhance our ability to confidently resolve signal from noise in studying the genetics of complex diseases and unveil the full potential of genetics research.
Investigating autoimmune risk variants
The techniques described so far in this Review can identify a relevant immune gene or locus and confirm the location of possible SNPs in a validated regulatory region. But to truly pinpoint and understand the mechanisms of these causal variants, further work is required. This work might include investigation of rare diseases to elucidate critical protein-coding variants and genes, computational fine-mapping studies to narrow down potential hits, traditional molecular biology approaches and genomic editing tools to directly assess variant function in cells of interest.
Rare diseases.
For some monogenic autoimmune disorders, researchers have identified causal variants in protein-coding regions, leading to a better understanding of variant and gene function. For example, single mutations in various genes are known to cause Aicardi–Goutières Syndrome, an autoimmune condition characterized by encephalitis with skin involvement, including mutations in TREX1, RNASEH2A, RNASEH2B and RNASEH2C. All these genes are involved in the regulation of nucleic acid degradation and contribute to excess inflammation66. As another example, the monogenic autoimmune disorder familial Mediterranean fever is typified by mutations in MEFV that result in aberrant activation of the pyrin inflammasome66. Mutations in WISP3 have also been linked to progressive pseudorheumatoid dysplasia, a progressive skeletal disorder that results in swelling and pain in multiple joints66. Other notable examples include autoimmune conditions resulting from mutations in AIRE, FAS and BACH2 (REFS21,67,68). Although these conditions are rare, they can provide considerable insight into disease pathophysiology and exemplify how a single variant can contribute to disease.
Computational approaches for prioritizing variants.
For non-coding variants, particularly common variants identified in GWAS, defining the causal variants can be more difficult than with monogenic disease-associated protein-coding variants. Within a single locus, multiple variants might be present in tight linkage with each other, some or all of which might explain the disease association. Additionally, multiple non-coding SNPs within a locus might work together to yield a particular phenotype, as exemplified by various non-coding SNPs in STAT4 that are associated with juvenile idiopathic arthritis69. Computational fine mapping is one way of identifying the most probable causal variants in complex polygenic rheumatic disorders. Fine mapping can refine the association signals in genotyping data via statistical methods to create credible sets of SNPs with the highest probability of being causal, which can then be investigated in further detail in functional studies70.
Another common approach to investigating the function of autoimmune risk alleles is expression quantitative trait loci (eQTL) analysis, which uses both genotyping and mRNA data to uncover DNA variants associated with altered gene expression. Large-scale studies of this kind have been conducted in cell lines and tissues to detect cell-specific and tissue-specific variants that might control gene transcription71-73. For example, in one eQTL analysis of whole blood, researchers could link 39% of 250 autoimmune disease-associated SNPs identified by GWAS to the expression levels of nearby genes74. Taking this approach a step further, another eQTL study found that some SNPs (12% of the eQTL SNPs analysed) had lineage-specific effects, highlighting the importance of studying the appropriate cell types and tissues75. Although eQTL studies can identify associations between variants and the expression of genes, linkage disequilibrium can make it difficult to disentangle disease-causing SNPs from incidental SNPs within an inherited block, limiting the interpretation of the results.
Together, fine mapping and eQTL analyses can provide a short list of potentially causal variants in autoimmune risk loci. However, experimental validation remains indispensable, both for proving causality and for deciphering the downstream mechanisms of action that contribute to disease.
Traditional approaches for functional validation.
Traditionally, functional validation of causal variants began with the study of individuals with a particular variant of interest. In this type of study, healthy individuals and patients with a particular disease and genotype at a variant of interest would be carefully selected and the relevant immune cells of these individuals profiled for differences in gene expression or other phenotypes76. Unfortunately, in this type of analysis, controlling for other genetic variants as well as differences in cell states is difficult, as such differences might be caused by environmental stimuli. Alternative molecular biology approaches are available that use synthesized oligonucleotides that encompass the variant of interest and the flanking genomic sequences to investigate functionality. These assays include electromobility shift assays that test the interactions of variants with nuclear material containing DNA-binding proteins, luciferase assays that test the ability of a variant to promote the expression of a reporter gene and affinity precipitation assays that similarly test the interaction of variants with DNA-binding proteins in nuclear material following magnetic pulldown77-79. Unfortunately, these techniques also have limitations, including a high level of technical noise and an inability to recapitulate the transcription factor interactions, histone modifications and chromatin organization found in the native cell state. To overcome these issues, researchers have developed sophisticated genome editing tools for investigating genetic variants, which can edit specific genomic regions within their native chromatin context (FIG. 3).
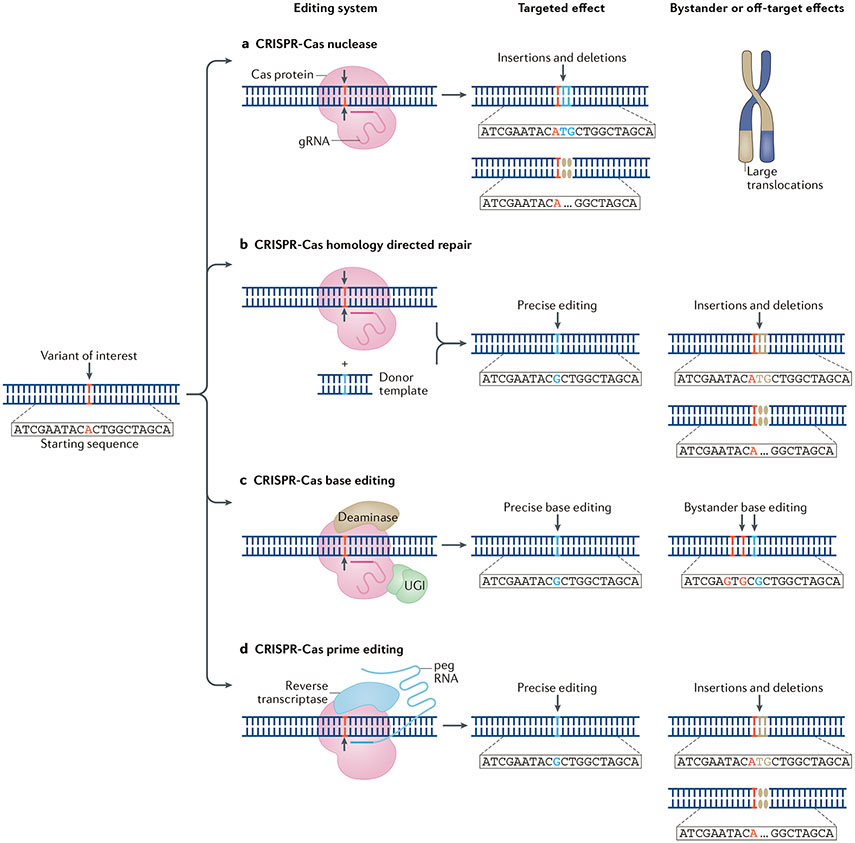
Fig. 3 ∣. Genomic editing to investigate variants of interest in rheumatic disease.
A variant of interest can be investigated using a number of genomic editing techniques. a ∣ The area of interest can be altered with small deletions or insertions using CRISPR–Cas nucleases. b ∣ Targeted homology-directed repair with CRISPR–Cas nucleases and a donor template can be used to precisely edit the variant of interest. c ∣ CRISPR–Cas base editors (comprising a nuclease-deactivated Cas9 protein (dCas9) or Cas nickase fused with an adenine or cytosine deaminase and a uracil DNA glycosylase inhibitor (UGI)) can be used to directly edit a base of interest. d ∣ CRISPR–Cas prime editors (comprising a Cas endonuclease or Cas nickase fused with a reverse transcriptase that is programmed with a prime editing guide RNA (pegRNA)) can be used for precise editing of the target site. For each approach, bystander or unwanted off-target effects can occur (as shown).
Genomic editing for functional variant validation.
The simplest application for CRISPR–Cas systems in the study of autoimmune risk variants is the generation of indels in close proximity to the SNP of interest. For example, by using such an approach to investigate six non-coding SNPs in close proximity to an HLA-DQB1 allele associated with type 1 diabetes, researchers could narrow down this list to a single SNP, rs71542466, which had an effect on gene and protein expression of HLA-DQB1 (REF.80). Similarly, CRISPR–Cas indels were used to investigate rs17622517, a variant in an enhancer region of a gene associated with autoimmune susceptibility, IRF1. Clonal selection of those cells with CRISPR–Cas-mediated deletions around the variant found that such deletions had a clear effect on IRF1 expression81. A similar approach was taken in the study of a BLK insertion variant associated with SLE, rs558245864; CRISPR–Cas9-mediated 6-bp and 18-bp deletions around this variant led to decreased accessibility of the site to the transcription factor CCCTC-binding factor (CFCF) and decreased BLK expression, highlighting the importance of this region and variant82.
Another major use of CRISPR–Cas genomic editing in variant validation is targeted editing. For this approach, CRISPR–Cas technology and exogenous template material can be used to induce double-strand breaks and HDR. Alternatively, CRISPR–Cas tools that incorporate modified base or prime editors, which have the capacity to directly mutate DNA without inducing double-stranded breaks, can be used.
For example, to validate the effects of the SNP rs71542466 on HLA-DQB1 expression, a CRISPR–Cas-guided HDR-based approach was used to mutate the variant from the reference G allele to the disease-associated C allele in a T cell line80. The clones with the resultant mutation had altered expression levels of HLA-DQB1, providing a causal link between the variant and the induced change80. In another study of the autoimmunity-linked gene TNAIP3, CRISPR–Cas-guided HDR repair was used to investigate a protein-coding variant associated with rheumatoid arthritis, rs2230926, in a monocyte cell line. Surprisingly, the mutation had no effect on NF-κB signalling but did increase the expression of PADI4 (REF.83). Importantly, CRISPR–Cas HDR methods can also be applied to primary human cells ex vivo. For example, non-viral CRISPR–Cas HDR methods have been used to correct coding mutations in IL2RA (encoding the soluble IL-2 receptor-α subunit, also known as CD25) in cells from a family of patients with a rare monogenic immune dysregulation disorder84. Amazingly, genomic editing of the primary CD4 T cells from the patients could restore the cell surface expression of CD25.
Base editors consist of Cas proteins fused with deaminases and enable a more precise way of mutating single nucleotides than CRISPR–Cas-mediated HDR. Instead of creating double-stranded breaks in the genome, these systems open the region of interest via Cas complex binding and then promote the mutation of selected nucleotides using the fused proteins. Current iterations of base editing enable both C to T and A to G transitions in a customizable editing window17,85-87. Base editors have been used to target pathogenic mutations in a number of disorders. For example, this tool has been used to target mutations in APOE4, MPDU1, HGB, HBB and DMD in cell lines, primary mouse cells and patient-derived fibroblasts85-88. The use of base editors has more recently been applied to human haematopoietic stem cells. For example, by using this approach on haematopoietic stem cells from a patient with sickle cell disease, researchers were able to edit the BCL11A enhancer and restore the expression of fetal haemoglobin in differentiated erythrocytes, thereby reducing the number of sickled cells89. Although not yet tested for autoimmune risk variants, base editors have also been applied to primary human T cells to introduce multiple simultaneous gene knockouts90.
Although incredibly effective and powerful, the current version of base editors can only induce a limited set of mutations, either C to T or A to G. To fill this gap and allow for investigation of other transitions, for instance C to G, an alternative approach has been developed, called prime editing. For this approach, Cas proteins are fused to a reverse transcriptase. The resulting complex is programmed with a prime editing guide RNA (pegRNA), which functions as both the targeting gRNA and the donor template for the reverse transcriptase enzyme. This technique has been used to repair pathogenic variants in HBB, HEXA, PRNP and DNMT1 in various cell lines including HEK293T cells, K562 cells, U2OS cells, HeLa S3 cells and mouse primary cortical neurons91. Using this technique, it might be possible to repair practically any variant in the human genome, including small insertions and deletions, although the editing efficiency can be low and variable91.
Resolving cell heterogeneity
Single-cell assays.
Another essential piece of the puzzle in unscrambling the genetics of autoimmune diseases is identifying critical cell populations and states that govern disease risk. Abundant evidence shows that immune cells are critical contributors to rheumatic disorders. In the past 10 years, genetic approaches aimed at identifying unique cell-type chromatin and transcription factor signatures, such as co-localizing GWAS hits with ATAC-seq, have been applied to decode which cell types and states might contribute to disease49,92-96. However, these genetic approaches rely on data collected from cell lines or bulk sorted samples and therefore fall short in capturing the true heterogeneity in the immune compartment of these disorders. Hence, more recently, large-scale unbiased single-cell investigations have begun to characterize relevant disease cells and states.
Single-cell technologies, such as droplet-based RNAseq and mass cytometiy, provide a comprehensive view of the main cellular states within diseased tissue and are often able to produce tens to thousands of readouts per cell97. In one single-cell analysis of synovial tissue from patients with rheumatoid arthritis or osteoarthritis, the heterogeneity of the CD4+ T cell compartment was resolved at a single-cell level, resulting in the discovery of a new lineage of cells, known as T peripheral helper cells, associated with disease98. In a parallel single-cell analysis of renal biopsy samples from patients with SLE, researchers were able to characterize an interferon and fibrotic mRNA signature in tubular cells that was associated with failure to respond to treatment96. A single-cell atlas of the human colon during ulcerative colitis has also been generated using colon mucosa biopsy samples, which identified 51 distinct cell types99. In this analysis, inflammatory fibroblasts, inflammatory monocytes, microfold-like cells and T cells that co-express CD8A and IL17A were all expanded in the colons of patients with ulcerative colitis compared with the colons of healthy individuals. Interestingly, mapping the various risk alleles associated with ulcerative colitis to particular cell types enabled the researchers to discover that many of the risk alleles were cell type-specific99. In general, the discovery of a cell population within a diseased tissue supports a potential role for that cell type in pathogenesis, and thereby implicates underlying genetic mechanisms in that cell type, although further experiments are required to confirm causality. Once such causal cell populations are determined, assessing the molecular phenotypes within these cell populations is an important next step.
With the ever-expanding scope and scale of scRNA-seq, single-cell eQTL analysis represent a promising approach to relate heterogeneous features of the immune cell compartment to variants of interest. In this regard, a new consortium was formed in 2020 (the single-cell eQTLGen consortium) that is aimed at identifying the cellular contexts in which disease-causing variants affect gene expression100.
Genomic editing in single cells.
Genomic editing can be paired with scRNA-seq to simultaneously assess the effects of perturbation on the heterogeneous immune cell compartments. As an example of this approach, researchers have used CRISPR–Cas screening in combination with scRNA-seq to assess the response of mouse bone marrow-derived dendritic cells to lipopolysaccharide at a single-cell level. By targeting 24 known transcription factors through CRISPR-based genetic perturbations, the researchers discovered four distinct groups of transcription factors that uniquely affected cell function and differentiation101. An extension of this approach paired CRISPR–Cas screening with ATAC-seq to measure the effects of CRISPR perturbation (using either CRISPR knockout or CRISPRi-based approaches) of transcription factors, chromatin-modifying factors, and non-coding RNAs on chromatin accessibility in human B cell lines and primary keratinocytes102. The researchers discovered how these transcription factors and other elements regulated B cell and keratinocyte cell states and might alter the chromatin accessibility of disease-associated variants. In a different example of paired CRISPR–Cas screening and scRNA-seq, this method was applied to TCR-stimulated Jurkat T cells to assess the function of six regulators and 23 transcription factors. The investigators discovered that CRISPR-mediated targeting of ETS1, RUNX1 and GATA3 reduced the viability of the cells and that CRISPR-mediated targeting of LCK and ZAP70 prevented TCR stimulation103. Finally, this approach has been cleverly modified and extended to primary human T cells through sequential use of gRNA-expressing lentiviruses and nucleofection of Cas proteins. Using this technique, the investigators characterized gene programmes controlled by important regulators of human T cell proliferation; notably, the ablation of some of these programmes in T cells results in enhanced killing of cancer cells in vitro34.
Although profiling total RNA or chromatin accessibility can be informative in elucidating changes in total gene expression in an unbiased fashion, this approach can also be costly. An alternative approach, hybridization of probes to RNA for sequencing (HyPR-seq), involves targeted scRNA quantification in combination with CRISPR–Cas screening. In this approach, a library of gRNA is transfected into cells that are then fixed and hybridized with RNA probes, before being processed into droplets for library generation and sequencing. By using targeted RNA probes, this approach allows for the highly sensitive detection of selected genes of interest and identification of the gRNA. Using HyPR-seq, researchers have been able to target regulatory regions around GATA1 and detect corresponding changes in gene expression104.
Finally, researchers have also developed a CRISPR knockin targeting approach that was combined with single-cell analysis to look at the effect of overexpressing various gene constructs in primary human T cells105. In this approach, a library of knock-in donor material was integrated into the genome with CRISPR–Cas to assess the effects of each knock-in DNA construct on T cell function. The correctly integrated gene constructs were identified by sequencing unique molecular barcodes introduced from the donor DNA, and the effects on T cell function could be analysed using single-cell RNA sequencing in the same cells. These experiments helped to discover a chimeric TGFBR2-41BB receptor that can promote clearance of a xenotransplant solid tumour model when knocked into human T cells along with an appropriate TCR. For rheumatic diseases, this approach can be used to overexpress candidate genes implicated in autoimmunity, complementing the previously described gene knockouts, to directly study function.
Overall, these single-cell approaches help to provide a full picture of the immune response in rheumatic disease. Paired with genomic editing, these techniques enable the investigation of rheumatic disease genes and variants at-scale in heterogeneous immune cell populations.
Future directions and limitations
Additional advances in genomic editing have helped to propel the potential and applicability of these tools. Cas proteins require particular protospacer adjacent motif (PAM) sites to initiate binding of the editing complex. Hence, several groups have focused on discovering and characterizing alternative Cas proteins with different PAM restrictions to broaden the range of possible targets106. Researchers have even been able to engineer nearly PAM-less Cas proteins through directed evolution107-111.
A promising alternative to DNA editing is RNA editing with Cas13 nucleases. Unlike the other variants of Cas, these proteins target RNA and not DNA, making it possible to target RNA molecules without potentially causing dangerous genomic mutations15,112,113. An interesting adaptation of this approach fuses deactivated Cas13 nucleases with adenosine deaminase acting on RNA type 2 (ADAR2), which enables direct mutagenesis of RNA, similar to the base editors discussed that target DNA. This approach has been applied in a cell line to correct two pathogenic G to A mutations in AVPR2 and FANCC, with modest efficiency112. Alternatively, CRISPR-free systems might represent a different route all together; indeed, a study in 2020 unveiled the newly developed CRISPR-free, transcription activator-like effector (TALE)-based editing system that was used to base-edit mitochondrial DNA for the first time114.
Genomic editing has various limitations; in particular, these tools can have off-target and bystander effects that might convolute results (TABLE 1). For example, cleavage by Cas nucleases induces a wide range of somewhat predictable mutations around the target site, but Cas binding and cleavage can also occur in off-target regions. Base and prime editors similarly have off-target effects but are also prone to both bystander-editing (editing of nearby non-target nucleotides) and, less frequently, unwanted insertions and deletions. As one group reported, the BE3 cytosine-targeted base editor, but not Cas9 or ABE7.10 adenosine-targeted base editors, was 20-fold more likely to induce random single nucleotide mutations in the genome than the spontaneous mutation rate115. Another important consideration is that base-editors edit a window of C or A nucleotides in the target site, which could make correcting a single nucleotide impossible, depending on the flanking sequences and the variant of interest. Finally, the discussed Cas13 nucleases that directly target RNA molecules are also reported to often induce transcriptome-wide off-target RNA mutations113. Optimization of Cas proteins, intelligent gRNA design and the use of multiple gRNA can offset some of these effects; however, the limitations and challenges of these technologies are important to keep in mind as we move towards their therapeutic application.
Table 1 ∣.
Limitations of CRISPR-Cas editing systems
| Application | Genomic editing technique | Advantages | Disadvantages |
|---|---|---|---|
| Identifying and investigating autoimmune-associated loci and genes | CRISPR–Cas-mediated genome wide screening30-34 | All annotated genes can be targeted | Limited to cell lines |
| Can discover cell type-specific effects | Requires a functional outcome (for example, proliferation) | ||
| CRISPR–Cas-mediated knockout of individual genes37,38 | Function can be assessed | Low throughput | |
| Amenable to primary immune cells | |||
| CRISPR–Cas-mediated activation (CRISPRa) or interference (CRISPRi) of gene expression61,64 | Easily multiplexable | Currently limited to cell lines | |
| Identifying autoimmune regulatory regions | CRISPR–Cas-mediated activation (CRISPRa) or interference (CRISPRi) of regulatory region60,62,63,65 | Can test any region of the genome | Effects might be context-dependent and require multiple cell lines for verification |
| CRISPR–Cas-mediated deletion or mutagenesis of regulatory regions55 | |||
| Identifying causal variants | CRISPR–Cas-mediated mutagenesis of causal variants58 | Amenable to primary immune cells | Induced deletions, insertions and substitutions are random and do not recapitulate variant changes |
| Some variants cannot be directly targeted | |||
| CRISPR–Cas-mediated homology directed repair80,83 | Can directly change reference allele to an alternative allele | Bystander mutations | |
| CRISPR–Cas-mediated base editing85-90 | Not all mutations are possible | ||
| CRISPR–Cas-mediated prime editing91 | Low efficiency | ||
| Linking variants to causal immune cell types | CRISPR–Cas-mediated editing paired with single-cell technologies101-105 | Can be used to directly identify cell populations of interest in rheumatic disorders | Expensive and prone to drop-out in gene expression |
| Can assess the function of a particular variant in a broad range of cell populations simultaneously |
Conclusions
The ease of use, customizability and accessibility of genomic editing technology has expedited its use in a number of applications, particularly for cell therapies in cancer and rare monogenic disorders. However, in the study of polygenic autoimmune diseases and associated variants, successful application remains scarce. The studies highlighted in this Review showcase how genomic editing can be used to identify and validate autoimmune disease-associated loci, genes and variants.
Practically, for rheumatologists and patients with rheumatic disease, genomic editing has potential in the development of new cellular therapies. Promising applications include the editing of stem cells to promote tissue regeneration and re-shape cytokine responses, the induction and strengthening of regulatory responses via autologous Treg cell therapies and the correction of pathogenic mutations in patients with monogenic diseases116,117.
As the field continues to grow, the applicability, scale and precision of genome editing technologies should continue to improve. All these advances in genomic editing will undoubtedly continue to resolve the causal genes, regions and SNPs responsible for complex polygenic autoimmune diseases. In the future, we anticipate that genomic editing directly in primary immune cells will become a major focus for proving causality and a powerful step towards defining associations between genotype and phenotype in immune cells of interest. Additionally, a renewed emphasis on multiplexed and multi-omic analyses should enable the simultaneous investigation and experimental validation of multiple variants or regions at the same time. Understanding how these variants interact individually and together to contribute to the development of complex and polygenic disorders represents the next frontier in genetics and genomic editing.
Overall, genomic editing has shown great promise in the study of the genetics of rheumatic diseases and remains the ideal approach for rapidly experimentally validating findings directly in primary human immune cells. One way or another, genomic editing tools are here to stay and will only become more accessible with time as researchers continue to adopt and expand these approaches in future studies.
Key points.
Hundreds of autoimmune risk loci have been discovered in coding and non-coding regions of the genome; however, their function and the causal alleles functioning within these loci have been difficult to discern.
Advances in genomic editing have made it possible to quickly and effectively investigate autoimmune disease-associated loci and variants using a number of approaches in both cell lines and primary cells.
CRISPR–Cas genomic editing can be used to induce insertions and deletions, correct precise mutations and induce epigenetic changes to investigate loci and variants associated with rheumatic diseases.
CRISPR–Cas screening approaches are effective tools for whole-genome investigation of autoimmune disease-related genes and detailed resolution of autoimmune risk regions.
Resolving the heterogeneity of cell types in rheumatic disorders with unbiased single-cell technologies is critical to understanding the genetics of disease.
Acknowledgements
S.R. is supported by funding from the National Institutes of Health (UH2AR067677, U01 HG009379, 1R01AR063759). Y.B. and S.R. are funded by the Broad Institute through the Scientific Projects to Accelerate Research (SPARC) programme and an investigator-initiated grant from Pfizer. P.A.N. is funded by National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) awards 2R01 AR065538, R01 AR075906, R01 AR073201, P30 AR070253, R21 AR076630 and NHLBI award R21 HL150575; the Fundación Bechara; the Arbuckle Family Fund for Arthritis Research; and the Samara Jan Turkel Center. A.M. is funded by NIH awards DP3DK111914-01 and R01DK1199979 and the National Multiple Sclerosis Society. A.M. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund and received the Lloyd Old STAR career award from the Cancer Research Institute (CRI). The Marson lab has received funding from the Innovative Genomics Institute (IGI), the Parker Institute for Cancer Immunotherapy (PICI), the Chan Zuckerberg Biohub and the Northern California JDRF Center of Excellence.
Glossary
- CUT&Tag
A technique that uses antibodies specific for DNA binding proteins to measure DNA regions bound by these proteins. The antibodies are tethered to a Tn5 transposase fusion protein and, following antibody-binding, activation of the transposase cleaves nearby DNA and generates fragment libraries for sequencing, the data of which are used to identify the bound regions.
- CUT&RUN
Similar to CUT&Tag, this technique analyses DNA regions bound by specific proteins using targeted antibodies. Unlike with CUT&Tag, the antibody is tethered to a micrococcal nuclease, which fragments nearby DNA elements.
- ATAC-seq
A technique used for assaying areas of open chromatin in the genome; the method relies on unguided Tn5 transposase-induced fragmentation of the genome.
- IMPACT
A computational genome annotation strategy that identifies regulatory elements defined by cell-state-specific transcription factor binding profiles.
- Massively parallel reporter assay
A technique used to identify regulatory regions of the genome in a high-throughput assay. Regions of interest are cloned into a minimal reporter with a unique barcode and a promoter to create a large pool of constructs. Constructs are expressed into cells and the RNA and DNA are sequenced to estimate the effects of each regulatory region on barcode gene expression, indicating regulatory capacity.
- Fluorescent in situ hybridization
A technique that measures RNA expression by flow cytometry using hybridization and amplification of fluorescent RNA probes.
- Single-cell RNA sequencing
An approach for measuring the expression of RNA in individual cells using droplet or plate-based technology.
- Computational fine mapping
A process by which a trait-associated region from a genome-wide association study is further analysed to identify genetic variants that are likely to causally influence the trait, usually through the integration of additional epigenetic or genomic data.
- Expression quantitative trait loci
Trait-associated regions that can explain a notable portion of the changes in expression of a gene.
- Electromobility shift assays
A molecular biology technique that measures the interaction of DNA and proteins on a protein-binding gel.
- Luciferase assays
A technique used to identify regions of the genome that can regulate gene expression. in these assays, the region of interest is cloned upstream or downstream of the gene encoding luciferase and the resultant plasmids are transfected into cells to measure the effect of the modification on luciferase expression.
- Affinity precipitation assays
A technique that is similar to electromobility shift assays, with the exception that bound complexes are magnetically pulled down prior to examination on a protein-binding gel.
- Droplet-based RNAseq
A single-cell RNA sequencing method that relies on droplet generation and encapsulation of individual cells.
- Mass cytometry
A type of single-cell analysis that tags cells with antibodies conjugated to heavy metals to then analyse staining intensity by time-of-flight mass spectrometry.
- HyPR-seq
A droplet-based targeted single-cell sequencing technique that involves hybridizing DNA probes to selected RNA to measure the expression of genes.
- Directed evolution
A process of protein engineering that mimics biological evolution. A library of mutated genes is expressed in cell lines and a phenotype is selected; the process is repeated with new mutations and harsher selection conditions until a desired outcome is achieved.
Footnotes
Competing interests
P.A.N. has been supported by investigator-initiated research grants from AbbVie, Bristol-Myers Squibb, Novartis, Pfizer, Sobi; consulting fees from Bristol-Myers Squibb (BMS), Cerecor, Miach Orthopedics, Novartis, Pfizer, Quench Bio, Sigilon, Simcere, Sobi, Exo Therapeutics and XBiotech; royalties from UpToDate Inc.; and salary support from the Childhood Arthritis and Rheumatology Research Alliance (CARRA). A.M. is a compensated co-founder, member of the boards of directors and a member of the scientific advisory boards of Spotlight Therapeutics and Arsenal Biosciences. A.M. was a compensated member of the scientific advisory board at PACT Pharma and was a compensated adviser to Juno Therapeutics and Trizell. A.M. owns stock in Arsenal Biosciences, Spotlight Therapeutics and PACT Pharma. A.M. has received honoraria from Merck and Vertex, a consulting fee from AlphaSights, and is an investor in and informal adviser to Offline Ventures. The Marson lab has received research support from Juno Therapeutics, Epinomics, Sanofi, GlaxoSmithKline, Gilead, and Anthem and reagents from Illumina. A.M. is an inventor on multiple inventions with the Whitehead Institute, UCSF and the Gladstone Institutes, including some that have been licensed. S.R. is a founder for Mestag, Inc, served in the past year as an adviser to Gilead, Biogen, Merck, Pfizer, Janssen and Abbvie. Y.B. and D.M. have no competing interests.
References
- 1.Cooper GS, Bynum MLK & Somers EC Recent insights in the epidemiology of autoimmune diseases: improved prevalence estimates and understanding of clustering of diseases. J. Autoimmun 33, 197–207 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogdanos DP et al. Twin studies in autoimmune disease: genetics, gender and environment. J. Autoimmun 38, J156–J169 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Plenge RM, Scolnick EM & Altshuler D Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov 12, 581–594 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Nelson MR et al. The support of human genetic evidence for approved drug indications. Nat. Genet 47, 856–860 (2015). [DOI] [PubMed] [Google Scholar]
- 5.King EA, Wade Davis J & Degner JF Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 15, e1008489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao C & Moult J GWAS and drug targets. BMC Genomics 15, S5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishigaki K et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat. Genet 52, 669–679 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shay T et al. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc. Natl Acad. Sci. USA 110, 2946–2951 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher MD & Chen-Plotkin AS The post-GWAS era: from association to function. Am. J. Hum. Genet 102, 717–730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Junhee Seok et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl Acad. Sci. USA 110, 3507–3512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Scheidt M et al. Applications and limitations of mouse models for understanding human atherosclerosis. Cell Metab. 25, 248–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue F & Ahituv N Decoding enhancers using massively parallel reporter assays. Genomics 106, 159–164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown CD, Mangravite LM & Engelhardt BE Integrative modeling of eQTLs and cis-regulatory elements suggests mechanisms underlying cell type specificity of eQTLs. PLoS Genet. 9, 1003649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson GJ & Yang M What rheumatologists need to know about CRISPR/Cas9. Nat. Rev. Rheumatol 13, 205–216 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Anzalone AV, Koblan LW & Liu DR Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol 38, 824–844 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Lee EG et al. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science 289, 2350–2354 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasegawa K et al. PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science 303, 685–689 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Kaplan MH, Sun YL, Hoey T & Grusby MJ Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature 382, 174–177 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Afzali B et al. BACH2 immunodeficiency illustrates an association between super-enhancers and haploinsufficiency. Nat. Immunol 18, 813–823 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siggs OM et al. Opposing functions of the T cell receptor kinase ZAP-70 in immunity and tolerance differentially titrate in response to nucleotide substitutions. Immunity 27, 912–926 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett CL et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IpEx) is caused by mutations of FOXP3. Nat. Genet 27, 20–21 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Morel L, Rudofsky UH, Longmate JA, Schiffenbauer J & Wakeland EK Polygenic control of susceptibility to murine systemic lupus erythematosus. Immunity 1, 219–229 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Todd JA et al. Genetic analysis of autoimmune type 1 diabetes mellitus in mice. Nature 351, 542–547 (1991). [DOI] [PubMed] [Google Scholar]
- 26.Ellinghaus D et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet 48, 510–518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li YR et al. Meta-analysis of shared genetic architecture across ten pediatric autoimmune diseases. Nat. Med 21, 1018–1027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy MI & Hirschhorn JN Genome-wide association studies: potential next steps on a genetic journey. Hum. Mol. Genet 17, R156 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bomba L, Walter K & Soranzo N The impact of rare and low-frequency genetic variants in common disease. Genome Biol. 18, 1–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang W et al. Genome-wide CRISPR screen identifies FAM49B as a key regulator of actin dynamics and T cell activation. Proc. Natl Acad. Sci. USA 115, E4051–E4060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parnas O et al. A Genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162, 675–686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henriksson J et al. Genome-wide CRISPR screens in T helper cells reveal pervasive crosstalk between activation and differentiation. Cell 176, 882–896.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cortez JT et al. CRISPR screen in regulatory T cells reveals modulators of Foxp3. Nature 582, 416 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shifrut E et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell 175, 1958–1971.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson W, Thorpe J, Long SA & Rawlings DJ Efficient CRISPR/Cas9 disruption of autoimmune-associated genes reveals key signaling programs in primary human T cells. J. Immunol 203, 3166–3178 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen DN et al. Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat. Biotechnol 38, 44–49 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumann K et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl Acad. Sci. USA 112, 10437–10442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sirvent S et al. Genomic programming of IRF4-expressing human Langerhans cells. Nat. Commun 11, 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu C-A et al. Genetic engineering in primary human B cells with CRISPR-Cas9 ribonucleoproteins. J. Immunol. Methods 457, 33–40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amariuta T et al. IMPACT: genomic annotation of cell-state-specific regulatory elements inferred from the epigenome of bound transcription factors. Am. J. Hum. Genet 104, 879–895 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maurano MT et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hindorff LA et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA 106, 9362–9367 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abascal F et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furey TS. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet 13, 840–852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Satpathy AT et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol 37, 925–936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skene PJ & Henikoff S An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaya-Okur HS et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun 10, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farh KK-H et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hrdlickova B et al. Expression profiles of long non-coding RNAs located in autoimmune disease-associated regions reveal immune cell-type specificity. Genome Med. 6, 88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J et al. Analysis of chromatin organization and gene expression in T cells identifies functional genes for rheumatoid arthritis. Nat. Commun 11, 4402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gustafsson M et al. A validated gene regulatory network and GWAS identifies early regulators of T cell-associated diseases. Sci. Transl. Med 7, 313ra178 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Hawkins RD et al. Global chromatin state analysis reveals lineage-specific enhancers during the initiation of human T helper 1 and T helper 2 cell polarization. Immunity 38, 1271–1284 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basak A & Sankaran VG Regulation of the fetal hemoglobin silencing factor BCL11A. Ann. N. Y. Acad. Sci 1368, 25–30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Canver MC et al. BCLIIA enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527, 192–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajagopal N et al. High-throughput mapping of regulatory DNA. Nat. Biotechnol 34, 167–174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korkmaz G et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnoi 34, 192–198 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Thynn HN et al. An allele-specific functional SNP associated with two systemic autoimmune diseases modulates IRF5 expression by long-range chromatin loop formation. J. Invest. Dermatol 140, 348–360. e11 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Doench JG Am i ready for CRISPR? A user’s guide to genetic screens. Nat. Rev. Genet 19, 67–80 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Simeonov DR et al. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature 549, 111–115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klann TS et al. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol 35, 561–568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ray JP et al. Prioritizing disease and trait causal variants at the TNFAIP3 locus using functional and genomic features. Nat. Commun 11, 1237 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fulco CP et al. Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat. Genet 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nasrallah R et al. A distal enhancer at risk locus 11q13.5 promotes suppression of colitis by Treg cells. Nature 583, 447–452 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gasperini M et al. A genome-wide framework for mapping gene regulation via cellular genetic screens in brief a highly multiplexed CRISPRi screen uncovers gene-enhancer relationships at scale. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell 176, 377–390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cho JH & Gregersen PK Genomics and the multifactorial nature of human autoimmune disease. N. Engl. J. Med 365, 1612–1623 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Husebye ES, Anderson MS & Kampe O Autoimmune polyendocrine syndromes. N. Engl. J. Med 378, 1132–1141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martínez-Feito A et al. Autoimmune lymphoproliferative syndrome due to somatic FAS mutation (ALPS-sFAS) combined with a germline caspase-10 (CASP10) variation. Immunobiology 221, 40–47 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Li G et al. High-throughput identification of noncoding functional SNPs via type IIS enzyme restriction. Nat. Genet 50, 1180–1188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wallace C et al. Dissection of a complex disease susceptibility region using a bayesian stochastic search approach to fine mapping. PLoS Genet. 11, 1–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lonsdale J et al. The genotype-tissue expression (GTEx) project. Nat. Genet 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tewhey R et al. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell 165, 1519–1529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen L et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell 167, 1398–1414.e24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ricaño-Ponce I et al. Refined mapping of autoimmune disease associated genetic variants with gene expression suggests an important role for non-coding RNAs. J. Autoimmun 68, 62–74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhernakova DV et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet 49, 139–145 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Gracey E et al. TYK2 inhibition reduces type 3 immunity and modifies disease progression in murine spondyloarthritis. J. Clin. Invest 130, 1863–1878 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Westra H-J et al. Fine-mapping and functional studies highlight potential causal variants for rheumatoid arthritis and type 1 diabetes. Nat. Genet 50, 1366–1374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hwang JS et al. NFAT1 and JunB cooperatively regulate IL-31 gene expression in CD4+ T cells in health and disease. J. Immunol 194, 1963–1974 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Tripathi SK et al. Genome-wide analysis of STAT3-mediated transcription during early human Th17 cell differentiation. Cell Rep. 19, 1888–1901 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Gutierrez-Arcelus M et al. Allele-specific expression changes dynamically during T cell activation in HLA and other autoimmune loci. Nat. Genet 52, 247–253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brandt M et al. An autoimmune disease risk variant has a trans master regulatory effect mediated by IRF1 under immune stimulation. bioRxiv 10.1101/2020.02.21.959734 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kumasaka N, Knights AJ & Gaffney DJ High-resolution genetic mapping of putative causal interactions between regions of open chromatin. Nat. Genet 51, 128–137 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Odqvist L et al. Genetic variations in A20 DUB domain provide a genetic link to citrullination and neutrophil extracellular traps in systemic lupus erythematosus. Ann. Rheum. Dis 78, 1363–1370 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roth TL et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559, 405–409 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gaudelli NM et al. Programmable base editing of T to G C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gehrke JM et al. An apobec3a-cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol 36, 977 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liang P et al. Correction of β-thalassemia mutant by base editor in human embryos. Protein Cell 8, 811–822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zeng J et al. Therapeutic base editing of human hematopoietic stem cells. Nat. Med. 26, 535–541 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Webber BR et al. Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nat. Commun 10, 5222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Anzalone AV et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Trynka G et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet 45, 124–130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Raj T et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gutierrez-Achury J et al. Functional implications of disease-specific variants in loci jointly associated with coeliac disease and rheumatoid arthritis. Hum. Mol. Genet 25, 180–190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thalayasingam N et al. CD4+ and B lymphocyte expression quantitative traits at rheumatoid arthritis risk loci in patients with untreated early arthritis: implications for causal gene identification. Arthritis Rheumatol. 70, 361–370 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Der E et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat. Immunol 20, 915–927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shalek AK & Benson M Single-cell analyses to tailor treatments. Sci. Transl. Med 9, 4730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rao DA et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 542, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smillie CS et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van der Wijst MGP et al. The single-cell eQTLGen consortium. eLife 9, e52155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dixit A et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rubin AJ et al. Coupled single-cell CRISPR screening and epigenomic profiling reveals causal gene regulatory etworks. Cell 176, 361–376.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Datlinger P et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marshall JL et al. HyPR-seq: Single-cell quantification of chosen RNAs via hybridization and sequencing of DNA probes. Proc. Natl Acad. Sci 117, 33404–33413 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roth TL et al. Pooled knockin targeting for genome engineering of cellular immunotherapies. Cell 181, 728–744.e21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grünewald J et al. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat. Biotechnol 37, 1041–1048 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Walton RT, Christie KA, Whittaker MN & Kleinstiver BP Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hu JH et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kleinstiver BP et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kleinstiver BP et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nishimasu H et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259–1262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cox DBT et al. RNA editing with CRISPR-Cas13. Science 358, 1019–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abudayyeh OO et al. A cytosine deaminase for programmable single-base RNA editing. Science 365, 382–386 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mok BY et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 583, 631–637 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zuo E et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Simeonov DR & Marson A CRISPR-based tools in immunity. Annu. Rev. Immunol 37, 571–597 (2019). [DOI] [PubMed] [Google Scholar]
- 117.Ewart DT, Peterson EJ & Steer CJ Gene editing for inflammatory disorders. Ann. Rheum. Dis 78, 6–15 (2019). [DOI] [PubMed] [Google Scholar]
- 118.Ben-David U et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 560, 325–330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ghandi M et al. Next-generation characterization of the cancer cell line encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]