Abstract
A plasmid able to transform and to be stably maintained both in Sulfolobus solfataricus and in Escherichia coli was constructed by insertion into an E. coli plasmid of the autonomously replicating sequence of the virus particle SSV1 and a suitable mutant of the hph (hygromycin phosphotransferase) gene as the transformation marker. The vector suffered no rearrangement and/or chromosome integration, and its copy number in Sulfolobus was increased by exposure of the cells to mitomycin C.
Among members of the archaeal domain of life, the extreme thermophiles exhibit intriguing and unique properties at both the physiological and molecular levels (4, 5). Nevertheless, the studies of these prokaryotes have so far been focused mainly on classical biochemistry, including the purification and characterization of individual proteins and low-molecular-weight compounds, and on the molecular cloning of structural genes (3), also used for determining the evolutionary relationships within members of the same and/or different kingdoms (1, 2, 11).
In contrast, recent studies on the molecular and cell biology of prokaryotes belonging to the domain Bacteria have progressed remarkably even for thermophilic representatives (14, 15, 22), because of their high similarity to Escherichia coli, the most extensively studied model system. The applicability of the experimental approaches of the E. coli model can also explain the very recent rapid and full development of molecular genetic techniques, such as the use of selective genetic markers and gene transfer, for halophilic members of the domain Archaea (9, 10).
Appropriate manipulative strategies for the thermophilic Archaea are still at a very early stage of setup, because of their notable diversity compared with their bacterial counterparts (11) and even among members of the same kingdom (12).
In this respect, the genus Sulfolobus, whose members are sulfur-metabolizing aerobes belonging to the archaeal kingdom Crenarchaeota (24), seems to be the most promising candidate for developing genetic systems since it has been investigated in greater physiologic and genetic detail than its relatives. In fact, several Sulfolobus species have been identified in very different areas on Earth, isolated, and characterized, and more interestingly, many of them have been shown to possess extrachromosomal genetic elements such as viruses (26) and plasmids, with a broad range of hosts (25). Sulfolobus solfataricus has been indicated as the most versatile recipient for both viruses, such as the SSV1 particle (20), isolated from the natural Sulfolobus shibatae host, and plasmids (21). Moreover, it can be plated as single colonies and cultivated both autotrophically and heterotrophically on different complex or simple nutrients (7).
Here, we present the construction of a shuttle vector, pEXSs, which is based on both the SSV1 viral autonomously replicating sequence (ARS) (18) and the pGEM5Zf(−) E. coli plasmid sequences, and the isolation of an antibiotic resistance gene marker for S. solfataricus, obtained by selection of a suitable mutant of the E. coli-derived hygromycin phosphotransferase (hph) gene (6).
The hph mutant was sequenced in order to identify the point mutation inserted. The corresponding plasmid carrying the selective determinant was shown to efficiently transform S. solfataricus and to be stably maintained as an autonomously replicating plasmid in this archaeon. This plasmid possesses shuttle capability since it can be transferred from S. solfataricus into E. coli and vice versa and propagated in both prokaryotes.
Sulfolobus growth conditions and screening for drug sensitivity.
Cells of S. solfataricus MT3 were kindly supplied by M. De Rosa (Istituto di Biochimica delle Macromolecole, II University of Naples), and S. shibatae B12 (DSM 5389) was provided by the Deutsche Sammlung von Mikroorganismen (DSM) (Braunschweig, Germany). All Sulfolobus cells were grown under medium, temperature, and pH conditions suggested by the DSM catalog of strains.
Different clones of S. solfataricus MT3 were purified by two subsequent plating cycles on Gelrite (Gellan gum; Sigma) (7) and characterized with respect to growth and nutritional requirements. Strain GΘ used in this study showed high reproducibility of plating efficiency with homogeneous colony sizes and grew relatively quickly in quite a wide range of temperatures (70 to 82°C) and pH values (2.5 to 5.0), with an optimum at 75°C and pH 3.8.
Inhibition of cell growth by several antibiotics and drugs at different concentrations was investigated, and the MIC was determined by monitoring the optical density at 600 nm (OD600) of liquid cultures and/or testing the cell viability by colony formation on Gelrite plates.
Results are shown in Table 1, together with the genes known to confer resistance to their natural hosts.
TABLE 1.
Substances tested for the growth inhibition of S. solfataricus GΘ
| Drug | MIC | Resistance genea | Source |
|---|---|---|---|
| Hygromycin B | 100 μg/ml | hph | E. coli |
| Geneticin (G418) | 150 μg/ml | neo | E. coli |
| Chloramphenicol | 50 μg/ml | cat | E. coli |
| Benzyl alcohol | 25 mM | Ssadh | S. solfataricus |
| adh-hT | B. stearothermophilus |
Gene determinants (resistance genes) conferring resistance to their natural hosts (indicated as sources).
Hygromycin B was chosen as the selective agent for transformation because it is very stable under the Sulfolobus growth conditions. In fact, it was still effective against E. coli at a concentration of 50 μg/ml after incubation for 1 week at 75°C of a 150-μg/ml stock solution in Brock’s basal medium buffered at pH 3.0. Moreover, a spontaneous resistance phenotype appeared at a very low frequency (10−9) in the Sulfolobus GΘ population.
Construction of the plasmid vector pEXSs.
Total genomic DNA, extracted from S. shibatae cells as described by Guagliardi et al. (8), was used as the template for the PCR amplification of a 1,700-bp region located between positions 4938 and 6617 of the SSV1 viral genome map, containing the putative ORI sequence for viral DNA replication, as suggested by Palm et al. (18). The oligonucleotides 5′-GTATGAATTCAGAGTTTGTGC-3′ and 5′-CTAACGTGAATTCTATTG-3′, both containing an EcoRI site (underlined in the sequences), were used as the 5′ and 3′ primers, respectively, for the amplification of the specific region with Pfu DNA polymerase.
The reaction was carried out for 30 cycles at a 45°C annealing temperature.
Aspartate aminotransferase gene promoter and terminator sequences were chosen for the heterologous gene expression of the E. coli hygromycin phosphotransferase gene (6). PCR amplification of the DNA regions in the plasmid pLV1, provided by Maria Luisa Tutino, Naples, Italy, using Pfu DNA polymerase and the specific oligonucleotides 5′-GATTTAGATAGGGCCCTAAGGATACC-3′, 5′-CGAGACCATGGGTGTATATGAAGAAC-3′, 5′-GCGGATATCGATTGATGAGCTAAACTC-3′, and 5′-CAAGGCTATTTGTCGACAAGAAGAGTG-3′ (for 30 cycles at 50°C annealing temperature) was used for the isolation of the specific sequences and adaptation of their termini for the in-frame insertion of the foreign gene. The ApaI and NcoI as well as EcoRV and SalI restriction sites on the 5′ and 3′ primers, respectively, are underlined. We also constructed a library of random mutant versions of the hph gene by using a PCR-mediated strategy, according to the method described by Leung et al. (13), by using dGTP and dCTP in two independent amplifications as the deoxynucleotide at the defective concentrations. The plasmid pHL1 carrying the E. coli hph gene linearized with SalI restriction enzyme was used as the template for the reaction. The oligonucleotides for the 40-cycle amplifications at a 50°C annealing temperature were 5′-GAGTCATGAAAAAGCCTGAACTCAC-3′ and 5′-CCGCATGCTATTCCTTTGCCCTCGG-3′, containing the restriction sites (BspHI and SphI, underlined in the sequences) for the appropriate insertion between the SsAspAT promoter and terminator sequences. The wild-type hph gene was also amplified by using Pfu DNA polymerase and the same oligonucleotides and annealing temperature, but under standard nonmutagenizing PCR conditions.
All these fragments were subcloned into the E. coli pGEM 5Zf(−) plasmid, resulting in the 6.2-kb vector, designated pEXSs, shown in Fig. 1. The control plasmid pEXSswt carrying the wild-type hph sequence was constructed similarly.
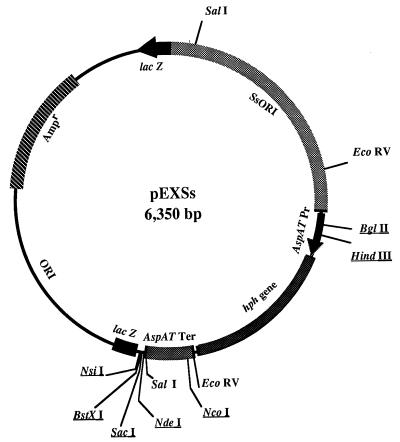
FIG. 1.
Plasmid map of the pEXSs shuttle vector. Unique restriction sites suitable for the insertion of foreign sequences are indicated. SsORI indicates the 1,700-bp fragment carrying the ARS of the S. shibatae SSV1 viral genome. AspAT Pr and AspAT Ter are the promoter and terminator sequences of the S. solfataricus aspartate aminotransferase gene, respectively. hph is the E. coli randomly mutagenized hygromycin phosphotransferase gene. The E. coli pGEM5Zf(−) plasmid moiety lies between the two lacZ gene fragments and comprises the sequences necessary for propagation (ORI) and transformant selection for ampicillin resistance (Ampr) in E. coli.
A total of 100 ng of the pEXSs DNA sublibrary was used to transform E. coli BO3301 by electroporation, and all the transformants were plated onto ampicillin selection plates.
Restriction analysis of plasmids isolated from independent clones showed the presence in two of five hph sequences of new sites, thus confirming the efficacy of the mutagenesis strategy applied.
Gene transfer experiments, selection, and characterization of Sulfolobus transformants at the DNA level.
S. solfataricus GΘ cells were grown in Brock’s basal salt medium containing 1 g of yeast extract per liter, 1 g of casein hydrolysate per liter, and 1 g of glucose per liter to a cell density of 0.3 OD600 unit. Preparation of cells competent for electroporation and the electroporation conditions tested were performed according to the procedure described by Schleper et al. for S. solfataricus infection by the SSV1 viral genome (20) with identical transformation efficiency. In the optimized experiments, 10 ng of the pEXSs plasmid sublibrary extracted from 105 independent clones was used.
The cells were mixed with 1 ml of 2× soft Gelrite and overlaid (7) onto solid medium containing 150 μg of hygromycin B per ml in polystyrene petri dishes. Plates were incubated at 75°C in a humidified chamber until the size of the colonies was about 2 mm (about 8 days). No colony growth was noted following a 15-day incubation of cells plated after electroporation with pGEM5Zf(−) DNA and a pEXSswt plasmid. This result confirms both the low frequency of the spontaneous resistance phenotype and the incapability of the wild-type hph gene to confer resistance to Sulfolobus cells.
Hygromycin B-resistant (HygBr) clones (5 per 10 ng of the pEXSs sublibrary) were picked up, grown for several generations in selective medium (150 μg of hygromycin B per ml), and harvested from the mid-log phase of growth; both extrachromosomal and total DNA were extracted in independent preparations.
The shuttle capability was ascertained by transformation of E. coli cells with the extrachromosomal DNA isolated from the Sulfolobus HygBr transformants and selection of the pEXSs-transformed E. coli clones on ampicillin selection plates.
Extrachromosomal DNA was prepared from 50-ml aliquots of the culture by using the alkaline lysis method and phenol extraction by following the procedure described by Sambrook et al. (19) for the E. coli plasmid minipreparations, with no lysozyme pretreatment step. DNA was resuspended in 50 μl of Tris-EDTA (TE) buffer, and 1 μl of the solutions was used to transform E. coli competent cells by electroporation.
The isolated plasmids were again transferred into Sulfolobus cells, and the efficiency of transformation, ranging from 102 to 104 clones per μg of plasmid DNA, was calculated in order to identify the most efficient shuttle vector among those isolated. All subsequent analyses and experiments were then performed on the most efficient plasmid, pEX2A, and on its host, clone 2A.
Total DNA was extracted from the HygBr Sulfolobus clone 2A as described above and characterized by restriction endonuclease-Southern blot analysis by using the 32P-radiolabelled pGEM5Zf(−) sequence as the probe under standard random priming labelling, blotting, and hybridization experimental conditions (19) (Fig. 2). In Southern analysis no difference in the restriction patterns was observed between the plasmid isolated from Sulfolobus and that propagated in E. coli prior to the transfer into Sulfolobus. This result indicated that no rearrangement or integration into the Sulfolobus genome occurred; namely, the vector could propagate autonomously in the Sulfolobus host cells. Moreover, the comparison of the pEX2A intensity signal with that derived by probing the same filter with the Ssadh gene (one copy per genome) allowed an estimation of 1 to 2 plasmid molecules per cell (data not shown).
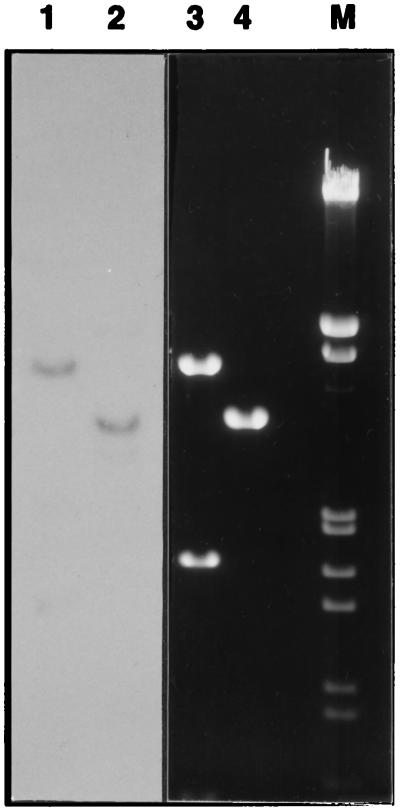
FIG. 2.
Test for the presence of pEXSs plasmid DNA in transformed S. solfataricus. Total DNA isolated from a clone, electroporated with the pEXSs2A plasmid, and plate selected for acquired resistance to hygromycin was cut with EcoRI (lane 1) and SalI (lane 2) and analyzed by filter hybridization with the pGEM5Zf(−) sequence as the probe. Lanes 3 and 4, ethidium bromide staining of the same restriction digests of the plasmid isolated from E. coli; lane M, λ DNA, EcoRI and HindIII digested as the DNA molecular weight marker.
Further proof that the SSV1 putative ORI sequence is able to drive autonomous replication in vivo was provided by the induction of the increase in the copy number of the plasmid vector constructed. Cells of Sulfolobus carrying the pEX2A plasmid vector were grown up to the late log phase and exposed to different nonlethal amounts of the DNA-damaging agent mitomycin C (0.5, 2.0, 5.0, and 10 μg/ml) (23). The drug was added to cultures with a cell density of 0.6 OD600 unit, and cells were grown for 24 and 48 h at 75°C and harvested. Extrachromosomal DNA was extracted, and an equal amount from the differently treated cells was used to transform E. coli cells. The number of E. coli transformants was used as an indirect measure of the relative amounts of plasmid in the different Sulfolobus DNA extracts in comparison with that extracted from untreated cells. The result shows that an increase of up to 10-fold could be obtained under the best conditions of drug concentration (5 μg/ml) and exposure time (24 h) tested here.
Sequence of the wild-type and mutant hygromycin phosphotransferase gene.
The coding sequence (hyg2A) of the hph gene from one of the Sulfolobus HygBr-transformed clones was determined by the dideoxy-chain termination method and compared to its wild-type counterpart. The mutant hph gene was sequenced on both strands and in duplicate by subcloning of suitable fragments and sequencing with the universal M13 primers and using the specific oligonucleotides 5′-GCAAAGTGCCGATAAAC-3′, 5′-GTTCTGCAGCCGGTCGC-3′, 5′-CGATTCCTTGCGGTCCG-3′, 5′-GTCCGGCACCTCGTGCA-3′, and 5′-GGGCGTATATGCTCCGC-3′, based on the hph gene for direct sequencing of the intact sequence in the pEXSs plasmid. Some silent (G348→T and G777→A) and two effective (G155→C and C156→G producing Ser52→Thr; G714→T resulting in Trp238→Cys) mutations could be located (numbers refer to the nucleotide sequence starting at the A of the ATG initiation codon); very interestingly, in addition to the nucleotide substitution inserted to construct the hybrid promoter-coding sequence, a nucleotide insertion was also located in the SsAspAT gene promoter between positions −13 and −14, upstream of the ATG start codon. The relationship between these nucleotide and/or amino acid replacements with the hypothetical thermal resistance of the mutated enzyme was not obvious. Currently under investigation are the contribution of the replacements in the coding sequence to thermal activity and stabilization of the mutant enzyme in comparison with the wild-type counterpart, as well as the possibility that the nucleotide insertion selected in the SsAspAT promoter sequence might increase hph gene transcription and, hence, enzyme levels.
Conclusions.
Genetic elements such as viruses and plasmids could represent powerful tools, as extensively demonstrated for Bacteria and Eucarya, to elucidate the molecular genetics of the still poorly understood domain Archaea.
The present paper describes the steps necessary to construct an E. coli-S. solfataricus shuttle vector that can be selected and maintained both in S. solfataricus and in E. coli.
It had been proven that the SSV1 virus particle of S. shibatae is able to also propagate as an extrachromosomal genetic element in S. solfataricus and that DNA-damaging agents, such as UV light or chemicals, can induce replication of the SSV1 genome and an increase of viral DNA copy number in the nonintegrated form (16, 17). These features rendered this genetic element a good vehicle for gene transfer into Sulfolobus. The present work shows that the putative SSV1 ARS is a real replicon unit able to drive autonomous propagation of hybrid Sulfolobus-E. coli DNA in the shuttle vector constructed.
The construction of the shuttle vector pEXSs was accompanied and hence made possible by the suitable modification of a specific enzyme of mesophilic source by the transferring of a gene able to confer an easily selectable phenotype after its random mutagenesis. Further tailoring of the 6.2-kb pEXSs shuttle vector is under way, with the replacement of the hph gene with other genetic markers, the insertion of different archaeal promoter and terminator sequences for gene regulation studies, and the expression of foreign mesophilic genes for thermal adaptation of the encoded proteins of interest.
Moreover, since the S. solfataricus genome sequencing project is in progress, the gene transfer system described here could be an effective strategy to express at reasonable levels potential coding regions as yet unknown or merely recognizable only by sequence similarity in order to identify and characterize genome products. The presence of a polycloning site in the newly constructed vector seems to be quite appropriate for these purposes.
Acknowledgments
This work was supported by a grant from the EC project “Extremophiles as Cell Factories,” contract BiO4-CT96-0488.
Special thanks are due to Wolfram Zillig for helpful scientific discussion.
REFERENCES
- 1.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot springs environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown J R, Doolittle W F. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci USA. 1995;92:2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciaramella M, Cannio R, Moracci M, Pisani F M, Rossi M. Molecular biology of extremophiles. World J Microbiol Biotechnol. 1995;11:71–84. doi: 10.1007/BF00339137. [DOI] [PubMed] [Google Scholar]
- 4.Cowan D A. Enzymes from thermophilic archaeabacteria: current and future application in biotechnology. Biochem Soc Symp. 1992;45:149–169. [PubMed] [Google Scholar]
- 5.Fontana A. How nature engineers proteins (thermo) stability. In: di Prisco G, editor. Life under extreme conditions: biochemical adaptation. Berlin, Germany: Springer-Verlag; 1991. pp. 89–113. [Google Scholar]
- 6.Gritz L, Davies J. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene. 1983;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- 7.Grogan D W. Phenotypic characterization of the archaeabacterial genus Sulfolobus: comparison of five wild-type strains. J Bacteriol. 1989;171:6710–6719. doi: 10.1128/jb.171.12.6710-6719.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guagliardi A, de Pascale D, Cannio R, Nobile V, Bartolucci S, Rossi M. The purification, cloning, and high level expression of a glutaredoxin-like protein from the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1995;270:3823–3827. doi: 10.1074/jbc.270.11.5748. [DOI] [PubMed] [Google Scholar]
- 9.Holmes M L, Dyall-Smith M L. A plasmid vector with a selectable marker for halophilic archaebacteria. J Bacteriol. 1990;172:756–761. doi: 10.1128/jb.172.2.756-761.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes M L, Nuttal S D, Dyall-Smith M L. Construction and use of halobacterial shuttle vectors and further studies on Haloferax DNA gyrase. J Bacteriol. 1991;173:3807–3813. doi: 10.1128/jb.173.12.3807-3813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keeling P J, Charlebois R L, Doolittle W F. Archaebacterial genomes: eubacterial form and eukaryotic content. Curr Opin Genet Dev. 1994;4:816–822. doi: 10.1016/0959-437x(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 12.Keeling P J, Doolittle W F. Archaea: narrowing the gap between prokaryotes and eukaryotes. Proc Natl Acad Sci USA. 1995;92:5761–5764. doi: 10.1073/pnas.92.13.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung D W, Chen E, Goeddel D V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 14.Liao H, McKenzie T, Hageman R. Isolation of a thermostable enzyme variant by cloning and selection in a thermophile. Proc Natl Acad Sci USA. 1986;83:576–580. doi: 10.1073/pnas.83.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mather M W, Fee J A. Development of plasmid cloning vectors for Thermus thermophilus HB8: expression of a heterologous, plasmid-borne kanamycin nucleotidyltransferase gene. Appl Environ Microbiol. 1992;58:421–425. doi: 10.1128/aem.58.1.421-425.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McEvoy J L, Murata H, Chatterjee A K. Genetic evidence for an activator required for induction of pectin lyase in Erwinia carotovora subsp. carotovora by DNA-damaging agents. J Bacteriol. 1992;174:5471–5474. doi: 10.1128/jb.174.16.5471-5474.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McEvoy J L, Murata H, Chatterjee A K. Molecular cloning and characterization of an Erwinia carotovora subsp. carotovora pectin lyase gene that responds to DNA-damaging agents. J Bacteriol. 1990;172:3284–3289. doi: 10.1128/jb.172.6.3284-3289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palm P, Schleper C, Grampp B, Yeats S, McWilliam P, Reiter W-D, Zillig W. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology. 1991;185:242–250. doi: 10.1016/0042-6822(91)90771-3. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Schleper C, Kubo K, Zillig W. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc Natl Acad Sci USA. 1992;89:7645–7649. doi: 10.1073/pnas.89.16.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schleper C, Holz I, Janekovic D, Murphy J, Zillig W. A multicopy plasmid of the extremely thermophilic archaeon Sulfolobus effects its transfer to recipients by mating. J Bacteriol. 1995;177:4417–4426. doi: 10.1128/jb.177.15.4417-4426.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber J M, Johnson S P, Vonstein V, Casadaban M J, Demirjian D C. A chromosome integration system for stable gene transfer into Thermus flavus. Bio/Technology. 1995;13:271–275. doi: 10.1038/nbt0395-271. [DOI] [PubMed] [Google Scholar]
- 23.Zillig, W. Personal communication.
- 24.Zillig W, Stetter K O, Wunderl S, Schulz W, Priess H, Scholz I. The Sulfolobus-“Caldariella” group: taxonomy on the basis of the structure of DNA-dependent RNA polymerases. Microbiology. 1980;125:259–269. [Google Scholar]
- 25.Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, Hain J, Lanzendörfer M, Kristjansson J K. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst Appl Microbiol. 1994;16:609–628. [Google Scholar]
- 26.Zillig W, Prangishvilli D, Schleper C, Elferink M, Holz I, Albers S, Janekovic D, Götz D. Viruses, plasmids and other genetic elements of thermophilic and hyperthermophilic Archaea. FEMS Microbiol Rev. 1996;18:225–236. doi: 10.1111/j.1574-6976.1996.tb00239.x. [DOI] [PubMed] [Google Scholar]