Abstract
Objectives:
The impact of rheumatoid arthritis (RA) on the shaping of the oral and gut microbiome raises the question of whether and how RA treatment modifies microbial communities. We examined changes in the oral and gut microbiota in a mouse model of antigen-induced arthritis (AIA) treated or not with methotrexate (MTX).
Methods:
Maxillae and stools were evaluated by the MiSeq platform of the V4 region of the 16S rRNA gene. Alveolar bone parameters were analysed by micro-computed tomography. Moreover, arthritis-induced changes in hyperalgesia and oedema were assessed, along with the impact on periodontal bone health.
Results:
Microbial communities in MTX-treated AIA mice revealed distinct clusters compared to the control and AIA groups. Overall, MTX impacted the richness and variability of microorganisms in the oral-gut axis microbiome at the phylum level. Regarding the oral microbiome, while in the control group the most dominant phylum was Firmicutes, in the AIA group there was a shift towards the predominance of Campilobacteriota and Bacteroidetes associated with the disease. MTX treatment led to greater dominance of the health-associated phylum Proteobacteria. In the gut microbiome, AIA induction resulted in increased abundance of the Verrucomicrobiota phylum, and MTX treatment restored its levels compared to control. Importantly, the MTX-treated AIA animals had significantly less periodontal bone loss, as well as decreased hyperalgesia and joint oedema compared to the AIA animals.
Conclusion:
Data suggest the benefit of MTX treatment in protecting alveolar bone, in addition to providing new insights on the drug-microbiome interaction in the course of RA.
Keywords: Alveolar bone loss, Antirheumatic agents, Gut microbiome, Methotrexate, Microbiota, Rheumatoid arthritis
1. Introduction
Rheumatoid arthritis (RA) is an autoimmune disease associated with inflammation and bone erosions in the joints [1]. Compelling evidence indicates that there is an interaction between the host and the microbiota in the presence of RA. This link suggests that RA-associated inflammation causes a dysbiosis in the oral and gut microbiota [2–6]. In turn, dysbiosis triggers a host response that enhances local and systemic inflammatory pathways and may contribute to joint damage [6]. The oral microbiota may further contribute to RA autoimmunity by expressing enzymes that induce host protein citrullination and, consequently, the production of antibodies to citrullinated proteins or peptides [7,8].
Previous studies have described the composition of the oral and gut microbiome in individuals with RA [2,3,5,9,10]; however, data on the concurrent effects of the oral-gut microbiota are sparse [3,11]. Importantly, some periodontopathic bacteria such as Tannerella and Treponema, were significantly higher in the oral microbiota of individuals with new-onset RA and tended to decrease with better-controlled established RA [2]. This result suggests that measures to improve RA by treatment impact the ecological adaptation of the oral microbial niche. However, studies investigating this issue are limited and have been conducted on relatively small numbers of subjects [2].
After almost 60 years of existence, the folic acid antagonist methotrexate (MTX) is the most widely used disease-modifying antirheumatic drug (DMARD), either as a single agent or in combination with other DMARDs [12]. Findings from a recent study have revealed that oral MTX can be metabolized by the human gut microbiota and have suggested that the gut microbiome may be of value in predicting the response to MTX in patients with RA [13]. Nevertheless, few studies have addressed whether treatment with MTX can reverse the impact of RA on the oral or gut microbiota [3,13,14]. In an experimental model, it has been suggested that MTX affects the abundance of some species in the gut and protects against periodontal bone loss associated with arthritis along with periodontal infection [15]. Herein, we analysed the impact of arthritis and MTX on the microbiome composition of the oral-gut axis in an antigen-induced arthritis (AIA) model. We hypothesized that MTX therapy partially reverses AIA-induced oral-gut axis dysbiosis and alveolar bone loss.
2. Material and methods
2.1. Animals and ethical clearance
The experiments were performed using male C57BL/6 mice weighing 20–25 g (6–8 weeks) obtained from the Animal Facility of the School of Medicine of Ribeirão Preto, University of São Paulo, Brazil. Mice were housed in temperature- and light-controlled rooms with ad libitum access to water and food in the animal facility of the Department of Pharmacology, School of Medicine of Ribeirão Preto, University of São Paulo, Brazil. Animal husbandry and procedures followed the guidelines of the Animal Ethics Committee of the School of Medicine of Ribeirão Preto (241/2018).
2.2. Induction of AIA in mice and MTX treatment
AIA was induced as previously described, with some modifications [16]. The mice were anesthetized with 2% isoflurane before immunization and challenge. Briefly, mice were sensitized with subcutaneous (s.c.) injections of 500 μg methylated bovine serum albumin (mBSA) in 0.2 mL of an emulsion containing 0.1 mL saline and 0.1 mL complete Freund’s adjuvant (CFA; 1 mg/mL of Mycobacterium tuberculosis) on day 0. The mice were boosted with the same preparation on day 7. Control mice received vehicle injections without the antigen (mBSA). Twenty-one days after the initial injection, arthritis was induced in the immunized animals by intra-articular (i.a.) injection of mBSA (100 μg/cavity) into the right knee joint using a sterile 33-gauge syringe. Control mice were injected with 10 μL of PBS alone. Mice received a second challenge on day 26 after the first immunization. On days 28, 35 and 42 after the first immunization, mice were challenged again in a similar fashion. In the arthritis group treated with MTX, mice received treatment with 2 mg/kg MTX (Tecnomet®, Zodiac; São Paulo, SP, Brazil), which was initially administered 2 weeks before day 0 (2 weeks before the first immunization), being repeated weekly by gavage until the week of euthanasia [17]. At the end of the experiment (day 43), a set of maxillae was collected for micro-computed tomography (micro-CT) and another set was collected for DNA extraction. Stools of all groups (control, AIA and AIA + MTX) were also collected for DNA extraction and stored at −70 °C.
2.3. Evaluation of articular hyperalgesia
Twenty-four hours after the last mBSA injection into the joint (day 43), knee joint thickness was measured in millimetres (mm) with a calliper. The results were expressed as means ± the difference between the diameter on day 0 (baseline) and after the last challenge. In parallel, articular hyperalgesia of the femur-tibial joint was determined as previously described [18]. An increasing perpendicular force was applied to the central area of the plantar surface of the hind paw challenged with mBSA in order to induce flexion of the femur-tibial joint followed by paw withdrawal. The pressure of the force applied when the paw was withdrawn was recorded with a pressure gauge. The mechanical threshold was expressed in grams and hyperalgesia was equated with a reduction in this threshold.
2.4. Micro-CT and data analysis
Maxillae were fixed in 10% neutral buffered formalin for 48 h and scanned using a micro-CT system (Skyscan 1174 X-ray microtomograph; Skyscan, Aartselaar, Belgium). Calibration was carried out with a known density calcium hydroxyapatite phantom (Skyscan; Aartselaar, Belgium). High-resolution scans were acquired using a 14.49 μm image pixel size (50 kV, 0.5 mm aluminium filter, 0.7° rotation angle). The images obtained were reconstructed using the NRecon software (Skyscan; Aartselaar, Belgium). Maxillae were positioned so as to standardize the analysed area using the Data-Viewer software (Chauvin Arnoux®; Asnières-Sur-Seine, France), and analysed using CTAn software (Skyscan; Aartselaar, Belgium). Alveolar bone crest (ABC) loss was measured by determining the area between the cemento-enamel junction (CEJ) and the ABC (ABC-CEJ, μm2) in three-dimensional images of the first, second and third molars (Fiji - National Institute of Health; Bethesda, MD, USA).
The GraphPad Prism software version 8.0 (GraphPad software, San Diego, CA, USA) was used for statistical analysis. One-Way ANOVA and the chi-square test were employed to evaluate the differences between groups. For all analyses, the level of significance was set at <0.05.
2.5. DNA extraction, 16S rRNA amplicon library preparation and sequencing
The DNA of the whole maxillae (i.e., bone, gingiva, and teeth) and stools was extracted using the DNeasy PowerSoil Pro kit (QIAGEN; Hilden, Germany). DNA concentration was assessed by spectrophotometry using a NanoDrop™ Spectrophotometer (Thermo Fisher; Waltham, MA, USA). The resulting purified DNA was stored at −20 °C until use. The V4 region of the 16S rRNA gene was amplified with region-specific primers that included the Illumina flowcell adapter sequences [19]. PCR was performed in 25 μL of a final volume containing 1X PCR Buffer, 0.2 mM dNTP, 2 mM MgSO4, 0.2 μM of primers, 1U Platinum™ Taq DNA Polymerase High Fidelity (Thermo Fisher; Waltham, MA, USA), and 20–30 ng of DNA. Cycle conditions were 1 cycle at 94 °C (3 min), then 30 each cycle at 94 °C (45 s), 57 °C (60 s), 68 °C (45 s), and a final extension at 68 °C (10 min). Libraries were purified using AMPure XP beads (Beckman Coulter; Brea, CA, USA) according to the Illumina 16S metagenomic sequencing library protocol. The size of the libraries was checked with the Bioanalyzer DNA 1000 Assay (Agilent; Santa Clara, CA, USA). Quantification was performed with the KAPA Library Quantification Kit Illumina (Roche; Basel, Switzerland) and further DNA libraries were normalized and pooled. The libraries were paired end sequenced (2 × 300) in a single run on the Miseq platform (Illumina, San Diego, CA USA).
2.6. Data processing and analysis
All raw data files were imported and processed in the R-environment (version 4.1.1; https://cran.r-project.org/) using inbuilt functions available in R. Data processing and raw reads were quality filtered and trimmed using the “filterAndTrim()” function from the Dada2 package [20]. An amplicon sequence variant (ASV) table was created and chimeras were removed using the “make-SequenceTable()” and “removeBimeraDenovo()” functions from the Dada2 package. Taxonomy was assigned using the Silva (version 138) database and “assignTaxonomy()” function [21]. Unassigned taxa and singletons were removed using the “subset_taxa ()” and “prune_taxa()” functions of the phyloseq package in R [22]. This data object was used for alpha/beta diversity analysis in order to calculate microbial abundances and to carry out other statistical tests using various functions in phyloseq and other packages.
2.7. Diversity analysis
Alpha diversity for the oral and gut microbial axis was computed using the Phyloseq R package [23] and was estimated by the Chao1 and Shannon diversity index. Statistical tests were performed after visual data inspection with histograms and normal data distribution was tested by the Shapiro-Wilk test for normality. Beta diversity was analysed using Bray-Curtis and unweighted UniFrac distances and visualized with PCoA. Sample groups were compared by permutational analysis using a dissimilarity matrix. The “adonis” function from the Vegan R package [24] was used to test differences in the composition of the community between sample groups (n. of permutation 999). Differential abundance analysis was performed by DESeq [25]. The taxa were considered to be differentially abundant if logFc |0| and p < 0.05.
2.8. Co-occurrence analysis
Co-occurrence analysis was performed by Spearman’s rank correlation at the genus level. The correlation threshold was ≥ 0.3 and the p value ≤ 0.05. The network was viewed through the Microbiome Analyst (https://www.microbiomeanalyst.ca/MicrobiomeAnalyst/).
2.9. Partial least squares discriminant analysis (PLS-DA)
To discriminate the microbial profiles between groups, PLS-DA was performed in the R package mixOmics framework for the analysis of mixMC microbial communities [26]. The loading plot displayed the microbial species contributing to the separation of PLS-DA scores between MTX, AIA and control groups.
3. Results
3.1. Treatment with MTX prevented AIA-induced alveolar bone loss, joint oedema and pain
Micro-CT revealed that MTX prevented the loss of alveolar bone crest, while greater loss of alveolar bone crest was observed in the AIA group compared to control (Fig. 1A–D). Arthritis parameters such as oedema and articular hyperalgesia mechanical threshold were analysed in parallel. The mechanical threshold was reduced in the AIA group. Treatment with MTX reduced the joint hyperalgesia induced by mBSA injected into immunized animals (Supplementary Fig. 1A). Likewise, MTX therapy reduced the oedema of treated animals compared to the AIA group (Supplementary Fig. 1B).
Fig. 1.
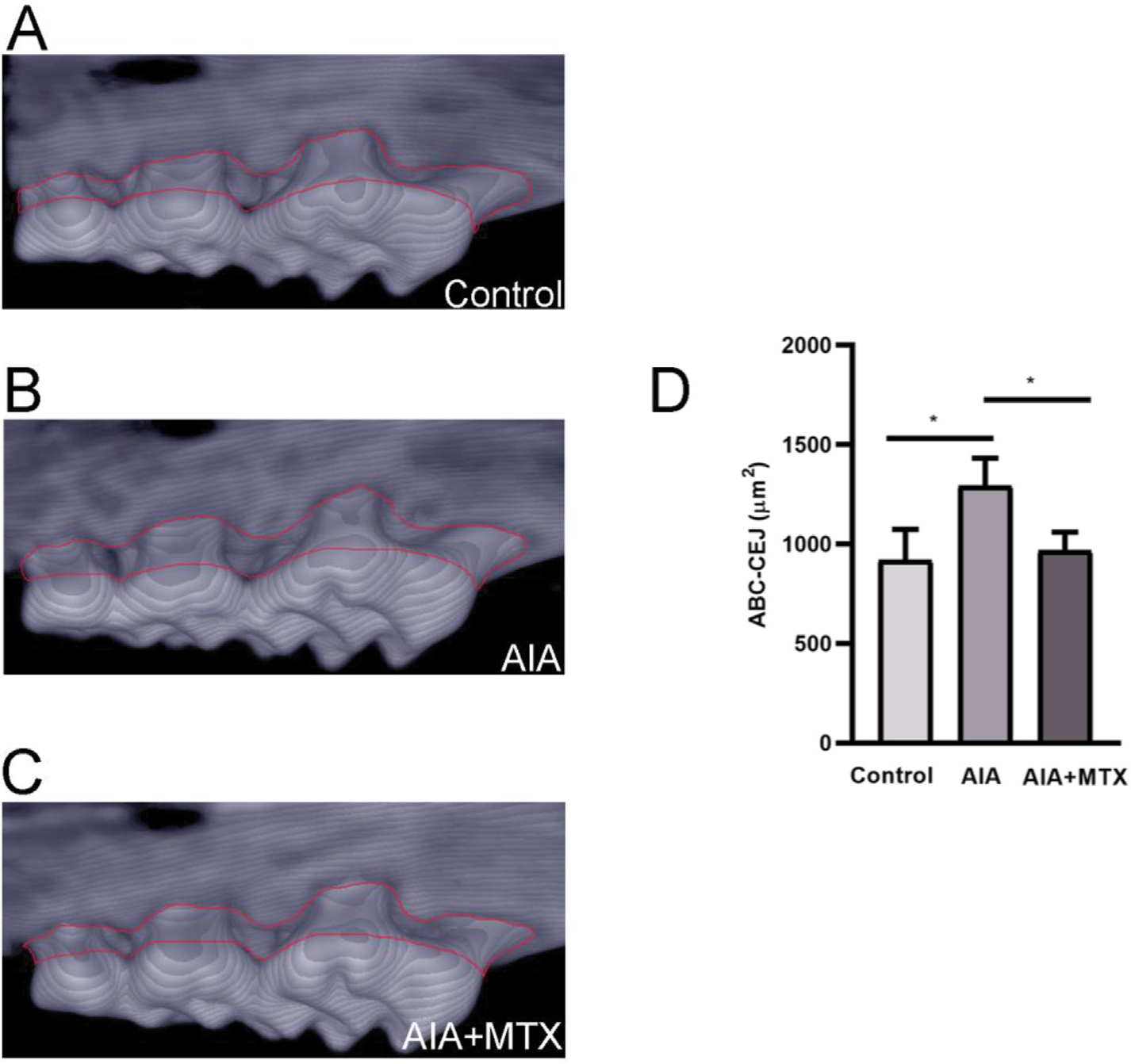
Representative 3D image of the alveolar bone crest of control mice (A), mice with antigen-induced arthritis (AIA) (B) and AIA mice treated with methotrexate (MTX) (C). Quantification of alveolar bone crest loss on the palatal surface of the maxillary first, second and third molar regions. The region analysed is circled in red (D). Note: n = 5 animals per group; One-Way ANOVA (p < 0.05).
3.2. Treatment with MTX affected the diversity of the oral and gut microbiota
In oral samples, AIA showed a shifting trend in alpha diversity measured by Chao1. In contrast, AIA + MTX significantly increased species richness (p < 0.05) compared to AIA measured by Chao1. When measured by Shannon diversity, AIA + MTX species were significantly different compared to untreated controls (p < 0.01), but not significantly different for the AIA group, with also a decrease in diversity compared to AIA and controls (Fig. 2A). To determine whether oral microbial communities differed among groups (beta-diversity), distances were calculated by unweighted UniFrac (p = 0.006) and Bray-Curtis (p = 0.02) statistics. No clear separation between the AIA and control groups was observed in most oral samples from the AIA group clustered separately from the control group; however, in the AIA + MTX group there was a clear distinction in relation to the control and AIA groups (Fig. 2B and C).
Fig. 2.
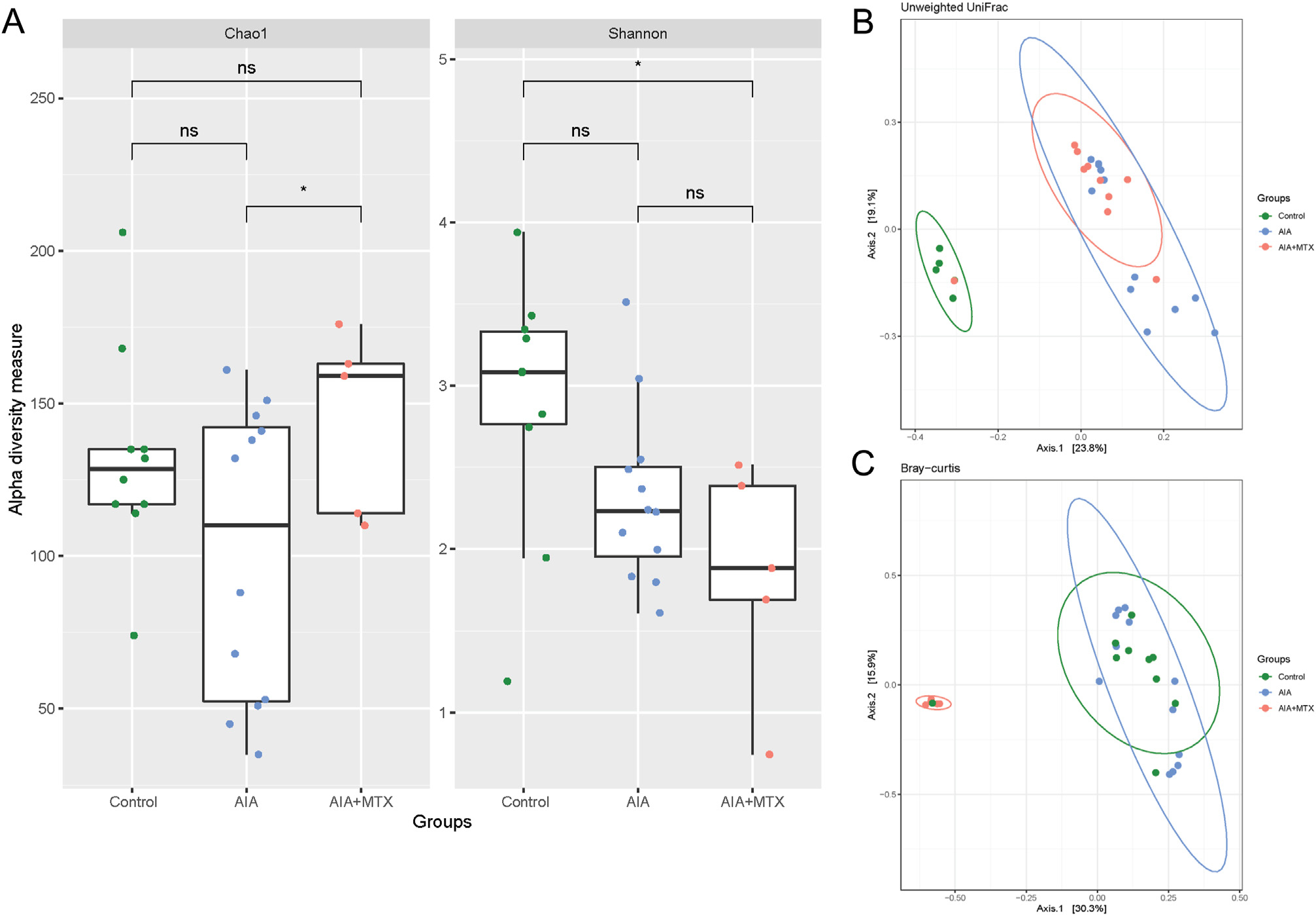
Diversity of oral microbiota. (A) Chao1 and Shannon alpha diversity index. (B) Microbiota separation on the principal coordinates calculated from unweighted and (C) weighted UniFrac distances. Control animals, arthritis induced group (AIA) and animals with arthritis treated with methotrexate (AIA + MTX). Note: n = 5 to 12 animals per group; ns: non-significant.
For the gut microbiome, the AIA groups tended to show increased diversity compared to control. However, MTX treatment caused a decrease in the diversity assessed by the Chao1 index (Fig. 3A). Similar to the oral microbiota, in the gut microbiome, most samples from the AIA group clustered separately from the control and the AIA + MTX group showed a clear difference in the gut microbiota composition (weighted UniFrac: p = 0.003; Bray-curtis: p = 0.005) (Fig. 3B and C).
Fig. 3.
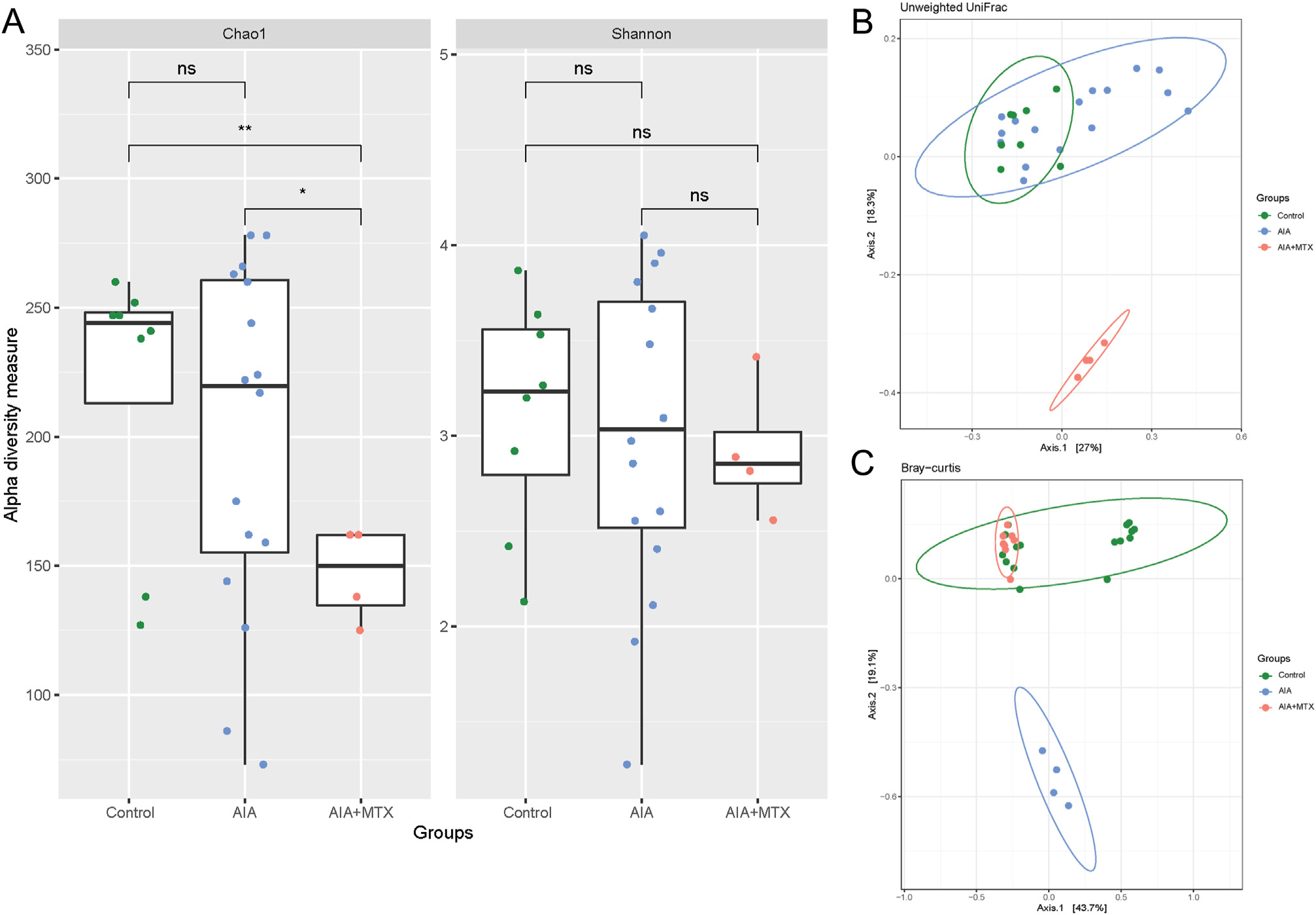
Diversity of gut microbiota. (A) Chao1 and Shannon alpha diversity index (B) Microbiota separation on the principal coordinates calculated from unweighted and (C) weighted UniFrac distances. Control animals, arthritis induced group (AIA) and animals with arthritis treated with methotrexate (AIA + MTX). Note: n = 4 to 16 animals per group; ns: non-significant.
3.3. Treatment with MTX modified the composition of the oral and gut microbiota by altering the dominant phyla
Regarding the differences in the abundance of the major phylum in the oral microbiota, in the control group, the most dominant phylum was Firmicutes, with many of its health-related members. In the AIA group, there was a shift toward the predominance of disease-associated Campilobacteriota and Bacteroidetes, and MTX treatment led to greater dominance of the health-associated phylum Proteobacteria. The abundance of Bacteroidota, Campilobacterota and Verrucomicrobiota increased in the AIA group compared to control, while MTX treatment reversed this increase and led to lower proportions of these bacteria than in the control group (Fig. 4A). In contrast, Firmicutes and Proteobacteria were reduced in AIA animals, with a pronounced increase in Proteobacteria after MTX treatment, making this phylum the dominant bacterial type in the MTX group (Fig. 4B).
Fig. 4.
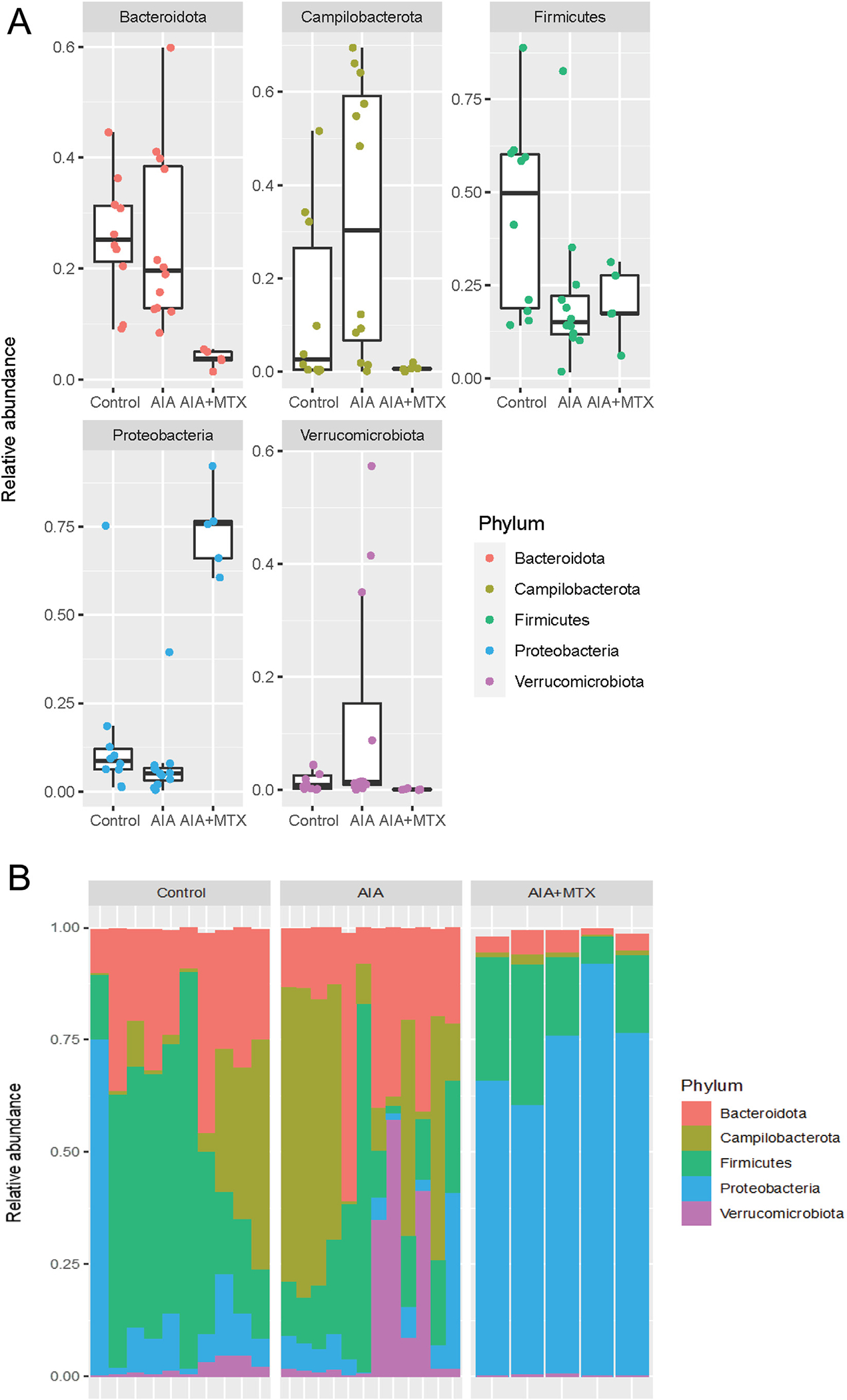
Relative abundance of oral microbiota composition at the phylum level. Note: n = 5 to 12 animals per group.
Most gut bacteria belonged to the phylum Bacteroidota, followed by Firmicutes. AIA had a diverse impact on these phyla, decreasing Bacteroidota and increasing Firmicutes. The Firmicutes/Bacteroidetes ratio is widely accepted to have an important influence on the maintenance of normal intestinal homeostasis. While MTX treatment partially reversed the AIA effect in Bacteroidota, an additional increase in Firmicutes was revealed in this group compared to the control and AIA groups (Fig. 5A). The Verrucomicrobiota phylum was the most impacted by the induction of AIA, with the highest proportion of these bacteria. Treatment with MTX restored the levels of Verrucomicrobiota in relation to the control. The Deferribacterota and Cyanobacteria phyla were significantly reduced in the AIA and AIA + MTX groups compared to control (Fig. 5B).
Fig. 5.
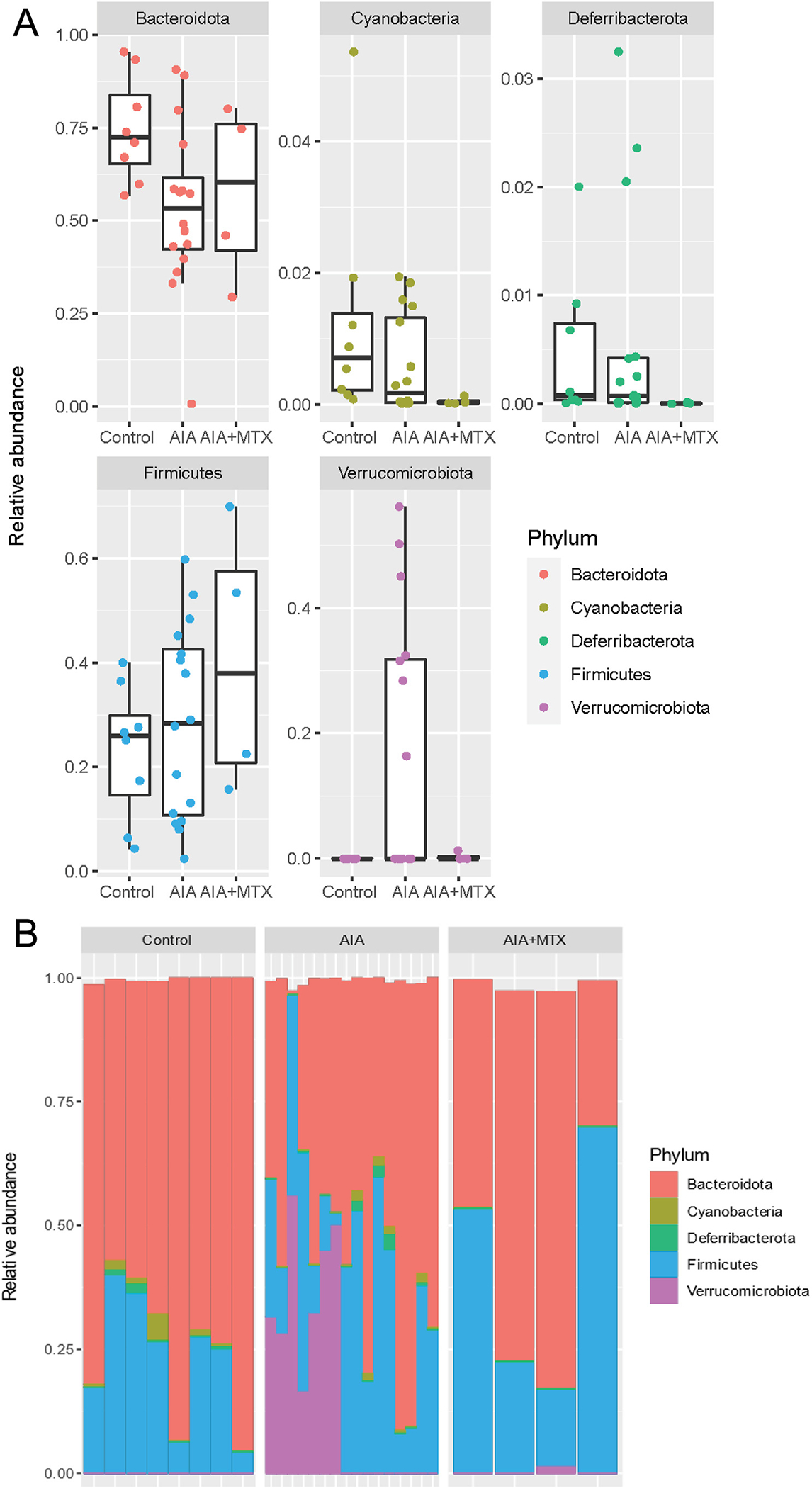
Relative abundance of gut microbiota composition at the phylum level. Note: n = 4 to 16 animals per group.
3.4. MTX reversed the increase of pathogenic species caused by the AIA shift
In the oral microbiota, the operational taxonomic units (OTU) differed considerably between the AIA and control groups. Many members of Lachnospirales were differentially affected by MTX, with some of them increasing in number and others decreasing compared to the AIA and control groups (Fig. 6A). When comparing the oral microbiota between the AIA and AIA + MTX groups, some differences were observed, such as an increased abundance of Rikenella and Streptococcus in the latter group (Fig. 6B). When comparing differentially abundant taxa at the species level, it was observed that Escheria coli increased in the AIA group compared to control. However, treatment with MTX drastically reduced the proportion of this species. Other species, including Enterococcus faecium, Akkermansia muciniphila, Bacteroides uniformis, were also reduced by MTX treatment. Conversely, Streptococcus respiraculi, Brevundimonas olei, Pseudomonas xanthomarina, and Rothia nasimurium numbers were lower in the control and AIA groups, but were significantly increased in the AIA + MTX group (Supplementary Fig. 2).
Fig. 6.
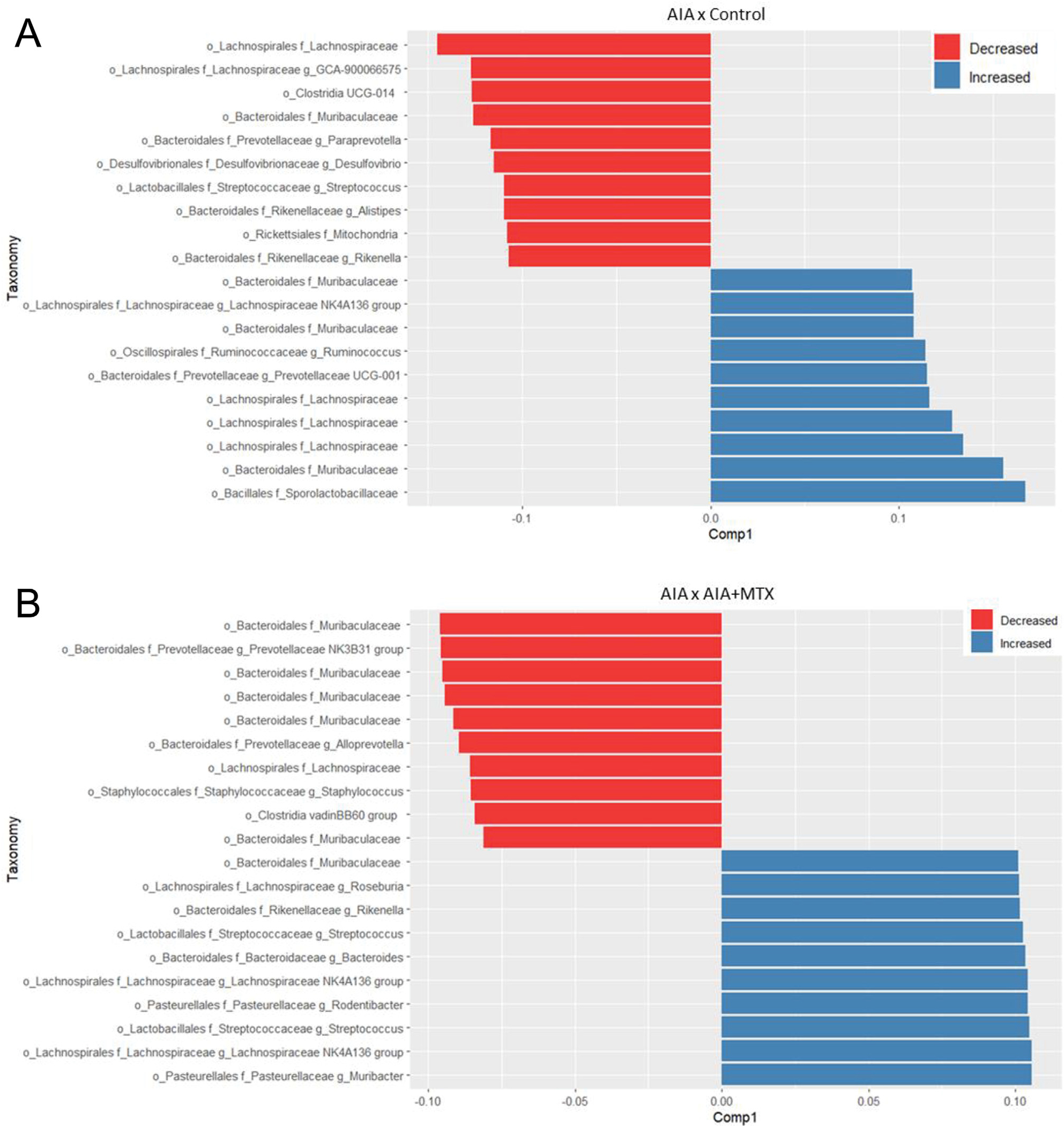
(A) Graph depicting oral operational taxonomic units (OTU) with differences in relative abundance in the AIA group compared to control animals based on partial least squares-discriminant analysis (PLSDA). (B) Graph depicting oral OTU with differences in relative abundance in the AIA + MTX group compared to AIA animals based on PLSDA results.
When comparing the gut microbiota between the control animals and the AIA group, differences were observed in the proportion of bacteria such as Roseburia and Ruminococcus, with a significant reduction in the AIA animals. Also, many OTU from the Muribaculaceae family were differentially abundant between groups. The proportions of some bacteria such as Bacteroides and Butyricimonas were decreased in the AIA group compared to the AIA + MTX group (Supplementary Fig. 3A). Other bacteria such as Rikenella and Odoribacter were increased in AIA animals (Supplementary Fig. 3B). Akkermansia muciniphila was increased in the AIA group, although it was reduced in the AIA + MTX group (Supplementary Fig. 4).
We next investigated the correlations among the microorganisms at the genus level in oral and gut samples. Negative correlations were found between Campylobacter and Rothia, and positive correlations between Campylobacter and Entereococcus, and Prevotellaceae and Staphylococcus (Supplementary Fig. 5A). In the gut microbiome, Akkermansia showed a negative correlation with Bacteroides, Escherichia Shigella and Lachnospiraceae and a positive correlation with Campylobacter and Enterorhabdus. Bacteroides was negatively correlated with Campylobacter and positively correlated with Butyricimonas (Supplementary Fig. 5B).
4. Discussion
We demonstrated that MTX was able to decrease joint oedema and prevent arthritis-induced alveolar bone loss in mice. These effects may be due to changes in the oral and gut microbiota since MTX impacted microbial communities. For the first time, our data collectively highlight the impact of MTX on the oral-gut axis microbiota and the protective activity of MTX against arthritis-induced alveolar bone loss.
It is known that MTX remains the anchor drug for the treatment of RA worldwide [12], although it may be ineffective for 30%–40% of individuals, with no indication about which patients may benefit from therapy [27]. In this scenario, contemporary studies have reported that the bioavailability and clinical efficacy of MTX in the treatment of RA can be modulated by the composition and function of gut microorganisms and their enzymatic products, leading to a decrease in host immune activation [3,11,13]. Furthermore, RA supposedly affects the oral microbiota [10]; in turn, the impact of MTX on the latter and on alveolar bone is unknown. Herein, we demonstrated that the response to MTX impacted the richness and variability of the oral and gut microbiome in an experimental model of arthritis. On this basis, it has recently been proposed that microbial diversity in the oral sample of individuals with RA is more likely to increase or to remain static in these individuals compared to healthy controls [10]. The most profound impact was on the gut microbiota, where AIA tended to increase diversity compared to control. However, treatment with MTX caused a decrease in diversity. Former studies have shown that MTX also has an impact on the gut microbiota in addition to decreasing alpha diversity in stool samples [3,11,28]. We also observed that most of the oral and gut samples from the AIA group clustered separately from the control group, whereas treatment with MTX clearly changed the beta diversity, as documented elsewhere [5,11,28].
In line with the alpha and beta diversity findings, we observed a series of changes in the oral and gut microbiota induced by MTX treatment both at the phylum and at the species level. Arthritis-induced alveolar bone loss has been associated with changes in the composition of the oral microbiota in the AIA model [29]. In our study, an in-depth analysis of the oral microbiome revealed similar findings. For instance, at the phylum level, the oral microbiota of control animals was dominated by Firmicutes, with a reduced proportion in the AIA group and partially restored with MTX treatment. Indeed, most members of the Firmicutes phylum are linked to healthy ones such as Streptococcus [10,30]. Also, the levels of the phyla Bacteroidota, Campilobacterota and Verrucomicrobiota were increased in the AIA group and were reduced by MTX treatment. Studies have shown that Bacteroidota are highly associated with disease [30,31]. On the other hand, our results showed a greater abundance of the Proteobacteria phylum after MTX treatment, consistent with previous studies showing higher Proteobacteria levels under healthy conditions [30,31]. Members of Lachnospirales were also differentially affected by MTX, with some increasing in abundance and others decreasing when compared to the AIA and control groups. Of note, we have previously documented that increased levels of some Gram-negative and anaerobe species, including Parvimonas micra, Veillonella parvula, and Selenomonas noxia in oral microbiota composition, were primarily associated with experimental AIA [29]. Another study, however, reported that oral pathobionts were associated with the synovial proteome in the RA model [32]. The authors hypothesized that this certainly occurred because periodontal bacteria shape the arthritis subtype and thereby set the groundwork for the ultimate response to therapy [32].
Our co-occurrence analysis regarding the oral microbioma indicated that the presence of pathogenic bacteria such as Escherichia shigella was positively correlated with other pathogenic bacteria such as Akkermansia [33] and Campylobacter [34]. Conversely, negative correlations were observed between Akkermansia and Haemophilus, and other members of the microbiome considered to be “healthy bacteria” such as Streptococcus and Rothia [34,35]. Notably, MTX satisfactorily prevented arthritis-induced bone loss, an effect possibly linked to the changes in bacterial communities. In this respect, we emphasize that it is uncertain how to differentiate what is bidirectional from changes in alveolar bone and oral microbiota caused by RA, MTX treatment, or interactions with commensal bacteria. In the case of reversal of alveolar bone loss by MTX treatment, it should be mentioned that, at least in experimental settings, bacteria in the oral cavity significantly affected the intestinal microbiome [6,36]. Nonetheless, further studies are needed to investigate specific bacteria in the oral-gut axis microbiota and their role in alveolar bone homeostasis. Recent literature revealed that MTX improved periodontal bone loss, although the authors did not investigate the isolated effects of MTX on the RA-induced alveolar bone [15]. The likelihood that MTX may also positively impact the dental supporting tissues of individuals with RA has been hypothesized [15]; however, in animals, MTX has been reported to promote osteoclast formation through increased levels of inflammatory cytokines [37].
An accumulating body of evidence supports a relationship between gut microbiota and joint inflammation in animals and humans [10,11,38,39]. Herein, we detected that, the induction of AIA led to a decrease in Bacteroidota in the gut microbiota, but treatment with MTX reversed this effect. The Firmicutes/Bacteroidetes ratio has been associated with several pathological conditions [40]. In this context, a study revealed that the population, diversity and major components of the gut microbiota of mice were dramatically modified in response to treatment with intraperitoneal MTX in a time-dependent manner. As a result, Bacteroidales exhibited the most distinct variation among all taxa [41]. Another study reported that oral administration of Porphyromonas gingivalis increases the Bacteriodetes and decreases the Firmicutes [36], while others showed that treatment with MTX increased the proportion of Bacteriodetes, Firmicutes and Verrucomicrobiota, in agreement with our findings [11].
Comparing differences at the species level, we found that Escherichia coli, a known pathogenic bacterium, was increased in the AIA group compared to control, and treatment with MTX drastically reduced the proportion of this species. Of interest, differences in colonization by pathogenic types of Escherichia coli were identified in individuals with inflammatory arthritis, who had lower joint scores and inflammatory markers although their IgA anti-Escherichia coli antibody responses were higher [42]. Enterococcus faecium, Akkermansia muciniphila [33] and, to a lesser extent, Bacteroides uniformis [11], are also bacteria that have been associated with some autoimmune rheumatic diseases and were reduced here by MTX treatment. Previous studies have demonstrated that the probiotic bacteria Escherichia coli and Enterococcus faecium had a beneficial effect on MTX treatment of adjuvant arthritis in murine models [43,44]. Dysbiosis of the gut microbiota has been detected in individuals with RA, as also observed for the oral microbiome, with some species being depleted (Haemophilus spp.) and others (Lactobacillus salivarius) being significantly increased. Curiously, these changes were partially recovered with treatment with DMARDs [3]. According to this premise, immunosuppressive agents that can restore gut microbial composition and immunologic balance may function as therapeutic drugs for RA [45]. Low-dose MTX-induced changes in the gut microbiome reduce host immune activation [11]. Specifically, alterations in the microflora can lead to the decrease of multiple populations of host cells, such as activated T cells, B cells, myeloid cells, among others [46]. In this line, antigen-specific regulatory T cells revealed a promising immunosuppressive key in an arthritis-prone mouse model [47]. Nonetheless, experimental studies are needed to unravel the induction of tolerogenic cell therapy in the microbiome content of the oral-gut axis in RA.
Taken together, our data support the idea that the oral and gut microbiome can experience dramatic changes after MTX therapy. These results underscore the importance of the changes in the oral-gut axis microbiome to monitor the response to MTX therapy. However, this study also has shortcomings that should be acknowledged. Despite the limitations that apply to animal models, especially in the context of recapitulation of the complex process of RA in humans, we chose the AIA because it has high reproducibility and allows the quantification of the response with quantitative parameters of the disease. Finally, the cellular and molecular mechanisms underlying MTX rescue in the oral-gut axis microbiota and the protective activity of MTX against arthritis-induced alveolar bone loss remain to be defined.
5. Conclusion
In summary, despite the significance of these results at the individual microorganism level, treatment with MTX clearly demonstrated a significant impact on the diversity and composition of the oral and gut microbiota while protecting alveolar bone loss. An orchestrated interaction between pathogenic microorganisms that can lead to dysbiosis in arthritis is a remarkable phenomenon. Our findings also provide new insights into drug–microbiome–host interactions during the course of RA.
Supplementary Material
Acknowledgments
Research supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (2013/08216–2). The authors thank Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Finance code 001) and Conselho Nacional de Desenvolvimento Científico e Tecnológico. D.T.G. was the recipient of grant R01DE017732 from the National Institute of Dental and Craniofacial Research (NIDCR). Mrs. E. Greene provided English editing of the manuscript.
Source of funding
Fundação de Amparo à Pesquisa do Estado de São Paulo (Centro de Pesquisa em Doenças Inflamatórias) (2013/08216–2) and National Institute of Dental and Craniofacial Research (R01DE017732).
Footnotes
Declaration of competing interest
The authors have no conflicts of interest relevant to this article.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.anaerobe.2022.102577.
References
- [1].Smolen JS, Aletaha D, McInnes IB, Rheumatoid arthritis, Lancet 388 (10055) (2016. Oct 22) 2023–2038, 10.1016/S0140-6736(16)30173-8. [DOI] [PubMed] [Google Scholar]
- [2].Scher JU, Ubeda C, Equinda M, Khanin R, Buischi Y, Viale A, Lipuma L, Attur M, Pillinger MH, Weissmann G, Littman DR, Pamer EG, Bretz WA, Abramson SB, Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis, Arthritis Rheum. 64 (10) (2012. Oct) 3083–3094, 10.1002/art.34539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, Lan Z, Chen B, Li Y, Zhong H, Xie H, Jie Z, Chen W, Tang S, Xu X, Wang X, Cai X, Liu S, Xia Y, Li J, Qiao X, Al-Aama JY, Chen H, Wang L, Wu QJ, Zhang F, Zheng W, Li Y, Zhang M, Luo G, Xue W, Xiao L, Li J, Chen W, Xu X, Yin Y, Yang H, Wang J, Kristiansen K, Liu L, Li T, Huang Q, Li Y, Wang J, The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment, Nat. Med. 21 (8) (2015. Aug) 895–905, 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- [4].Lopez-Oliva I, Paropkari AD, Saraswat S, Serban S, Yonel Z, Sharma P, de Pablo P, Raza K, Filer A, Chapple I, Dietrich T, Grant MM, Kumar PS, Dysbiotic subgingival microbial communities in periodontally healthy patients with rheumatoid arthritis, Arthritis Rheumatol. 70 (7) (2018. Jul) 1008–1013, 10.1002/art.40485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Corrêa JD,Fernandes ^GR, Calderaro DC, Mendonça SMS, Silva JM,Albiero ML, Cunha FQ, Xiao E, Ferreira GA, Teixeira AL, Mukherjee C, Leys EJ, Silva TA, Graves DT, Oral microbial dysbiosis linked to worsened periodontal condition in rheumatoid arthritis patients, Sci. Rep. 9 (1) (2019. Jun 10) 8379, 10.1038/s41598-019-44674-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hamamoto Y, Ouhara K, Munenaga S, Shoji M, Ozawa T, Hisatsune J, Kado I, Kajiya M, Matsuda S, Kawai T, Mizuno N, Fujita T, Hirata S, Tanimoto K, Nakayama K, Kishi H, Sugiyama E, Kurihara H, Effect of Porphyromonas gingivalis infection on gut dysbiosis and resultant arthritis exacerbation in mouse model, Arthritis Res. Ther. 22 (1) (2020. Oct 19) 249, 10.1186/s13075-020-02348-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sato K, Takahashi N, Kato T, Matsuda Y, Yokoji M, Yamada M, Nakajima T, Kondo N, Endo N, Yamamoto R, Noiri Y, Ohno H, Yamazaki K, Aggravation of collagen-induced arthritis by orally administered Porphyromonas gingivalis through modulation of the gut microbiota and gut immune system, Sci. Rep. 7 (1) (2017. Jul 31) 6955, 10.1038/s41598-017-07196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cheng Z, Do T, Mankia K, Meade J, Hunt L, Clerehugh V, Speirs A, Tugnait A, Emery P, Devine D, Dysbiosis in the oral microbiomes of anti-CCP positive individuals at risk of developing rheumatoid arthritis, Ann. Rheum. Dis. 80 (2) (2021. Feb) 162–168, 10.1136/annrheumdis-2020-216972. [DOI] [PubMed] [Google Scholar]
- [9].Eriksson K, Fei G, Lundmark A, Benchimol D, Lee L, Hu YOO, Kats A, Saevarsdottir S, Catrina AI, Klinge B, Andersson AF, Klareskog L, Lundberg K, Jansson L, Yucel-Lindberg T, Periodontal health and oral microbiota in patients with rheumatoid arthritis, J. Clin. Med. 8 (5) (2019. May 8) 630, 10.3390/jcm8050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chu XJ, Cao NW, Zhou HY, Meng X, Guo B, Zhang HY, Li BZ, The oral and gut microbiome in rheumatoid arthritis patients: a systematic review, Rheumatology 60 (3) (2021. Mar 2) 1054e1066, 10.1093/rheumatology/keaa835. [DOI] [PubMed] [Google Scholar]
- [11].Nayak RR, Alexander M, Deshpande I, Stapleton-Gray K, Rimal B, Patterson AD, Ubeda C, Scher JU, Turnbaugh PJ, Methotrexate impacts conserved pathways in diverse human gut bacteria leading to decreased host immune activation, Cell Host Microbe 29 (3) (2021. Mar 10) 362–377, 10.1016/j.chom.2020.12.008, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lopez-Olivo MA, Siddhanamatha HR, Shea B, Tugwell P, Wells GA, Suarez-Almazor ME, Methotrexate for treating rheumatoid arthritis, Cochrane Database Syst. Rev. 2014 (6) (2014. Jun 10), CD000957, 10.1002/14651858.CD000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Artacho A, Isaac S, Nayak R, Flor-Duro A, Alexander M, Koo I, Manasson J, Smith PB, Rosenthal P, Homsi Y, Gulko P, Pons J, Puchades-Carrasco L, Izmirly P, Patterson A, Abramson SB, Pineda-Lucena A, Turnbaugh PJ, Ubeda C, Scher JU, The pretreatment gut microbiome is associated with lack of response to methotrexate in new-onset rheumatoid arthritis, Arthritis Rheumatol. 73 (6) (2021. Jun) 931–942, 10.1002/art.41622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Romero-Sanchez C, Rodríguez C, Santos-Moreno P, Mesa AM, Lafaurie GI, Giraldo QS, De-Avila J, Castillo DM, Duran M, Chalem PC, Bello Gualtero JM, Valle-Oñate R, Is the treatment with biological or non-biological DMARDS a modifier of periodontal condition in patients with rheumatoid arthritis? Curr. Rheumatol. Rev. 13 (2) (2017) 139–151, 10.2174/1573397113666170407161520. [DOI] [PubMed] [Google Scholar]
- [15].Lübcke PM, Ebbers MNB, Volzke J, Bull J, Kneitz S, Engelmann R, Lang H, Kreikemeyer B, Müller-Hilke B, Periodontal treatment prevents arthritis in mice and methotrexate ameliorates periodontal bone loss, Sci. Rep. 9 (1) (2019. May 31) 8128, 10.1038/s41598-019-44512-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pinto LG, Talbot J, Peres RS, Franca RF, Ferreira SH, Ryffel B, Aves-Filho JC, Figueiredo F, Cunha TM, Cunha FQ, Joint production of IL-22 participates in the initial phase of antigen-induced arthritis through IL-1β production, Arthritis Res. Ther. 17 (1) (2015. Sep 2) 235, 10.1186/s13075-015-0759-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Peres RS, Liew FY, Talbot J, Carregaro V, Oliveira RD, Almeida SL, França RF, Donate PB, Pinto LG, Ferreira FI, Costa DL, Demarque DP, Gouvea DR, Lopes NP, Queiroz RH, Silva JS, Figueiredo F, Alves-Filho JC, Cunha TM, Ferreira SH, Louzada-Junior P, Cunha FQ, Low expression of CD39 on regulatory T cells as a biomarker for resistance to methotrexate therapy in rheumatoid arthritis, Proc. Natl. Acad. Sci. U. S. A. 112 (8) (2015. Feb 24) 2509–2514, 10.1073/pnas.1424792112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Schneider AH, Machado CC, Veras FP, Maganin AGM, de Souza FFL, Barroso LC, de Oliveira RDR, Alves-Filho JC, Cunha TM, Fukada SY, Louzada-Júnior P, da Silva TA, Cunha FQ, Neutrophil extracellular traps mediate joint hyperalgesia induced by immune inflammation, Rheumatology 60 (7) (2021. Jul 1) 3461–3473, 10.1093/rheumatology/keaa794. [DOI] [PubMed] [Google Scholar]
- [19].Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R, Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms, ISME J. 6 (8) (2012. Aug) 1621–1624, 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP, DADA2: high-resolution sample inference from Illumina amplicon data, Nat. Methods 13 (7) (2016. Jul) 581–583, 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McLaren MR, Callahan BJ, Silva 138.1 Prokaryotic SSU Taxonomic Training Data Formatted for DADA2 [Data Set], Zenodo, 2021, 10.5281/zenodo.4587955. [DOI] [Google Scholar]
- [22].McMurdie PJ, Holmes S, phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data, PLoS One 8 (4) (2013. Apr 22), e61217, 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP, Bioconductor workflow for microbiome data analysis: from raw reads to community analyses, F1000Res 5 (2016. Jun 24) 1492, 10.12688/f1000research.8986.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Oksanen JF, Blanchet G, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H, vegan: community Ecology Package. R package version 2.5–7. https://CRAN.R-project.org/package=vegan, 2020. [Google Scholar]
- [25].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol. 15 (12) (2014) 550, 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lê Cao KA, Costello ME, Lakis VA, Bartolo F, Chua XY, Brazeilles R, Rondeau P, MixMC: a multivariate statistical framework to gain insight into microbial communities, PLoS One 11 (8) (2016. Aug 11), e0160169, 10.1371/journal.pone.0160169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Maciejewski M, Sands C, Nair N, Ling S, Verstappen S, Hyrich K, Barton A, Ziemek D, Lewis MR, Plant D, Prediction of response of methotrexate in patients with rheumatoid arthritis using serum lipidomics, Sci. Rep. 11 (1) (2021. Mar 31) 7266, 10.1038/s41598-021-86729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Letertre MPM, Munjoma N, Wolfer K, Pechlivanis A, McDonald JAK, Hardwick RN, Cherrington NJ, Coen M, Nicholson JK, Hoyles L, Swann JR, Wilson ID, A two-way interaction between methotrexate and the gut microbiota of male Sprague-Dawley rats, J. Proteome Res. 19 (8) (2020. Aug 7) 3326–3339, 10.1021/acs.jproteome.0c00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Corrêa JD, Saraiva AM, Queiroz-Junior CM, Madeira MF, Duarte PM, Teixeira MM, Souza DG, da Silva TA, Arthritis-induced alveolar bone loss is associated with changes in the composition of oral microbiota, Anaerobe 39 (2016. Jun) 91–96, 10.1016/j.anaerobe.2016.03.006. [DOI] [PubMed] [Google Scholar]
- [30].Park OJ, Yi H, Jeon JH, Kang SS, Koo KT, Kum KY, Chun J, Yun CH, Han SH, Pyrosequencing analysis of subgingival microbiota in distinct periodontal conditions, J. Dent. Res. 94 (7) (2015. Jul) 921–927, 10.1177/0022034515583531. [DOI] [PubMed] [Google Scholar]
- [31].Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ, Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing, ISME J. 6 (6) (2012. Jun) 1176–1185, 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Buschhart AL, Bolten L, Volzke J, Ekat K, Kneitz S, Mikkat S, Kreikemeyer B, Müller-Hilke B, Periodontal pathogens alter the synovial proteome. Periodontal pathogens do not exacerbate macroscopic arthritis but alter the synovial proteome in mice, PLoS One 15 (12) (2020. Dec 31), e0242868, 10.1371/journal.pone.0242868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Coit P, Sawalha AH, The human microbiome in rheumatic autoimmune diseases: a comprehensive review, Clin. Immunol. 170 (2016. Sep) 70–79, 10.1016/j.clim.2016.07.026. [DOI] [PubMed] [Google Scholar]
- [34].Joshi V, Matthews C, Aspiras M, de Jager M, Ward M, Kumar P, Smoking decreases structural and functional resilience in the subgingival ecosystem, J. Clin. Periodontol. 41 (11) (2014. Nov) 1037–1047, 10.1111/jcpe.12300. [DOI] [PubMed] [Google Scholar]
- [35].Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J, Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples, Genome Biol. 13 (6) (2012. Jun 14) R42, 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Arimatsu K, Yamada H, Miyazawa H, Minagawa T, Nakajima M, Ryder MI, Gotoh K, Motooka D, Nakamura S, Iida T, Yamazaki K, Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota, Sci. Rep. 4 (2014. May 6) 4828, 10.1038/srep04828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].King TJ, Georgiou KR, Cool JC, Scherer MA, Ang ES, Foster BK, Xu J, Xian CJ, Methotrexate chemotherapy promotes osteoclast formation in the long bone of rats via increased pro-inflammatory cytokines and enhanced NF-κB activation, Am. J. Pathol. 181 (1) (2012. Jul) 121–129, 10.1016/j.ajpath.2012.03.037. [DOI] [PubMed] [Google Scholar]
- [38].Rogier R, Evans-Marin H, Manasson J, van der Kraan PM, Walgreen B, Helsen MM, van den Bersselaar LA, van de Loo FA, van Lent PL, Abramson SB, van den Berg WB, Koenders MI, Scher JU, Abdollahi-Roodsaz S, Alteration of the intestinal microbiome characterizes preclinical inflammatory arthritis in mice and its modulation attenuates established arthritis, Sci. Rep. 7 (1) (2017. Nov 15) 15613, 10.1038/s41598-017-15802-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zaiss MM, Joyce Wu HJ, Mauro D, Schett G, Ciccia F, The gut-joint axis in rheumatoid arthritis, Nat. Rev. Rheumatol. 17 (4) (2021. Apr) 224–237, 10.1038/s41584-021-00585-3. [DOI] [PubMed] [Google Scholar]
- [40].Magne F, Gotteland M, Gauthier L, Zazueta A, Pesoa S, Navarrete P, Balamurugan R, The Firmicutes/Bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients? Nutrients 12 (5) (2020. May 19) 1474, 10.3390/nu12051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhou B, Xia X, Wang P, Chen S, Yu C, Huang R, Zhang R, Wang Y, Lu L, Yuan F, Tian Y, Fan Y, Zhang X, Shu Y, Zhang S, Bai D, Wu L, Xu H, Yang L, Induction and amelioration of methotrexate-induced gastrointestinal toxicity are related to immune response and gut microbiota, EBioMedicine 33 (2018. Jul) 122–133, 10.1016/j.ebiom.2018.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Newkirk MM, Zbar A, Baron M, Manges AR, Distinct bacterial colonization patterns of Escherichia coli subtypes associate with rheumatoid factor status in early inflammatory arthritis, Rheumatology 49 (7) (2010. Jul) 1311–1316, 10.1093/rheumatology/keq088. [DOI] [PubMed] [Google Scholar]
- [43].Rovenský J, Stancíková M, Svík K, Utesený J, Bauerov a K, Jurcovicová J, Treatment of adjuvant-induced arthritis with the combination of methotrexate and probiotic bacteria Escherichia coli O83 (Colinfant), Folia Microbiol. 54 (4) (2009) 359–363, 10.1007/s12223-009-0045-2. [DOI] [PubMed] [Google Scholar]
- [44].Rovenský J, Svík K, Matha V, Istok R, Kamarád V, Ebringer L, Ferencík M, Stancíková M, Combination treatment of rat adjuvant-induced arthritis with methotrexate, probiotic bacteria Enterococcus faecium, and selenium, Ann. N. Y. Acad. Sci. 1051 (2005. Jun) 570–581, 10.1196/annals.1361.101. [DOI] [PubMed] [Google Scholar]
- [45].Xu H, Zhao H, Fan D, Liu M, Cao J, Xia Y, Ju D, Xiao C, Guan Q, Interactions between gut microbiota and immunomodulatory cells in rheumatoid arthritis, Mediat. Inflamm. 2020 (2020. Sep 9), 1430605, 10.1155/2020/1430605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Yan H, Su R, Xue H, Gao C, Li X, Wang C, Pharmacomicrobiology of methotrexate in rheumatoid arthritis: gut microbiome as predictor of therapeutic response, Front. Immunol. 12 (2021. Dec 16), 789334, 10.3389/fimmu.2021.789334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Farooq SM, Ashour HM, Type II collagen-specific B cells induce immune tolerance in Th1-Skewed, Th2-Skewed, and arthritis-prone strains of mice, Cells 10 (4) (2021. Apr 12) 870, 10.3390/cells10040870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


