ABSTRACT
Ambrosia gall midges (AGMs) are mostly host plant-specific. In their galls, they harbor fungal symbionts on which they feed. Therefore, they represent unique steps in the evolution of the gall-forming Cecidomyiidae (Diptera). Gall-associated fungi have been studied predominantly by cultivations, and potential larval endosymbionts have been completely neglected. Using ITS2 rRNA metabarcoding, we characterized the mycobiomes of individual gall compartments (gall surface, gall interior, and larva) of six species from two phylogenetically separated tribes (Asphondyliini and Lasiopterini). Compared to the gall surface and interior, the larvae harbored significantly higher fungal richness and taxonomic diversity, and a larger pool of indicator taxa. Larval mycobiome composition was more species-specific; however, the fungal genera Fusarium, Filobasidium, Tilletiopsis, Alternaria, and Aureobasidium were indicator taxa shared among species. Overall, the larvae harbored 29% of unique taxa that can play a functional role in the host (e.g., initiation of gall development or selection of the mycelia composition). The mycobiome of the gall interior was assembled least stochastically, and its composition was the least species-specific, being dominated by Botryosphaeria dothidea (except for Lasioptera arundinis). Therefore, the interior of ambrosia galls offers a unique environment that supports the growth of similar fungi, regardless of the host plant species and the phylogenetic distance between the AGM tribes. Our study illustrates a range of fungal microorganisms indicative of individual gall compartments, but their potential function, especially in larvae, remains to be solved.
IMPORTANCE
Ambrosia gall midges are endophagous insect herbivores whose larvae live enclosed within a single gall for their entire development period. They may exhibit phytomycetophagy, a remarkable feeding mode that involves the consumption of plant biomass and mycelia of their cultivated gall symbionts. Thus, AGMs are ideal model organisms for studying the role of microorganisms in the evolution of host specificity in insects. However, compared to other fungus-farming insects, insect–fungus mutualism in AGMs has been neglected. Our study is the first to use DNA metabarcoding to characterize the complete mycobiome of the entire system of the gall-forming insects as we profiled gall surfaces, nutritive mycelia, and larvae. Interestingly, larval mycobiomes were significantly different from their nutritive mycelia, although Botryosphaeria dothidea dominated the nutritive mycelia, regardless of the evolutionary separation of the tribes studied. Therefore, we confirmed a long-time hypothesized paradigm for the important evolutionary association of this fungus with AGMs.
KEYWORDS: ambrosia gall midge, Cecidomyiidae, fungiculture, Asphondylia, Lasioptera, larval mycobiome, metabarcoding, nutritive mycelium, phytomycetophagy
INTRODUCTION
Ambrosia gall midges (AGMs) are mostly host plant-specific and feed on the mycelia of the cultivated fungal symbiont(s). Due to their mixed diet (phytomycetophagy), this group represents a unique step in the evolution of gall-forming Cecidomyiidae (1). Most AGMs belong to two taxonomically separated tribes of the subfamily Cecidomyiinae: Asphondyliini, and Lasiopterini (2 – 5). Fungiculture, which has evolved independently at least six times in insects (6), has been intensively studied in bark and ambrosia beetles (7), woodwasps (8, 9), leaf-cutting ants (10), and fungus-growing termites (11); however, in AGMs, this interaction has received far less attention.
Based on current knowledge, Asphondyliini may be associated with specific fungal symbionts, primarily Botryosphaeria dothidea (2, 12 – 15). However, this fungus has also been found in other AGM-induced galls, including Lasiopterini (13, 16 – 18), and also in the unrelated Cynipidae (Hymenoptera) (19, sequence accession KT823763), where fungi as a diet are not expected. Moreover, a variety of other fungi have been found in the galls of Asphondylia (2, 12, 20, 21), Lasioptera (22, 23), and other AGM genera (13). However, due to the lack of experimental design in most studies, these fungi have been reported to be “associated” rather than “mutualists” and sometimes even as incidentally captured saprotrophs (17, 24). Therefore, the specificity and richness of fungal symbionts in ambrosia galls remain largely speculative.
Although insect fungicultures are usually complex (6), fungal associates of AGMs have been studied predominantly by cultivation, sometimes complemented by standard DNA barcoding (12, 13, 17, 18, 20, 22, 25 – 28). In addition to capturing only a fragment of diversity, gall-based cultivations are prone to contamination (29) and fail frequently, especially for dominant B. dothidea (13, 16). Some fungi present in ambrosia galls may be only saprophytes, representing opportunistic colonization rather than strict mutualism (12). These species frequently reported as gall associates may conceal the real symbiont(s) during the isolation procedure (27). These shortcomings can be overcome by the application of culture-independent methods and the inclusion of control samples, for example, tissues of gall surfaces or ungalled plant parts.
In AGMs, mycobiome studies have focused primarily on nutritive mycelia (14, 18). On occasion, fungal samples were collected from the external surfaces of the galls (30), ungalled plant parts (20), eggs (17), and larval surfaces (13). However, a comparison of the respective gall compartments (i.e., gall surface, interior, and larvae) has never been made. Despite the urgency to detect fungal DNA from larvae (13), this has never been done, and it remains an open question whether there is a distinct mycobiome of the individuals that live permanently inside the gall enclosed by their nutritive mycelia. In herbivorous or detritivorous insects, fungi represent an important part of the microbiome, often forming communities distinct from those present in the feeding substrates (31 – 35). Paradoxically, in fungus-feeding insects, only the bacterial component of the gut has been studied (36 – 40), except for ambrosia beetles (41, 42). Therefore, the potential role of fungi in AGM larvae remains unknown.
In this study, we sampled galls from six AGM species from two phylogenetically separated Cecidomyiidae tribes (Asphondyliini and Lasiopterini). We used DNA metabarcoding to profile the fungal communities of each compartment of the whole AGM system: external gall surface (plant tissue), nutritive mycelia (gall interior), and larvae. We aimed to (i) investigate the effect of AGM taxonomy on mycobiome diversity and composition; (ii) compare the mycobiome of individual gall compartments and investigate the involvement of neutral processes in fungal community assembly; and (iii) discuss the potential functional significance of the revealed fungal symbionts. We hypothesized that larvae would harbor fungal microbes different from nutritive mycelia, in analogy to detritophagous and herbivorous insects (33, 34, 43). Based on this assumption, presumed symbiosis with B. dothidea, and the active suppression of unsuitable taxa by larval symbionts (44), we expected a lower involvement of neutral processes in the gall interior and larval microbiome compared to the gall surface. Our sampling design enabled us to distinguish random fungal associates (plant endophytes and epiphytes) from real symbionts.
RESULTS
From 31 triplets, 795 086 fungal reads were obtained. After discarding the contaminant and unassigned reads, 793 200 fungal reads (μ = 8529.03; 95% of them ranging from 1007 to 18 159) were assigned to 645 amplicon sequence variants (ASVs), which we classified into 184 species (gall surfaces = 112; gall interiors = 73; larvae = 127). On average, we observed 20.61 fungal species per sample (μ gall surfaces = 14.97 ± 5.12; μ interiors = 18.03 ± 5.19; μ larvae = 28.84 ± 7.05). For most samples, the sequencing depth was sufficient, as rarefaction curves at the ASV level reached their asymptotes (Fig. S1; Supplementary Material 1).
Composition and diversity of mycobiomes
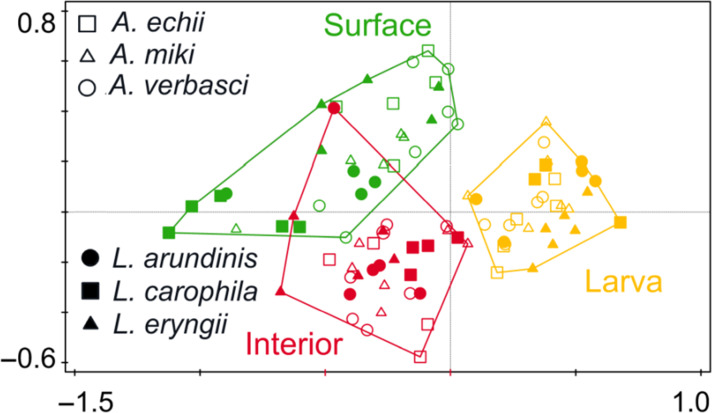
The gall compartment (i.e., gall surface, gall interior, and larva) best explained the mycobiome composition (13.86% of variability; df = 90, F = 10.03, P = 0.001), followed by AGM species (16.28%; df = 85, F = 4.71, P = 0.001) and their interaction (18.05%, df = 75, F = 2.61, P = 0.001; Fig. 1), while the genus did not decrease Akaike information criterion (AICc). The results of this analysis at the ASV level were similar (Fig. S2; Supplementary Material 1). Analysis of each gall compartment separately revealed the greatest difference among individual species at the larvae level (AGM species explaining 47.68% of variability, df = 25, F = 4.56, Padj = 0.001), followed by the surface of the gall (that is, different host plants, 40.89% of variability, df = 25, F = 3.46, Padj = 0.001), while the lowest but still significant differences between AGM species were detected in the gall interior (30.73% of variability, df = 25, F = 2.22, Padj = 0.001). β-Diversity did not differ between the gall compartments (df = 88, F = 2.44, P = 0.093).
Fig 1.
Principal coordinates analysis plot based on partial-canonical correspondence analysis (F = 3.80, P = 0.001) showing dissimilarity in the composition of fungal species among mycobiomes of gall surface, interior, and larvae of Asphondylia spp. and Lasioptera spp.
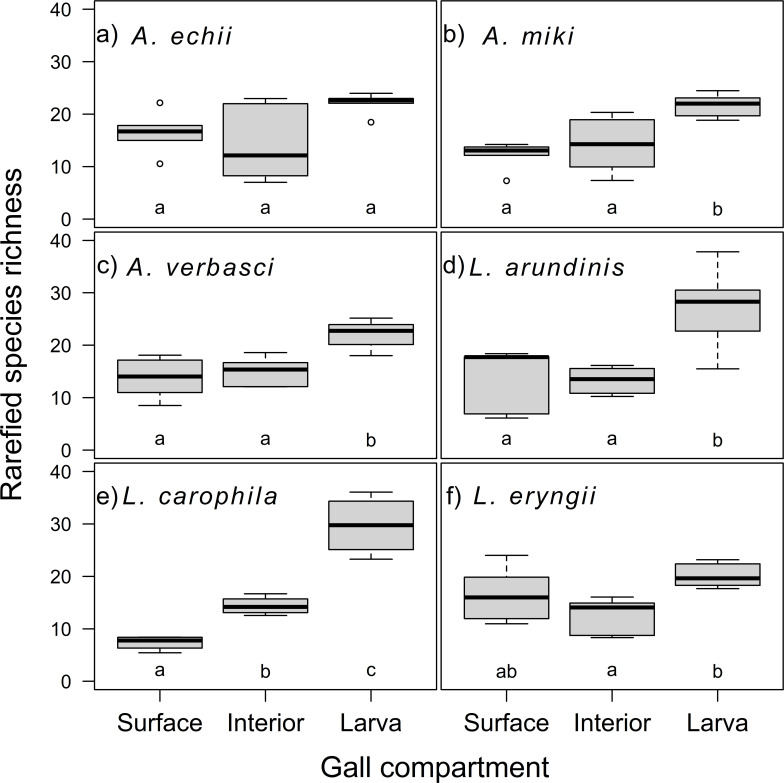
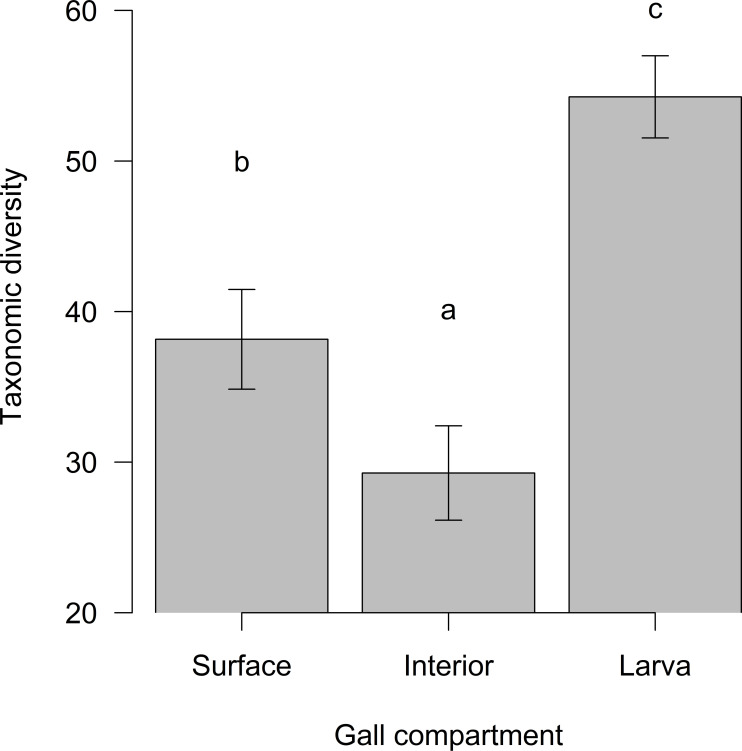
The gall compartment was the only significant explanatory variable for the fungal richness (42.70% variability; df = 90, F = 35.55, P < 0.001); neither the AGM genus (df = 89, F = 0.36, P = 0.552) nor the species (df = 85, F = 0.82, P = 0.516) further decreased AIC. The highest richness was associated with larvae (Fig. 2). Compared to the gall surface, the richness of the gall interior did not differ (df = 90, t = 0.50, P = 0.620), while the larval richness differed significantly (df = 90, t = 6.93, P < 0.001), as indicated also by the species accumulation curves (Fig. S3; Supplementary Material 1). The ASV-based analysis of richness yielded similar results (Fig. S4; Supplementary Material 1). The taxonomic diversity of the mycobiome differed significantly between the gall compartments (df = 90, F = 13.43, P < 0.001), with gall interior having lower (t = −2.21, P = 0.030) and larvae higher (t = 2.94, P = 0.004) taxonomic diversity compared to surface mycobiome (Fig. 3).
Fig 2.
Fungal species richness standardized by rarefaction for gall mycobiomes of individual gall parts for each gall midge species: (A) Asphondylia echii, (B) A. miki, (C) A. verbasci, (D) Lasioptera arundinis, (E) L. carophila, and (F) L. eryngii. Significant differences are indicated by different letters.
Fig 3.
Taxonomic diversity (Δ) of fungal species in individual gall compartments (mean ± SE). Significant differences are indicated by different letters.
Involvement of neutral processes in community assembly
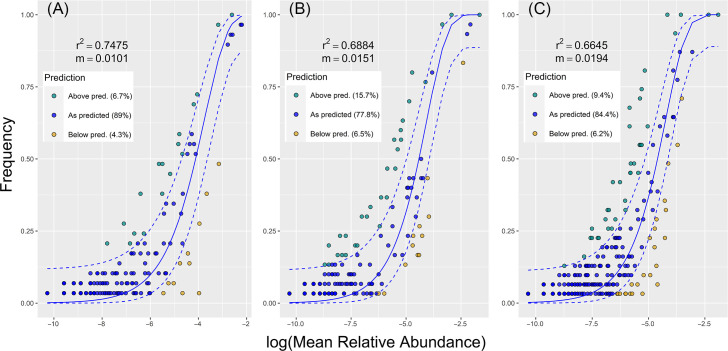
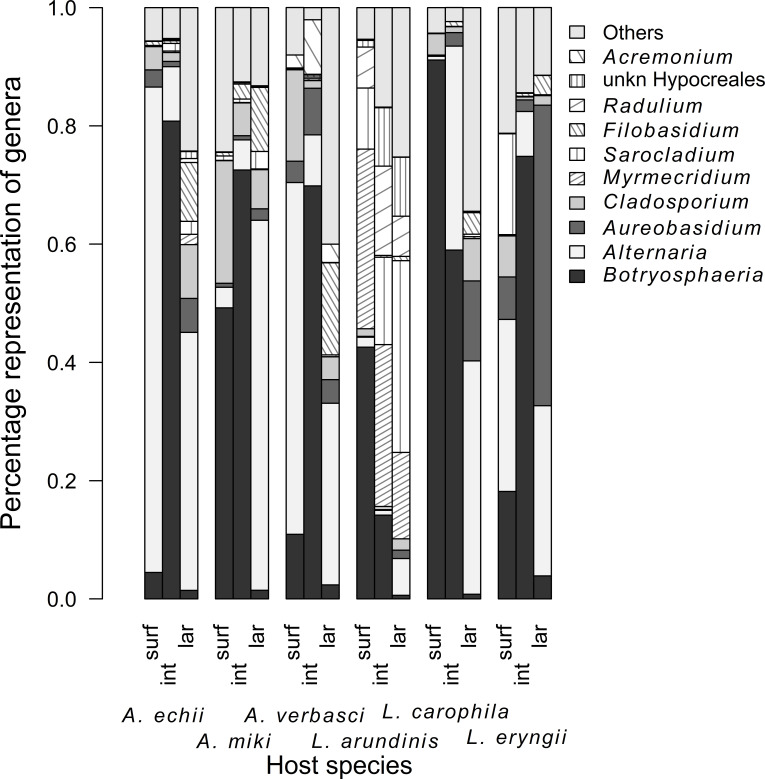
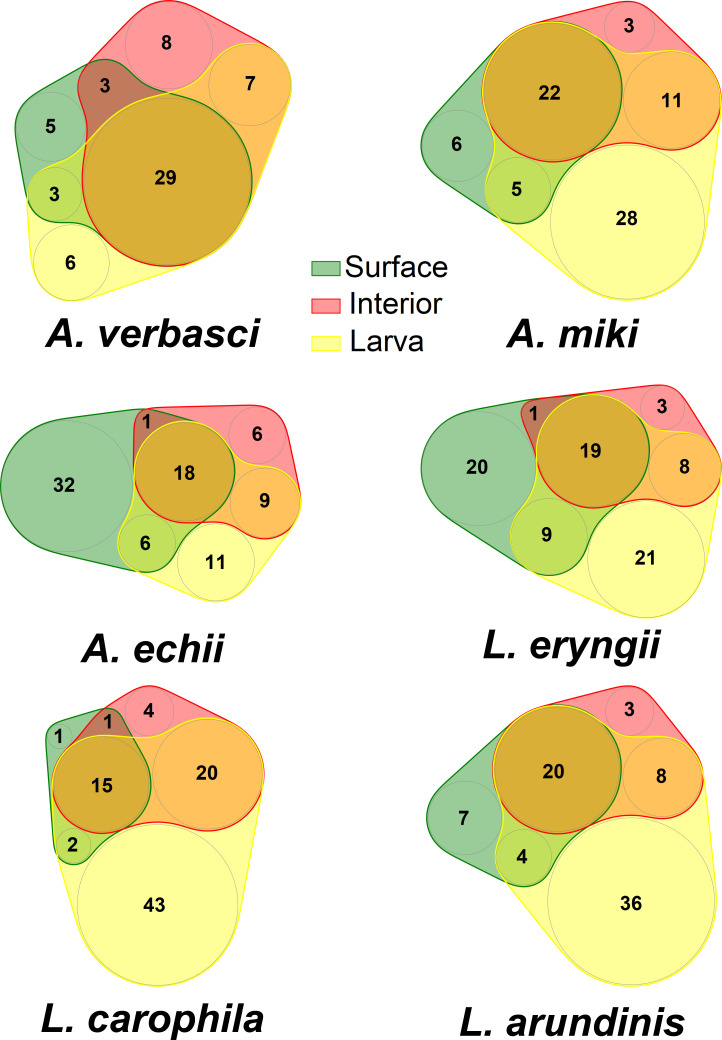
The mycobiome was assembled most stochastically on the gall surface (89.00% of the ASVs fitted the prediction of the neutral model), while it was assembled least stochastically in the gall interior (77.80% of ASVs fitted the model prediction), and the result for the larvae was intermediate (84.40%, Fig. 4). We found 25 fungal taxa significantly indicative of larvae of at least one AGM species, six of which were common to three or more AGM species: Fusarium sporotrichioides (5 AGM species), Filobasidium oeirense (5), Tilletiopsis washingtonensis (3), Alternaria sp. (A. alternata species complex) (3), unidentified Alternaria (3), and Aureobasidium pullulans (3). Only three fungal species were indicative of the gall interior: Botryosphaeria dothidea (4 AGM species), Cercospora sp. (C. beticola species complex) (1), and unidentified Myrmecridium species (1). Botryosphaeria dothidea was eudominant (>10%) in A. echii (80.80% of reads), A. miki (72.54%), A. verbasci (69.84%), and L. eryngii (74.84%), while in L. carophila (58.99%) it was accompanied by Alternaria sp. (A. alternata species complex) (31.26%). In L. arundinis, we found four eudominant species (>10%): unidentified Myrmecridium sp. (27.39%), Radulidium subulatum (15.11%), B. dothidea (14.16%), and Sarocladium strictum (11.01%). Five fungal species were indicative of gall surfaces and none of these were common among AGM species (Table 1; Fig. 5). The ITS2 barcode did not provide sufficient discrimination power for some taxa, and the identities of significant taxa are provided in Supplementary Material 2B. Precise barcoding of the pure cultures confirmed the presence of B. dothidea and identified other dominant taxa, such as various species of Akanthomyces, Aureobasidium, Alternaria, Cladosporium, Filobasidium, Fusarium, Heterophoma, Mucor, Papiliotrema, Rhodosporidium, Rhodosporidiobolus, and Sarocladium (Supplementary Material 2C). As confirmed by the Venn diagrams, the pool of fungi associated purely with larvae formed in a total of 27.87% of taxa and was the largest subset in four of six gall midge species (Fig. 6).
Fig 4.
Fit of the neutral community model (NCM) of fungal community assembly for (A) gall surface, (B) gall interior, and (C) larvae predicted occurrence frequencies for x and y. Solid blue lines indicate the best fit for the NCM according to Sloan et al. (45), and the dashed blue lines represent 95% confidence intervals (CIs) around the model prediction. Fungal species that occur more or less frequently than predicted by the NCM are shown in different colors. m = metacommunity size ×immigration; r2 = the goodness of fit of this model (coefficient of determination).
TABLE 1.
Fungal species indicative of gall surfaces, gall interiors, and larvae for individual ambrosia gall midge (AGM) species mycobiomes based on MC statistics for multi-level pattern analysis (P < 0.050) a
| AGM species | Gall surfaces | Gall interiors | AGM larvae |
|---|---|---|---|
| Asphondylia echii | Alternaria alternata species complex | Botryosphaeria dothidea | Filobasidium oeirense, Fusarium sporotrichioides, Fusarium avenaceum species complex) |
| Asphondylia miki | B. dothidea |
A. alternata species complex, Alternaria sp., Fil. oeirense, Fus. sporotrichioides,
Neosetophoma sp., Periconia byssoides, Tilletiopsis washingtonensis |
|
| Asphondylia verbasci | Botryosphaeriaceae sp., Cladosporium sp. 1, Cladosporium sp. 2 | B. dothidea, Myrmecridium sp. | Fil. oeirense, Fil. wieringae, Aureobasidium pullulans, Ramularia sp., Rhodotorula glutinis |
| Lasioptera arundinis | Cercospora. beticola species complex | A. alternata species complex, Alternaria sp., Apenidiella sp. 1, Apenidiella sp. 2, Aureobasidium pullulans, Fil. oeirense, Fus. sporotrichioides, A. pullulans, Mucor fragilis, Pseudopithomyces rosae, T. washingtonensis | |
| Lasioptera carophila | B. dothidea | Botrytis cinerea, Fus. sporotrichioides, Myrmecridium sp., Papiliotrema frias, T. washingtonensis | |
| Lasioptera eryngii | B. dothidea |
A. alternata species complex, Alteranaria sp., Aur. pullulans, Cercospora zebrina, Cladosporium sp. 2, Fil. oeirense, Fil. wieringae, Fus. sporotrichioides,
A. pullulans, Periconia sp., Pse. rosae, Mycosphaerellaceae sp. |
Full taxa names are listed in Supplementary Material 2B.
Fig 5.
Composition of mycobiomes at the genera level associated with the gall surfaces (S), gall interiors (I), and larvae (L) for individual Asphondylia and Lasioptera species.
Fig 6.
Venn diagrams showing the overlap between mycobiome of gall surface (green), gall interior (red), and gall midge larvae (yellow) of individual Asphondylia and Lasioptera species.
DISCUSSION
Larval mycobiomes differed significantly in composition from the surface, and gall interior mycobiomes were the most species-rich and taxonomically diverse. They hosted the largest pool of significantly associated and species-specific taxa. These results indicate that the larval interior is a prolific environment that supports the growth of diverse and host-specific fungal consortia. In accordance with our hypothesis, the mycobiome of the gall surfaces was assembled most stochastically, while the involvement of neutral processes was the lowest in the mycobiomes of the gall interiors, which also had the lowest taxonomic diversity. The fungal composition in nutritive mycelia tended to be more similar across AGM species than larval and gall surface mycobiomes, suggesting that the interior of ambrosia galls offers a unique environment that supports the growth of similar fungal groups, despite the phylogenetic distances between the host plant species and between the AGM tribes Asphondyliini and Lasiopterini.
The higher similarity of the mycobiomes of the gall interiors between species was due to the dominance of Botryosphaeria (except in L. arundinis). In our study, the sequenced ITS2 regions assigned to Botryosphaeria were identical to the type strain of B. dothidea and conspecific isolates from gall midges (12, 13, 20, 27, 46). This marker is not specific to distinguish it from related species such as B. auasmontanum, B. fabicerciana, B. scharifii, B. ramosa, and B. fusispora. All these species were not reported as AGM symbionts and were known only from outside Europe (47) (Table S1; Supplementary Material 2A). Finally, our taxonomic analysis using ITS rDNA and TEF1α barcode on pure cultures confirmed its identity as B. dothidea.
The dominance of B. dothidea as an AGM symbiont aligns with Bisset and Borkent (2), who suggested that there might be fewer species of fungi than cecidomyiids involved in ambrosia galls, with several midges utilizing the same dominant fungus (12). Asphondyliini may depend on one principal fungus, primarily B. dothidea, sharing specific clonal strains among several species, supplied by some nonspecific fungi (2, 12 – 15, 18, 48, 49). Botryosphaeria dothidea can also be found in other AGMs, including Lasiopterini (50) and Alycaulini (12), but the specificity in Lasiopterini has received much less attention. As Asphondyliini and Lasiopterini form distant clades of Cecidomyiidae, a long evolutionary association with gall midges is likely for B. dothidea (2, 26).
In L. arundinis and L. carophila, B. dothidea was more prevalent on the gall surfaces than in the gall interiors (42.56% and 91.13% of reads, respectively). Botryosphaeria dothidea is a ubiquitous endophyte and pathogen that occurs in multiple plants worldwide (51). Facilitated colonization of endophytes in galled plant tissue is known for non-AGMs (52, 53). It is possible that in AGMs, endophytes may have become fungal symbionts, being genetically identical to free-living populations (17). However, the midge Asteromyia carbonifera (Osten Sacken 1862; tribe Alycaulini, supertribe Lasiopteridi) appears to be associated with a single lineage of B. dothidea despite the supposed lack of vertical transmission. This symbiosis exhibits an unusually high level of specificity for ectosymbiotic associations (12), which are prone to invasion and replacement of symbionts (17, 54). Therefore, B. dothidea may be vertically transmitted by mycangia (17) or acquired environmentally by horizontal transfer in each generation, as is known in fungal symbionts of termites and some wood wasps (55, 56). In such cases, microbial competition among potential symbionts may sustain the specificity of symbiosis through competition-based selection, potentially because hosts provide a specific environment to selectively cultivate favorable symbionts (57). AGM larvae have been shown to inhibit the growth of competing fungi (15, 58) or regulate the metabolism of B. dothidea because the virulence of pathogenic B. dothidea is suppressed in the galls (18, 48). Botryosphaeria dothidea itself may also produce metabolites with antimicrobial or antifungal activities (59); therefore, B. dothidea and larvae may protect each other from unwanted invaders of the gall. Competition-based selection or active larval selection can be corroborated by a reduced role of neutral processes in the gall interior compared to other gall compartments.
In the interior of L. arundinis galls, the dominant fungal species were Myrmecridium sp., R. subulatum, and Sarocladium strictum. Furthermore, a significantly indicative taxon was assigned to the genus Cercospora. The association between L. arundinis and R. subulatum, previously classified as Ramichloridium sp., is already recognized (2, 60). A closely related gall midge, L. hungarica, exhibits a similar relationship with a fungus named Sporotrix sp., probably also conspecific with R. subulatum (61). Cercospora is a plant pathogen, endophyte, and saprobe. The same applies to Sarocladium (62 – 64), which was also found in the galls of Lasioptera donacis (22). Species from the genus Myrmecridium can be found as saprobes (64). Other dominant fungi in the gall interior were Alternaria and Aureobasidium. Alternaria has been reported in mycangia or galls of various Asphondylia species, sometimes also as a prominent fungus (12, 20, 28, 65). Aureobasidium covers the interior of the galls of some Asphondylia (24, 66) or Lasioptera (67). Both Alternaria and Aureobasidium are known as plant endophytes (68, 69) and may also have a nutritive role (21, 70), and both were predominant in larval mycobiomes.
The larval mycobiome was likely composed mainly of the taxa present in the gut. It is not clear whether the fungi occurred in the internal organs or the hemolymph, but the effect of surface contamination was likely very low given the distinct composition of the mycobiome of the larvae and the gall interior and the fact that the larvae were surface-sterilized. In Cecidomyiidae, data on the bacterial part of the intestinome are only available (36, 39), although in fungus-feeding insects, gut microbiomes may help digest fungal biomass (37, 71). In our study, the AGM gut mycobiomes differed substantially from the gall interior; moreover, there was a significant effect of AGM species on the fungal community composition. The significant effect of the host species on the mycobiome composition is contradictory to the patterns found in larval leaf miners (also concealed within the plant tissue but not fungal feeders (43), but similar to those found in leaf-chewing Lepidoptera (33). As the extent to which AGM larvae feed on plants, except nutritive mycelia, remains almost unknown (72, 73), it is possible that these associates are acquired from the host plant and subsequently filtered by the gut environment, which is usually species-specific (33).
Based on previous studies, the core mycobiome of AGM galls is largely composed of saprotrophs (27) and sometimes pathogenic fungi widespread in plants (12, 46), growing epiphytically or endophytically (27, 74). These taxa are good biomass degraders and are tolerant to extreme conditions and stress, they grow rapidly and sporulate. This may also apply to certain taxa indicative of larvae in our study, such as several Fusarium species. In addition to being pathogenic to plants (75), Fusarium can play a role in the defense of insects against pathogens (76), as many strains produce a wide variety of mycotoxins (77), many of which have antibacterial activity (78, 79). Insect vectoring of plant pathogens after ingestion can evolve into mutualism if the insect benefits from the plant infection (80). On the other hand, Fusarium spp. are often mentioned as entomopathogens whose mycotoxins can have a noxious effect on the larvae rather than protecting them from infections (81).
A similar spectrum of genera found in the larvae in this study (Alternaria, Fusarium, Cladosporium, and various yeasts, including Aureobasidium) has been found in the guts of other plant biomass feeders. Among the taxa recorded in this study as significantly indicative of larvae, T. washingtonensis was found in the gut mycobiomes of herbivorous beetles (82), Fil. oeirense and Pseudopithomyces occurred in the gut mycobiomes of Tephritidae fruit flies (83, 84) and Filobasidium and Didymella in the larval gut mycobiomes of ambrosia beetles (41, 42). Interestingly, T. washingtonensis and A. pullulans are probably capable of inducing gall formation (67, 85), and Didymellaceae and Filobasidium have been reported from various galls and gall formers (85 – 87). Neosetophoma is an anamorph of Didymella, which is known to infect the feeding sites of some Cecidomyiidae (88), and Fusarium has been recorded in the galls of some Daphnephila (Cecidomyiidae: Asphondyliini) (14, 48) and Asphondylia (20) species. The secretions produced by AGM larvae are believed to be responsible for gall development (2, 5), and the aforementioned fungi may be involved in the formation of these secretions. Galling insects have been proposed to mediate gall induction by endosymbiotic microorganisms or gall-inducing genes acquired from microbial symbionts through horizontal gene transfer (89, 90).
The larvae likely affect the growth of the fungi in nutritive mycelia because the main fungal development and changes in color and rigidity are observed when larvae stop feeding, die, or are attacked by a parasitoid (13, 15, 16, 23, 91). It is possible that the enzymes secreted by larvae can help modify the composition of the nutritive mycelia. We hypothesized that some of the fungi indicative of larvae might play a role in producing such enzymes. Antibiotic-producing microbes that defend fungal gardens from antagonistic organisms occur in other fungus-growing insects (92 – 97), and the convergent evolution paradigm may suggest the presence of particular antibiotic-producing microbes in gall midges (98). Microbes with symbiotic functions were found in the bacteriocyte-like structure of eggs transmitted maternally in distantly related midges (99). Thus, larval fungal symbionts may be included in further research on potential endosymbiotic microorganisms in AGM species.
Conclusion
For the first time in gall-forming insects, we present a complete characterization of the mycobiomes of the whole system as we profiled gall surfaces, nutritive mycelia, and larvae. Our study suggests a spectrum of fungal microorganisms indicative of individual gall compartments. We have discovered that the most diverse and unique communities are associated with hitherto unstudied intestinal mycobiomes of larvae. However, in AGM larvae, the specificity and role of these fungi remain unresolved. As antibiotic and antifungal properties may be found in these endosymbionts during further research, congruent with the convergent evolution paradigm, AGM larvae could have considerable biochemical potential.
MATERIALS AND METHODS
Sampling and processing of larvae and plant tissues
Galls were sampled in the Cerová vrchovina Protected Landscape Area (48.219N, 19.967E, Slovakia) in August 2019. We sampled six AGM species: Asphondyliini: Asphondylia echii (Loew 1850) from Echium vulgare L., A. miki Wachtl, 1880 from Medicago falcata L., A. verbasci (Vallot 1827) from Verbascum sp. L.; Lasiopterini: Lasioptera arundinis Schiner (1854) from Phragmites australis (Cav.) Trin. ex Steud., L. carophila Löw (1874) from Daucus carota L., and L. eryngii (Vallot 1829) from Eryngium campestre L. The galls were placed in separate plastic containers using sterilized tweezers to avoid contamination. To obtain the microbiota of the gall surface (plant tissue), the surface tissue of the galls was scratched with a sterilized razor blade (approx. thickness 0.1 mm). The galls were then washed by vortexing in a 30-mL centrifuge tube with a 20-mL sterile solution of phosphate-buffered saline (PBS) 1% with Tween 80 (Sigma-Aldrich, Saint Louis, MI, USA) at 2,100 rpm for 45 s to minimize contamination during dissection (100). The galls were dissected on paraffin wax, which was previously sterilized by pouring ethanol and igniting. For subsequent experiments, we used galls with one living larva inside for Asphondylia and at least one living larva inside for Lasioptera (empty galls and galls with pupae, dead larvae, or larvae with parasitoids were discarded). A total of 31 galls were used (5–6 galls from each species): A. echii = 5, A. miki = 6, A. verbasci = 6, L. arundinis = 5, L. carophila = 4, L. eryngii = 5. We scratched their interior and separated one larva. The larvae were washed in the same manner as the galls to minimize contamination by the gall interior. The gall surface, gall interior, and larva represented triplets in subsequent analyses (n = 31 × 3 gall compartments).
Metabarcoding of mycobiomes
Total microbial DNA was extracted from the samples using the NucleoSpin Tissue DNA Isolation Kit, following the manufacturer’s protocol with minor modifications. For a complete lysis of the samples and higher DNA yields, we crushed the samples multiple times in 1.5 mL tubes using pestles and liquid nitrogen before the cell lysis step. To ensure the broad recovery of fungal diversity and to significantly reduce the recovery of chloroplasts, we used highly degenerate primers to amplify the ITS2 rDNA variable gene regions. All PCRs were performed in triplicate. We used the primers ITS3_KYO2 5′–GATGAAGAACGYAGYRAA–3′ (forward) and ITS4_KYO3 5′–CTBTTVCCKCTTCACTCG–3′ (reverse) (101), with barcodes added to the 5′ end of both primers, enabling us to identify each sample. Amplification was performed as described by Toju et al. (101), with minor modifications consisting of initial denaturation at 95°C for 3 min; 35 cycles at 94°C for 30 s, 55°C for 60 s, and 72°C for 60 s; and a final extension at 72°C for 10 min. Each PCR (25 µL) consisted of 9.4 µL molecular biology grade water (New England BioLabs), 0.5 U KAPA2G Robust HotStart DNA Polymerase, 5 µL 5 × KAPA2G Buffer B, 5 µL 5 × KAPA2G Enhancer (all Kapa Biosystems), 0.5 µL 10 mM dNTP Mix (Thermo Fisher Scientific), 0.8 µM of each primer, and 2 µL genomic DNA. On the plate (n = 96), negative controls (n = 3) (mastermix +water + primers) were placed evenly. All PCR products were checked on a 1.5% agarose gel. Subsequently, we pooled triplicate PCRs within each sample, measured DNA concentration using the Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific), and equalized concentrations within all samples. Furthermore, the samples were pooled to create a library. Amplicons of specific length were excised from the 2% agarose gel and purified using QIAquick Gel Extraction Kit (Qiagen) and subjected to DNA ligation of sequencing adapters and library-unique multiplex identifiers necessary for demultiplexing the reads using the KAPA Hyper Prep Kit (Kapa Biosystems) following the manufacturer’s instructions. The ligated library was quantified using the KAPA Library Quantification Kit (Kapa Biosystems) and diluted to create a final sequencing library at 7.5 ng/µL. The library was subjected to paired-end sequencing on an Illumina MiSeq instrument at the Genomics Core Facility, CEITEC (Masaryk University, Brno, Czech Republic), producing 2 × 300 bp long reads (four runs in total). Sequences dedicated to this study represented 4.68% of the total sequencing output.
Metabarcoding data processing
Sequencing data were processed using QIIME 2.0 2020.2 (102). Raw paired-end reads were demultiplexed and quality filtered, including extraction of the ITS region using the q2-ITSxpress plugin (103). Reads were denoized using the DADA2 algorithm (104). Taxonomy was assigned using the q2-feature-classifier classify-sklearn (105) using a trained naïve Bayes classifier against the reference sequences in the UNITE QIIME release for Fungi version 8.0 (106). Information on the read counts for all amplicon sequence variants (ASVs) from all samples together with taxonomic information was compiled into the ASV table.
Statistical analysis
All analyses were performed in R 4.2.1 (107) and Canoco 5.01 (108). We identified the contaminant ASVs based on negative controls using the library “decontam” (109) and discarded them. We then pooled the identified ASVs at the level of fungal species to perform the analyses, but the major analyses were repeated also at the level of ASVs to validate the patterns with a more detailed resolution (Figs S1, S2, and S4; Supplementary Material 1). We analyzed the differences in composition by Permutation Multivariate Analysis of Variance (PERMANOVA) with Bray-Curtis distance matrices and tested 999 permutations, using the library “vegan” (110). As potential explanatory variables, we used the triplet ID (if this was selected based on the AICc, it would be used as a random term in the final model), gall compartment, and taxonomy of AGM (species or genus). We built the final model by forward selection based on the AICc. We accompanied this analysis with canonical correspondence analysis (CCA) using the gall compartment as an explanatory variable and gall species as a covariate and testing the significance using the Monte Carlo test with 999 permutations. As the results of PERMANOVA can be compromised in case of an unbalanced number of samples, we added the PERMDISP2 procedure for the analysis of multivariate homogeneity of group dispersions (variances) based on the Bray–Curtis distance, measuring the distance to the group centroids (111). Differences in β-diversity among gall compartments were tested using ANOVA and Tukey HSD post hoc test. Based on our results, we repeated the PERMANOVA analysis for each gall compartment separately, with AGM species as an explanatory variable.
To analyze which explanatory variables best explained the richness, we used a generalized linear model (GLM) with Gamma distribution with forward selection based on AICc. Before analysis, we standardized the species richness of all samples by rarefying/extrapolating the read counts to a uniform value (n = 1,000) using the library “iNEXT” (112, 113). To analyze taxonomic diversity, we calculated an index developed by Clarke and Warwick (114, 115) (Δ; the average taxonomic distance between any two organisms, randomly chosen from the sample). Six levels of taxonomic resolution were used for index calculations: phylum, class, order, family, genus, and species. For analysis of taxonomic distance, we built GLM with Gamma distribution with forward selection based on AICc from previously mentioned explanatory variables. We accompanied the analyses with species accumulation curves (116) to assess the sufficiency of the sampling effort and with Venn diagrams created using a simulation-based nVenn algorithm from the library “nVennR” (117) for a simple display of the similarity of the gall compartments.
To quantify the involvement of neutral processes in the fungal community assembly, we fitted neutral models at the ASV level for each gall compartment according to Sloan et al. (45) using libraries “reltools” (118), “phyloseq” (119), and “GUniFrac” (120). First, we rarefied samples to the same sequence depth, i.e., 1,000 reads. Then, we fitted the neutral models and extracted information about taxa fitting the null model, being above or below prediction. At the level of fungal species, we identified the indicator species for each gall compartment by IndVal, the indicator value relating to the frequency and relative abundance of the reads (121), and multilevel pattern analysis from libraries “indicspecies” and “labdsv” (122, 123).
Cultivation and identification of fungi
Because in some fungal taxa, ITS2 generated by metabarcoding does not allow accurate taxonomic determination, we directly cultivated fungi from the galls and larvae of A. echii, A. miki, A. pruniperda, A. verbasci, Lasioptera artemisiae, L. arundinis, L. carophila, L. eryngii, and L. rubi. Small fragments of ambrosial fungal mat (1 × 1 mm) and individual larvae were resuspended in 1 mL of 1% PBS solution and crushed with a sterile stamen. One hundred thirty-two micorliters of the suspension (undiluted and 10-fold diluted) was subsequently spread evenly on agar plates with Malt Extract Agar (HiMedia, Mumbai, India). Agar plates were cultivated at 25°C for one week in the dark. After this period, colonies were morphotyped, and morphologically unique cultures were identified. Fungi were identified by ITS rDNA region (primers ITS1 and ITS4) and TEF1α region (primers EF 526F and 986R) barcode according to Kolařík et al. (124). Sequences were compared with data deposited in the NCBI database using the BLASTn tool, with a preference for data obtained from type cultures or reliable taxonomic studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank all colleagues from the Laboratory of Insect Trophic Strategies at the University of Ostrava for laboratory processing. We thank Soňa Kajzrová and Kateřina Křížková for assistance with fungal cultivation and identification.
This work was supported by the Czech Science Foundation (GA23-07026S).
P.D., P.P., and M.Š. designed the research; P.P. and J.Š. sampled the data; P.P., D.V., and M.Š. performed the laboratory work and databased the data; M.Kos. and M.Kol. processed the data bioinformatically; P.P. performed the statistical analysis; P.P. and H.Š. wrote the manuscript; and M.Š., M.Kol., J.Š., and P.D. edited the manuscript and provided additional comments.
Contributor Information
Petr Pyszko, Email: petr.pyszko@osu.cz.
Christina A. Cuomo, Broad Institute, Cambridge, Massachusetts, USA
Shu Benshui, Zhongkai University of Agriculture and Engineering, Guangzhou, China .
Rosario Nicoletti, Università di Napoli Federico II, Naples, Italy .
DATA AVAILABILITY
Raw demultiplexed sequencing data with sample annotations are available under the accession number PRJNA693163 at the NCBI Bioproject website.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.02830-23.
Taxonomic identity of ITS2 sequences ascribable to the Botryosphaeria genus.
Taxonomic identity of cultures isolated from gall interior and larvae.
Figures S1 to S4.
Taxonomic identity of species significantly indicative for gall surfaces, gall interiors, and AGM larvae.
An accounting of the reviewer comments and feedback.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Mamaev BM. 1975. Evolution of gall forming insects, gall midges. British Library Lending Division, Wetherby, UK. [Google Scholar]
- 2. Bissett J, Borkent A. 1988. Ambrosia galls: the significance of fungal nutrition in the evolution of the cecidomyiidae (Diptera), p 203–226. In Pirozynski KA, Hawksworth DL (ed), Coevolution of fungi with plants and animals. Academic Press, London. [Google Scholar]
- 3. Dorchin N, Harris KM, Stireman JO. 2019. Phylogeny of the gall midges (Diptera, Cecidomyiidae, Cecidomyiinae): systematics, evolution of feeding modes and diversification rates. Mol Phylogenet Evol 140:106602. doi: 10.1016/j.ympev.2019.106602 [DOI] [PubMed] [Google Scholar]
- 4. Sikora T, Jaschhof M, Mantič M, Kaspřák D, ševčík J. 2019. Considerable congruence, enlightening conflict: molecular analysis largely supports morphology-based hypotheses on Cecidomyiidae (Diptera) phylogeny. Zool J Linn Soc 185:98–110. doi: 10.1093/zoolinnean/zly029 [DOI] [Google Scholar]
- 5. Yukawa J, Rohfritsch O. 2005. Biology and ecology of gall-inducing Cecidomyiidae (Diptera), p 273–304. In Raman A (ed), Biology, ecology, and evolution of gall-inducing arthropods. Science Publishers, Inc, Enfield, USA. [Google Scholar]
- 6. Dentinger BT, Bills C. 2018. Fungal cultivation by insects, p 1–9. In eLS [Google Scholar]
- 7. Beaver RA. 1989. Insect-fungus relationships in the bark and ambrosia beetles, p 121–137. In Wilding N, Collins NM, Hammond PM, Webber JF (ed), Insect-fungus interactions. Academic Press, London, UK. [Google Scholar]
- 8. Gilbertson RL. 1984. Relationships between insects and wood-rotting Basidiomycetes, p 130–165. In Wheeler Q, Blackwell M (ed), Fungus-insect relationships, perspectives in ecology and evolution. Columbia University Press, New York. [Google Scholar]
- 9. Morgan FD. 1968. Bionomics of Siricidae. Annu Rev Entomol 13:239–256. doi: 10.1146/annurev.en.13.010168.001323 [DOI] [Google Scholar]
- 10. Cherrett JM, Powell RJ, Strandling DJ. 1989. The mutualism between leaf-cutting ants and their fungus, p 93–116. In Wilding N, Collins NM, Hammond PM, Webber JF (ed), Insect-fungus interactions. Academic Press, London, UK. [Google Scholar]
- 11. Wood TG, Thomas RJ. 1989. The mutualistic association between Macrotermitinae and Termitomyces, p 69–88. In Wilding N, Collins NM, Hammond PM, Webber JF (ed), Insect-fungus interactions. Academic Press, London, UK. [Google Scholar]
- 12. Adair RJ, Burgess T, Serdani M, Barber P. 2009. Fungal associations in Asphondylia (Diptera: Cecidomyiidae) galls from Australia and South Africa: implications for biological control of invasive acacias. Fungal Ecol 2:121–134. doi: 10.1016/j.funeco.2009.02.003 [DOI] [Google Scholar]
- 13. Kobune S, Kajimura H, Masuya H, Kubono T. 2012. Symbiotic fungal flora in leaf galls induced by Illiciomyia yukawai (Diptera: Cecidomyiidae) and in its mycangia. Microb Ecol 63:619–627. doi: 10.1007/s00248-011-9962-0 [DOI] [PubMed] [Google Scholar]
- 14. Pan L-Y, Chen W-N, Chiu S-T, Raman A, Chiang T-C, Yang M-M. 2015. Is a gall an extended phenotype of the inducing insect? A comparative study of selected morphological and physiological traits of leaf and stem galls on Machilus thunbergii (Lauraceae) induced by five species of Daphnephila (Diptera: Cecidomyiidae) in northeastern Taiwan. Zoolog Sci 32:314–321. doi: 10.2108/zs140244 [DOI] [PubMed] [Google Scholar]
- 15. Rohfritsch O. 2008. Plants, gall midges, and fungi: a three-component system. Entomologia Exp Applicata 128:208–216. doi: 10.1111/j.1570-7458.2008.00726.x [DOI] [Google Scholar]
- 16. Heath JJ, Stireman III JO. 2010. Dissecting the association between a gall midge, Asteromyia carbonifera, and its symbiotic fungus, Botryosphaeria dothidea. Entomologia Exp Applicata 137:36–49. doi: 10.1111/j.1570-7458.2010.01040.x [DOI] [Google Scholar]
- 17. Janson EM, Peeden ER, Stireman III JO, Abbot P. 2010. Symbiont-mediated phenotypic variation without co-evolution in an insect–fungus association. J Evol Biol 23:2212–2228. doi: 10.1111/j.1420-9101.2010.02082.x [DOI] [PubMed] [Google Scholar]
- 18. Park I, Sanogo S, Hanson SF, Thompson DC. 2019. Molecular identification of Botryosphaeria dothidea as a fungal associate of the gall midge Asphondylia prosopidis on mesquite in the United States. BioControl 64:209–219. doi: 10.1007/s10526-019-09924-6 [DOI] [Google Scholar]
- 19. Meyer JB, Gallien L, Prospero S. 2015. Interaction between two invasive organisms on the European chestnut: does the chestnut blight fungus benefit from the presence of the gall wasp? FEMS Microbiol Ecol 91:fiv122. doi: 10.1093/femsec/fiv122 [DOI] [PubMed] [Google Scholar]
- 20. Bernardo U, Nugnes F, Gualtieri L, Nicoletti R, Varricchio P, Sasso R, Viggiani G. 2018. A new gall midge species of Asphondylia (Diptera: Cecidomyiidae) inducing flower galls on Clinopodium nepeta (Lamiaceae) from Europe, its phenology, and associated fungi. Environ Entomol 47:609–622. doi: 10.1093/ee/nvy028 [DOI] [PubMed] [Google Scholar]
- 21. Te Strake D, Keagy AH, Stiling PD. 2006. Fungi associated with Borrichia frutescens (Asteraceae): insect galls and endophytes. SIDA Contrib Bot 22:755–763. [Google Scholar]
- 22. Bon M-C, Guermache F, Simone D de, Cristofaro M, Vacek A, Goolsby J. 2018. Insights into the microbes and nematodes hosted by Pupae of the Arundo leaf miner, Lasioptera donacis (Diptera: Cecidomyiidae). Florida Entomologist 101:505–507. doi: 10.1653/024.101.0309 [DOI] [Google Scholar]
- 23. Rohfritsch O. 1992. A fungus associated gall midge, Lasioptera arundinis (Schiner), on Phragmites australis (Cav.) Trin. Bull Société Bot Fr Lett Bot 139:45–49. doi: 10.1080/01811797.1992.10824942 [DOI] [Google Scholar]
- 24. Batra LR, Lichtwardt RW. 1963. Association of fungi with some insect galls. J Kans Entomol Soc 36:262–278. [Google Scholar]
- 25. Sugiura S, Yamazaki K. 2009. Gall-attacking behavior in phytophagous insects, with emphasis on Coleoptera and Lepidoptera. Terr Arthropod Rev 2:41–61. doi: 10.1163/187498309X435658 [DOI] [Google Scholar]
- 26. Veenstra AA, Lebel T, Milne J, Kolesik P. 2019. Two new species of Dactylasioptera (Diptera: Cecidomyiidae) inducing stem galls on Maireana (Chenopodiaceae). Austral Entomol 58:220–234. doi: 10.1111/aen.12363 [DOI] [Google Scholar]
- 27. Zimowska B, Okoń S, Becchimanzi A, Krol ED, Nicoletti R. 2020. Phylogenetic characterization of Botryosphaeria strains associated with Asphondylia galls on species of Lamiaceae. Diversity 12:41. doi: 10.3390/d12020041 [DOI] [Google Scholar]
- 28. Zimowska B, Viggiani G, Nicoletti R, Furmańczyk A, Becchimanzi A, Kot I. 2017. First report of the gall midge Asphondylia serpylli on thyme (Thymus vulgaris), and identification of the associated fungal symbiont. Ann Appl Biol 171:89–94. doi: 10.1111/aab.12360 [DOI] [Google Scholar]
- 29. Amann RI, Ludwig W, Schleifer KH. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169. doi: 10.1128/mr.59.1.143-169.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nagle FS, Casamatta DA, Rossi A. 2019. Genetic analysis of the fungal community resident in Asphondylia borrichiae (Diptera: Cecidomyiidae) galls. St. Louis, MO: ESA; [Google Scholar]
- 31. Gurung K, Wertheim B, Falcao Salles J. 2019. The microbiome of pest insects: it is not just bacteria. Entomol Exp Appli 167:156–170. doi: 10.1111/eea.12768 [DOI] [Google Scholar]
- 32. Ravenscraft A, Berry M, Hammer T, Peay K, Boggs C. 2019. Structure and function of the bacterial and fungal gut microbiota of Neotropical butterflies. Ecol Monogr 89. doi: 10.1002/ecm.1346 [DOI] [Google Scholar]
- 33. Šigut M, Pyszko P, Šigutová H, Višňovská D, Kostovčík M, Kotásková N, Dorňák O, Kolařík M, Drozd P. 2022. Fungi are more transient than bacteria in caterpillar gut microbiomes. Sci Rep 12:15552. doi: 10.1038/s41598-022-19855-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Větrovský T, Soukup P, Stiblik P, Votýpková K, Chakraborty A, Larrañaga IO, Sillam-Dussès D, Lo N, Bourguignon T, Baldrian P, Šobotník J, Kolařík M. 2020. Termites host specific fungal communities that differ from those in their ambient environments. Fungal Ecol 48:100991. doi: 10.1016/j.funeco.2020.100991 [DOI] [Google Scholar]
- 35. Višňovská D, Pyszko P, Šigut M, Kostovčík M, Kolařík M, Kotásková N, Drozd P. 2020. Caterpillar gut and host plant phylloplane mycobiomes differ: a new perspective on fungal involvement in insect guts. FEMS Microbiol Ecol 96:fiaa116. doi: 10.1093/femsec/fiaa116 [DOI] [PubMed] [Google Scholar]
- 36. Bansal R, Hulbert SH, Reese JC, Whitworth RJ, Stuart JJ, Chen M-S. 2014. Pyrosequencing reveals the predominance of pseudomonadaceae in gut microbiome of a gall midge. Pathogens 3:459–472. doi: 10.3390/pathogens3020459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hu H, da Costa RR, Pilgaard B, Schiøtt M, Lange L, Poulsen M. 2019. Fungiculture in termites is associated with a mycolytic gut bacterial community. mSphere 4:e00165-19. doi: 10.1128/mSphere.00165-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Long Y-H, Xie L, Liu N, Yan X, Li M-H, Fan M-Z, Wang Q. 2010. Comparison of gut-associated and nest-associated microbial communities of a fungus-growing termite (Odontotermes yunnanensis). Insect Sci 17:265–276. doi: 10.1111/j.1744-7917.2010.01327.x [DOI] [Google Scholar]
- 39. Ojha A, Sinha DK, Padmakumari AP, Bentur JS, Nair S. 2017. Bacterial community structure in the Asian rice gall midge reveals a varied microbiome rich in proteobacteria. Sci Rep 7:9424. doi: 10.1038/s41598-017-09791-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Richards C, Otani S, Mikaelyan A, Poulsen M. 2017. Pycnoscelus surinamensis cockroach gut microbiota respond consistently to a fungal diet without mirroring those of fungus-farming termites. PLoS One 12:e0185745. doi: 10.1371/journal.pone.0185745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ceriani-Nakamurakare E, Mc Cargo P, Gonzalez-Audino P, Ramos S, Carmarán C. 2020. New insights into fungal diversity associated with Megaplatypus mutatus: gut mycobiota. Symbiosis 81:127–137. doi: 10.1007/s13199-020-00687-8 [DOI] [Google Scholar]
- 42. Gao G, Gao J, Hao C, Dai L, Chen H. 2018. Biodiversity and activity of gut fungal communities across the life history of Trypophloeus klimeschi (Coleoptera: Curculionidae: Scolytinae). Int J Mol Sci 19:2010. doi: 10.3390/ijms19072010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Šigutová H, Šigut M, Pyszko P, Kostovčík M, Kolařík M, Drozd P. 2023. Seasonal shifts in bacterial and fungal microbiomes of leaves and associated leaf-mining larvae reveal persistence of core taxa regardless of diet. Microbiol Spectr 11:e0316022. doi: 10.1128/spectrum.03160-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rodrigues A, Cable RN, Mueller UG, Bacci M, Pagnocca FC. 2009. Antagonistic interactions between garden yeasts and microfungal garden pathogens of leaf-cutting ants. Antonie Van Leeuwenhoek 96:331–342. doi: 10.1007/s10482-009-9350-7 [DOI] [PubMed] [Google Scholar]
- 45. Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP. 2006. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol 8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x [DOI] [PubMed] [Google Scholar]
- 46. Lebel T, Peele C, Veenstra A. 2012. Fungi associated with Asphondylia (Diptera: Cecidomyiidae) galls on Sarcocornia quinqueflora and Tecticornia arbuscula (Chenopodiaceae). Fungal Divers 55:143–154. doi: 10.1007/s13225-012-0157-x [DOI] [Google Scholar]
- 47. Phillips AJL, Alves A, Abdollahzadeh J, Slippers B, Wingfield MJ, Groenewald JZ, Crous PW. 2013. The Botryosphaeriaceae: genera and species known from culture. Stud Mycol 76:51–167. doi: 10.3114/sim0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Raman A, Suryanarayanan TS. 2017. Fungus–plant interaction influences plant-feeding insects. Fungal Ecol 29:123–132. doi: 10.1016/j.funeco.2017.06.004 [DOI] [Google Scholar]
- 49. Tokuda M, Yukawa J. 2007. Biogeography and evolution of gall midges (Diptera: Cecidomyiidae) inhabiting broad-leaved evergreen forests in Oriental and eastern Palearctic regions. Oriental Insects 41:121–139. doi: 10.1080/00305316.2007.10417502 [DOI] [Google Scholar]
- 50. Latinović J, Hrnčić S, Perović T, Latinović N. 2014. Botryosphaeria dothidea - causal agent of olive fruit rot - pathogen of wounds or not? Botryosphaeria dothidea - causal agent olive fruit rot. Pathog Wounds Not 108:35–38. doi: 10.1016/j.cropro.2013.02.004 [DOI] [Google Scholar]
- 51. Marsberg A, Kemler M, Jami F, Nagel JH, Postma-Smidt A, Naidoo S, Wingfield MJ, Crous PW, Spatafora JW, Hesse CN, Robbertse B, Slippers B. 2017. Botryosphaeria dothidea: a latent pathogen of global importance to woody plant health. Mol Plant Pathol 18:477–488. doi: 10.1111/mpp.12495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carroll GC. 1986. The biology of endophytism in plants with particular reference to woody perennials, p 203–222. In Microbiol Phyllosphere [Google Scholar]
- 53. Petrini LE, Petrini O, Laflamme G. 1989. Recovery of endophytes of Abies balsamea from needles and galls of Paradiplosis tumifex. Phytoprotection 70:97–103. [Google Scholar]
- 54. Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience Publishers, New York. https://agris.fao.org/agris-search/search.do?recordID=US201300597419. [Google Scholar]
- 55. Aanen DK, Ros VID, de Fine Licht HH, Mitchell J, de Beer ZW, Slippers B, Rouland-Lefèvre C, Boomsma JJ. 2007. Patterns of interaction specificity of fungus-growing termites and Termitomyces symbionts in South Africa. BMC Evol Biol 7:115. doi: 10.1186/1471-2148-7-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pažoutová S, Šrůtka P, Holuša J, Chudíčková M, Kolařík M. 2010. Diversity of xylariaceous symbionts in Xiphydria woodwasps: role of vector and a host tree. Fungal Ecol 3:392–401. doi: 10.1016/j.funeco.2010.07.002 [DOI] [Google Scholar]
- 57. Itoh H, Jang S, Takeshita K, Ohbayashi T, Ohnishi N, Meng X-Y, Mitani Y, Kikuchi Y. 2019. Host-symbiont specificity determined by microbe-microbe competition in an insect gut. Proc Natl Acad Sci U S A 116:22673–22682. doi: 10.1073/pnas.1912397116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bronner N. 1992. The role of nutritive cells in the nutrition of cynipids and cecidomyiids, p 118–140. In Biology of insect-induced galls. Oxford University Press, Oxford. [Google Scholar]
- 59. Xiao J, Zhang Q, Gao Y-Q, Tang J-J, Zhang A-L, Gao J-M. 2014. Secondary metabolites from the endophytic Botryosphaeria dothidea of Melia azedarach and their antifungal, antibacterial, antioxidant, and cytotoxic activities. J Agric Food Chem 62:3584–3590. doi: 10.1021/jf500054f [DOI] [PubMed] [Google Scholar]
- 60. Rohfritsch O. 1997. Morphological and behavioural adaptations of the gall midge Lasioptera arundinis (Schiner) (Diptera, Cecidomyiidae) to collect and transport conidia of its fungal symbiont. Tijdschr Voor Entomol 140:59–66. [Google Scholar]
- 61. Ryckegem G. 2005. Fungi on common reed (phragmites australis): fungal diversity, community structure and decomposition processes / Gunther Van Ryckegem Dissertation thesis, Universiteit Ghent, Ghent, Belgium: [Google Scholar]
- 62. Giraldo A, Gené J, Sutton DA, Madrid H, de Hoog GS, Cano J, Decock C, Crous PW, Guarro J. 2015. Phylogeny of Sarocladium (Hypocreales). Persoonia 34:10–24. doi: 10.3767/003158515X685364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rashmi M, Kushveer J, Sarma V. 2019. A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere 10:798–1079. doi: 10.5943/mycosphere/10/1/19 [DOI] [Google Scholar]
- 64. Wijayawardene NN, Hyde KD, Rajeshkumar KC, Hawksworth DL, Madrid H, Kirk PM, Braun U, Singh RV, Crous PW, Kukwa M, et al. 2017. Notes for genera: ascomycota. Fungal Divers 86:1–594. doi: 10.1007/s13225-017-0386-0 [DOI] [Google Scholar]
- 65. Borkent A, Bissett J. 1985. Gall midges (Diptera: Cecidomyiidae) are vectors for their fungal symbionts. Symbiosis 1:185–194. [Google Scholar]
- 66. Malagaris P. 2011. Biology and ecology of Asphondylia coridothymi (Diptera: Cecidomyiidae) inducing galls on Coridothymus capitatus on the island of Samos, Greece. Acta Soc Zool Bohemicae 75:239–251. [Google Scholar]
- 67. Herman RP, Bynum HG, Alexander AB. 1993. Interaction between the black yeast Aureobasidium pullulans and the gall midge Lasioptera ephedricola in gall formation on the desert shrub Ephedra trifurca. Ecography 16:261–268. doi: 10.1111/j.1600-0587.1993.tb00215.x [DOI] [Google Scholar]
- 68. Schena L, Nigro F, Pentimone I, Ligorio A, Ippolito A. 2003. Control of postharvest rots of sweet cherries and table grapes with endophytic isolates of Aureobasidium pullulans. Postharvest Biol Technol 30:209–220. doi: 10.1016/S0925-5214(03)00111-X [DOI] [Google Scholar]
- 69. Wang Y, Yang M-H, Wang X-B, Li T-X, Kong L-Y. 2014. Bioactive metabolites from the endophytic fungus Alternaria alternata. Fitoterapia 99:153–158. doi: 10.1016/j.fitote.2014.09.015 [DOI] [PubMed] [Google Scholar]
- 70. McLeod JM. 1969. On the habits of a jack pine needle-miner, Eucordylea canusella (Lepidoptera : Gelechiidae), with special reference to its association with a fungus, Aureobasidium pullulans (Maniliales (Deuteromycetes) dematiaceae). Can Entomol 101:166–179. doi: 10.4039/Ent101166-2 [DOI] [Google Scholar]
- 71. Barcoto MO, Carlos-Shanley C, Fan H, Ferro M, Nagamoto NS, Bacci M, Currie CR, Rodrigues A. 2020. Fungus-growing insects host a distinctive microbiota apparently adapted to the fungiculture environment. Sci Rep 10:12384. doi: 10.1038/s41598-020-68448-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Janson EM, Grebenok RJ, Behmer ST, Abbot P. 2009. Same host-plant, different sterols: variation in sterol metabolism in an insect herbivore community. J Chem Ecol 35:1309–1319. doi: 10.1007/s10886-009-9713-6 [DOI] [PubMed] [Google Scholar]
- 73. Stireman JO, Devlin H, Carr TG, Abbot P. 2010. Evolutionary diversification of the gall midge genus Asteromyia (Cecidomyiidae) in a multitrophic ecological context. Mol Phylogenet Evol 54:194–210. doi: 10.1016/j.ympev.2009.09.010 [DOI] [PubMed] [Google Scholar]
- 74. Hammon KE, Faeth SH. 1992. Ecology of plant-herbivore communities: a fungal component? Nat Toxins 1:197–208. doi: 10.1002/nt.2620010307 [DOI] [PubMed] [Google Scholar]
- 75. Ma L-J, Geiser DM, Proctor RH, Rooney AP, O’Donnell K, Trail F, Gardiner DM, Manners JM, Kazan K. 2013. Fusarium pathogenomics. Annu Rev Microbiol 67:399–416. doi: 10.1146/annurev-micro-092412-155650 [DOI] [PubMed] [Google Scholar]
- 76. Pyszko P, Višňovská D, Drgová M, Šigut M, Drozd P. 2020. Effect of bacterial and fungal microbiota removal on the survival and development of bryophagous beetles. Environ Entomol 49:902–911. doi: 10.1093/ee/nvaa060 [DOI] [PubMed] [Google Scholar]
- 77. Fotso J, Leslie JF, Smith JS. 2002. Production of beauvericin, moniliformin, fusaproliferin, and fumonisins B1, B2, and B3 by fifteen ex-type strains of Fusarium species. Appl Environ Microbiol 68:5195–5197. doi: 10.1128/AEM.68.10.5195-5197.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ingle A, Gade A, Pierrat S, Sonnichsen C, Rai M. 2008. Mycosynthesis of silver nanoparticles using the fungus Fusarium acuminatum and its activity against some human pathogenic bacteria. Curr Nanosci 4:141–144. doi: 10.2174/157341308784340804 [DOI] [Google Scholar]
- 79. Wang Q, Xu L. 2012. Beauvericin, a bioactive compound produced by fungi: a short review. Molecules 17:2367–2377. doi: 10.3390/molecules17032367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Frago E, Dicke M, Godfray HCJ. 2012. Insect symbionts as hidden players in insect–plant interactions. Trends Ecol Evol 27:705–711. doi: 10.1016/j.tree.2012.08.013 [DOI] [PubMed] [Google Scholar]
- 81. Santos AC da S, Diniz AG, Tiago PV, de Oliveira NT. 2020. Entomopathogenic Fusarium species: a review of their potential for the biological control of insects, implications and prospects. Fungal Biology Reviews 34:41–57. doi: 10.1016/j.fbr.2019.12.002 [DOI] [Google Scholar]
- 82. Molnár O, Wuczkowski M, Prillinger H. 2008. Yeast biodiversity in the guts of several pests on maize; comparison of three methods: classical isolation, cloning and DGGE. Mycol Prog 7:111–123. doi: 10.1007/s11557-008-0558-0 [DOI] [Google Scholar]
- 83. Majumder R, Sutcliffe B, Taylor PW, Chapman TA. 2020. Microbiome of the Queensland fruit fly through metamorphosis. Microorganisms 8:795. doi: 10.3390/microorganisms8060795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Vitanović E, Aldrich JR, Boundy-Mills K, Čagalj M, Ebeler SE, Burrack H, Zalom FG. 2020. Olive fruit fly, Bactrocera oleae (Diptera: Tephritidae), attraction to volatile compounds produced by host and insect-associated yeast strains. J Econ Entomol 113:752–759. doi: 10.1093/jee/toz341 [DOI] [PubMed] [Google Scholar]
- 85. Glushakova AM, Kachalkin AV. 2017. Endophytic yeasts in Malus domestica and Pyrus communis fruits under anthropogenic impact. Microbiology 86:114–122. doi: 10.1134/S0026261716060102 [DOI] [PubMed] [Google Scholar]
- 86. Garas LS, Uzabakiriho JD, Chimwamurombe PM. 2012. Isolation and identification of fungal species associated with gall formation on Acacia mellifera in the western Windhoek. J Pure Appl Microbiol 6:713–716. [Google Scholar]
- 87. Morales-Rodriguez C, Sferrazza I, Aleandri M, Dalla Valle M, Mazzetto T, Speranza S, Contarini M, Vannini A. 2019. Fungal community associated with adults of the chestnut gall wasp Dryocosmus kuriphilus after emergence from galls: taxonomy and functional ecology. Fungal Biol 123:905–912. doi: 10.1016/j.funbio.2019.09.009 [DOI] [PubMed] [Google Scholar]
- 88. Tóth P, Tóthová M, Váňová M. 2006. First records of Resseliella theobaldi (Diptera, Cecidomyiidae), an important pest of raspberry from Slovakia. Biologia 61:239–240. doi: 10.2478/s11756-006-0044-6 [DOI] [Google Scholar]
- 89. Bartlett L, Connor EF. 2014. Exogenous phytohormones and the induction of plant galls by insects. Arthropod-Plant Interact 8:339–348. doi: 10.1007/s11829-014-9309-0 [DOI] [Google Scholar]
- 90. Giron D, Glevarec G. 2014. Cytokinin-induced phenotypes in plant-insect interactions: learning from the bacterial world. J Chem Ecol 40:826–835. doi: 10.1007/s10886-014-0466-5 [DOI] [PubMed] [Google Scholar]
- 91. Arduin M, Kraus JE. 2001. Anatomy of Ambrosia leaf galls in Baccharis concinna and Baccharis dracunculifolia (Asteraceae). Braz J Bot 24:63–72. doi: 10.1590/S0100-84042001000100007 [DOI] [Google Scholar]
- 92. Barke J, Seipke RF, Grüschow S, Heavens D, Drou N, Bibb MJ, Goss RJM, Yu DW, Hutchings MI. 2010. A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol 8:109. doi: 10.1186/1741-7007-8-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Cafaro MJ, Currie CR. 2005. Phylogenetic analysis of mutualistic filamentous bacteria associated with fungus-growing ants. Can J Microbiol 51:441–446. doi: 10.1139/w05-023 [DOI] [PubMed] [Google Scholar]
- 94. Cardoza YJ, Vasanthakumar A, Suazo A, Raffa KF. 2009. Survey and phylogenetic analysis of culturable microbes in the oral secretions of three bark beetle species. Entomol Exp Appl 131:138–147. doi: 10.1111/j.1570-7458.2009.00844.x [DOI] [Google Scholar]
- 95. Cardoza YJ, Klepzig KD, Raffa KF. 2006. Bacteria in oral secretions of an endophytic insect inhibit antagonistic fungi. Ecol Entomol 31:636–645. doi: 10.1111/j.1365-2311.2006.00829.x [DOI] [Google Scholar]
- 96. Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. 2006. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science 311:81–83. doi: 10.1126/science.1119744 [DOI] [PubMed] [Google Scholar]
- 97. Um S, Fraimout A, Sapountzis P, Oh D-C, Poulsen M. 2013. The fungus-growing termite Macrotermes natalensis harbors bacillaene-producing Bacillus sp. that inhibit potentially antagonistic fungi. Sci Rep 3:3250. doi: 10.1038/srep03250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Bittleston LS, Pierce NE, Ellison AM, Pringle A. 2016. Convergence in multispecies interactions. Trends Ecol Evol 31:269–280. doi: 10.1016/j.tree.2016.01.006 [DOI] [PubMed] [Google Scholar]
- 99. Bansal R, Hulbert S, Schemerhorn B, Reese JC, Whitworth RJ, Stuart JJ, Chen M-S, Ho PL. 2011. Hessian fly-associated bacteria: transmission, essentiality, and composition. PLoS One 6:e23170. doi: 10.1371/journal.pone.0023170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bateman C, Šigut M, Skelton J, Smith KE, Hulcr J. 2016. Fungal associates of the Xylosandrus compactus (Coleoptera: Curculionidae, Scolytinae) are spatially segregated on the insect body. Environ Entomol 45:883–890. doi: 10.1093/ee/nvw070 [DOI] [PubMed] [Google Scholar]
- 101. Toju H, Tanabe AS, Yamamoto S, Sato H. 2012. High-coverage ITS primers for the DNA-based identification of Ascomycetes and Basidiomycetes in environmental samples. PLoS One 7:e40863. doi: 10.1371/journal.pone.0040863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet C, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. 2018. QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science. PeerJ 6:e27295v2. doi: 10.7287/peerj.preprints.27295v2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Rivers AR, Weber KC, Gardner TG, Liu S, Armstrong SD. 2018. ITSxpress: software to rapidly trim internally transcribed spacer sequences with quality scores for marker gene analysis. F1000Res 7:1418. doi: 10.12688/f1000research.15704.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J. 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Nilsson RH, Larsson K-H, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L, Saar I, Kõljalg U, Abarenkov K. 2019. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res 47:D259–D264. doi: 10.1093/nar/gky1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. R Core Team . 2022. R: a language and environment for statistical computing (4.2.1). R foundation for statistical computing. Vienna, Austria. [Google Scholar]
- 108. Ter Braak CJF, Šmilauer P. 2012. CANOCO reference manual and CanoDraw for windows user’s guide: software for Canonical community ordination (version 5.01). Microcomputer Power, Ithaca, NY. [Google Scholar]
- 109. Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. 2018. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6:226. doi: 10.1186/s40168-018-0605-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens M, Szoecs E, Wagner H. 2019. vegan: community ecology package. R package version 2.4-5
- 111. Anderson MJ. 2006. Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62:245–253. doi: 10.1111/j.1541-0420.2005.00440.x [DOI] [PubMed] [Google Scholar]
- 112. Chao A, Gotelli NJ, Hsieh TC, Sander EL, Ma KH, Colwell RK, Ellison AM. 2014. Rarefaction and extrapolation with Hill numbers: a framework for sampling and estimation in species diversity studies. Ecol Monogr 84:45–67. doi: 10.1890/13-0133.1 [DOI] [Google Scholar]
- 113. Hsieh TC, Ma KH, Chao A. 2022. iNEXT: iNterpolation and EXTrapolation for species diversity. R package version 3.0.0
- 114. Clarke KR, Warwick RM. 2001. A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Mar Ecol Prog Ser 216:265–278. doi: 10.3354/meps216265 [DOI] [Google Scholar]
- 115. Clarke KR, Warwick RM. 1998. A taxonomic distinctness index and its statistical properties. J Appl Ecol 35:523–531. doi: 10.1046/j.1365-2664.1998.3540523.x [DOI] [Google Scholar]
- 116. Coleman BD, Mares MA, Willig MR, Hsieh Y-H. 1982. Randomness, area, and species richness. Ecology 63:1121–1133. doi: 10.2307/1937249 [DOI] [Google Scholar]
- 117. Pérez-Silva JG, Araujo-Voces M, Quesada V. 2018. nVenn: generalized, quasi-proportional Venn and Euler diagrams. Bioinformatics 34:2322–2324. doi: 10.1093/bioinformatics/bty109 [DOI] [PubMed] [Google Scholar]
- 118. Sprockett D. 2022. reltools: microbiome amplicon analysis and visualization. R package version 0.1.0
- 119. McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Chen J, Zhang X, Yang L. 2022. GUniFrac: generalized Unifrac distances, distance-based multivariate methods and feature-based univariate methods for microbiome data analysis. R package version 1.7
- 121. Dufrêne M, Legendre P. 1997. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr 67:345–366. doi: 10.1890/0012-9615(1997)067[0345:SAAIST]2.0.CO;2 [DOI] [Google Scholar]
- 122. De Cáceres M, Legendre P. 2009. Associations between species and groups of sites: indices and statistical inference. Ecology 90:3566–3574. doi: 10.1890/08-1823.1 [DOI] [PubMed] [Google Scholar]
- 123. Roberts DW. 2019. Ordination and multivariate analysis for ecology. R package version 2.0-1
- 124. Kolařík M, Stępniewska H, Jankowiak R. 2021. Taxonomic revision of the acidophilic genus Acidiella (Dothideomycetes, Capnodiales) with a description of new species from Poland. Plant Syst Evol 307:38. doi: 10.1007/s00606-021-01753-4 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Taxonomic identity of ITS2 sequences ascribable to the Botryosphaeria genus.
Taxonomic identity of cultures isolated from gall interior and larvae.
Figures S1 to S4.
Taxonomic identity of species significantly indicative for gall surfaces, gall interiors, and AGM larvae.
An accounting of the reviewer comments and feedback.
Data Availability Statement
Raw demultiplexed sequencing data with sample annotations are available under the accession number PRJNA693163 at the NCBI Bioproject website.