ABSTRACT
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein is a highly immunogenic and mutable protein that is the target of vaccine prevention and antibody therapeutics. This makes the encoding S-gene an important sequencing target. The SARS-CoV-2 sequencing community overwhelmingly adopted tiling amplicon-based strategies for sequencing the entire genome. As the virus evolved, primer mismatches inevitably led to amplicon dropout. Given the exposure of the spike protein to host antibodies, mutation occurred here most rapidly, leading to amplicon failure over the most insightful region of the genome. To mitigate this, we developed a targeted method to amplify and sequence the S-gene. We evaluated 20 distinct primer designs through iterative in silico and in vitro testing to select the optimal primer pairs and run conditions. Once selected, periodic in silico analysis monitors primer conservation as SARS-CoV-2 evolves. Despite being designed during the beta wave, the selected primers remain >99% conserved through Omicron as of 19 October 2023. To validate the final design, we compared targeted S-gene data to National SARS-CoV-2 Strain Surveillance whole-genome data for 321 matching samples. Consensus sequences for the two methods were highly identical (99.998%) across the S-gene. This method can serve as a complement to whole-genome surveillance or can be leveraged where only S-gene sequencing is of interest.
IMPORTANCE
The COVID-19 pandemic was accompanied by an unprecedented surveillance effort. The resulting data were and will continue to be critical for surveillance and control of SARS-CoV-2. However, some genomic surveillance methods experienced challenges as the virus evolved, resulting in incomplete and poor quality data. Complete and quality coverage, especially of the S-gene, is important for supporting the selection of vaccine candidates. As such, we developed a robust method to target the S-gene for amplification and sequencing. By focusing on the S-gene and imposing strict coverage and quality metrics, we hope to increase the quality of surveillance data for this continually evolving gene. Our technique is currently being deployed globally to partner laboratories, and public health representatives from 79 countries have received hands-on training and support. Expanding access to quality surveillance methods will undoubtedly lead to earlier detection of novel variants and better inform vaccine strain selection.
KEYWORDS: gene sequencing, SARS-CoV-2, surveillance studies
INTRODUCTION
In December 2019, an outbreak of pneumonia of unknown cause began in Wuhan, China (1). This illness (COVID-19) was found to be caused by a novel betacoronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2 or SC2) (2). The virus quickly spread around the world, and in March 2020, the World Health Organization officially declared COVID-19 a pandemic (3). As of May 2023, COVID-19 has caused roughly 677 million infections and 6.9 million deaths (4).
As the COVID-19 pandemic progressed, waves of new variants spread (5), and mutations within the surface glycoprotein (spike) accumulated (6, 7). The spike protein is key to the viral replication cycle as its binding to the human angiotensin-converting enzyme 2 receptor initiates cellular entry of the virus (8). It also bears clinical significance as it is the target of vaccine prevention (9) and antibody therapeutics (10). The continual evolution of SARS-CoV-2 to evade immune pressures has led to a plethora of spike mutations that have been deleterious to vaccine effectiveness (11) and antibody neutralization (12 – 14). Importantly, many of these mutations are located within the receptor-binding domain (RBD) (15), where 90% of neutralizing antibodies target SARS-CoV-2 (16). As such, the spike protein encoding S-gene is an important sequencing target, and complete and accurate data for the S-gene are paramount for high-quality surveillance information.
Genomic tools, such as the widely used Artic SARS-CoV-2 primer set, have required numerous updates to remain effective against new variants (17 – 20) (https://github.com/artic-network/artic-ncov2019/tree/master/primer_schemes/nCoV-2019). It can be challenging for surveillance laboratories, which are likely operating at surge capacity, to examine available alternative methods and validate revisions to the method for use. When overlooked, these limitations can lead to overt sequencing gaps or areas of low coverage, usually within the S-gene (21). This issue has occurred multiple times during the COVID-19 pandemic with major variant transitions (origin strain to Alpha, Alpha to Delta, and Delta to Omicron). However, this problem is not limited to major variant shifts. Variability in sequencing protocols and the design of sequencing primers in highly mutable regions of the SARS-CoV-2 spike protein causes intermittent sequencing dropouts, even with moderate amounts of variation. This is complicated further by the variability of organization for countrywide sequencing. Some countries have centralized health systems with more direct control of sequencing protocols and communication, whereas other countries contract out sequencing to private laboratories, which can yield higher data volume but have more variability in sequencing methods and directness of communication. This can lead to protocol issues that are very slow to address, causing blind spots to critical regions of the spike protein as evolution occurs. As an illustration, we examined global surveillance data across the SARS-CoV-2 RBD over different time periods (Figure S1 at https://figshare.com/articles/data set/Supplemental_Material/22762076). During the transition from Delta to Omicron, this critical region was missing a significant amount of data. Moreover, the shape of those missing data resembles the amplicon 76 dropout known to affect the Artic SARS-CoV-2 primer set during the emergence of Omicron (19). The coverage across this region has since improved, but this does illustrate the issues of using mutation-sensitive amplification methods through a highly mutable region of a highly mutable virus.
Efforts have been made to focus surveillance to the S-gene; however, these methods have serious limitations. One such effort is to use eight overlapping amplicons (342–979 bp) and Sanger sequencing to bring SARS-CoV-2 surveillance to low-resource areas (22). Unfortunately, the need for eight reverse transcription PCRs (RT-PCRs) per sample and the use of Sanger sequencing are costly, labor intensive, and seriously limit throughput potential. In an effort to improve the throughput of S-gene-only sequencing, a modified version of Artic V3 SARS-CoV-2 primer set, HiSpike, was developed (23). HiSpike retains many of the limitations of the Artic SARS-CoV-2 protocol, most notably, the use of small amplicons (~400 bp) and the need for many primers to bind within the spike coding region. Using many primers to generate many small overlapping amplicons is not ideally suited to the surveillance of a rapidly evolving RNA virus and will likely again lead to sequencing dropouts due to primer mismatches. Indeed, multiple primers from both studies have conservation issues.
Because of these challenges, it was critical to develop a robust method for obtaining rapid sequence information, specifically for the S-gene. For this purpose, we developed a targeted method to amplify and sequence the S-gene (Fig. 1). This method uses four carefully selected and highly conserved primers (Table 1) to produce two overlapping amplicons that yield full coverage of the S-gene (Fig. 2). Our method is suitable as a complement to whole-genome sequencing (WGS) for research purposes where only the S-gene is of interest, as a stand-alone surveillance method, and as a means to expand surveillance to laboratories that may be new to surveillance. The technique is currently being deployed globally to partner laboratories in an effort to expand access to quality surveillance methods, detect novel variants earlier, and better inform vaccine strain selection.
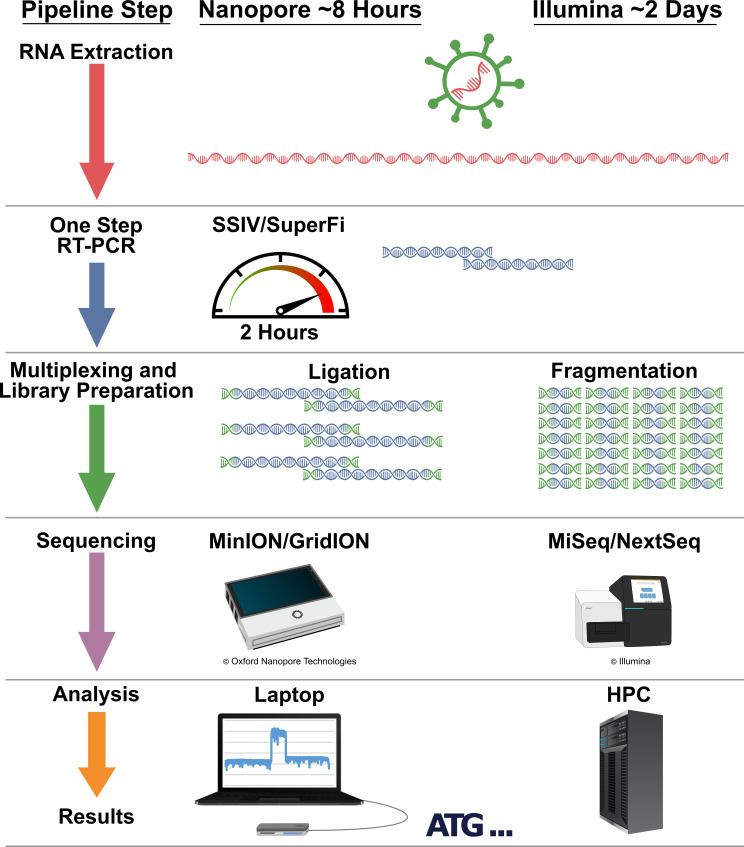
Fig 1.
Sequencing workflow. Targeted S-gene sequencing is presented here as RT-PCR amplification and Nanopore sequencing. The workflow is designed to be flexible as the amplicons can be diverted to other sequencing platforms.
TABLE 1.
Primers: targeted S-gene amplification primer sequences and working stock concentration
| Oligo | Sequence 5′−3′ | µM in pool |
|---|---|---|
| S1 primer pool | ||
| S1F_21358 | ACAAATCCAATTCAGTTGTCTTCCTATTC | 5 |
| S1R_23813 | TGCTGCATTCAGTTGAATCACC | 5 |
| S2 primer pool | ||
| S2F_23288 | GTCCGTGATCCACAGACACTT | 5 |
| S2R_25460 | GCATCCTTGATTTCACCTTGCTTC | 5 |
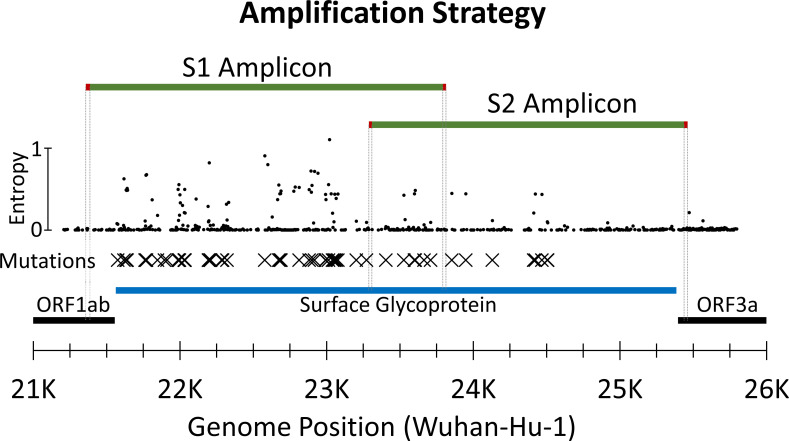
Fig 2 .
Amplification strategy. Amplicons are green with primer locations highlighted in red and traced down to the open reading frames (ORFs). Diversity (entropy) across the region is plotted with small circles. Detected amino acid mutations are maked with Xs. SARS-CoV-2 ORFs are black with the surface glycoprotein ORF highlighted in blue. Separate one-step RT-PCRs generate overlapping S1 and S2 amplicons that are 2.2 and 2.5 kb, respectively. These amplicons extend beyond the coding region and overlap across the S1-S2 subunit cleavage site.
RESULTS
Primer selection and validation
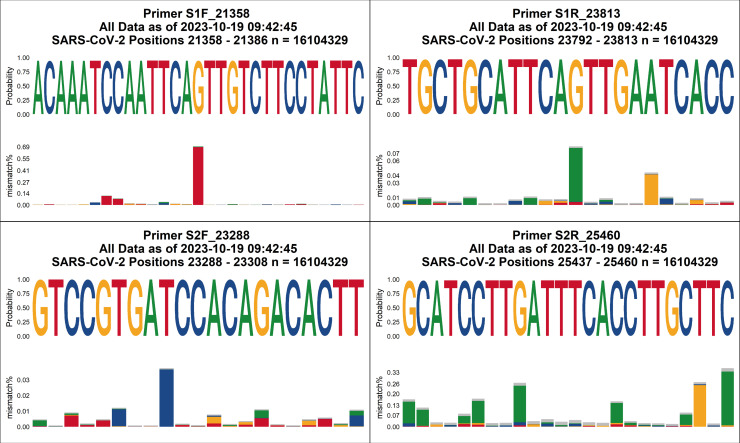
We used the conservation of all available SARS-CoV-2 sequences to identify (Table S1 at https://figshare.com/articles/data set/Supplemental_Material/22762076) and evaluate (Table S2 at https://figshare.com/articles/data set/Supplemental_Material/22762076) candidate primers. We identified three candidates for each of the four needed primers (S1F, S1R, S2F, and S2R) with additional candidates for S2R, where SARS-CoV-2 (Wuhan-hu-1, NC_045512.2) and SARS-CoV-1 (NC_004718.3) shared identity. We eliminated those with <95% conservation for all available SARS-CoV-2 sequences. By testing candidate primer combinations across an annealing temperature gradient (Table S3; Fig. S2 and S3 at https://figshare.com/articles/data set/Supplemental_Material/22762076), we were able to simultaneously eliminate possible combinations with poor performance and select 60°C as the annealing temperature. Finally, a limit of detection (LOD) assay (Tables S3 and S4; Fig. S4 at https://figshare.com/articles/data set/Supplemental_Material/22762076) was used to select S1F_21358, S1R_23813, S2F_23288, and S2R_25460 as the final primers (Table 1; Seq S1 at https://figshare.com/articles/data set/Supplemental_Material/22762076).
We periodically monitor the conservation of these primers, and as of 19 October 2023, the selected primers remain highly conserved against SARS-CoV-2 using 3 months of US data, 3 months of global data, and all global data (Fig. 3; Fig. S5 to S16 at https://figshare.com/articles/data set/Supplemental_Material/22762076). Our primers also show some conservation against related coronaviruses (Seq S2; Fig. S17 at https://figshare.com/articles/data set/Supplemental_Material/22762076). If a particular subvariant is of concern, we can perform a more focused conservation analysis. Such was the case with Omicron XBB, XBB.1.5, and derivatives. Analysis against those subvariants, as of 18 January 2023, demonstrated that our primers remained conserved (Table S5 at https://figshare.com/articles/data set/Supplemental_Material/22762076).
Fig 3.
Visualization of primer conservation for all global SARS-CoV-2 surveillance data. Data as of 19 October 2023 indicate >99% conservation at every position.
Our design results in an amplification strategy where two overlapping amplicons, each in their own RT-PCR reaction, span the entire gene. The four selected primers, which will generally be known as S1F, S1R, S2F, and S2R, avoid known mutations and regions of high diversity (Fig. 2 and 3).
Sequencing runs
We performed 14 Nanopore sequencing runs to validate and characterize our method. We prepared three Nanopore libraries from samples spanning SARS-CoV-2 diversity through the Delta variant (RNA Plate01–Plate03). From those libraries, three sequencing runs (Spike01_MIN–Spike03_MIN) validated the method using standard MinION flow cells. Using those same libraries, three sequencing runs (Spike01_FLG–Spike03_FLG) characterized the yield of disposable Flongle flow cells. Sequencing runs Spike04_MIN and Spike05_MIN validated the method against priority Omicron samples. Sequencing runs Spike06_MIN and Spike07_FLG validated the method against Omicron isolates. Four sequencing runs (LOD01_MIN, LOD02_Bulk_MIN, LOD03_Bulk_FLG, and LOD04-05_Mixed_Dried_MIN) characterized the limit of detection. A summary of these runs is available in the supplemental materials (Table S6 at https://figshare.com/articles/data set/Supplemental_Material/22762076).
Sensitivity and specificity
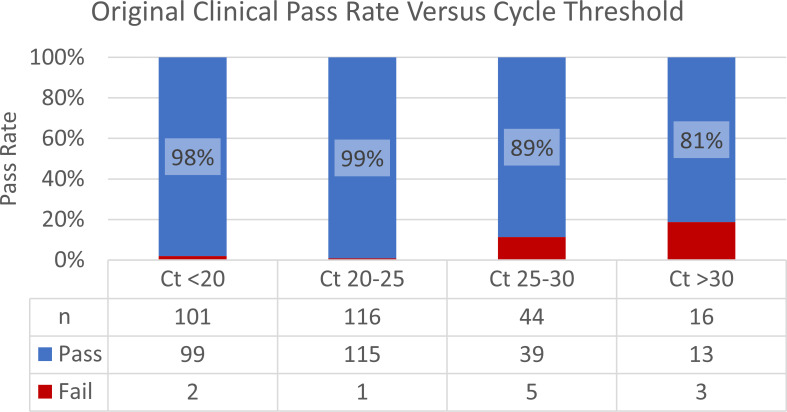
The LOD via MinION flow cell sequencing was ~100 copies/µL with a Ct value of 30 (Table S7; Fig. S18 at https://figshare.com/articles/data set/Supplemental_Material/22762076). Via Flongle flow cell sequencing, the LOD was ~100 copies/µL with a Ct value of 27 (Table S8; Fig. S19 at https://figshare.com/articles/data set/Supplemental_Material/22762076). These runs also contained 84 no template controls (NTCs), and no reads from these mapped to SARS-CoV-2 (Tables S9 and S10; Fig. S20 and S21 at https://figshare.com/articles/data set/Supplemental_Material/22762076). From these LOD runs, we determined that the LOD, which is largely a property of the RT-PCR and not the the sequencers, was 100 copies/µL with a Ct value of 27. More important is the performance on a clinical material versus Ct values within the context of the distribution of those Ct values. To that end, we tested a subset of 277 clinical specimens from the NS3 project. By comparing Ct values to coverage results (Table S11 at https://figshare.com/articles/data set/Supplemental_Material/22762076), we found that 98%–99% of samples with a Ct value less than 25 (n = 217) passed the coverage threshold of requiring ≥50× coverage at every position. For samples with Ct values between 25 and 30 (n = 44), 89% of the samples passed. Moreover, for samples with Ct values over 30 (n = 16), 81% of the samples passed (Fig. 4). Notably, 78% of the samples had a Ct value below 25.
Fig 4.
Original clinical pass rate versus cycle threshold. We compared the coverage results and Ct values for 277 clinical specimens. Samples were grouped by Ct range and assigned a pass if they met the coverage threshold of requiring ≥50x coverage at every position. For samples with a Ct value less than 25, 98-99% of samples passed. For samples with Ct values between 25 and 30, 89% of the samples passed. For samples with Ct values over 30, 81% of the samples passed.
Method validation
We tested 377 samples for a pairwise comparison to National SARS-CoV-2 Strain Surveillance (NS3) WGS data. Of those, 321 samples passed targeted sequencing (Seq S4 at https://figshare.com/articles/data set/Supplemental_Material/22762076) and WGS (Seq S5 at https://figshare.com/articles/data set/Supplemental_Material/22762076) to be carried forward for further analysis. The S-gene consensus sequences were highly identical with 1,225,156 identities out of 1,225,185 positions (99.998% identical). Analyzing S-gene-only data via Nextclade or Pangolin is limited by some group defining mutation residing outside of the S-gene. Still, with the widespread use of these analytical tools, we wanted to characterize the Nextclade results of S-gene sequences in comparison to WGS data. Of the 281 samples that had a variant assignment (e.g., Delta or Omicron), Nextclade assignment of these variants was 100% concordant between S-gene and WGS data. These assignments included the variants Alpha, Beta, Gamma, Delta, Epsilon, Eta, Iota, Lambda, Mu, and Omicron (24, 25). Identical clades were assigned for 91% of the samples. As expected, clades with identical S-gene sequences, such as clades 21A (Delta) and 21J (Delta), were often conflated. Clades 21K (Omicron) and 21L (Omicron) were accurately assigned due to the S-gene diversity between those clades. Identical Nextclade_pango lineages were assigned for 72% of the samples. Similar to clade identification, the resolution of S-gene-only lineage identification is limited by a great number of named lineages and their identification being based off mutations outside the S-gene (Table S12 at https://figshare.com/articles/data set/Supplemental_Material/22762076).
Importantly, our method identified 4,428 mutations which include all 4,422 spike protein mutations identified by WGS. For six samples, we identified one additional mutation each (Table S12 at https://figshare.com/articles/data set/Supplemental_Material/22762076). Further investigation of raw read data confirmed that these additional mutations were due to minor subpopulations at >20% frequency amplified at variable proportions due to separate rounds of PCR between targeted S-gene amplification and NS3. In any case, correctly identifying all 4,422 presumably true spike protein mutations reflects a high degree of accuracy that is more than sufficient for surveillance purposes (26).
For a subset of 277 clinical specimens, we split the three Nanopore libraries for loading on standard MinION flow cells (FLO-MIN106) for 72 hours and disposable Flongle flow cells (FLO-FLG001) for 24 hours. These flow cell types are known to have disparate sequencing yields, and indeed the Flongle flow cells produced just ~1% of the average coverage compared to the MinION flow cells. However, the coverage thresholds only require that a full assembly be made and ≥50× coverage at every position. Using those requirements, MinION flow cell sequencing passed 267 of the 277 samples (96%), and Flongle flow cell sequencing passed 241 of the 277 samples (87%). In other words, ~1% of the average coverage from Flongle flow cells passed 90% (241 of 267) of as many samples with respect to MinION flow cells (Table S13; Fig. S22 at https://figshare.com/articles/data set/Supplemental_Material/22762076).
For this same subset of 277 clinical specimens, we diverted a portion of the spike amplicons to Illumina sequencing, and 251 samples passed both techniques. We compared consensus level identity between spike amplicons sequenced via Nanopore (MIN) to those same amplicons sequenced via Illumina (ILL, Seq S6 at https://figshare.com/articles/data set/Supplemental_Material/22762076). For 251 samples, consensus sequences were highly identical with 958,236 identities out of 958,239 positions (99.9997% identical). This was expanded to a three-way comparison that includes the corresponding NS3 generated S-gene sequences (Table S14 at https://figshare.com/articles/data set/Supplemental_Material/22762076). The MINvNS3 consensus sequences were 99.9972% (958,212 of 958,239) identical, and the ILLvNS3 consensus sequences were 99.9969% (958,210 of 958,240) identical. All 251 samples had 100% identity between at least two of the three methods, and 20 samples had discrepant results. Because at least two of the methods always agreed, the discrepant results always appeared in pairs, were of identical magnitude, and shared a common method. For example, the three-way blast results of sample 3002648260 for MINvILL are 100% identical, whereas MINvNS3 and ILLvNS3 are both 99.974% identical. This indicates that the discrepancy lies with the NS3-derived S-gene consensus for sample 3002648260. Of the 20 samples with discrepant results, 18 are due to discrepancies with the NS3-derived S-gene consensus, and 2 are due to discrepancies with the Illumina sequenced spike amplicons. This distribution of discrepancies is expected as the NS3 samples were independently amplified, processed, and analyzed. Ultimately though, these discrepancies are very minor and more than acceptable for surveillance purposes.
Phylogenetics
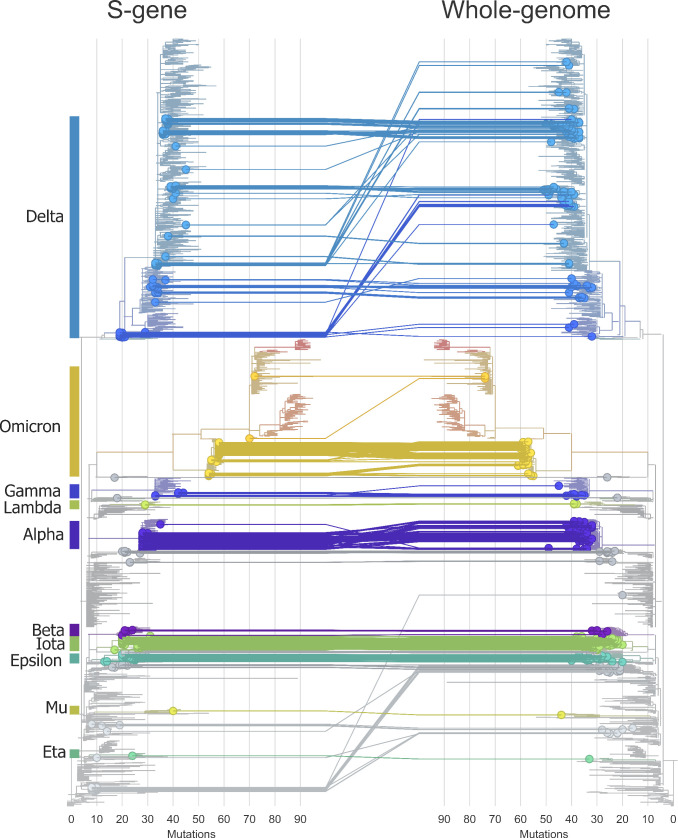
We visualized the Nextclade results in auspice to generate a tanglegram (Fig. 5) of matching samples (n = 321) that passed both S-gene and WGS. As detailed in Table S12 at https://figshare.com/articles/data set/Supplemental_Material/22762076, variant assignment was 100% concordant, and clade assignment was highly concordant with ambiguities appearing for different clades with identical S-gene sequences.
Fig 5.
Pairwise phylogenetics of matching samples (n = 321) that passed both S-gene (circles left) and whole-genome sequencing (circles right). Phylogenetics were exported from Nextclade and visualized using Auspice.
DISCUSSION
We have developed and validated a robust method for amplifying and sequencing the SARS-CoV-2 surface glycoprotein. The length of the S-gene necessitated internal primers and separate overlapping RT-PCRs. Still, we were able to limit the total number of primers to four and the number of primers within the coding region of the S-gene to two. With so few primers required, we were able to evaluate many candidates for conservation and efficacy. We also ensured that the primer-binding sites avoid any major structural/functional elements. This design and process of primer selection gives the best odds at avoiding mutations that might affect primer annealing. Indeed, despite being originally designed during the beta wave, the selected primers have remained highly conserved through Omicron as of 19 October 2023. This strategy does require longer amplicons compared to other methods, which raises concerns over sensitivity; however, we have demonstrated a high degree of sensitivity within the clinically relevant range of viral abundance.
We validated this method against hundreds of clinical specimens collected for genomic surveillance by the NS3 project. We compared S-gene data to the available WGS data to confirm that our method accurately represents the S-gene amino acid mutations. Having only one amplicon in each reaction and two amplicons in total, quality control (QC) via electrophoresis can immediately reveal dropouts. This, combined with strict coverage requirements, ensures only complete and high-quality S-gene sequencing data are reported.
As the pandemic progressed, the methods used by NS3 required several revisions and, occasionally, relied upon our method for sequence completion (Fig. S23 at https://figshare.com/articles/data set/Supplemental_Material/22762076) or confirmation of recombination (27). The speed of targeted S-gene sequencing has also been proven to be useful during the floods of high-priority samples associated with the appearance of new variants. In one case, our method was used to confirm the presence of Omicron in the US Virgin Islands, which allowed the territory to acquire antibody therapeutics best suited at the time of infection with the Omicron variant.
Targeted S-gene sequencing can serve as a complement to WGS data with S-gene coverage gaps, be leveraged as a tool for projects in which only S-gene sequencing is of interest, and stand alone as a means of surveillance. Our method was evaluated and approved by the Centers for Disease Control and Prevention (CDC) Infectious Disease Test Review Board and is currently being deployed to partner laboratories. In collaboration with the World Health Organization, we are hosting intensive week-long regional trainings where country representatives will gain hands-on experience and receive reagents, consumables, and an Mk1C sequencing device sufficient to perform a year of SARS-CoV-2 S-gene (and influenza A virus) surveillance. These regional trainings dedicate a great deal of time on foundational knowledge about the viruses themselves, the fundamentals of a quality surveillance effort, the importance of each step of data analysis and curation, and critically evaluating all step of the surveillance pipeline to ensure only quality data are submitted to public databases. While it is tempting to simply ship out point-and-click solutions, we want to develop a strong foundational knowledge about surveillance and data curation when training and equipping laboratories and countries new to next-generation sequencing (NGS) surveillance. This not only ensures the best use of resources but also gives those laboratories and countries the best chance at being successful in generating quality data and participating in this global surveillance effort. Moreover, because this method targets a portion of the genome, a given amount of surveillance capacity could cover several times more samples compared to WGS on the same platform. The Nanopore platform used by our method is compatible with low to moderate throughputs, and its simplicity better enables users to achieve accurate results, even in low-resource settings. Finally, the relatively low capital expenditure makes this strategy an ideal starting point for public health laboratories new to NGS surveillance. As of August 2023, public health representatives from 79 countries have received this training. One limitation has been a lower adoption rate of this method versus the companion influenza A virus sequencing. This was expected as some laboratories had established WGS for SARS-CoV-2 but not for influenza A virus. Despite the advantages, this method still requires two RT-PCR reactions per sample, and some laboratories chose not to incorporate this method into their surveillance work.
WGS by a variety of methods will remain an integral part of SARS-CoV-2 surveillance, and we are not intending our method to simply be a replacement. WGS is the only way to properly assign phylogenetic relationships or monitor for amino acid mutations outside of the S-gene that can, for example, affect viral replication and pathogenesis (28). Moreover, quality WGS data are necessary to monitor primer conservation for any targeted amplification strategy.
Targeted S-gene sequencing represents a refocusing on essential information needed from surveillance data. Whole-genome surveillance of SARS-CoV-2 has occasionally and unfortunately prioritized getting any result at the expense of sequence completeness and quality. As an example, eagerness to define new clades/lineages based on trivial differences has convoluted the classification of SARS-CoV-2 viruses and obscured the relationships between similar or disparate S-gene mutations that carry clinical significances. By focusing on the S-gene, imposing strict coverage and quality metrics, and applying lessons learned through surveillance of the diverse RNA influenza viruses, we hope to supplement SARS-CoV-2 surveillance with complete and quality reporting on the rapidly mutating S-gene.
MATERIALS AND METHODS
Molecular workflow
To amplify the S-gene, we produced overlapping amplicons (S1 and S2) via separate SuperScript IV One-Step RT-PCR System (Thermo Fischer Scientific, USA) reactions. The RT-PCR mixture contained 4.25-µL nuclease-free water, 12.5-µL super script IV (SSIV) 2× reaction mix, 0.25-µL SSIV RT Mix, 5-µL S1 or S2 primer pairs, and 3 µL of RNA. The RT-PCR conditions are as follows: 10 minutes at 50°C, 2 minutes at 98°C, 40 cycles of 10 seconds at 98°C, 10 seconds at 60°C, and 1 minute 15 seconds at 72°C, a final elongation of 5 minutes at 72°C, and a hold at 4°C. Electrophoresis quality control was performed on individual RT-PCRs. After QC, corresponding S1 and S2 amplicons were combined, cleaned via SPRI beads (1×) with ethanol washes, and eluted into 15 µL of nuclease-free water.
Nanopore libraries were prepared using SQK-LSK109 and EXP-NBD196 and sequenced on GridION (Oxford Nanopore Technologies, UK) using FLO-MIN106 or FLO-FLG001 flow cells.
Laboratory procedures for RT-PCR and library preparation are available in the supplemental material (Text S1 and Text S2 at https://figshare.com/articles/data set/Supplemental_Material/22762076).
For Illumina sequencing, a portion of the cleaned amplicons were taken and prepared using the Nextera XT sample preparation kit. Since the size of the SARS-CoV-2 S-gene amplicons is similar to that of the influenza virus amplicons, they were processed via the standard influenza surveillance pipeline used by the CDC Genomics and Diagnostics Team (29, 30).
Sequencing data analysis
During the sequencing run, we used the GridION MinKNOW to perform super-accuracy basecalling live (ont-guppy-for-gridion 5.0.17 or 5.1.13) to trim the barcodes and to filter the reads. We trimmed primers using BBDuk (31), restricted the trimming using restrictleft = 50 and restrictright = 50, and referred to the primer sequences (Seq S1 at https://figshare.com/articles/data set/Supplemental_Material/22762076). We assembled reads using IRMA (https://wonder.cdc.gov/amd/flu/irma/irma.html) with the CoV-s-gene module (IRMA version 1.0.3, https://wonder.cdc.gov/amd/flu/irma/release_notes.html) and mapped to the S-gene reference (29). For a sample to pass, it must meet coverage and quality metrics. Specifically, it must have a complete S-gene assembly, have at least 50× coverage at every position, and be free of frameshift mutations. Mutations were identified using Nextclade Web version 2.6.1 (https://clades.nextstrain.org, accessed 30 September 2022) SARS-CoV-2 without recombinants (24).
Updated laboratory procedures and analysis tools are available online https://cdcgov.github.io/MIRA (32).
Primer selection and validation
We selected four primer target regions where S1F and S2R would lie outside of the S-gene coding region, and S1R and S2F would be on opposite sides of the S1/S2 cleavage site and avoid major structural elements. We identified multiple sets of candidate primers for each S1F, S1R, S2F, and S2R. For S2R, we also evaluated an area where SARS-CoV-2 (Wuhan-hu-1, NC_045512.2) and SARS-CoV-1 (NC_004718.3) shared identity (Table S1 at https://figshare.com/articles/data set/Supplemental_Material/22762076). During the Beta wave (March 2021), we evaluated the conservation of primer candidates against 476,466 SARS-CoV-2 genomes (Table S2 at https://figshare.com/articles/data set/Supplemental_Material/22762076). Twenty primer combinations were tested (Table S3 at https://figshare.com/articles/data set/Supplemental_Material/22762076). We initially screened the candidate primer pairs across a temperature gradient using RNA from B.1.351 (Beta) with a Ct value of 25 as determined by the flu SC2 multiplex assay (33). We used an LOD of B.1.351 (Beta) from a Ct value of 14–30 (846,000 to 16 copies/µL; Table S4 at https://figshare.com/articles/data set/Supplemental_Material/22762076) to finalize the primer selection. The presence of amplicons was determined using a QIAxcel HT fragment analyzer.
We monitored the conservation of the primers via data downloading from Global Initiative on Sharing All Influenza Data (GISAID). Downloaded genomic data were aligned to the Wuhan-Hu-1 reference (NCBI accession MN908947.3) genome using striped Smith-Waterman algorithm (34). Aligned genome primer regions were regularly compared for mismatches against each individual primer sequence. This information was used to highlight potential assay issues with new emerging variants. We downloaded diversity (entropy) data from Nextstrain (https://nextstrain.org/ncov/gisaid/global/6m, accessed 6 March 2023) (24).
Sensitivity and specificity
To measure the absolute limit of detection, we used a custom synthetic RNA fragment from Twist Bioscience (San Francisco, CA, USA) based on the Delta lineage virus hCoV-19/USA/CO-CDC-MMB09467199/2021. The sequence for this fragment (TwistDeltaFragment_4276451.fasta) is available in the supplemental materials (Seq S3 at https://figshare.com/articles/data set/Supplemental_Material/22762076). The 4,626-nucleotide fragment spans the S-gene and extends into neighboring genes. The synthetic fragment aliquots were delivered at 629,000 copies/µL as determined via manufacturer ddPCR. To measure the viral limit of detection, we used a propagated isolate of Delta SARS-CoV-2 and measured the Ct value of the serial dilutions using the flu SC2 multiplex assay (33). The limit of detection was determined by the most dilute sample to pass coverage and quality thresholds for all the replicates.
We prepared RT-PCR master mixes for triplicate limit of detection assays using both synthetic and viral materials. The dilution series were a fivefold serial dilution through seven steps with a water NTC as the eighth step. LOD amplicons were split at the end-prep stages for sequencing on both MinION and Flongle flow cells. For sequencing on MinION flow cells, we included 48 additional water NTCs. For sequencing on Flongle flow cells, we included 24 additional A549 RNA (Rp Ct 22) NTCs.
Method validation
To validate this method, we tested a total of 377 specimens from the NS3 project. We started with a retrospective analysis of 277 clinical specimens that were collected from March to August 2021 and that captured the diversity of SARS-CoV-2 into the Delta wave. During the omicron wave, we continued validation concurrently with NS3. These additional 100 samples were collected from November 2021 to January 2022. Of these 377 samples, 321 passed targeted S-gene sequencing and WGS to be carried forward for further analysis (Seq S4 and S5, respectively, at https://figshare.com/articles/data set/Supplemental_Material/22762076).
We compared matching samples (n = 321) that passed both S-gene and WGS using ncbi-blast+/2.9.0 (35) and Nextclade Web version 2.6.1 (https://clades.nextstrain.org, accessed 30 September 2022) SARS-CoV-2 without recombinants (24). Using the output of Nextclade, we evaluated the concordance of variant, clade, and lineage assignment. We also compared the reported S-gene amino acid mutations for complete matches of corresponding samples and by counting individual mutations for corresponding samples (Table S12 at https://figshare.com/articles/data set/Supplemental_Material/22762076).
A subset of 277 samples through Delta was used to compare Ct values to coverage Nanopore sequencing yield on two flow cell types (FLO-MIN106 versus FLO-FLG001) and Nanopore sequencing accuracy to Illumina sequencing. Each time the RNA was thawed, we tested it with the flu SC2 multiplex assay (33) to determine the Ct value and amplified the S-gene using the methods presented here. For samples with an undefined Ct value (n = 2), a Ct value of 40 was assigned. We then split the spike amplicons to both Illumina and Nanopore sequencing methods. For Nanopore sequencing, we prepared libraries using the methods described here and loaded both standard MinION flow cells (FLO-MIN106) for 72 hours and disposable Flongle flow cells (FLO-FLG001) for 24 hours.
All 277 samples from this subset (pass or fail) were used to assess the relative pass rates of standard MinION flow cells (FLO-MIN106) versus Ct value (Table S11 at https://figshare.com/articles/data set/Supplemental_Material/22762076 and Fig. 4) and versus disposable Flongle flow cells (FLO-FLG001; Table S13 and Fig. S22 at https://figshare.com/articles/data set/Supplemental_Material/22762076).
From that subset of 277 samples, 251 samples passed both Nanopore (FLO-MIN106) and Illumina sequencing of the S-gene (Seq S6 at https://figshare.com/articles/data set/Supplemental_Material/22762076). For each of these 251 samples, we used ncbi-blast+/2.9.0 (35) to generate a three-way comparison between S-gene amplification and Nanopore sequencing (MIN), Illumina sequencing of those same amplicons (ILL), and NS3 surveillance results for the S-gene (NS3, Table S14 at https://figshare.com/articles/data set/Supplemental_Material/22762076).
Phylogenetics
We compared matching samples (n = 321) that passed both S-gene and WGS using Nextclade Web version 2.6.1 (https://clades.nextstrain.org; accessed September 30, 2022) SARS-CoV-2 without recombinants (24). From this analysis, we exported the phylogenetics and visualized them with Auspice (https://auspice.us; accessed October 6, 2022). We added a metadata sheet to label and highlight added sequences above the backbone sequences.
Primer kit manufacturing
CDC’s Division of Scientific Resources manufactured the primers used in this study and distributed to public health laboratories. The Oligo Synthesis Laboratory synthesized the primers, purified via high-performance liquid chromatography and verified by mass spectrophotometry. Following initial synthesis and purification, we received three QC aliquots for limit of detection analysis and excess material for use in this study. The remaining material (5 mmol each primer) was then transferred to the Diagnostic Manufacturing Laboratory for stochiometric mixing of forward and reverse primers, dispensing, drying, and kit assembly. We received three aliquots for QC testing.
Supplemental material
Supplemental material for this article may be found at https://figshare.com/articles/dataset/Supplemental_Material/22762076. Supplemental legends are available in supplemental Text S3 at https://figshare.com/articles/dataset/Supplemental_Material/22762076.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Centers for Disease Control and Prevention/National Center for Emerging and Zoonotic Infectious Diseases (CDC/NCEZID), Division of Scientific Resources/Biotechnology Core Facility Branch/Oligo Synthesis Laboratory for synthesizing the primers used in this study. We thank the CDC/NCEZID/Division of Scientific Resources/Reagent and Diagnostic Services Branch/Diagnostic Manufacturing Laboratory for manufacturing the primer kits.
Contributor Information
Matthew W. Keller, Email: nqp3@cdc.gov.
John R. Barnes, Email: fzq9@cdc.gov.
Day-Yu Chao, National Chung Hsing University, Taichung, Taiwan .
Michael Owusu, Kumasi Centre for Collaborative Research in Tropical Medicine (KCCR), Kumasi, Ghana .
DATA AVAILABILITY
Corresponding S-gene consensus sequences (Nanopore sequencing) and NS3 whole-genome consensus sequences are available in the supplemental materials (n = 321 each, Seq S04-S05). S-gene amplification and Illumina sequencing-derived S-gene consensus sequences (n = 251) are available in the supplemental materials (Seq S06). FASTQ reads (that BLAST matched to IRMA reference) are available online at NCBI under BioProject: PRJNA999712 (Sequence Read Archive SRS18443091–SRS18443900). The BioSamples (n=810) include the 321 primary validation samples (320 FLO-MIN106 and 1 FLO-FLG001), the 238 Flongle yield replicates that passed, and 251 Illumina accuracy replicates that passed. Consolidated metadata, biosample, and SRA identifiers associated with that raw sequencing data are available in the supplemental materials (Table S15).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.02982-23.
An accounting of the reviewer comments and feedback.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. WHO . 2020. Pneumonia of Unknown Cause – China. Available from: https://www.who.int/emergencies/disease-outbreak-news/item/2020-DON229. Retrieved 4 Oct 2022.
- 2. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cucinotta D, Vanelli M. 2020. WHO declares COVID-19 a pandemic. Acta Biomed 91:157–160. doi: 10.23750/abm.v91i1.9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dong E, Du H, Gardner L. 2020. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 20:533–534. doi: 10.1016/S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lambrou AS, Shirk P, Steele MK, Paul P, Paden CR, Cadwell B, Reese HE, Aoki Y, Hassell N, Zheng XY, et al. 2022. Genomic surveillance for SARS-CoV-2 variants: predominance of the Delta (B.1.617.2) and Omicron (B.1.1.529) variants - United States, June 2021-January 2022. MMWR Morb Mortal Wkly Rep 71:206–211. doi: 10.15585/mmwr.mm7106a4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Frampton D, Rampling T, Cross A, Bailey H, Heaney J, Byott M, Scott R, Sconza R, Price J, Margaritis M. 2021. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B. Lancet Infect Dis 21:1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dhar MS, Marwal R, Vs R, Ponnusamy K, Jolly B, Bhoyar RC, Sardana V, Naushin S, Rophina M, Mellan TA, et al. 2021. Genomic characterization and epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. Science 374:995–999. doi: 10.1126/science.abj9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. 2020. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367:1444–1448. doi: 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thompson MG, Burgess JL, Naleway AL, Tyner H, Yoon SK, Meece J, Olsho LEW, Caban-Martinez AJ, Fowlkes AL, Lutrick K, et al. 2021. Prevention of COVID-19 with the BNT162b2 and mRNA-1273 vaccines. reply. N Engl J Med 385:320–329. doi: 10.1056/NEJMc2113575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen RE, Winkler ES, Case JB, Aziati ID, Bricker TL, Joshi A, Darling TL, Ying B, Errico JM, Shrihari S, et al. 2021. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature 596:103–108. doi: 10.1038/s41586-021-03720-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, COVID-19 Genomics UK (COG-UK) Consortium, Peacock SJ, Robertson DL. 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. doi: 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rees-Spear C, Muir L, Griffith SA, Heaney J, Aldon Y, Snitselaar JL, Thomas P, Graham C, Seow J, Lee N, Rosa A, Roustan C, Houlihan CF, Sanders RW, Gupta RK, Cherepanov P, Stauss HJ, Nastouli E, Doores KJ, van Gils MJ, McCoy LE. 2021. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep 34:108890. doi: 10.1016/j.celrep.2021.108890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. VanBlargan LA, Errico JM, Halfmann PJ, Zost SJ, Crowe JE, Purcell LA, Kawaoka Y, Corti D, Fremont DH, Diamond MS. 2022. An infectious SARS-CoV-2 B.1.1.529 omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med 28:490–495. doi: 10.1038/s41591-021-01678-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe JE, Bloom JD. 2021. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 29:44–57. doi: 10.1016/j.chom.2020.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, Huang W, Li Q, Wang P, An R, et al. 2022. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602:657–663. doi: 10.1038/s41586-021-04385-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Piccoli L, Park YJ, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M, Silacci-Fregni C, Pinto D, Rosen LE, Bowen JE, et al. 2020. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell 183:1024–1042. doi: 10.1016/j.cell.2020.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Itokawa K, Sekizuka T, Hashino M, Tanaka R, Kuroda M. 2020. Disentangling primer interactions improves SARS-CoV-2 genome sequencing by multiplex tiling PCR. PLoS One 15:e0239403. doi: 10.1371/journal.pone.0239403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davis JJ, Long SW, Christensen PA, Olsen RJ, Olson R, Shukla M, Subedi S, Stevens R, Musser JM. 2021. Analysis of the ARTIC version 3 and version 4 SARS-CoV-2 primers and their impact on the detection of the G142D amino acid substitution in the spike protein. Microbiol Spectr 9:e0180321. doi: 10.1128/Spectrum.01803-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arctic Network . 2021. SARS-CoV-2 V4.1 update for Omicron variant. Available from: https://community.artic.network/t/sars-cov-2-v4-1-update-for-omicron-variant/342. Retrieved 3 Mar 2022.
- 20. Tyson JR, James P, Stoddart D, Sparks N, Wickenhagen A, Hall G, Choi JH, Lapointe H, Kamelian K, Smith AD, Prystajecky N, Goodfellow I, Wilson SJ, Harrigan R, Snutch TP, Loman NJ, Quick J. 2020. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv:2020.09.04.283077. doi: 10.1101/2020.09.04.283077 [DOI] [Google Scholar]
- 21. Sanderson T, Barrett JC. 2021. Variation at spike position 142 in SARS-CoV-2 delta genomes is a technical artifact caused by dropout of a sequencing amplicon. Wellcome Open Res 6:305. doi: 10.12688/wellcomeopenres.17295.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Salles TS, Cavalcanti AC, da Costa FB, Dias VZ, de Souza LM, de Meneses MDF, da Silva JAS, Amaral CD, Felix JR, Pereira DA, Boatto S, Guimarães MAAM, Ferreira DF, Azevedo RC, Ito E. 2022. Genomic surveillance of SARS-CoV-2 spike gene by sanger sequencing. PLoS ONE 17:e0262170. doi: 10.1371/journal.pone.0262170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fass E, Zizelski Valenci G, Rubinstein M, Freidlin PJ, Rosencwaig S, Kutikov I, Werner R, Ben-Tovim N, Bucris E, Erster O, Zuckerman NS, Mor O, Mendelson E, Dveyrin Z, Rorman E, Nissan I. 2021. Hispike method for high-throughput cost effective sequencing of the SARS-CoV-2 spike gene. Front Med (Lausanne) 8:798130. doi: 10.3389/fmed.2021.798130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aksamentov I, Roemer C, Hodcroft EB, Neher RA. 2021. Nextclade: clade assignment, mutation calling and quality control for viral genomes. JOSS 6:3773. doi: 10.21105/joss.03773 [DOI] [Google Scholar]
- 25. Rambaut A, Holmes EC, O’Toole Á, Hill V, McCrone JT, Ruis C, du Plessis L, Pybus OG. 2020. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5:1403–1407. doi: 10.1038/s41564-020-0770-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Organization WH. 2021. Genomic sequencing of SARS-CoV-2: a guide to implementation for maximum impact on public health. Retrieved 8 Jan 2021.
- 27. Lacek KA, Rambo-Martin B, Batra D, Keller MW, Wilson M, Sheth M, Davis M, Burroughs M, Gerhart J, Hassell N, Lee J, Shepard SS, Cook PW, Wentworth DE, Barnes JR, Kondor R, Paden CR, Peacock TPR, Sakaguchi H. 2022. Identification of a novel SARS-CoV-2 delta-omicron recombinant virus in the United States. bioRxiv:2022.03.19.484981. doi: 10.1101/2022.03.19.484981 [DOI] [Google Scholar]
- 28. Johnson BA, Zhou Y, Lokugamage KG, Vu MN, Bopp N, Crocquet-Valdes PA, Kalveram B, Schindewolf C, Liu Y, Scharton D, Plante JA, Xie X, Aguilar P, Weaver SC, Shi P-Y, Walker DH, Routh AL, Plante KS, Menachery VD, Shih SR. 2022. Nucleocapsid mutations in SARS-CoV-2 augment replication and pathogenesis. PLoS Pathog 18:e1010627. doi: 10.1371/journal.ppat.1010627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shepard SS, Meno S, Bahl J, Wilson MM, Barnes J, Neuhaus E. 2016. Erratum to: viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler. BMC Genomics 17:801. doi: 10.1186/s12864-016-3138-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rambo-Martin BL, Keller MW, Wilson MM, Nolting JM, Anderson TK, Vincent AL, Bagal UR, Jang Y, Neuhaus EB, Davis CT, Bowman AS, Wentworth DE, Barnes JR. 2020. Influenza A virus field surveillance at a swine-human interface. mSphere 5:e00822-19. doi: 10.1128/mSphere.00822-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bushnell B. 2014. Bbmap: A fast, accurate, splice-aware Aligner. Berkeley, CA (United States) Lawrence Berkeley National Lab.(LBNL) [Google Scholar]
- 32. Rambo-Martin BL, Lacek KA, Chau R. 2023. MIRA: an interactive dashboard for influenza genome and SARS-CoV-2 spike-gene assembly and curation. Available from: https://cdcgov.github.io/MIRA/index.html
- 33. Shu B, Kirby MK, Davis WG, Warnes C, Liddell J, Liu J, Wu K-H, Hassell N, Benitez AJ, Wilson MM, Keller MW, Rambo-Martin BL, Camara Y, Winter J, Kondor RJ, Zhou B, Spies S, Rose LE, Winchell JM, Limbago BM, Wentworth DE, Barnes JR. 2021. Multiplex real-time reverse transcription PCR for influenza A virus, influenza B virus, and severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 27:1821–1830. doi: 10.3201/eid2707.210462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao M, Lee WP, Garrison EP, Marth GT. 2013. SSW library: an SIMD Smith-Waterman C/C++ library for use in genomic applications. PLoS One 8:e82138. doi: 10.1371/journal.pone.0082138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An accounting of the reviewer comments and feedback.
Data Availability Statement
Corresponding S-gene consensus sequences (Nanopore sequencing) and NS3 whole-genome consensus sequences are available in the supplemental materials (n = 321 each, Seq S04-S05). S-gene amplification and Illumina sequencing-derived S-gene consensus sequences (n = 251) are available in the supplemental materials (Seq S06). FASTQ reads (that BLAST matched to IRMA reference) are available online at NCBI under BioProject: PRJNA999712 (Sequence Read Archive SRS18443091–SRS18443900). The BioSamples (n=810) include the 321 primary validation samples (320 FLO-MIN106 and 1 FLO-FLG001), the 238 Flongle yield replicates that passed, and 251 Illumina accuracy replicates that passed. Consolidated metadata, biosample, and SRA identifiers associated with that raw sequencing data are available in the supplemental materials (Table S15).