Abstract
The study of protein complexes and protein-protein interactions is of great importance due to their fundamental roles in cellular function. Proximity labeling, often coupled with mass spectrometry, has become a powerful and versatile tool for studying protein-protein interactions by enriching and identifying proteins in the vicinity of a specified protein-of-interest. Here, we describe and compare traditional approaches to investigate protein-protein interactions to current day state-of-the-art proximity labeling methods. We focus on the wide array of proximity labeling strategies and underscore studies using diverse model systems to address numerous biological questions. In addition, we highlight current advances in mass spectrometry-based technology that exhibit promise in improving the depth and breadth of the data acquired in proximity labeling experiments. In all, we show the diversity of proximity labeling strategies and emphasize the broad range of applications and biological inquiries that can be addressed using this technology.
1. Introduction
Protein-protein interactions play critical roles in all aspect of cellular life. From cell structure to complex signaling networks, and from stable to transient interactions, developing deeper knowledge of the specific proteins that interact and the factors that drive their association, dissociation, and duration of interaction is of great importance. These interactions are essential for the proper function of the processes of life, and are often blocked, perturbed, or altered in disease.
Protein-protein interactions are also very complex. It is estimated that the human body contains 200 different cell types and, at a minimum, 20,000 nonmodified proteins (Ponomarenko et al., 2016). Each cell type has a different complement of proteins with changing levels of protein expression that are organized into spatially distinct subcellular compartments. Furthermore, proteins can be found in different proteoforms arising from alternative splicing during expression, single amino acid polymorphisms, and posttranslational modifications. All of these different factors have effects on protein-protein interactions. For example, the role and interacting partners of a protein in a subcompartment of one cell type may be quite different than the role and interacting partners of that same protein in a different subcompartment of a different cell type. Also, as seen often with proteins involved in intercellular signaling pathways, reversible posttranslational modifications, such as phosphorylation, can dictate association/dissociation of different interactors to a given protein.
In the last two decades, different methods involving proximity labeling have been developed to study protein-protein interactions. In this review, we highlight the breadth and depth of these methods for the study of known protein-protein interactions as well as for the discovery of new protein interactors. Our aim is to provide the reader a clear comparison among proximity labeling methods as well as classical nonproximity labeling methods. We also highlight advances in mass spectrometry technology to enhance data acquisition which can be coupled to proximity labeling strategies. Of note, many of the proximity labeling techniques discussed herein have also been adapted for the study of protein-RNA or protein-DNA interactions to which we direct readers to other excellent reviews on the topic (Qin, Cho, Cavanagh, & Ting, 2021; Trinkle-Mulcahy, 2019; Ummethum & Hamperl, 2020; Zhou & Zou, 2021).
2. Nonproximity labeling based methods for studying protein-protein interactions
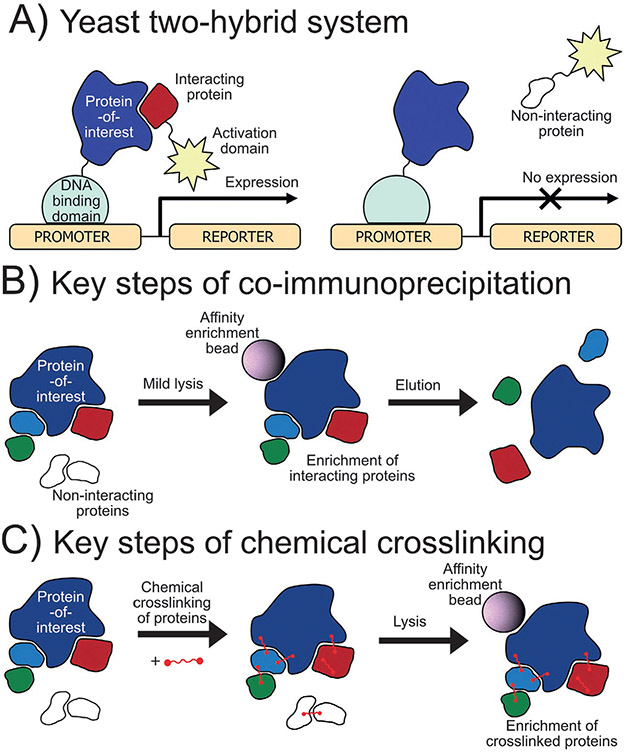
One common nonproximity based method to identify protein-protein interactors is the yeast two-hybrid (Y2H) system which was reported first in 1989 (Fields & Song, 1989). Different variations of this method have been developed but in general, the protein-of-interest and potential binding partners are expressed in genetically modified yeast as fusions to the DNA binding domain and the activation domain of a transcription factor, respectively (Fig. 1A). Upon interaction of the two proteins, the activation and DNA binding domains of the transcription factor are reconstituted upstream to a reporter gene that often allows yeast to grow in a selective media or induces a color change for detection. Advantages such as cost, parallel experimentation, and no requirement to purify large amounts of protein has led to widespread use of this method including recently in a high-throughput screen to identify approximately 53,000 protein-protein interactions (Luck et al., 2020). However, drawbacks such as low expression proteins-of-interest, misfolding in yeast, lack of or abnormal posttranslational modifications (such as phosphorylation) that modulate protein-protein interactions, and the inability to determine multiprotein complexes also limit the scope of this technique.
FIG. 1.
Traditional methods for studying protein-protein interactions. (A) Yeast two-hybrid method to determine protein-protein interactors: A protein-of-interest is fused to the DNA binding domain while the potential interactor is fused to the activation domain of a functional transcription factor. If the proteins interact, the transcription factor is activated producing a color or allowing growth in selective media. (B) Coimmunoprecipitation of protein interactors: A protein-of-interest is purified by affinity chromatography by antibody or affinity tag bringing along interactors. (C) Chemical crosslinking of protein interactors: Bifunctional linkers are used to covalently crosslink protein interactors that are then purified by affinity chromatography.
Another popular method to study protein-protein interactions is coimmunoprecipitation (co-IP). In this method, cells expressing the protein-of-interest are lysed under mild conditions and the protein-of-interest is enriched along with interacting partners by affinity chromatography using either a primary antibody or an enrichment tag fused to the protein-of-interest (Fig. 1B). This approach is often used as a verification method for protein-protein interactions discovered by other methods using immunoblotting for detection. This affinity purification approach has also been coupled to mass spectrometry (AP-MS) and used widely in studies aiming to discover novel interactors, such as the large scale BioPlex project (Huttlin et al., 2021, 2017, 2015). The advantages of this technique include high sensitivity, detection of multiprotein complexes, and the ability to detect and quantify posttranslational modifications that may affect the protein-protein interactions. Limitations of AP-MS include the requirement of a high-quality primary antibody or proper fusion to an enrichment tag, high background from nonspecific protein binding during enrichment, and the loss of weak or transient protein interactors during purification.
Chemical crosslinking mass spectrometry (XL-MS) was developed to address the difficulty of identifying weak or transient protein-protein interactions and allow for harsher washing to remove nonspecific binders. In this technique, a chemical reagent is added to crosslink nearby proteins through covalent bonds (Fig. 1C) or, alternatively, unnatural amino acids may be incorporated into proteins to facilitate crosslinking. Often, these unnatural amino acids have side chain functionality that is photosensitive and generates a radical on the side chain that can react with interacting proteins forming a direct covalent bond. Once linked, the proteins can be enriched by affinity chromatography and processed for mass spectrometry analysis. The covalent linking of the proteins traps even transient protein-protein interactions and permits stronger washing during enrichment to improve signal-to-noise (Sinz, 2006, 2010). Many crosslinking reagents are available that vary in length, reactivity, cleavability, and other properties (Yakovlev, 2009). This technique has been used recently to map 6439 interactions among 2484 proteins in HEK293 cells (Wheat et al., 2021). One limitation of this method has been the difficulty of mass spectrometry data processing due to the increased and more complex search space, however, today many software tools have been and continue to be developed to address this challenge (Mintseris & Gygi, 2020; Yılmaz et al., 2018).
In addition, pairing chromatographic separation of a native complex and mass spectrometry may facilitate a high-throughput means of studying global protein-protein interactions. Protein correlation profiling (Foster et al., 2006) couples native protein cofractionation of complexes with global mass spectrometry-based analysis to correlate chromatographic profiles and infer protein-protein interactions. The complexes are generally separated according to their hydrodynamic radius by size-exclusion chromatography (Bosch, Chen, & Perrimon, 2021). In such an analysis, hundreds of complexes that represent thousands of interactions can be present in each fraction (Bludau et al., 2020; Heusel et al., 2019). Protein-protein interactions and protein complex composition can be inferred as protein subunits of the same complex should have consistent size-exclusion chromatography profiles (Fossati et al., 2021). These data based on chromatographic coelution and associated protein-protein interactions and protein complexes can be used to both qualitatively and quantitatively compare across biological conditions, thereby facilitating the discovery of condition-specific differences (Heusel et al., 2020). The coupling of protein correlation profiling with advanced mass spectrometry technology holds much promise to elucidate protein-protein interactions on a global scale.
The value of the methods discussed above cannot be understated, however, the drawbacks of these methods have driven a search for a complementary general method that can be (1) used in a more endogenous environment, (2) used in the study of multiprotein complexes, (3) capture even the most transient interactions, and (4) have a rapid on/off mechanism for temporal control. Proximity labeling methods that conjugate a “reporter” to proteins near or in direct contact with a protein-of-interest have been developed to achieve this goal. Though outside of the scope of this review, a DNA proximity labeling method is often cited as the inspiration for many proximity-based methods to study protein-protein interactions. In 2000, van Steensel and Henikoff introduced DamID, a technique that fuses a DNA adenine methyltransferase (Dam) from Escherichia coli to a chromatin protein-of-interest resulting in the local methylation of N6-position of adenine in the DNA sequence GATC. The sites of methylation can then be determined by antibodies and quantified by PCR. This technique was used to map the target loci of heterochromatin protein 1 (HP1) in Drosophila cells and whole flies identifying positions that previously had no direct evidence of HP1 interaction (van Steensel & Henikoff, 2000). This general approach—fusion of a protein or part of a protein that drives labeling to or near a protein-of-interest—is a key concept to many of the proximity labeling techniques discussed.
3. Proximity labeling methods for studying protein-protein interactions
3.1. Binary interaction methods
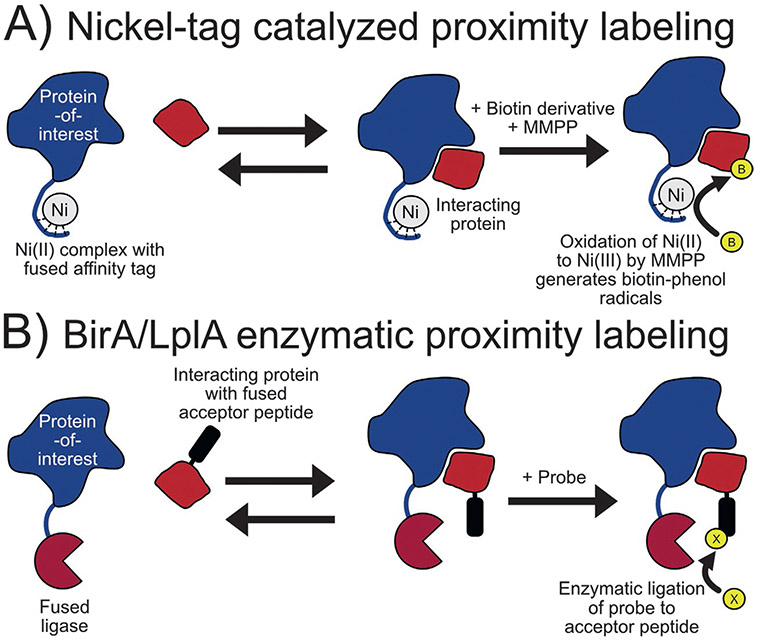
First used for protein crosslinking (Fancy, Melcher, Johnston, & Kodadek, 1996), chemically induced proximity biotinylation using nickel-tag complex can be used for the quantitative characterization of protein-protein and protein-peptide interactors. Using purified proteins, nickel is coordinated to the hexahistidine (His6) or N-terminal glycine-glycine-histidine (NH2-GGH)-tag of one protein before adding the interacting partner at various concentrations in the presence of biotin tyramide. Proximity labeling is initiated by the addition of a peroxy acid, magnesium monoperoxyphthalate hexahydrate (MMPP) (Fig. 2A). The peroxy acid oxidizes the Ni(II) that is localized to the protein tag to a Ni(III) species then mediates biotinylation of nearby electron-rich amino acids by a radical mechanism. Labeling is then quenched and the extent of biotinylation of the protein partner was determined by immunoblotting (Amini, Kodadek, & Brown, 2002). Using this technique, Amini and coworkers determined the stoichiometry and KD of known interactors TATA binding protein/Gal4 activation domain fusion protein and Rad23/ubiquitin with values in good agreement to the values determined by other techniques (Amini, Denison, Lin, Kuo, & Kodadek, 2003).
FIG. 2.
Binary interaction proximity labeling methods. (A) Proximity labeling by nickel (Ni) catalyst: Nickel is coordinated to His6 or NH2-GGH tag then oxidized to catalyze the radical activation of biotin tyramide. (B) Mechanism of proximity labeling using the BirA/biotin acceptor peptide and mutant lipoic acid ligase/ligase acceptor peptide techniques: The fused enzyme links a probe to the specific acceptor peptide if the two proteins are interacting.
BirA is a 35 kDa protein expressed in E. coli that functions as a biotin ligase or biotin-operon repressor in response to the cellular level of biotin (Barker & Campbell, 1981a, 1981b). As a biotin ligase, BirA is highly specific for its endogenous substrate: a subunit of acetyl-CoA carboxylases. An unnatural minimal peptide substrate, called biotin acceptor peptide, was created during the course of investigating BirA substrate specificity (Beckett, Kovaleva, & Schatz, 1999; Schatz, 1993) and was later expressed fused to a protein allowing quantitative in vivo biotinylation for protein purification and assay development (Smith et al., 1998). In 2008, this enzyme-substrate pair was adapted further for studying protein-protein interactors in vitro and in mammalian cell culture. Here, a protein-of-interest is expressed as a fusion with BirA while another known partner is expressed tagged with a truncated derivative of the biotin acceptor peptide. Exogenous biotin addition then promotes biotinylation of the biotin acceptor peptide by BirA should the two proteins be in close proximity (Fig. 2B). The efficacy of this technique was shown in the study of FKBP/FRB and Cdc25C phosphatase/14-3-3ε protein interacting pairs (Fernández-Suárez, Chen, & Ting, 2008). Further adaptation of this technique optimized the biotin acceptor peptide sequence for detection by mass spectrometry after proteolysis and allowed for multiplexing analysis (Kulyyassov et al., 2011). Named PUB-ChIP, this technique was later used to study the effect of RAB18 in close proximity to chromatin, finding that this potential interaction has an effect on the level and pattern of H4 acylation (Shoaib et al., 2013).
Similar to the BirA-BAP strategy, Interaction-Dependent PRobe Incorporation Mediated by Enzymes or ID-PRIME uses a mutant lipoic acid ligase, LplA* (W37A, T57I, F147L, H267R), from E. coli and ligase acceptor peptide in a manner analogous to the BirA-biotin acceptor peptide pair but using biotin, lipoic acid, or picolyl azide as the labeling probe (Liu, Loh, Lam, White, & Ting, 2013). This method was then utilized with a click chemistry detection strategy (Uttamapinant et al., 2012) to image neurexin and neuroligin interactions in HEK, mixed HEK-neuron, and pure neuron cell cultures.
These proximity labeling methods for the study of binary protein-protein interactions offer useful options, however, these methods are limited to the study of the association/dissociation of known protein-protein interactors. In the following sections we will discuss more promiscuous proximity labeling methods that allow for the study of multiprotein complexes and the discovery of previously unknown protein-protein interactors.
3.2. Ligase-based methods
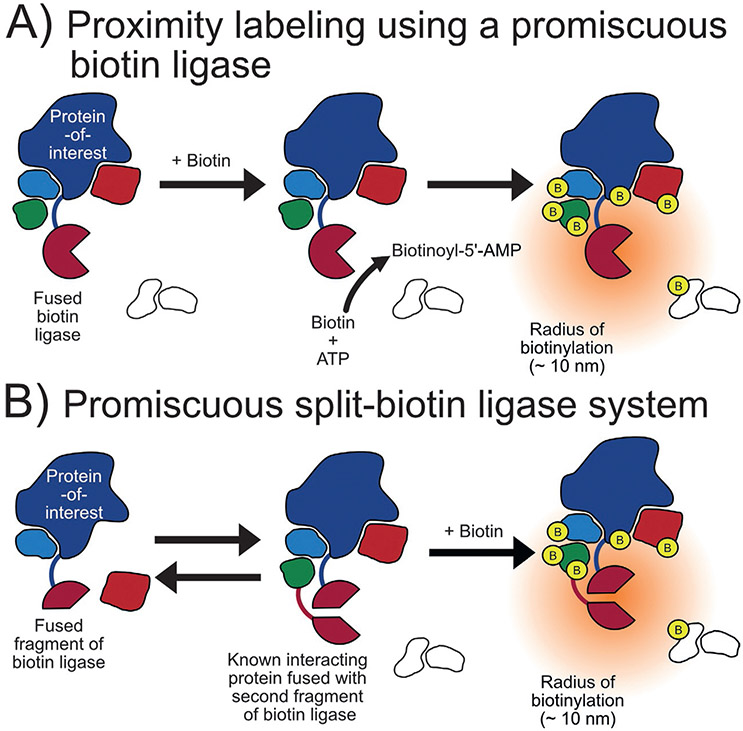
Biochemical profiling of BirA identified a number of mutations that effect enzymatic activity by altering substrate, biotin, and intermediate biotinoyl-5′-AMP binding. One of these mutants, R118GBirA, results in a 400-fold greater dissociation constant for biotinoyl-5′-AMP indicating that it leaks this intermediate into its vicinity (Kwon & Beckett, 2000). Once leaked, this reactive biotin derivate enables R118GBirA to promiscuously label nearby proteins through biotinylation of lysine residues (Choi-Rhee, Schulman, & Cronan, 2004) (Fig. 3A). Inspired by DamID, Roux and coworkers leveraged the promiscuity of R118GBirA to develop BioID and highlighted the technique by identifying the proteins in proximity of human lamin A. Lamin A is a filament protein that is a main constituent of the inner nuclear membrane that faces the interior of the nucleus and is insoluble making conventional methods to investigate molecular interactors difficult. BioID enabled the identification of known components of the nuclear membrane as well as previously unknown proteins, including a novel peripheral membrane protein (Roux, Kim, Raida, & Burke, 2012). BioID was later determined to have an approximate labeling radius of 10nm while investigating the architecture of the nuclear pore complex (Kim et al., 2014). BioID has been used in vivo to identify putative c-MYC interactors in xenograft tumors in mice (Dingar et al., 2015) and to discover new inhibitory post-synaptic proteins in murine brain tissue (Uezu et al., 2016). BioID has also been used widely in many other in vivo studies including in Toxoplasma gondii in which a protein interaction network analysis revealed conoid protein hub 1 (CPH1) functions as a hub linking key motor and structural proteins for microtubule organization (Long, Anthony, Drewry, & Sibley, 2017).
FIG. 3.
Biotin ligase-based proximity labeling methods. (A) Mechanism of promiscuous biotin ligases such as BioID: The biotin ligase is fused to a protein-of-interest which then activates biotin as a biotinoyl-5′-AMP and labels the lysine residues of nearby proteins. (B) The split-biotin ligase system: The biotin ligase is split into two parts that can only catalyze proximity labeling only once brought together.
A smaller biotin ligase from Aquifex aeolicus (27kDa) was determined to be functionally comparable to R118GBirA when used for proximity labeling of interactors of lamin A, as done with BioID. Called BioID2, this method uses a ligase containing a R40G mutation which is analogous to the R118G mutation of BirA in BioID. This study also determined that BioID2 is less disruptive due to its smaller size and sustains high levels of biotinylation at low biotin concentrations (>3.2μM) when compared to BioID (50μM). Also of note, it was found that both enzymes show reduced activity at temperatures below 37°C and that the radius of proximity labeling of BioID2 can be increased when a longer linker is placed between the two regions of the fusion protein (Kim et al., 2016). More recently, a BioID2 knock-in mouse model was developed that identified potentially novel cardiac dyad proteins (Feng et al., 2020) and was used in zebrafish to profile interactors of ErbB2 in cardiomyocytes (Pronobis, Zheng, Singh, Goldman, & Poss, 2021).
Three versions of a reporter-fragment complementation assay utilizing BioID have been reported in the literature. In reporter-fragment complementation assays, a reporter is split into two parts that only become active when close enough to reform the intact reporter (Morell, Ventura, & Avilés, 2009) (Fig. 3B). This arrangement adds more spatiotemporal control to labeling as the reporter, in this case a biotin ligase, is only active when reformed. In 2017, De Munter and coworkers split R118GBirA at E140/Q141 and fused one part to protein phosphatase PP1 (PP1) and the other to two known interactors of PP1, nuclear inhibitor of PP1 or RepoMan. In addition to identifying known interactors of each protein, this approach allowed for the determination of apparent interactors of the PP1-NIPP1 complex to a much greater extent than the PPI-RepoMan complex (De Munter et al., 2017). Schopp and coworkers also developed a split-BioID in 2017 by splitting the ligase at E256/G257 and used this system to investigate the miRNA silencing pathway. Each fragment was fused to Ago2, a known member of two functionally distant complexes in the silencing pathway, and the other part was fused to a specific component of each complex. Experiments using each pair, Ago2-TNRC6C and Ago2-Dicer, identified distinct proteins reflecting Ago2s different role in each complex (Schopp et al., 2017). More recently, Kwak and coworkers introduced a third split-BioID system, called Contact-ID, that split R118GBirA at G78/G79 after having considered B factors (a calculation of the atomic flexibility in the crystalline state) to choose the split sites investigated. This system was then used to map the contact site of the endoplasmic reticulum (ER) and mitochondria by conjugating each piece to SEC61B (part of the ER cytosolic membrane) and TOM20 (part of the outer mitochondrial membrane) and identified 115 proteins that implied that steroidogenesis may be an important function of the mitochondria-associated membrane (Kwak et al., 2020).
In 2018, TurboID (35kDa) and miniTurboID (28kDa) were introduced as more efficient alternatives to BioID and BioID2 that reduced the necessary labeling time from >16h to 10 min. Each enzyme was engineered from R118SBirA using a yeast display-based directed evolution method. TurboID was found to be slightly more active than miniTurboID (1.5–2-fold) but promoted more labeling prior the delivery of exogenous biotin. Both new enzymes were then compared to BioID in various experiments in cell culture, flies, worms, and yeast concluding that, compared to BioID, both show rapid kinetics, are more functional at lower temperatures, and can be more useful for in vivo studies with the precautions outlined by the authors (Branon et al., 2018). For instance, the protein network and associating proteins of plant transcription factors, FAMA, have been investigated in Arabidopsis and Nicotiana benthamiana using TurboID and miniTurboID (Mair, Xu, Branon, Ting, & Bergmann, 2019).
The same laboratory then developed a split-TurboID system in 2020, which operates as described for the split-BioID versions discussed previously, however, requires a much-reduced labeling time (4h). Screening 14 different split sites determined that TurboID split at L73/G74 gave the desired characteristics and was used to examine ER-mitochondria contacts by conjugating one piece of split-TurboID to Cb5 (part of the ER cytosolic membrane) and the other to TOM20 (part of the outer mitochondrial membrane). The resulting dataset identified 17 proteins that had not been reported previously at the ER-mitochondria interface, including two (FUNDC2 and MTFR1) that appear to have tethering functions at the interface (Cho et al., 2020). This technology has also been applied in vivo, such as for identifying proximity partners of lamin A in Zebrafish embryos (Rosenthal et al., 2021) and to determine interaction networks for Cavin4 and Cavin1 proteins in Zebrafish muscle cells (Xiong et al., 2021).
A de novo designed BirA enzyme was recently constructed using an ancestral reconstruction algorithm analysis of metagenome data for use in proximity labeling. This novel enzyme, called AirID, was then characterized and compared to BioID and TurboID in various experiments for perspective. It was then successfully applied in a proximity labeling experiment that identified interactors of IκBα in mammalian cells (Kido et al., 2020).
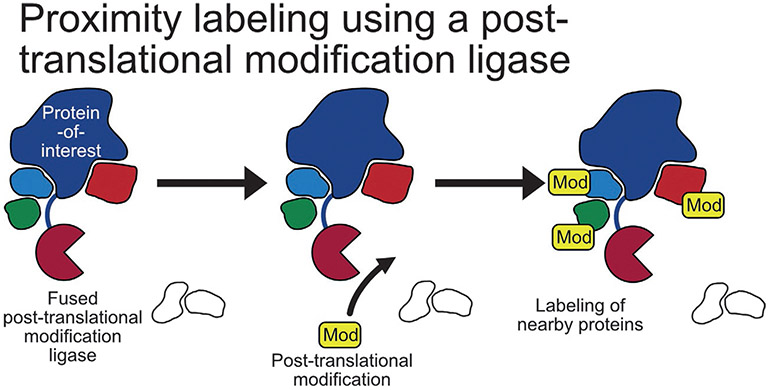
A posttranslation modification-based proximity labeling approach that leveraged a NEDD8 E2-conjugating enzyme, Ubc12, fused to a protein-of-interest was reported in 2013. Called NEDDylator, this system uses Ubc12 to attach NEDD8, a stable ubiquitin homolog, to lysine residues of proteins near the protein-of-interest which can subsequently be enriched by a histidine-biotin (HB) tag on NEDD8 (Fig. 4). This technology was then used in cell lysates to identify potential substrates of two RING-type E3 ubiquitin transferases, XIAP and cIAP1, and confirming one of the substrates found, PGAM5, is proapoptotic (Zhuang, Guan, Wang, Burlingame, & Wells, 2013). Interestingly, this method has also be used to identify the protein targets of a small molecule (Hill, Pollock, Zhuang, & Wells, 2016).
FIG. 4.
Posttranslational modification proximity labeling methods. Mechanism of posttranslational modification proximity labeling: A posttranslational modification ligase is fused to a protein-of-interest that links a modification to nearby proteins that can later be used as an enrichment handle.
PUP-IT, a proximity labeling method similar to NEDDylator uses a ligase, PafA, of a small protein, Pup, both from Corynebacterium glutamicum. In this method, a protein-of-interest fused to PafA is then coexpressed with a modified version of Pup that itself is fused to a bacteria-derived carboxylase domain that becomes biotinylated in cells. Coexpression results in the conjugation of the modified Pup to lysine residues of proteins near the protein-of-interest that can subsequently be enriched by affinity chromatography (Fig. 4). This method was then used to identify interactors of CD28, including >200 possible interactors of the C-terminal tail region (Liu et al., 2018).
Ligase-based methods for proximity labeling to investigate protein-protein interactions have been utilized widely in the literature, especially BioID and TurboID. Unlike the proximity labeling methods to study binary interactions, these methods allow for the straightforward discovery of potentially unknown interacting partners to a protein-of-interest. Furthermore, simply requiring only the addition of nontoxic, exogenous biotin has allowed many of these methods to be used in animal models. On drawback of using ligase-based methods, however, is that at least 10 min are required for effective proximity labeling which has spawned the development of peroxidase-based methods in experiments requiring faster kinetics.
3.3. Peroxidase-based methods
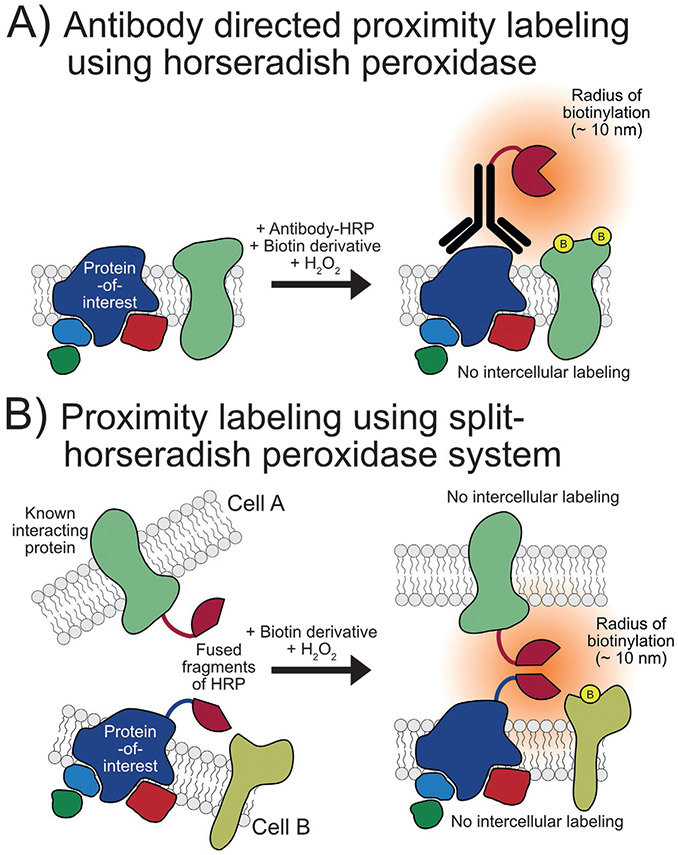
In 2008, Kotani and coworkers reported the discovery of an aryl azide-biotin reagent conversion to an active radical species by horseradish peroxidase (HRP), termed “enzyme-mediated activation of radical source” or “EMARS” reaction. This reaction was then leveraged to visualize molecular clusters on the surface of living cells by first treating the cells with a primary antibody to each of β1 integrin, EGFR, IGF1R, and EphA2 followed by an HRP-conjugated secondary antibody and finally aryl azide-biotin resulting in the radical biotinylation of electron-rich amino acids (primarily tyrosine) of nearby proteins on the cell surface. The level of biotinylation of receptor tyrosine kinases (RTKs) in proximity of each protein-of-interest was then visualized using an antibody array kit showing that different cellular clusters can be investigated using EMARS (Kotani et al., 2008). This technique was used later in combination with proteomics to identify 30 membrane and secreted proteins that potentially cocluster with ganglioside GM1, a lipid raft marker (Jiang et al., 2012).
In 2014, Li and coworkers developed a method for mapping cell surface interactors called selective proteomic proximity labeling using tyramide (SPPLAT). Similar to EMARS, this method uses biotin tyramide chemistry catalyzed by exogenous hydrogen peroxide to label the proteins near a protein-of-interest targeted by HRP-conjugated antibody recognition (Fig. 5A). This technique was then used for the proximity labeling of proteins within IgM-class B cell receptor clusters that identified 12 proteins of high significance, half of which had not been reported previously as possible components of the cluster (Li et al., 2014).
FIG. 5.
Horseradish peroxidase-based proximity labeling methods. (A) Use of HRP-linked antibody for the proximity labeling of proteins near a protein-of-interest on a cell surface. (B) Use of a split-HRP system for the proximity labeling at a cell-cell interface.
In 2016, a split-HRP (sHRP) was engineered by Martell and coworkers using yeast display to improve evolution of a reporter-fragment complementation assay for the first time (Fig. 5B). After screening 17 cuts to determine 213/214 was the best split site, a yeast display-based directed evolution was utilized to improve signal yielding a sHRP which has six mutations compared to HRP: T21I, P78S, R93G, N175S, N255D, and L299R. The authors then used sHRP to visualize synapses between specific sets of neurons (Martell et al., 2016). Although, to our knowledge, sHRP has not been used in a proteomics experiment to date, it has potential to be utilized in future experiments.
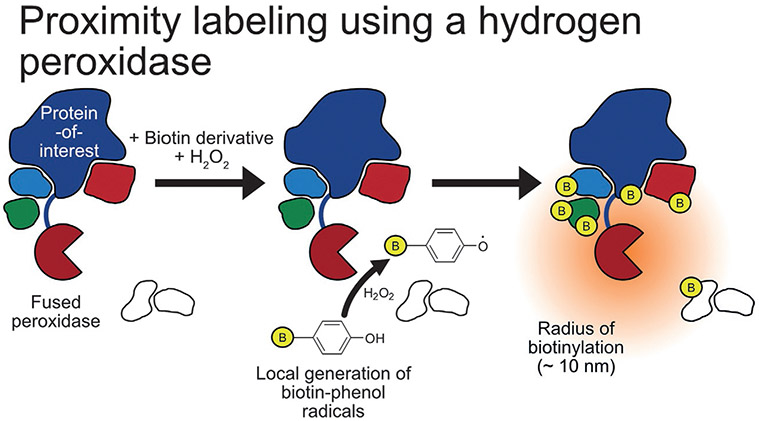
An engineered ascorbate peroxidase (APEX) was developed in 2012 for application in electron microscopy as an alternative to HRP, in part due to the inactivity of the latter in the cytosol (Martell et al., 2012). The following year, APEX (28kDa) was used for proximity labeling and proteomics to map the proteins in the mitochondrial matrix. Cells expressing mito-APEX were first preincubated with a biotin-phenol derivative and then hydrogen peroxide was added to promote robust biotinylation for only 1 min before the reaction was quenched and samples prepared for downstream analysis (Fig. 6). This resulted in the detection of 495 proteins with high spatial- and temporal-control and aided the reassignment of the cellular location of two catalysts in heme biosynthesis, PPOX and CPOX. Though not experimentally determined, APEX is considered to have a labeling radius of <20nm due to the short half-life of phenoxyl radicals (Rhee et al., 2013). Interestingly, synthetic modification of the biotin-phenol probe can introduce a cleavable portion for use during affinity enrichment (Chang et al., 2017; Li et al., 2014) or be used to alter the probe’s reactivity and selectivity for proximity labeling (Ke et al., 2021; Zhou et al., 2019). It must also be noted that such derivatization may also effect probe solubility, toxicity, and the rate of cellular uptake/distribution. Additionally, alkyne-based probes compatible with the proximity labeling chemistry have also been developed (Li, Tian, et al., 2020). APEX has been utilized to map the mitochondrial matrix proteome of Drosophila (Chen et al., 2015) and to map proteomes in Caenorhabditis elegans in a tissue- and subcellular-specific manner (Reinke, Mak, Troemel, & Bennett, 2017).
FIG. 6.
Peroxidase-based proximity labeling methods. A peroxidase, such as APEX2, is fused to a protein-of-interest with catalyzes the radical addition of biotin-phenol to nearby proteins using hydrogen peroxide.
Using a yeast display-based directed evolution method, A134PAPEX was found to have considerably increased activity compared to APEX allowing lower levels of expression to provide sufficient biotinylation for different applications. Called APEX2, Lam and coworkers compared APEX and APEX2 by targeting each to the mitochondrial outer membrane or ER membrane and inducing biotinylation. Immunoblotting then showed APEX2 had a more substantial increase of known outer membrane proteins Tom20 and Tom70 and ER membrane protein BCAP31 when each enzyme was targeted to each location (Lam et al., 2015). This increased activity has been utilized in the study of rapidly changing protein-protein interactions, such as those involved in G-protein coupled receptor signaling (Lobingier et al., 2017; Paek et al., 2017). The use of APEX2 is not limited to in vitro assays as this technology has been used recently to visualize induced pluripotent stem cell-derived cardiomyocytes in mouse hearts (Hatani et al., 2018). Moreover, APEX2 forms the basis of the integrative DNA And Protein Tagging (iDAPT) approach which has been used to investigate transcription factor access to sites of gene regulation (Lee et al., 2021).
In addition, a split-APEX2 method was developed by Xue and coworkers in 2017 by screening different split sites leading to 201/202 being deemed as the site that yielded the highest enzymatic activity and lowest background signal. This split-APEX2 method was then used to detect STIM1 and Orial1 homodimers as well as the location of biotinylation on STIM1 following site-specific mass spectrometry analysis (Xue et al., 2017). Another split-APEX2 system (sAPEX) was developed in 2019 using yeast display-based directed evolution after identifying 200/201 as the best split site, resulting in 9 mutations of the APEX2 fragments to produce a functional sAPEX2 (Han et al., 2019).
Also in 2019, it was found that C32SAPEX2 improved expression in mammalian cells when fused to four SLC protein family members (SLC1A5, SLC6A5, ALC6A14, and SLC7A1) compared to APEX2, potentially caused by mismatched disulfide bond formation. C32SAPEX2 was found to exhibit comparable enzymatic activity to APEX2, allow correct SLC fusion protein localization, and provide consistent levels of biotinylation using biotin tyramide concluding that this cysteine-free mutant of APEX2 offers a good surrogate of APEX2 should poor fusion expression be found (Huang et al., 2019).
The use of peroxidase-based methods, especially APEX, has been utilized widely for the study of protein-protein interactions, due in large part, to the improved labeling kinetics compared to ligase-based methods. The fast labeling kinetics of APEX2 has been particularly useful the study of G-protein coupled receptors as first reported in 2017 (Lobingier et al., 2017; Paek et al., 2017) and used by our labs more recently (Pfeiffer et al., 2021). APEX2 proximity labeling was used to identify membrane protein LMBRD2 as novel modulator of beta-2 adrenergic receptor and discriminate the different rates of internalization between beta-2 adrenergic receptor and another receptor of a different G-protein coupled receptor subclass, angiotensin II type I receptor (Paek et al., 2017). Recently, we have used this technique to investigate the effect of different “biased” agonists of angiotensin II type I receptor. Our data found that ligands of the same biased classification had similar proximity labeling profiles while ligands from different classes had very different labeling profiles and suggests differences in internalization rate and receptor trafficking and degradation (Pfeiffer et al., 2021). However, the use of toxic hydrogen peroxide may limit the general use of peroxidase-based methods. In an effort to bring together the fast labeling kinetics of peroxidase-based methods and the low toxicity of the ligase-based methods, the field has also looked to develop photochemical and chemical-based methods that may offer new options for proximity labeling.
3.4. Photochemical- and chemical-based methods
Inspired again by DamID, Beck and coworkers utilized a photoactivatable probe that is recruited to a protein-of-interest by genetic tag fusion for the proximity labeling of other proteins and RNA in 2014. A fusion protein of EZH2, the catalytic subunit a chromatin-modifying protein complex, and monomeric streptavidin was expressed in mammalian cell culture before addition of the same aryl azide-biotin probe used in EMARS. This probe is recruited to the protein-of-interest through biotin-streptavidin interaction and subsequently a 365nm UV light source was used to generate an active radical species that can attach to other nearby molecules. Harsh lysis conditions and the lower affinity of monomeric streptavidin to biotin then allow the noncovalent interaction to be broken and molecules covalently labeled during the experiment can be enriched for analysis. Using this technique, three known protein interactors of EZH2 were identified by mass spectrometry. Separate experimentation using a SNRNP70-monomeric streptavidin fusion protein successfully labeled spliceosomal U1 small nuclear RNA as determined by deep sequencing (Beck et al., 2014).
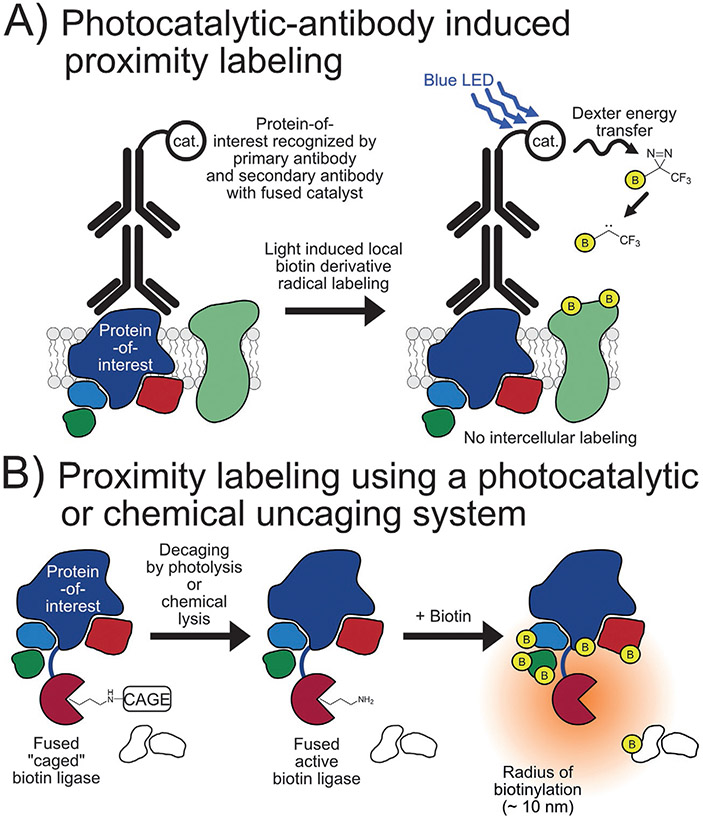
In 2020, Geri and coworkers reported the development of a photocatalytic-antibody method to identify protein-protein interactors on cell membranes, named μMap. An optimized organometallic photocatalyst was developed that can drive N2 elimination from diazirines to generate reactive carbenes in the presence of blue light irradiation through a short-range Dexter energy transfer mechanism. This catalyst was then conjugated to a secondary antibody and utilized in a manner similar to EMARS and SPPLAT (Fig. 7A). The effectiveness of this approach was highlighted in conjunction with proteomics to map the protein neighbors of PD-L1, identifying potentially new interactors (Geri et al., 2020).
FIG. 7.
Photochemical- and chemical-based proximity labeling methods. (A) The mechanism of blue light/Dexter energy transfer catalyzed proximity labeling of proteins on a cell surface near a protein-of-interest. (B) The photo- or chemical-based decaging of a biotin ligase to initiate proximity labeling.
A photocatalytic uncaging system for proximity labeling with rapid turn-on kinetics that minimizes cytotoxicity from background labeling was introduced in 2021. Named subcellular-specific uncaging-assisted biotinylation and mapping of the phosphoproteome, or SubMAPP, this approach uses TurboID but genetically replaces the active site lysine (K182) with a caged lysine analog through genetic code expansion, thus inhibiting enzymatic activity until uncaging by photolysis at 365nm (photoTurbo) (Fig. 7B). This technique was then used to investigate ER proteome and phosphoproteome dynamics under thapsigargin-induced ER stress identifying 18 proteins that are enriched compared to nonstressed samples and suggesting increased chaperone/enzyme activity to maintain ER calcium homeostasis. Of great note, this uncaging approach was also extended into living mice by modifying the nature of the cage from a photocleavage analog to a chemical-cleavage analog (chemoTurbo) (Liu, Zeng, et al., 2021).
4. Coupling mass spectrometry to proximity labeling
The most powerful applications using proximity labeling in the study of protein-protein interactions has been when coupled to mass spectrometry. In this case, proximity labeling is used to attach a chemical enrichment tag, most often biotin, to proteins near a protein-of-interest which then allows for affinity-based enrichment before enzymatic digestion and mass spectrometry analysis. A benefit of MS is that proteins cannot only be identified but can be quantified, posttranslational modifications such as phosphorylation can be investigated, and site-specific labeling can be used to support conformational predictions. For identification, tandem MS (MS/MS) is used to generate peptide fragmentation spectra that are computationally matched to known peptide sequences from a database of proteins. MS quantification can be achieved using label-free, stable isotope labeling of amino acids in cell culture (SILAC), or by tandem mass tag (TMT) methods (Pappireddi, Martin, & Wühr, 2019). In label-free MS, each sample is run individually on the mass spectrometer and spectral counting or spectrometric signal intensity is used to determine the relative amount of each peptide detected. As the name suggests, SILAC involves culturing cells with different isotopes of amino acids (typically arginine and lysine) which then allows up to four different samples to be analyzed in the same MS run, reducing run-to-run variability and improving quantification. TMT quantification involves labeling digested peptides with isotopically different tags allowing up to 18 different samples to be quantified in a single MS run (Li, Van Vranken, et al., 2020). Additionally the site-specific identification and quantification of posttranslational modifications, such as phosphorylation or glycosylation, can be used to investigate another layer of signaling complexity (Doll & Burlingame, 2015). This approach can also be used to analyze the site-specific extent of proximity labeling as a marker of site accessibility to infer protein conformation (Kim et al., 2018; Minde, Ramakrishna, & Lilley, 2020; Nierves & Lange, 2021).
Analysis of proximity labeling-based experiments is possible on virtually any modern mass spectrometer able to perform other proteomics-based data acquisition. As such, proximity labeling may leverage many emerging mass spectrometry-based technologies, particularly ion mobility, real-time database searching, and targeted proteomics methods. Ion mobility separates gas phase ions in the presence of an electric field based on their mobility through an inert buffer gas. The separation is a function of the shape, charge, and size of the ionized peptide (Jiang, Chung, May, McLean, & Robinson, 2019). Recently, ion mobility has grown in appeal as it has been integrated seamlessly into routine mass spectrometry applications. More specifically, High-Field Asymmetric waveform Ion Mobility Spectrometry (FAIMS) has been implemented through a FAIMS Pro interface and used seamlessly on Orbitrap-based mass spectrometers (Pfammatter et al., 2020, 2018; Pfammatter, Bonneil, & Thibault, 2016; Schweppe et al., 2019; Schweppe, Rusin, Gygi, & Paulo, 2020). This technology has shown an increase in depth and quantitative accuracy in both label-free and isobaric-tag based applications focusing on both whole proteome and posttranslational modifications (Bridon, Bonneil, Muratore-Schroeder, Caron-Lizotte, & Thibault, 2012; Pfammatter et al., 2018; Pfammatter, Bonneil, McManus, & Thibault, 2019; Schweppe et al., 2019). Like FAIMS, real-time database searching (RTS) enhances the depth and accuracy of global proteomics applications. RTS is applied to isobaric tagging applications using SPS-MS3-based data acquisition strategies in which peptide identification and quantification are decoupled. The usage of RTS results in higher accuracy compared to traditional MS2-only methods (Gygi et al., 2020, 2019; Paulo, Navarrete-Perea, Guha Thakurta, & Gygi, 2019; Paulo, O’Connell, & Gygi, 2016). More specifically, fragment ions from the MS2 stage are isolated, fragmented further to release reporter ions, and a time-consuming high resolution Orbitrap scan is performed. Traditional SPS-MS3 suffers from long acquisition duty cycles that are detrimental to proteome depth. RTS offers a solution by which a database search is performed on a single MS2 spectrum (requiring only 5–15ms) and the instrument decides whether an MS3 scan should be acquired (Schweppe et al., 2020). As such, this additional time is used for the analysis of new precursors rather than unsuitable and lengthy MS3 scans. Moreover, if specific proteins or peptides are sought, targeted acquisition strategies, such as Tomahto or TOMAHAQ could be implemented (Erickson et al., 2019; Yu et al., 2020). Such targeted methods require knowledge of known proximal protein targets and prevent the discovery of new targets. Along with the increased multiplexing capability attributed to novel isobaric tags (currently up to 18-plex) (Li et al., 2021; Li, Van Vranken, et al., 2020), these innovative mass spectrometry tools will enhance the depth of proximity label-based data analysis.
As sample numbers increase, the need for automated sample processing becomes increasingly apparent. Robotics and automation, such liquid handlers and magnetic particle transporters can carry out repetitive and potentially error-prone sample preparation procedures, thereby increasing throughput and reproducibility (Ippoliti et al., 2016; Ruelcke, Loo, & Hill, 2016; Zhao et al., 2017). The use of magnetic particles throughout a proximity labeling experiment would facilitate automation. Precipitation on magnetic nanoparticles is often used in lieu of variability-prone chemical-based precipitation methods, such as trichloroacetic acid (TCA) or methanol-chloroform precipitation (Hughes et al., 2014; Hughes, Moggridge, et al., 2019; Hughes, Sorensen, & Morin, 2019; Moggridge, Sorensen, Morin, & Hughes, 2018). In addition, streptavidin-coated magnetic nanoparticles are available to precipitate biotin-conjugated proteins. In all, these magnetic nanoparticles offer the advantage of automatability throughout sample processing. Automated liquid handlers, such as the opentrons OT-2 (Liu, Gygi, & Paulo, 2021), Thermo King-Fisher Flex (Leutert, Rodríguez-Mias, Fukuda, & Villén, 2019), or Hamilton Vantage (Gaun et al., 2021) systems, include magnetic modules which can accommodate nanoparticle-based precipitation in a 96-well format and have been used for proteomics applications. In all, improvements in mass spectrometry will only enhance data quality, depth, and throughput for proximity labeling experiments.
5. Advantages and disadvantages of the different methods
Traditional studies of protein-protein interactions have shown great success over the course of many years. For instance, Y2H screening is a powerful method that is rapid and scalable for the detection of binary protein-protein interactions. However, problems with protein expression, posttranslational modifications, and the difficulty of investigating proteins in a nonnative environment limit its use for certain approaches. In part to address these issues, two-hybrid assays have been developed in mammalian cells (Luo, Batalao, Zhou, & Zhu, 1997), bacteria (Karimova, Pidoux, Ullmann, & Ladant, 1998), plants (Ehlert et al., 2006), and insect cells (Mon et al., 2009). However, unlike proximity labeling, the two-hybrid system cannot be used to study multiprotein complexes. Co-IP is another important method for the study of protein-protein interactions, however, when used for the discovery of new interactors, this technique may miss weak or transient interactions that would be identified using proximity labeling. Conversely, co-IP identifies only direct interactors to the protein-of-interest whereas proximity labeling also identifies noninteracting proteins nearby that can indicate subcellular location. Therefore, co-IP is often used in conjunction with proximity labeling studies for validation of possibly new direct protein interactors. Chemical crosslinking is another useful technique; however, crosslinking efficiency is restricted by linker length, may require the use of genetic code expansion, and the data processing of XL-MS experiments often necessitates more complex analysis. Other techniques not discussed here in detail have similar limitations, for example, cofractionation mass spectrometry relies on protein interactors to coelute during chromatographic separation which may miss weak or transient interactors.
The proximity labeling approaches discussed here offer unique options for the study of protein-protein interactions. Proximity labeling and immunoblotting can be used to determine the KD of a known protein-protein interactions using microgram quantities of purified protein (Amini et al., 2003). This approach offers an alternative to commonly used methods, though at lower throughput and reduced sensitivity, to other techniques used to determine KD such as surface plasmon resonance, isothermal titration calorimetry, or fluorescence resonance energy transfer (FRET) that may require expensive instrumentation, larger amounts of protein, or other experimental modifications that can effect binding (Lakey & Raggett, 1998). Proximity labeling can also be used to study a specific protein-protein interaction in cells using highly specific enzyme-protein pairs such as BirA/biotin acceptor peptide and LplA*/ligase acceptor peptide and subsequent staining for imaging (Liu et al., 2013).
Choosing which proximity method will be the best fit depends on the experimental question(s) being asked, as each method has their own set of strengths and limitations. Table 1 summarizes the similarities and differences among the most commonly used promiscuous proximity labeling methods discussed here. For example, the biotin ligase methods discussed earlier do not require the addition of hydrogen peroxide as used in the peroxidase-based methods, making them much more amendable for use in animal models. Conversely, the peroxidase methods are more reactive and therefore have much shorter labeling times (30–60s) than ligase-based method (10+min) making them much more suitable for the study of very dynamic protein-protein interactions, such as those involved in biased G-protein coupled receptor signaling (Paek et al., 2017; Pfeiffer et al., 2021). More recent developments in photo-based methods such as μMap (Geri et al., 2020) and photoTurbo (Liu, Zeng, et al., 2021) can also achieve a high level of activity, however, will be limited by the difficulty in light penetration in tissue or animal models. Also of consideration, μMap and HRP-based methods (Kotani et al., 2008; Li et al., 2014; Martell et al., 2016) can only be used in extracellular experiments. Furthermore, most ligase-based methods react with lysine side chains whereas the peroxidase methods label tyrosine, meaning each family of techniques could miss certain proteins should that protein not contain the accessible amino acid required. Each method discussed also has similar limitations in that the use of any type of fusion protein may have an effect on protein expression, folding, cellular location, or interactions with other proteins.
Table 1.
Characteristics of the most commonly used proximity labeling methods.
| BioID | TurboID | APEX | |
|---|---|---|---|
| Enzyme | Biotin ligase | Biotin ligase | Peroxidase |
| Amino acid target | Lysine | Lysine | Mainly tyrosine |
| Labeling substrate | Biotin | Biotin | Biotin-phenol |
| Time of incubation with substrate | 12–24 h | 10 min | 30–60 min |
| Time of labeling | 12–24 h | 10 min | 1 min |
Recently, various enrichment techniques have been combined with proximity labeling to study different protein-protein interactions. For example, as described above, SubMAPP utilized enrichment both for proximity labeling at the protein-level and subsequently phosphorylation at the peptide-level to investigate subcellular phosphoproteome dynamics (Liu, Zeng, et al., 2021). Liu and coworkers combined proximity labeling and chemical crosslinking, named PL/XL-MS, to study protein-protein interactions at the nuclear envelope (Liu et al., 2020). This approach uses proximity labeling to enrich for both direct interactors and nearby proteins while chemical crosslinking is then utilized as part of the experimental workflow to distinguish only the direct interactors. Qin and coworkers paired proximity labeling with each of phosphoprotein enrichment, O-GlcNAcylated protein enrichment, and RNA-protein crosslinking with orthogonal organic phase enrichment, the latter of which, named APEX-PS, was then used to identify 15 RNA binding protein that are candidates for recruiting and/or retaining mitochondrial mRNAs at the outer mitochondrial membrane (Qin, Myers, Carey, Carr, & Ting, 2021). With such a broad range of experimental strategies, the specific technique selected must depend on the biological question being addressed or the hypothesis being tested.
6. Summary
Over the past two decades proximity labeling has become a vital tool for the study of known protein-protein interactions as well as for the discovery of new interactors. Many different proximity labeling methods have been developed, each with its own unique strengths and limitations that should be considered for use in different types of experiments. In this review, we highlight the different methods that have been developed. Furthermore, we discuss how they compare with each other and other common methods used in the study of protein-protein interactions. In summary, we provide both an entry level introduction to the field and a guide to those seeking to use proximity labeling in their own research.
Disclosures
This work was supported by National Institutes of Health Grant HL056687 to H.A.R., R01 GM132129 to J.A.P., GM67945 to S.P.G., and an institutional NIH training grant T32HL007101 to C.T.P. Dr. H.A.R. is one of the scientific founders of Trevena, Inc.
References
- Amini F, Denison C, Lin H-J, Kuo L, & Kodadek T (2003). Using oxidative crosslinking and proximity labeling to quantitatively characterize protein-protein and protein-peptide complexes. Chemistry & Biology, 10(11), 1115–1127. 10.1016/j.chembiol.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Amini F, Kodadek T, & Brown KC (2002). Protein affinity labeling mediated by genetically encoded peptide tags. Angewandte Chemie International Edition, 41(2), 356–359. . [DOI] [PubMed] [Google Scholar]
- Barker DF, & Campbell AM (1981a). Genetic and biochemical characterization of the birA gene and its product: Evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin operon in Escherichia coli. Journal of Molecular Biology, 146(4), 469–492. 10.1016/0022-2836(81)90043-7. [DOI] [PubMed] [Google Scholar]
- Barker DF, & Campbell AM (1981b). The birA gene of Escherichia coli encodes a biotin holoenzyme synthetase. Journal of Molecular Biology, 146(4), 451–467. 10.1016/0022-2836(81)90042-5. [DOI] [PubMed] [Google Scholar]
- Beck DB, Narendra V, Drury WJ, Casey R, Jansen PWTC, Yuan Z-F, et al. (2014). In vivo proximity labeling for the detection of protein–protein and protein–RNA interactions. Journal of Proteome Research, 13(12), 6135–6143. 10.1021/pr500196b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D, Kovaleva E, & Schatz PJ (1999). A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Science: A Publication of the Protein Society, 8(4), 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bludau I, Heusel M, Frank M, Rosenberger G, Hafen R, Banaei-Esfahani A, et al. (2020). Complex-centric proteome profiling by SEC-SWATH-MS for the parallel detection of hundreds of protein complexes. Nature Protocols, 15(8), 2341–2386. 10.1038/s41596-020-0332-6. [DOI] [PubMed] [Google Scholar]
- Bosch JA, Chen C-L, & Perrimon N (2021). Proximity-dependent labeling methods for proteomic profiling in living cells: An update. Wiley Interdisciplinary Reviews. Developmental Biology, 10(1), e392. 10.1002/wdev.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, et al. (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nature Biotechnology, 36(9), 880–887. 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridon G, Bonneil E, Muratore-Schroeder T, Caron-Lizotte O, & Thibault P (2012). Improvement of phosphoproteome analyses using FAIMS and decision tree fragmentation. Application to the insulin signaling pathway in Drosophila melanogaster S2 cells. Journal of Proteome Research, 11(2), 927–940. 10.1021/pr200722s. [DOI] [PubMed] [Google Scholar]
- Chang L, Chen Y-J, Fan C-Y, Tang C-J, Chen Y-H, Low P-Y, et al. (2017). Identification of Siglec ligands using a proximity labeling method. Journal of Proteome Research, 16(10), 3929–3941. 10.1021/acs.jproteome.7b00625. [DOI] [PubMed] [Google Scholar]
- Chen C-L, Hu Y, Udeshi ND, Lau TY, Wirtz-Peitz F, He L, et al. (2015). Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proceedings of the National Academy of Sciences of the United States of America, 112(39), 12093–12098. 10.1073/pnas.1515623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KF, Branon TC, Rajeev S, Svinkina T, Udeshi ND, Thoudam T, et al. (2020). Split-TurboID enables contact-dependent proximity labeling in cells. Proceedings of the National Academy of Sciences of the United States of America, 117(22), 12143–12154. 10.1073/pnas.1919528117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi-Rhee E, Schulman H, & Cronan JE (2004). Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Science: A Publication of the Protein Society, 13(11), 3043–3050. 10.1110/ps.04911804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Munter SD, Görnemann J, Derua R, Lesage B, Qian J, Heroes E, et al. (2017). Split-BioID: A proximity biotinylation assay for dimerization-dependent protein interactions. FEBS Letters, 591(2), 415–424. 10.1002/1873-3468.12548. [DOI] [PubMed] [Google Scholar]
- Dingar D, Kalkat M, Chan P-K, Srikumar T, Bailey SD, Tu WB, et al. (2015). BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. Journal of Proteomics, 118, 95–111. 10.1016/j.jprot.2014.09.029. [DOI] [PubMed] [Google Scholar]
- Doll S, & Burlingame AL (2015). Mass spectrometry-based detection and assignment of protein posttranslational modifications. ACS Chemical Biology, 10(1), 63–71. 10.1021/cb500904b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert A, Weltmeier F, Wang X, Mayer CS, Smeekens S, Vicente-Carbajosa J, et al. (2006). Two-hybrid protein–protein interaction analysis in Arabidopsis protoplasts: Establishment of a heterodimerization map of group C and group S bZIP transcription factors. The Plant Journal, 46(5), 890–900. 10.1111/j.1365-313X.2006.02731.x. [DOI] [PubMed] [Google Scholar]
- Erickson BK, Mintseris J, Schweppe DK, Navarrete-Perea J, Erickson AR, Nusinow DP, et al. (2019). Active instrument engagement combined with a real-time database search for improved performance of sample multiplexing workflows. Journal of Proteome Research, 18(3), 1299–1306. 10.1021/acs.jproteome.8b00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy DA, Melcher K, Johnston SA, & Kodadek T (1996). New chemistry for the study of multiprotein complexes: The six-histidine tag as a receptor for a protein crosslinking reagent. Chemistry & Biology, 3(7), 551–559. 10.1016/S1074-5521(96)90146-5. [DOI] [PubMed] [Google Scholar]
- Feng W, Liu C, Spinozzi S, Wang L, Evans SM, & Chen J (2020). Identifying the cardiac dyad proteome in vivo by a BioID2 knock-in strategy. Circulation, 141(11), 940–942. 10.1161/CIRCULATIONAHA.119.043434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Suárez M, Chen TS, & Ting AY (2008). Protein-protein interaction detection in vitro and in cells by proximity biotinylation. Journal of the American Chemical Society, 130(29), 9251–9253. 10.1021/ja801445p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, & Song O (1989). A novel genetic system to detect protein-protein interactions. Nature, 340(6230), 245–246. 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fossati A, Frommelt F, Uliana F, Martelli C, Vizovisek M, Gillet L, et al. (2021). System-wide profiling of protein complexes via size exclusion chromatography-mass spectrometry (SEC-MS). Methods in Molecular Biology (Clifton, N.J.), 2259, 269–294. 10.1007/978-1-0716-1178-4_18. [DOI] [PubMed] [Google Scholar]
- Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, et al. (2006). A mammalian organelle map by protein correlation profiling. Cell, 125(1), 187–199. 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Gaun A, Lewis Hardell KN, Olsson N, O’Brien JJ, Gollapudi S, Smith M, et al. (2021). Automated 16-plex plasma proteomics with real-time search and ion mobility mass spectrometry enables large-scale profiling in naked mole-rats and mice. Journal of Proteome Research, 20(2), 1280–1295. 10.1021/acs.jproteome.0c00681. [DOI] [PubMed] [Google Scholar]
- Geri JB, Oakley JV, Reyes-Robles T, Wang T, McCarver SJ, White CH, et al. (2020). Microenvironment mapping via Dexter energy transfer on immune cells. Science, 367(6482), 1091–1097. 10.1126/science.aay4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi JP, Rad R, Navarrete-Perea J, Younesi S, Gygi SP, & Paulo JA (2020). A triple knockout isobaric-labeling quality control platform with an integrated online database search. Journal of the American Society for Mass Spectrometry, 31(7), 1344–1349. 10.1021/jasms.0c00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi JP, Yu Q, Navarrete-Perea J, Rad R, Gygi SP, & Paulo JA (2019). Web-based search tool for visualizing instrument performance using the triple knockout (TKO) proteome standard. Journal of Proteome Research, 18(2), 687–693. 10.1021/acs.jproteome.8b00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Branon TC, Martell JD, Boassa D, Shechner D, Ellisman MH, et al. (2019). Directed evolution of split APEX2 peroxidase. ACS Chemical Biology, 14(4), 619–635. 10.1021/acschembio.8b00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatani T, Funakoshi S, Deerinck TJ, Bushong EA, Kimura T, Takeshima H, et al. (2018). Nano-structural analysis of engrafted human induced pluripotent stem cell-derived cardiomyocytes in mouse hearts using a genetic-probe APEX2. Biochemical and Biophysical Research Communications, 505(4), 1251–1256. 10.1016/j.bbrc.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusel M, Bludau I, Rosenberger G, Hafen R, Frank M, Banaei-Esfahani A, et al. (2019). Complex-centric proteome profiling by SEC-SWATH-MS. Molecular Systems Biology, 15(1), e8438. 10.15252/msb.20188438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusel M, Frank M, Köhler M, Amon S, Frommelt F, Rosenberger G, et al. (2020). A global screen for assembly state changes of the mitotic proteome by SEC-SWATH-MS. Cell Systems, 10(2). 133–155.e6 10.1016/j.cels.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill ZB, Pollock SB, Zhuang M, & Wells JA (2016). Direct proximity tagging of small molecule protein targets using an engineered NEDD8 ligase. Journal of the American Chemical Society, 138(40), 13123–13126. 10.1021/jacs.6b06828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M-S, Lin W-C, Chang J-H, Cheng C-H, Wang HY, & Mou KY (2019). The cysteine-free single mutant C32S of APEX2 is a highly expressed and active fusion tag for proximity labeling applications. Protein Science: A Publication of the Protein Society, 28(9), 1703–1712. 10.1002/pro.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CS, Foehr S, Garfield DA, Furlong EE, Steinmetz LM, & Krijgsveld J (2014). Ultrasensitive proteome analysis using paramagnetic bead technology. Molecular Systems Biology, 10, 757. 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CS, Moggridge S, Müller T, Sorensen PH, Morin GB, & Krijgsveld J (2019). Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nature Protocols, 14(1), 68–85. 10.1038/s41596-018-0082-x. [DOI] [PubMed] [Google Scholar]
- Hughes CS, Sorensen PH, & Morin GB (2019). A standardized and reproducible proteomics protocol for bottom-up quantitative analysis of protein samples using SP3 and mass spectrometry. Methods in Molecular Biology (Clifton, N.J.), 1959, 65–87. 10.1007/978-1-4939-9164-8_5. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Bruckner RJ, Navarrete-Perea J, Cannon JR, Baltier K, Gebreab F, et al. (2021). Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell, 184(11). 3022–3040.e28 10.1016/j.cell.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, et al. (2017). Architecture of the human interactome defines protein communities and disease networks. Nature, 545(7655), 505–509. 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, et al. (2015). The BioPlex network: A systematic exploration of the human interactome. Cell, 162(2), 425–440. 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippoliti PJ, Kuhn E, Mani DR, Fagbami L, Keshishian H, Burgess MW, et al. (2016). Automated microchromatography enables multiplexing of immunoaffinity enrichment of peptides to greater than 150 for targeted MS-based assays. Analytical Chemistry, 88(15), 7548–7555. 10.1021/acs.analchem.6b00946. [DOI] [PubMed] [Google Scholar]
- Jiang W, Chung NA, May JC, McLean JA, & Robinson RAS (2019). Ion mobility–mass spectrometry. In Encyclopedia of analytical chemistry (pp. 1–34). 10.1002/9780470027318.a9292.pub2. [DOI] [Google Scholar]
- Jiang S, Kotani N, Ohnishi T, Miyagawa-Yamguchi A, Tsuda M, Yamashita R, et al. (2012). A proteomics approach to the cell-surface interactome using the enzyme-mediated activation of radical sources reaction. Proteomics, 12(1), 54–62. 10.1002/pmic.201100551. [DOI] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, & Ladant D (1998). A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proceedings of the National Academy of Sciences of the United States of America, 95(10), 5752–5756. 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke M, Yuan X, He A, Yu P, Chen W, Shi Y, et al. (2021). Spatiotemporal profiling of cytosolic signaling complexes in living cells by selective proximity proteomics. Nature Communications, 12(1), 71. 10.1038/s41467-020-20367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido K, Yamanaka S, Nakano S, Motani K, Shinohara S, Nozawa A, et al. (2020). AirID, a novel proximity biotinylation enzyme, for analysis of protein–protein interactions. eLife, 9, e54983. 10.7554/eLife.54983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Cutler JA, Na CH, Reckel S, Renuse S, Madugundu AK, et al. (2018). BioSITe: A method for direct detection and quantitation of site-specific biotinylation. Journal of Proteome Research, 17(2), 759–769. 10.1021/acs.jproteome.7b00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Jensen SC, Noble KA, KC B, Roux KH, Motamedchaboki K, et al. (2016). An improved smaller biotin ligase for BioID proximity labeling. Molecular Biology of the Cell, 27(8), 1188–1196. 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, KC B, Zhu W, Motamedchaboki K, Doye V, & Roux KJ (2014). Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proceedings of the National Academy of Sciences of the United States of America, 111(24), E2453–E2461. 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani N, Gu J, Isaji T, Udaka K, Taniguchi N, & Honke K (2008). Biochemical visualization of cell surface molecular clustering in living cells. Proceedings of the National Academy of Sciences of the United States of America, 105(21), 7405–7409. 10.1073/pnas.0710346105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulyyassov A, Shoaib M, Pichugin A, Kannouche P, Ramanculov E, Lipinski M, et al. (2011). PUB-MS: A mass spectrometry-based method to monitor protein–protein proximity in vivo. Journal of Proteome Research, 10(10), 4416–4427. 10.1021/pr200189p. [DOI] [PubMed] [Google Scholar]
- Kwak C, Shin S, Park J-S, Jung M, Nhung TTM, Kang M-G, et al. (2020). Contact-ID, a tool for profiling organelle contact sites, reveals regulatory proteins of mitochondrial-associated membrane formation. Proceedings of the National Academy of Sciences of the United States of America, 117(22), 12109–12120. 10.1073/pnas.1916584117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K, & Beckett D (2000). Function of a conserved sequence motif in biotin holoenzyme synthetases. Protein Science: A Publication of the Protein Society, 9(8), 1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakey JH, & Raggett EM (1998). Measuring protein–protein interactions. Current Opinion in Structural Biology, 8(1), 119–123. 10.1016/S0959-440X(98)80019-5. [DOI] [PubMed] [Google Scholar]
- Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, et al. (2015). Directed evolution of APEX2 for electron microscopy and proximity labeling. Nature Methods, 12(1), 51–54. 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JD, Paulo JA, Posey RR, Mugoni V, Kong NR, Cheloni G, et al. (2021). Dual DNA and protein tagging of open chromatin unveils dynamics of epigenomic landscapes in leukemia. Nature Methods, 18(3), 293–302. 10.1038/s41592-021-01077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutert M, Rodríguez-Mias RA, Fukuda NK, & Villén J (2019). R2-P2 rapid-robotic phosphoproteomics enables multidimensional cell signaling studies. Molecular Systems Biology, 15(12), e9021. 10.15252/msb.20199021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Cai Z, Bomgarden RD, Pike I, Kuhn K, Rogers JC, et al. (2021). TMTpro-18plex: The expanded and complete set of TMTpro reagents for sample multiplexing. Journal of Proteome Research, 20(5), 2964–2972. 10.1021/acs.jproteome.1c00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-W, Rees JS, Xue P, Zhang H, Hamaia SW, Sanderson B, et al. (2014). New insights into the DT40 B cell receptor cluster using a proteomic proximity labeling assay. Journal of Biological Chemistry, 289(21), 14434–14447. 10.1074/jbc.M113.529578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tian C, Liu K, Zhou Y, Yang J, & Zou P (2020). A clickable APEX probe for proximity-dependent proteomic profiling in yeast. Cell Chemical Biology, 27(7). 858–865. e8 10.1016/j.chembiol.2020.05.006. [DOI] [PubMed] [Google Scholar]
- Li J, Van Vranken JG, Pontano Vaites L, Schweppe DK, Huttlin EL, Etienne C, et al. (2020). TMTpro reagents: A set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nature Methods, 17(4), 399–404. 10.1038/s41592-020-0781-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-H, Chien M-J, Chang Y-C, Cheng Y-H, Li F-A, & Mou KY (2020). Combining proximity labeling and cross-linking mass spectrometry for proteomic dissection of nuclear envelope interactome. Journal of Proteome Research, 19(3), 1109–1118. 10.1021/acs.jproteome.9b00609. [DOI] [PubMed] [Google Scholar]
- Liu X, Gygi SP, & Paulo JA (2021). A semiautomated paramagnetic bead-based platform for isobaric tag sample preparation. Journal of the American Society for Mass Spectrometry, 32(6), 1519–1529. 10.1021/jasms.1c00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DS, Loh KH, Lam SS, White KA, & Ting AY (2013). Imaging trans-cellular neurexin-neuroligin interactions by enzymatic probe ligation. PLoS One, 8(2), e52823. 10.1371/journal.pone.0052823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zeng R, Wang R, Weng Y, Wang R, Zou P, et al. (2021). Spatiotemporally resolved subcellular phosphoproteomics. Proceedings of the National Academy of Sciences of the United States of America, 118(25). 10.1073/pnas.2025299118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zheng J, Sun W, Huo Y, Zhang L, Hao P, et al. (2018). A proximity-tagging system to identify membrane protein–protein interactions. Nature Methods, 15(9), 715–722. 10.1038/s41592-018-0100-5. [DOI] [PubMed] [Google Scholar]
- Lobingier BT, Hüttenhain R, Eichel K, Miller KB, Ting AY, von Zastrow M, et al. (2017). An approach to spatiotemporally resolve protein interaction networks in living cells. Cell, 169(2). 350–360.e12. 10.1016/j.cell.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S, Anthony B, Drewry LL, & Sibley LD (2017). A conserved ankyrin repeat-containing protein regulates conoid stability, motility and cell invasion in Toxoplasma gondii. Nature Communications, 8, 2236. 10.1038/s41467-017-02341-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck K, Kim D-K, Lambourne L, Spirohn K, Begg BE, Bian W, et al. (2020). A reference map of the human binary protein interactome. Nature, 580(7803), 402–408. 10.1038/s41586-020-2188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Batalao A, Zhou H, & Zhu L (1997). Mammalian two-hybrid system: A complementary approach to the yeast two-hybrid system. BioTechniques, 22(2), 350–352. 10.2144/97222pf02. [DOI] [PubMed] [Google Scholar]
- Mair A, Xu S-L, Branon TC, Ting AY, & Bergmann DC (2019). Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID. eLife, 8, e47864. 10.7554/eLife.47864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, et al. (2012). Engineered ascorbate peroxidase as a genetically-encoded reporter for electron microscopy. Nature Biotechnology, 30(11), 1143–1148. 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell JD, Yamagata M, Deerinck TJ, Phan S, Kwa CG, Ellisman MH, et al. (2016). A split horseradish peroxidase for detection of intercellular protein-protein interactions and sensitive visualization of synapses. Nature Biotechnology, 34(7), 774–780. 10.1038/nbt.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minde D-P, Ramakrishna M, & Lilley KS (2020). Biotin proximity tagging favours unfolded proteins and enables the study of intrinsically disordered regions. Communications Biology, 3(1), 1–13. 10.1038/s42003-020-0758-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintseris J, & Gygi SP (2020). High-density chemical cross-linking for modeling protein interactions. Proceedings of the National Academy of Sciences of the United States of America, 117(1), 93–102. 10.1073/pnas.1902931116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moggridge S, Sorensen PH, Morin GB, & Hughes CS (2018). Extending the compatibility of the SP3 paramagnetic bead processing approach for proteomics. Journal of Proteome Research, 17(4), 1730–1740. 10.1021/acs.jproteome.7b00913. [DOI] [PubMed] [Google Scholar]
- Mon H, Sugahara R, Hong S-M, Lee J-M, Kamachi Y, Kawaguchi Y, et al. (2009). Analysis of protein interactions with two-hybrid system in cultured insect cells. Analytical Biochemistry, 392(2), 180–182. 10.1016/j.ab.2009.05.033. [DOI] [PubMed] [Google Scholar]
- Morell M, Ventura S, & Avilés FX (2009). Protein complementation assays: Approaches for the in vivo analysis of protein interactions. FEBS Letters, 583(11), 1684–1691. 10.1016/j.febslet.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Nierves L, & Lange PF (2021). Detectability of biotin tags by LC–MS/MS. Journal of Proteome Research, 20(5), 3002–3008. 10.1021/acs.jproteome.0c01049. [DOI] [PubMed] [Google Scholar]
- Paek J, Kalocsay M, Staus DP, Wingler L, Pascolutti R, Paulo JA, et al. (2017). Multidimensional tracking of GPCR signaling via peroxidase-catalyzed proximity labeling. Cell, 169(2). 338–349.e11. 10.1016/j.cell.2017.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappireddi N, Martin L, & Wühr M (2019). A review on quantitative multiplexed proteomics. Chembiochem: A European Journal of Chemical Biology, 20(10), 1210–1224. 10.1002/cbic.201800650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo JA, Navarrete-Perea J, Guha Thakurta S, & Gygi SP (2019). TKO6: A peptide standard to assess interference for unit-resolved isobaric labeling platforms. Journal of Proteome Research, 18(1), 565–570. 10.1021/acs.jproteome.8b00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo JA, O’Connell JD, & Gygi SP (2016). A triple knockout (TKO) proteomics standard for diagnosing ion interference in isobaric labeling experiments. Journal of the American Society for Mass Spectrometry, 27(10), 1620–1625. 10.1007/s13361-016-1434-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfammatter S, Bonneil E, Lanoix J, Vincent K, Hardy M-P, Courcelles M, et al. (2020). Extending the comprehensiveness of immunopeptidome analyses using isobaric peptide labeling. Analytical Chemistry, 92(13), 9194–9204. 10.1021/acs.analchem.0c01545. [DOI] [PubMed] [Google Scholar]
- Pfammatter S, Bonneil E, McManus FP, Prasad S, Bailey DJ, Belford M, et al. (2018). A novel differential ion mobility device expands the depth of proteome coverage and the sensitivity of multiplex proteomic measurements. Molecular & Cellular Proteomics: MCP, 17(10), 2051–2067. 10.1074/mcp.TIR118.000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfammatter S, Bonneil E, McManus FP, & Thibault P (2019). Accurate quantitative proteomic analyses using metabolic labeling and high field asymmetric waveform ion mobility spectrometry (FAIMS). Journal of Proteome Research, 18(5), 2129–2138. 10.1021/acs.jproteome.9b00021. [DOI] [PubMed] [Google Scholar]
- Pfammatter S, Bonneil E, & Thibault P (2016). Improvement of quantitative measurements in multiplex proteomics using high-field asymmetric waveform spectrometry. Journal of Proteome Research, 15(12), 4653–4665. 10.1021/acs.jproteome.6b00745. [DOI] [PubMed] [Google Scholar]
- Pfeiffer CT, Wang J, Paulo JA, Jiang X, Gygi SP, & Rockman HA (2021). Mapping angiotensin II type 1 receptor-biased signaling using proximity labeling and proteomics identifies diverse actions of biased agonists. Journal of Proteome Research, 20(6), 3256–3267. 10.1021/acs.jproteome.1c00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarenko EA, Poverennaya EV, Ilgisonis EV, Pyatnitskiy MA, Kopylov AT, Zgoda VG, et al. (2016). The size of the human proteome: The width and depth. International Journal of Analytical Chemistry, 2016, 7436849. 10.1155/2016/7436849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronobis MI, Zheng S, Singh SP, Goldman JA, & Poss KD (2021). In vivo proximity labeling identifies cardiomyocyte protein networks during zebrafish heart regeneration. eLife, 10, e66079. 10.7554/eLife.66079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Cho KF, Cavanagh PE, & Ting AY (2021). Deciphering molecular interactions by proximity labeling. Nature Methods, 18(2), 133–143. 10.1038/s41592-020-01010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Myers SA, Carey DK, Carr SA, & Ting AY (2021). Spatiotemporally-resolved mapping of RNA binding proteins via functional proximity labeling reveals a mitochondrial mRNA anchor promoting stress recovery. Nature Communications, 12(1), 4980. 10.1038/s41467-021-25259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke AW, Mak R, Troemel ER, & Bennett EJ (2017). In vivo mapping of tissue- and subcellular-specific proteomes in Caenorhabditis elegans. Science Advances, 3(5), e1602426. 10.1126/sciadv.1602426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H-W, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, et al. (2013). Proteomic mapping of mitochondria in living cells via spatially-restricted enzymatic tagging. Science (New York, N.Y.), 339(6125), 1328–1331. 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal SM, Misra T, Abdouni H, Branon TC, Ting AY, Scott IC, et al. (2021). A toolbox for efficient proximity-dependent biotinylation in zebrafish embryos. Molecular & Cellular Proteomics, 20, 100128. 10.1016/j.mcpro.2021.100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, & Burke B (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. The Journal of Cell Biology, 196(6), 801–810. 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelcke JE, Loo D, & Hill MM (2016). Reducing the cost of semi-automated in-gel tryptic digestion and GeLC sample preparation for high-throughput proteomics. Journal of Proteomics, 149, 3–6. 10.1016/j.jprot.2016.03.025. [DOI] [PubMed] [Google Scholar]
- Schatz PJ (1993). Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: A 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology, 11(10), 1138–1143. 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Schopp IM, Amaya Ramirez CC, Debeljak J, Kreibich E, Skribbe M, Wild K, et al. (2017). Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nature Communications, 8. 10.1038/ncomms15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweppe DK, Prasad S, Belford MW, Navarrete-Perea J, Bailey DJ, Huguet R, et al. (2019). Characterization and optimization of multiplexed quantitative analyses using high-field asymmetric-waveform ion mobility mass spectrometry. Analytical Chemistry, 91(6), 4010–4016. 10.1021/acs.analchem.8b05399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweppe DK, Rusin SF, Gygi SP, & Paulo JA (2020). Optimized workflow for multiplexed phosphorylation analysis of TMT-labeled peptides using high-field asymmetric waveform ion mobility spectrometry. Journal of Proteome Research, 19(1), 554–560. 10.1021/acs.jproteome.9b00759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoaib M, Kalypso A, Robin C, Winczura K, Tarlykov P, Despas E, et al. (2013). PUB-NChIP—“In vivo biotinylation” approach to study chromatin in proximity to a protein of interest. Genome Research, 23(2), 331–340. 10.1101/gr.134874.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinz A. (2006). Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein–protein interactions. Mass Spectrometry Reviews, 25(4), 663–682. 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- Sinz A. (2010). Investigation of protein–protein interactions in living cells by chemical cross-linking and mass spectrometry. Analytical and Bioanalytical Chemistry, 397(8), 3433–3440. 10.1007/s00216-009-3405-5. [DOI] [PubMed] [Google Scholar]
- Smith PA, Tripp BC, DiBlasio-Smith EA, Lu Z, LaVallie ER, & McCoy JM (1998). A plasmid expression system for quantitative in vivo biotinylation of thioredoxin fusion proteins in Escherichia coli. Nucleic Acids Research, 26(6), 1414–1420. 10.1093/nar/26.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L. (2019). Recent advances in proximity-based labeling methods for interactome mapping. F1000Research, 8. 10.12688/f1000research.16903.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezu A, Kanak DJ, Bradshaw TWA, Soderblom EJ, Catavero CM, Burette AC, et al. (2016). Identification of an elaborate complex mediating post-synaptic inhibition. Science, 353(6304), 1123–1129. 10.1126/science.aag0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ummethum H, & Hamperl S (2020). Proximity labeling techniques to study chromatin. Frontiers in Genetics, 11. 10.3389/fgene.2020.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttamapinant C, Tangpeerachaikul A, Grecian S, Clarke S, Singh U, Slade P, et al. (2012). Fast, cell-compatible click chemistry with copper-chelating azides for biomolecular labeling. Angewandte Chemie (International Ed. in English), 51(24), 5852–5856. 10.1002/anie.201108181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, & Henikoff S (2000). Identification of in vivo DNA targets of chromatin proteins using tethered Dam methyltransferase. Nature Biotechnology, 18(4), 424–428. 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- Wheat A, Yu C, Wang X, Burke AM, Chemmama IE, Kaake RM, et al. (2021). Protein interaction landscapes revealed by advanced in vivo cross-linking–mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America, 118(32), e2023360118. 10.1073/pnas.2023360118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Lo HP, McMahon K-A, Martel N, Jones A, Hill MM, et al. (2021). In vivo proteomic mapping through GFP-directed proximity-dependent biotin labelling in zebrafish. eLife, 10, e64631. 10.7554/eLife.64631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Hou J, Wang L, Cheng D, Lu J, Zheng L, et al. (2017). Optimizing the fragment complementation of APEX2 for detection of specific protein-protein interactions in live cells. Scientific Reports, 7(1), 12039. 10.1038/s41598-017-12365-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev AA (2009). Crosslinkers and their utilization for studies of intermolecular interactions. Neurochemical Journal, 3(2), 139–144. 10.1134/S181971240902010X. [DOI] [Google Scholar]
- Yılmaz Ş, Shiferaw GA, Rayo J, Economou A, Martens L, & Vandermarliere E (2018). Cross-linked peptide identification: A computational forest of algorithms. Mass Spectrometry Reviews, 37(6), 738–749. 10.1002/mas.21559. [DOI] [PubMed] [Google Scholar]
- Yu Q, Xiao H, Jedrychowski MP, Schweppe DK, Navarrete-Perea J, Knott J, et al. (2020). Sample multiplexing for targeted pathway proteomics in aging mice. Proceedings of the National Academy of Sciences of the United States of America, 117(18), 9723–9732. 10.1073/pnas.1919410117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Liu G, Zambito FC, Zhang YJ, DeSilva BS, Kozhich AT, et al. (2017). A multiplexed immunocapture liquid chromatography tandem mass spectrometry assay for the simultaneous measurement of myostatin and GDF-11 in rat serum using an automated sample preparation platform. Analytica Chimica Acta, 979, 36–44. 10.1016/j.aca.2017.04.028. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Wang G, Wang P, Li Z, Yue T, Wang J, et al. (2019). Expanding APEX2 substrates for proximity-dependent labeling of nucleic acids and proteins in living cells. Angewandte Chemie International Edition, 58(34), 11763–11767. 10.1002/anie.201905949. [DOI] [PubMed] [Google Scholar]
- Zhou Y, & Zou P (2021). The evolving capabilities of enzyme-mediated proximity labeling. Current Opinion in Chemical Biology, 60, 30–38. 10.1016/j.cbpa.2020.06.013. [DOI] [PubMed] [Google Scholar]
- Zhuang M, Guan S, Wang H, Burlingame AL, & Wells JA (2013). Substrates of IAP ubiquitin ligases identified with a designed orthogonal E3 ligase, the NEDDylator. Molecular Cell, 49(2), 273–282. 10.1016/j.molcel.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]