Abstract
Streptomyces griseus protease B, a member of the chymotrypsin superfamily, is encoded by a gene that expresses a pre-pro-mature protein. During secretion the precursor protein is processed into a mature, fully folded protease. In this study, we constructed a family of genes which encode deletions at the amino-terminal end of the propeptide. The secretion of active protease B was seen to decrease in an exponential manner according to the length of the deletion. The results underscore the intimate relationship between folding and secretion in bacterial protease expression. They further suggest that the propeptide segment of the zymogen stabilizes the folding of the mature enzyme through many small binding interactions over the entire surface of the peptide rather than through a few specific contacts.
Secreted proteins of prokaryotes and eukaryotes are commonly expressed in precursor forms. These are subsequently processed into mature enzymes by the action of one or more peptide hydrolases. For example, the chymotrypsin-like proteases of Streptomyces griseus are translated in the form of pre-pro-mature precursors, whereas only the mature segments of the polypeptides appear in culture supernatants.
Precursor polypeptides have signal sequences attached at either amino-terminal (in which case they are referred to as “pre”) or carboxyl-terminal ends of the protein. Signal sequences are responsible for directing the nascent proteins to the cell surface. The role of the propeptide in the precursor is less clear. Recent studies suggest roles for propeptides in protein folding, protein secretion, and inhibition of enzyme activity. The propeptides of certain serine proteases are thought to catalyze a rate-determining step in protein folding (1). For example, the propeptide of the α-lytic protease was shown to lower the activation barrier between active and inactive conformations of this enzyme by as much as 27 kcal · mol−1 (3). In this example the propeptide appears to function as the intramolecular equivalent of the chaperonin. During α-lytic protease secretion the propeptide is cleaved from the precursor protein and degraded; consequently, the enzyme becomes trapped in a conformational well.
Some propeptides are reported to be potent inhibitors of the activity of the mature enzymes (2, 15, 26). It is possible that inhibition is a major biological role for the pro regions, ensuring that the enzyme is inactive until it has been secreted from the cell. Alternatively, enzyme inhibition may be an artifact of the primary function of the propeptide, to reduce the activation energy between the folded and unfolded states of the protein (23). In either case, some propeptides appear to possess both folding and inhibitory functions.
Despite a common catalytic mechanism, it is possible to categorize serine proteases into distinct families or clans of distant evolutionary origin (4). Subtilisin from Bacillus subtilis (11), α-lytic protease from Lysobacter enzymogenes (23), and carboxypeptidase Y from Saccharomyces cerevisiae (26) are representative enzymes from these evolutionarily distant families. Yet, remarkably, the three nascent proteases each contain propeptides implicated in folding and maturation, appearing to have converged on a similar propeptide-dependent folding pathway.
The bacterial proteases rely on the activity of a signal peptidase to cleave the bond at the junction between the pre and pro domains of the zymogen, whereas processing of the pro-mature junction is autocatalytic. It has not been established, however, whether this autocatalytic processing event is intramolecular (one molecule self-processing) or intermolecular (one mature enzyme processing another zymogen) or if both intramolecular and intermolecular processing takes place. There are, to date, no three-dimensional structures of bacterial chymotrypsin-like proteases that include propeptides. Consequently, the details of interaction between propeptides and mature peptides are still very much in question.
Extensive propeptides are present in proteases SGPA, SGPB (10), SGPC (22), SGPD (21), and SGPE (20) of S. griseus and in the related α-lytic protease of L. enzymogenes (24). Although the mature enzymes are similar in size, sequence, and three-dimensional structure (where it is known), the propeptides are extremely diverse (21). Considering the substantial variations in the sizes and sequences of propeptides in the bacterial chymotrypsin-like proteases, it is reasonable to question whether the folding function of the propeptide involves interactions over its entire length or whether a few key residues are involved in the folding activity. In a previous study, Fujishige et al. (9) reported that the deletion of as few as 5 residues from the amino terminus of the propeptide of α-lytic protease abolished the secretion and generation of active enzyme. Similarly, a 14-amino-acid deletion in the propeptide of subtilisin led to failure to generate active protease in vivo (11). A series of deletions in carboxypeptidase Y resulted in a reduction of intracellular enzyme activity (16).
In this report we demonstrate (i) that deletions to the 76-amino-acid propeptide of SGPB cause an incremental loss in the production of mature enzyme and (ii) that the propeptide of the related enzyme SGPA can catalyze the maturation and secretion of mature SGPB.
Effects of amino-terminal propeptide deletions on SGPB production in vivo.
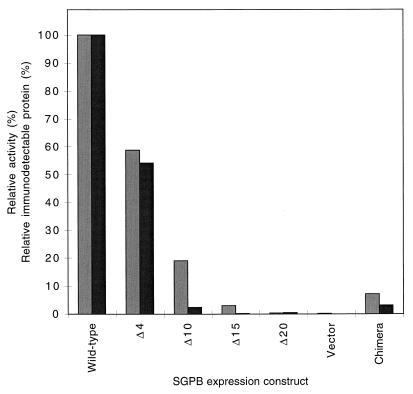
To analyze the role of the propeptide in the production of active SGPB, deletions were made to that part of the sprB gene (the gene encoding SGPB) that encodes the amino terminus of the propeptide. Plasmid pEB-B8 is derived from the Bacillus expression vector pEB11 (20). pEB-B8 encodes pro-mature SGPB driven from the Bacillus amyloliquefaciens subtilisin BPN′ promoter and is fused to the pre regions of subtilisin. The plasmids pEB-BPΔ4 and pEB-BPΔ15, encoding 4 and 15 amino acid deletions, respectively, were constructed from pEB-B8 by using synthetic oligonucleotide primers and standard PCR techniques. Plasmids pEB-BPΔ10 and pEB-BPΔ20, which encode 10 and 20 amino acid deletions, respectively, were constructed by standard restriction digestions. The constructs were verified by DNA sequence and restriction enzyme analyses. B. subtilis DB104 was transformed (12) with each of the above vectors and with pEB-B8. Transformants were streaked onto yeast extract-tryptone (YT) agar (13) containing 1.5% skim milk powder and 50 μg of kanamycin/ml. Broth cultures of YT (2 ml each) supplemented with kanamycin (50 μg/ml) were inoculated with single colonies and grown for approximately 12 h at 30°C, with shaking at 200 rpm. A series of 125-ml Erlenmeyer flasks, each containing 50 ml of YT media plus antibiotic, were then inoculated to 0.1%. The cultures were grown in duplicate at 30°C with shaking at 200 rpm for 100 h. At various time points, cell growth was determined spectrophotometrically by measuring the absorbance at 600 nm (A600). Aliquots from each culture were taken, and cells were precipitated by microcentrifugation. The protease activity of culture supernatants was then determined by measuring the release of p-nitroanilide at 412 nm from the chromogenic substrate N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide (14) at 25°C in 50 mM Tris (pH 8.0). Figure 1 shows the maximum activities observed from the various deletion mutants. Values were normalized for cell density and expressed as percentages of the wild-type clone’s activity. The activity of culture supernatants was seen to decrease exponentially with increasing deletions to the propeptide of SGPB.
FIG. 1.
Relative production of SGPB from mutant sprB genes. Expression constructs encoding SGPB with different numbers of amino acids (shown following Δ) deleted from the N terminus of the propeptide were used. Protease activities (░⃞) and immunodetectable protein (▪) are shown relative to those of the wild type (encoded by pEB-B8). The vector (encoded by pEB-11) contains no sprB insert. The chimera (encoded by pEB-PAmB) is the proA-mature B fusion.
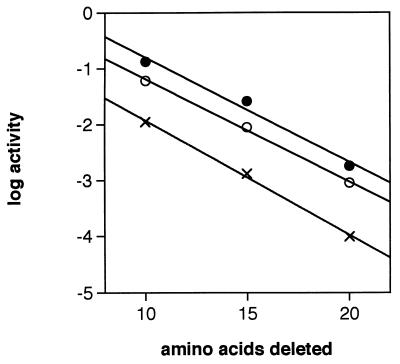
To determine whether this relationship was temperature dependent, the mutant cultures were grown at 21, 30, and 37°C. When activity was plotted as a log function of the number of amino acids deleted from the propeptide amino terminus, parallel lines were observed for the pEB-BPΔ10, pEB-BPΔ15, and pEB-BPΔ20 cultures at each temperature (Fig. 2). This suggests that the phenomenon is not merely an artifact of the growth conditions of B. subtilis. Furthermore, Northern blot analysis of the sprB deletion mutants indicated that the genes were being transcribed at the same levels as wild-type sprB; therefore, the decrease in activity in the mutants was not a transcriptional artifact (data not shown).
FIG. 2.
Effect of temperature on mutant SGPB activity. B. subtilis cultures harboring pEB-BPΔ10, pEB-BPΔ15, pEB-BPΔ20, or pEB-11 were grown for 59 h at 30°C. Activity is plotted as a log function of the number of amino acids deleted from the propeptide for cultures grown at 21 (•), 30 (○), and 37°C (×).
We made the assumption that the decreased activities measured in culture supernatants of the deletion mutants corresponded to a decrease in concentration of active, mature protease, as opposed to incompletely processed or inactive forms of the enzyme. In order to determine this, a polyclonal antibody was raised against SGPB. An immunogenic surface loop, corresponding to amino acids 109 through 126 of SGPB, was predicted by using the program PC Gene (Intelligenetics). The peptide, conjugated to keyhole limpet hemocyanin, was synthesized by the Alberta Peptide Institute. Rabbit polyclonal antibodies were raised against the peptide-keyhole limpet hemocyanin conjugate by using standard procedures (19).
Polypeptides in culture supernatants and in whole-cell lysates were separated by 0.1% sodium dodecyl sulfate–15% polyacrylamide gel electrophoresis (19) and transferred to supported nitrocellulose (Bio-Rad). Blots were then probed with rabbit anti-SGPB serum by using donkey anti-rabbit immunoglobulin G-horseradish peroxidase conjugate (DAR-HRP) as the secondary antibody. Adsorbed DAR-HRP was detected by using the enhanced chemiluminescence kit (Amersham) according to the directions of the manufacturer. It can be seen that only mature forms of the enzyme were detected in culture supernatants and that the decrease in protease activity corresponded directly to a decrease in mature enzyme (Fig. 3). No SGPB could be detected in whole-cell fractions (data not shown). The high-molecular-weight species which cross-reacted with the anti-SGPB was not a variant form of SGPB, since it was also present in supernatants of B. subtilis harboring pEB-11, which does not contain the sprB gene. No SGPB was detected from pEB-BPΔ15 and pEB-BPΔ20, presumably because the amounts of protein (based on activities of about 3 and 0.3% of wild-type activity, respectively) were below the detectable level.
FIG. 3.
Identification of extracellular SGPB. A Western blot probed with anti-SGPB is shown. B. subtilis expression constructs encoding different numbers of amino acids (shown following Δ) deleted from the N terminus of the propeptide of SGPB were used. B, purified SGPB. The chimera (encoded by pEB-PAmB) is the proA-mature B fusion. The wild type is encoded by pEB-B8. The vector (pEB-11) contains no sprB insert.
To further confirm that the reductions in protease activity corresponded to a reduction in mature SGPB, we quantitated SGPB by enzyme-linked immunosorbent assay using anti-SGPB. Supernatants (100 μl) were incubated at 100°C for 5 min and applied to Immulon-2 microtiter plates (Dynatech Laboratories); the wells were then blocked with 0.5% ovalbumin in phosphate-buffered saline (19). The wells were probed with anti-SGPB diluted 1:1,000, followed by DAR-HRP (diluted 1:4,000). Three washes, each with 300 μl of water, were performed between each step to remove unadsorbed protein and antibodies. Adsorbed DAR-HRP was detected by reaction with o-phenylenediamine (Sigma); the reaction was stopped with 20% H2SO4, and the difference between the A490 and A630 was recorded. It is clear that the decrease in activities observed with the deletion mutants corresponds with a decrease in immunodetectable protein (Fig. 1).
Our observation that the protease activity in culture supernatants corresponds only to the presence of native enzyme is consistent with studies which show that folding and cellular transport (including secretion) are intimately linked (8, 9, 16). If the propeptide is missing, protein (native or denatured) is not secreted. The connection between propeptide-catalyzed folding and secretion is unclear, but it may be that the propeptide maintains the enzyme in a metastable (denatured) conformational state so that it can interact with the secretion machinery (7). Once outside the cell, the propeptide is cleaved from the enzyme and degraded, thereby trapping the enzyme in a stable, native conformation.
Related S. griseus proteases.
We wanted to establish whether the observed reductions in activity with the propeptide deletion mutants of SGPB could be extended to related proteases from S. griseus. Expression constructs encoding deletions in the N termini of the propeptide-encoding regions of SGPC, SGPD, and SGPE were also produced by PCR and restriction enzyme digestions. When constructs pEB-CPΔ11 and pEB-CPΔ16, encoding 11 and 16 amino acid deletions in the N terminus of the propeptide of SGPC, respectively, were expressed in B. subtilis DB104, maximum protease activities of 2.3 and 0.05% of the maximum wild-type activity were observed. Similarly, deletions of 20 and 26 amino acids to the N termini of the propeptide-encoding regions of SGPD and SGPE, respectively, resulted in the production of 3.4 and 1.7% of the maximum wild-type activities produced by pEB-D8 and pEB-E. Therefore, these propeptide deletion mutants produced reduced but detectable levels of activity, similar to those of the sprB deletion mutants.
proA-mature B chimeric protease.
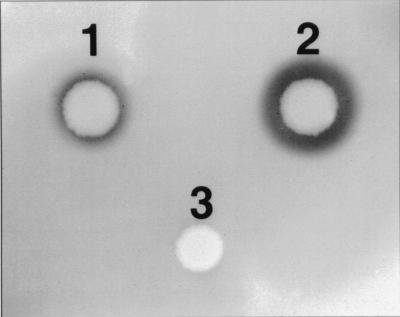
The propeptide region of SGPA is 43% identical in amino acid sequence to that of SGPB. The mature regions of the same enzymes are 181 and 185 residues long, respectively, and show 61% identity in amino acid sequence. Furthermore, the α-carbon positions of the two enzymes are 85% equivalent topologically, based on their crystal structures (6). In order to address the question of whether the propeptide of one protease is able to catalyze the folding of another related enzyme, a chimeric gene construct was prepared in which the propeptide of SGPA was fused to the mature domain of SGPB. The secretion vector pEB-PAmB, encoding the chimeric pro-mature protease, was expressed in B. subtilis DB104 along with pEB-B8 and pEB-11. Zones of clearing, indicating the secretion of active protease, were visible with both the pEB-PAmB and pEB-B8 transformants on YT agar containing 1.5% skim milk (25), 50 μg of kanamycin/ml, and 80 μM isopropyl-β-d-thiogalactopyranoside (IPTG), suggesting the production of active protease (Fig. 4). It was evident from Northern blotting experiments that the chimeric gene was as efficiently transcribed as the gene encoding wild-type SGPB (data not shown). After 73 h of growth at 30°C, the chimeric gene produced 7% of the activity produced by wild-type sprB (Fig. 1). Therefore, the propeptide of SGPA can catalyze the maturation and secretion of mature SGPB, although not as efficiently as does the propeptide of SGPB.
FIG. 4.
Recombinant expression by chimeric proA-mature B protease. B. subtilis DB104 transformants harboring pEB-pAmB (chimera) (1), pEB-B8 (wild type) (2), or pEB-11 (vector control) (3) were grown on YT-kanamycin agar supplemented with 1.5% skim milk and incubated for 18 h at 30°C. The diameter of each zone of clearing is proportional to the level of proteolytic activity.
Hypothetical folding mechanism.
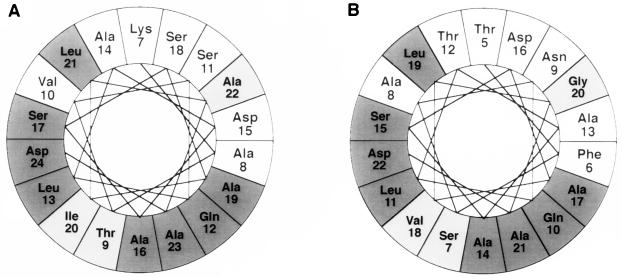
As indicated, SGPA and SGPB have a high degree of structural homology. We have shown that the propeptide of SGPA will, to some extent, catalyze the folding of SGPB. It follows, therefore, that the folding pathways of both proteases would be very close. A secondary-structure analysis using the method of Chou and Fasman (5) and Rose (17) predicts that the propeptides of SGPA and SGPB are predominantly helical. This is also a characteristic of the propeptide of carboxypeptidase Y (26) and of some heat shock chaperonin proteins (18). In contrast, crystal structures reveal the mature regions of SGPA and SGPB to be predominantly β-sheet (6). Most of the deletions made to the propeptide of SGPB fall within the first putative amino-terminal α-helix (residues 5 to 22). An analysis of this α-helix and the corresponding α-helix in SGPA (residues 7 to 24), performed with the computer program MacDNASIS Pro (Hitachi), revealed a conserved and a variable face (Fig. 5). It is tempting to speculate that the propeptide is aligned so that the conserved face interacts with the mature domain during folding. In essence, the propeptide could act as a scaffold to stabilize the formation of β-sheet structure, thereby reducing the activation energy barrier to the native protease.
FIG. 5.
Helical wheel representations of amino acids 7 to 24 of the propeptide of SGPA (A) and the homologous amino acids 5 to 22 of the propeptide of SGPB (B) (8). The analysis was generated with the computer program MacDNASIS Pro (Hitachi), with 3.6 amino acids per turn. Dark shading, chemically identical amino acids; light shading, chemically similar amino acids.
In summary, we have shown that propeptides can mediate the folding of related enzymes. Observations of the deletion mutants in this study indicate that propeptides likely have multiple interactions with the mature region during folding. The data suggest that these interactions are distributed evenly across the amino-terminal region of the propeptide rather than being limited to a few key residues. These results substantiate the observation that the processes of folding and secretion are intimately linked. We are now studying internal deletions in the propeptide in order to further define the interactions between the pro and mature regions of SGPB.
Acknowledgments
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada. J.E. is a recipient of a Medical Research Council of Canada Studentship.
REFERENCES
- 1.Baker D, Shiau A K, Agard D A. The role of pro regions in protein folding. Curr Opin Cell Biol. 1993;5:966–970. doi: 10.1016/0955-0674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 2.Baker D, Silen J L, Agard D A. Protease pro region required for folding is a potent inhibitor of the mature enzyme. Proteins Struct Funct Genet. 1992;12:339–344. doi: 10.1002/prot.340120406. [DOI] [PubMed] [Google Scholar]
- 3.Baker D, Sohl J L, Agard D A. A protein-folding reaction under kinetic control. Nature. 1992;356:263–265. doi: 10.1038/356263a0. [DOI] [PubMed] [Google Scholar]
- 4.Barrett A J, Rawlings N D. Families and clans of serine proteases. Arch Biochem Biophys. 1995;318:247–250. doi: 10.1006/abbi.1995.1227. [DOI] [PubMed] [Google Scholar]
- 5.Chou P, Fasman G D. Prediction of the secondary structures of proteins from their amino acid sequence. Adv Enzymol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 6.Delbaere L T J, Brayer G D, James M N G. The 2.8Å resolution structure of Streptomyces griseus protease B and its homology with α-chymotrypsin and Streptomyces griseus protease A. Can J Biochem. 1979;57:135–144. doi: 10.1139/o79-017. [DOI] [PubMed] [Google Scholar]
- 7.Eder J, Fersht A R. Pro-sequence-assisted folding. Mol Microbiol. 1995;16:609–614. doi: 10.1111/j.1365-2958.1995.tb02423.x. [DOI] [PubMed] [Google Scholar]
- 8.Eilers M, Schatz G. Binding of a specific ligand inhibits import of a purified precursor protein into the mitochondria. Nature. 1986;322:228–232. doi: 10.1038/322228a0. [DOI] [PubMed] [Google Scholar]
- 9.Fujishige A, Smith K R, Silen J L, Agard D A. Correct folding of α-lytic protease is required for its extracellular secretion from Escherichia coli. J Cell Biol. 1992;118:33–42. doi: 10.1083/jcb.118.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson G, Krygsman P, Liu C J, Davey C C, Malek L T. Characterization and structure of genes for proteases A and B from Streptomyces griseus. J Bacteriol. 1987;169:3778–3784. doi: 10.1128/jb.169.8.3778-3784.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikemura H, Takagi H, Inouye M. Requirement of pro-sequence for the production of active subtilisin E in Escherichia coli. J Biol Chem. 1987;262:7859–7864. [PubMed] [Google Scholar]
- 12.Kawamura F, Doi R H. Construction of a Bacillus subtilisdouble mutant deficient in extracellular alkaline and neutral proteases. J Bacteriol. 1984;160:442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 14.Nakajima K, Powers J C, Ashe B M, Zimmerman M. Mapping the extended substrate binding site of cathepsin G and human leukocyte elastase. J Biol Chem. 1979;254:4027–4032. [PubMed] [Google Scholar]
- 15.Ohta Y, Inouye M. Pro-subtilisin E: purification and characterization of its autoprocessing to active subtilisin E in vitro. Mol Microbiol. 1990;4:295–304. doi: 10.1111/j.1365-2958.1990.tb00596.x. [DOI] [PubMed] [Google Scholar]
- 16.Ramos C, Winther J R, Kielland-Brandt M C. Requirement of the propeptide for the in vivo formation of active yeast carboxypeptidase Y. J Biol Chem. 1994;269:7006–7012. [PubMed] [Google Scholar]
- 17.Rose G D. Prediction of chain turns in globular proteins on a hydrophobic basis. Nature. 1978;272:586–590. doi: 10.1038/272586a0. [DOI] [PubMed] [Google Scholar]
- 18.Sadis S, Raghavendra K, Hightower L E. Secondary structure of the mammalian 70 kilodalton heat shock protein analysed by circular dichroism spectroscopy and secondary structure prediction. Biochemistry. 1990;29:8199–8206. doi: 10.1021/bi00488a001. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Sidhu S, Kalmar G, Borgford T. Characterization of the gene encoding the glutamic acid-specific protease of Streptomyces griseus. Biochem Cell Biol. 1993;71:454–461. doi: 10.1139/o93-067. [DOI] [PubMed] [Google Scholar]
- 21.Sidhu S S, Kalmar G B, Willis L, Borgford T J. Protease evolution in Streptomyces griseus: discovery of a dimeric serine protease. J Biol Chem. 1995;270:7594–7600. doi: 10.1074/jbc.270.13.7594. [DOI] [PubMed] [Google Scholar]
- 22.Sidhu S S, Kalmar G B, Willis L, Borgford T J. Streptomyces griseusprotease C: a novel enzyme of the chymotrypsin superfamily. J Biol Chem. 1994;269:20167–20171. [PubMed] [Google Scholar]
- 23.Silen J L, Agard D A. The α-lytic protease pro-region does not require a physical linkage to activate the protease domain in vivo. Nature. 1989;341:462–464. doi: 10.1038/341462a0. [DOI] [PubMed] [Google Scholar]
- 24.Silen J L, McGrath C N, Smith K R, Agard D A. Molecular analysis of the gene encoding α-lytic protease: evidence for a preproenzyme. Gene. 1988;69:237–244. doi: 10.1016/0378-1119(88)90434-9. [DOI] [PubMed] [Google Scholar]
- 25.Wells J A, Ferrari E, Henner D J, Estell D A, Chen E Y. Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtilisin in Bacillus subtilis. Nucleic Acids Res. 1983;11:7911–7925. doi: 10.1093/nar/11.22.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winther J R, Sorensen P. Propeptide of carboxypeptidase Y provides a chaperone-like function as well as inhibition of the enzymatic activity. Proc Natl Acad Sci USA. 1991;88:9330–9334. doi: 10.1073/pnas.88.20.9330. [DOI] [PMC free article] [PubMed] [Google Scholar]