Abstract
The products of in-frame overlapping genes II and X carried by the filamentous phage f1 genome are proteins with required but opposing functions in phage DNA replication. Their normal relative levels are important for continuous production of phage DNA without killing infected Escherichia coli hosts. Here we identify several factors responsible for determining the relative levels of pII and pX and that, if perturbed, alter the normal distribution of phage DNA species in infected hosts. Translation of the two proteins is essentially relegated to separate mRNAs. The mRNAs encoding genes II and X are also differentially sensitive to cleavage dependent on rne, the gene encoding the only E. coli endo-RNase known to have a global role in mRNA stability. Whereas pII levels are limited at the level of mRNA stability, normal pX levels require transcription in sufficient amounts from the promoter for the smaller mRNA encoding only pX.
Three proteins encoded by the Ff filamentous phage (f1, M13, and fd) are required for the stages of phage DNA replication (Fig. 1) that occur after a complement to the incoming circular (+) strand has been synthesized by Escherichia coli host enzymes (18). pII and pX arise from in-frame overlapping genes II and X. Gene X is contained entirely within gene II (Fig. 1), and yields a 13-kDa protein identical to the C-terminal third of the 46-kDa pII (30). The single-stranded DNA binding protein pV is encoded by gene V. pII functions early in infection to produce replicative forms (RF). It initiates synthesis by site-specific cleavage on supercoiled RFI, participates in the synthesis reaction, and terminates each round by cleaving and ligating the newly displaced strand. pX is needed later for synthesis of single-stranded viral DNA (ssDNA), and although less well understood biochemically than pII, pX probably acts as an inhibitor of pII function (5). pV, once it reaches a sufficient concentration, promotes synthesis of viral strands by sequestering ssDNA in a flexible rod away from the replication proteins. pV also turns down synthesis of pII and pX late in infection by repressing translation on the mRNA for both gene II and gene X (4, 17, 31).
FIG. 1.
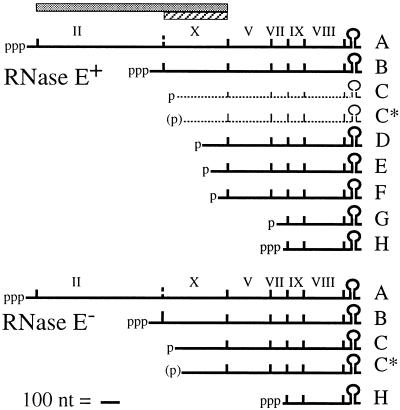
Bacteriophage f1 mRNA for RNase E+ and RNase E− E. coli hosts. The extents of gene II (shaded bar) and gene X (hatched bar) are shown. The mRNA diagram for the RNase E+ hosts is based on the work of many authors (reviewed in reference 18), and that for the RNase E− hosts is as described in reference 10. The f1 mRNAs, scaled as indicated, range in size from 370 to 2,000 nt. Coding regions are demarcated by vertical lines. Primary transcripts from strong constitutive promoters and posttranscriptional cleavage products are indicated by known or probable (in parentheses) phosphorylation status. RNAs C and C* are short-lived species observed at very low levels in wild-type hosts (1, 2) but higher levels in RNase E− hosts (10). The f1 mRNAs have a common 3′ end generated by rho-independent termination.
The phage DNA replication proteins map in a transcription unit expressed in wild-type hosts as a series of abundant polycistronic mRNAs (Fig. 1). Only the two large primary transcripts, RNAs A and B, include genes II and X. RNAs C to G are the 3′ products of endonuclease cleavage (reviewed in reference 18). A number of the cleavages appear to involve RNase E, a key endonuclease in the processing and decay of mRNA, the 9S precursor to 5S rRNA, and the antisense regulator of plasmid ColE1 replication (3). At the nonpermissive temperature (42°C) in a host bearing a temperature-sensitive rne-1 mutation in the gene encoding RNase E, the f1 mRNA processing pattern was shown to be markedly different (10, 28). The steady-state levels of RNA B, RNA C, and RNA C* increased, whereas the other processed RNAs were virtually absent. Because the probe used for S1 nuclease mapping did not visualize RNA A, the effects on this RNA of inactivation of RNase E were not clear. The results suggested that the f1 mRNA processing pathway is comprised minimally of rne-independent cleavages at the C and C* sites and rne-dependent steps at distal sites. Importantly, the same block in processing occurred in the rne-1 host at 37°C, a temperature permissive for cell growth and phage infection.
Evidence from Fulford and Model (5) has indicated that the relative levels of pII and pX are critical to maintaining a normal replication cycle. Using plasmids to deliver an excess dose of pII and/or pX in infected hosts, they observed an increase in RFs at the expense of ssDNA synthesis whenever a higher-than-normal ratio of pII to pX occurred. Their findings suggested that one or more mechanisms exist to ensure that expression of genes II and X yields appropriate relative levels of pII and pX. To determine how the normal relative levels are achieved, we have exploited the effectiveness of the block in processing in rne-1 hosts at permissive temperatures to study expression of f1 genes II and X from RNAs A and B. To verify observations for the rne+ and rne-1 hosts, the mRNA and protein levels were also examined in wild-type hosts infected with phage mutants containing progressively weaker promoters for RNA B.
Quantitation of phage primary transcripts A and B in an RNase E-deficient host.
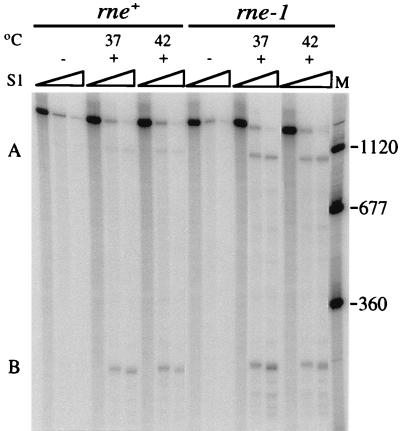
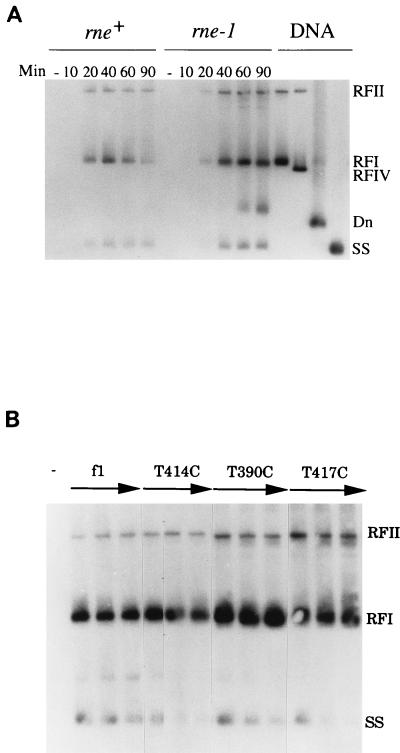
To determine whether inactivation of RNase E affects the steady-state levels of RNAs A and B, the levels present in isogenic rne+ and rne-1 hosts at permissive and nonpermissive temperatures were quantified by S1 nuclease protection methods carried out at probe excess (10). RNA was isolated from cultures incubated at 37 or 42°C after infection with wild-type f1 at 34°C (10). RNAs A and B were detected with an ssDNA probe made by annealing a 5′-end-labeled primer to a position on f1 (+) strand DNA ∼250 nucleotides (nt) downstream from the RNA B start and extending it to a length (1,515 nt) that would detect both RNAs (Fig. 2). In the sets of lanes containing RNA from infected cultures, products were present that had appropriate mobilities (1,110 and 250 nt) for fragments protected by RNAs A and B. Quantitation revealed that in the rne+ host, approximately fivefold less RNA A was present than RNA B at 37 and 42°C. In the rne-1 host, the amount of RNA A relative to RNA B increased. At 37°C, the level of RNA A was only twofold lower than that of RNA B, and at 42°C, the levels of the two RNAs were nearly equivalent. The nature of the rne-1 defect and the fact that the changes in mRNA levels were exacerbated as the mutant phenotype became more severe made it likely that turnover of RNA A had decreased. The results suggested that inactivation of the rne-dependent RNA cleavage pathway stabilized RNA A more than RNA B, an interpretation confirmed by the sixfold versus threefold increases in RNAs A and B seen for the rne-1 host on Northern blots (6). This suggests that RNA A serves as a better substrate than RNA B for rne-dependent cleavage. While the basis is not known, a likely possibility is that as yet unidentified sites of rate-limiting cleavage are located between the two transcription start points. Another explanation would be that structural distinctions between RNAs A and B make RNA A a more effective target for recognition by the complex containing RNase E (3, 16, 23).
FIG. 2.
Primary transcripts A and B in phage-infected rne+ and rne-1 hosts. DS211 (F′Tn10/rne+ λ−) and DS212 (F′Tn10/rne-1 λ−) were grown at 34°C to 2 × 108 cells/ml in Luria-Bertani broth (27) containing 12 μg of tetracycline per ml and infected (multiplicity of infection of 50) with wild-type f1 (+) or left uninfected (−). They were shifted to the indicated temperatures at 15 min after infection. Samples (5 ml) were removed immediately prior to infection or 60 min after the temperature shift. The 5′-32P-labeled probe and size standards (nucleotides) (lane M) were generated in a similar manner by primer extension and purification on alkaline gels. Each lane contained the products of analysis of 2 μg (DS211) or 1 μg (DS212) of RNA and 0.1 pmol of probe. The S1 nuclease concentrations for each set of lanes were none or 0.1 and 0.35 U/μg of RNA. Samples were electrophoresed on 6% polyacrylamide sequencing gels containing a gradient of 0.5× to 2.5× TBE (89 mM Tris-borate, 2.5 mM EDTA [pH 8.3]).
pII and pX production in the RNase E-deficient host.
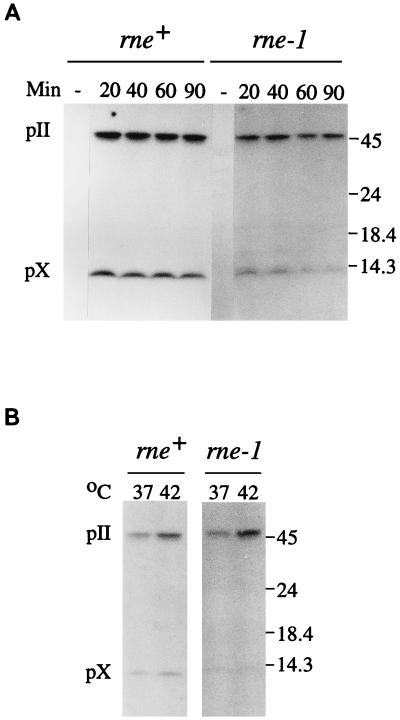
To determine if the altered relative levels of RNAs A and B were reflected in gene II and X expression, the amounts of pII and pX at times after phage infection were determined by Western blot analysis (Fig. 3A). Polyclonal antiserum raised against pX was used so that the same epitopes would be recognized in both proteins and give quantitative estimates for pII and pX. At 37°C, the ratio of pII to pX in the rne+ host was 1.7 to 1.9 throughout infection, whereas in the rne-1 host, the relative amount of pII increased to more than threefold over pX as early as 20 min and was sevenfold greater by 90 min. The increase was more pronounced at 42°C (Fig. 3B), with approximately sevenfold more pII than pX as early as 60 min after infection. Thus, a change in the relative amounts of pII and pX does accompany the increase in the relative amount of RNA A in the rne-1 host. The increase in the relative amount of pII was in fact greater than expected if RNA A functions as an efficient mRNA template for both pII and pX. This raised the possibility that little or no pX is made from RNA A and that RNA B is the major template for translation of pX.
FIG. 3.
Immunoblot analysis of pII and pX. The rne+ and rne-1 hosts were infected with wild-type f1 (multiplicity of infection of 300) or left uninfected (−) and shifted to 37°C (A) or 37 and 42°C (B). Samples removed immediately prior to infection or at the indicated times in minutes after the temperature shift were chilled in 0.5 ml of a combination of 10 mM (each) Tris-HCl (pH 7.5), EDTA, NaCl, and NaN3. Following centrifugation, cells were resuspended in sample buffer, incubated at 100°C for 5 min, and analyzed on sodium dodecyl sulfate-polyacrylamide gels containing gradients of 10 to 20% acrylamide and 0.26 to 1.0% bisacrylamide. Electrophoretic transfer was to 0.2-μm-pore-size BA83 nitrocellulose membranes (24, 30). Each lane contained an amount of extract equivalent to 108 cells. pII and pX were detected with anti-pX immunoglobulin G (100 μg) and 125I-labeled protein A (0.5 μCi). Antiserum against pX was made from protein overexpressed in strain K561 (7) containing a plasmid with the pII coding region under control of the tacI promoter. To a culture (50 ml) grown at 37°C to 2 × 108 cells/ml in Luria-Bertani broth containing 0.1 mg of ampicillin per ml, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to 2 mM. Ampicillin was replenished at the time of induction and 1 h later. Bacteria were harvested by centrifugation after 2 h and resuspended in 5 ml of sample buffer (30), and samples were fractionated on sodium dodecyl sulfate-polyacrylamide gels (13). pX was eluted electrophoretically. Antiserum was prepared by Pocono Rabbit Farm and Laboratory, Inc., and immunoglobulin G fractions were isolated by chromatography on protein A-Sepharose. The positions of pII, pX, and molecular mass standards (in kilodaltons) are indicated to the right.
Altered pII and pX production from phage bearing mutations in the promoter for RNA B.
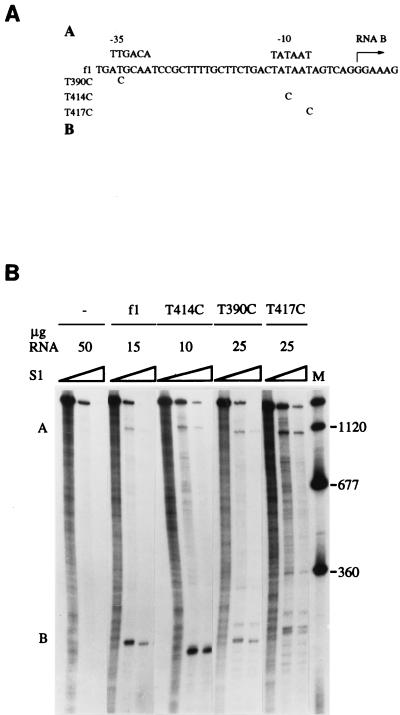
Although the changes in the relative levels of the gene II and X products appeared to arise from the differences in the steady-state levels of the mRNAs, the changes could have represented pleiotropic effects of the rne-1 mutation rather than direct consequences of the mRNA processing defect. To rule this out, the relative levels of RNAs A and B were varied by a different means in a wild-type rne+ host grown at 37°C. Mutations aimed at decreasing the strength of the promoter for RNA B were introduced into the phage (Fig. 4A). The mutants, designated by nucleotide position in f1 DNA (8), contained a T→C change in the −35 or −10 region. The base substitutions were made in the third codon position so as not to change any amino acids within the larger protein, pII. Based on the effects of other mutations at various positions in the −35 and −10 hexamers (19), the −10 change in T414C was expected to reduce promoter activity by <50%, whereas the −35 and −7 changes in T390C and T417C were expected to decrease activity by >90%. S1 nuclease protection assays revealed mRNA levels generally consistent with these expectations (Fig. 4B). The level of RNA A in S26r1eλ− infected with wild-type f1 was five- to sixfold lower than that of RNA B, the same as that observed for the rne+ host, DS211. These relative levels changed as the strength of the promoter for RNA B decreased and the relative level of RNA A increased. The −10 change (T414C) had little effect, giving RNA A levels 4.5- to 5-fold lower than RNA B levels. The −35 change (T390C) had a greater effect, giving RNA A levels only twofold lower than the RNA B levels. The −7 change (T417C) showed the most severe effect. RNA A levels were normal, but RNA B levels were drastically reduced, with some evidence for ambiguity in start site selection. The low yield of protected fragments representing RNA B made quantitation difficult, but RNA A appeared to be about twice as abundant as RNA B. The mutants provided a range of promoter activities decreasing the steady-state levels of RNA B.
FIG. 4.
Primary transcripts A and B in a wild-type host infected with phage bearing mutations in the promoter for RNA B. (A) Location of the point mutations in the f1 DNA sequence (8). Consensus sequences for the −35 and −10 regions of sigma 70 promoters and the probable start point for transcription of RNA B (+1) are shown. Oligonucleotide mutagenesis (12) used uracil-containing f1 (+) strand DNA generated in CJ236 (dut ung mutant). Following extension of the annealed primers and closure of new strands, the double-stranded circular molecules were introduced into JM109 (29) and incubated overnight at 37°C in Luria-Bertani top agar in a lawn of JM109. The desired mutations were identified by dideoxy sequencing (25). (B) S1 nuclease protection analysis of RNAs A and B following infection of S26r1eλ− (Hfr Cavalli phoA4(Am) serU132 λ−) with no phage (−), wild-type phage (f1), or the indicated mutants. Assays, performed with samples isolated 50 min after infection, contained the indicated quantities of RNA and 5′-end-labeled probe (0.05 pmol). The final S1 nuclease concentrations in each set of lanes were none or 0.33 and 1.0 U/μg of RNA. The panel is a composite that presents one RNA concentration of several tested for each phage strain. Sizes are shown to the right in nucleotides.
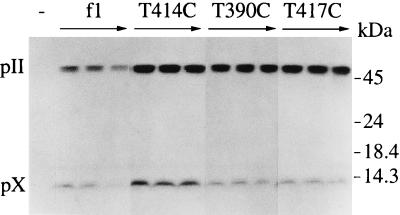
Western blots quantified pII and pX (Fig. 5). Over the time course of infection, wild-type f1 showed the typical accumulation of twofold more pII than pX. T414C showed a small but detectable increase (20%) in the amount of pII relative to pX by 1 h after infection. T390C, which reduced promoter activity more significantly, resulted in approximately fivefold more pII than pX throughout the course of infection. The increases in the relative levels of RNA A and pII for this promoter mutant were very similar to those observed for the rne-1 host compared to its wild-type counterpart (Fig. 2 [37°C lanes] and 3A). T417C, the most severe down mutation, resulted in the most marked change in protein levels, with 8- to 10-fold more pII than pX. Since decreasing synthesis of RNA B in these experiments had effects on pII and pX levels similar to those observed in the rne-1 host, the altered levels of pII and pX in the rne-1 host can be attributed directly to the differences in relative mRNA levels brought about by the processing defect.
FIG. 5.
Immunoblot analysis of pII and pX produced by the phage promoter mutants. Cell extracts from S26r1eλ− left uninfected (−) or infected with the indicated phage strains were prepared immediately prior to infection or 30, 60, and 90 min after infection.
The data for the promoter mutants in addition demonstrate the importance of RNA B as the major template for translation of pX. The T417C mutant at position −7 provided the clearest indication that the proteins are synthesized from separate transcripts. RNA B levels dropped more than 10-fold, and pX levels decreased sharply, despite the fact that synthesis of RNA A was not changed. Since only RNA A encodes gene II and the low levels of RNA B could account for the pX observed, the bulk of translation in the II-X region of RNA A appears to be devoted to producing pII. The results thus answer a question first raised when overlapping genes II and X were discovered (30): whether pX is synthesized from one or both of the mRNAs that include the coding region. The results may also have implications for other pairs of overlapping genes, a number of which contain dual promoters (21). Two possible mechanisms could explain why little or no pX arises from RNA A. Elongating ribosomes within the gene II mRNA coding region could block internal initiation, or rapidly forming secondary structure between ribosomes could render the initiator region largely inaccessible.
Effect of altered gene II and X expression on phage DNA replication.
Phage containing the down mutations in the promoter for RNA B were detected exclusively as very small plaques, generally a reliable indicator of reduced phage production. This observation, considered with evidence that a specific ratio of pII to pX is required for a normal pattern of DNA replication (5), raised the possibility that the increased relative levels of pII had detectable effects on replication. Thus, the distribution of phage DNA species from the rne+ and rne-1 hosts infected at 37°C with wild-type f1 was examined on Southern blots (Fig. 6A). The distribution of DNA species was normal in the rne+ host, with RF production at early times and later revealing a shift toward ssDNA synthesis. In contrast, the DNA samples from the rne-1 host showed a higher-than-normal accumulation of RF species relative to ssDNA. The identity of the additional species migrating between RFI and ssDNA remains to be resolved, but as judged from standards, it does not represent relaxed RFIV or denatured DNA. A similar pattern was observed when DNA samples from S26r1eλ− infected with wild-type f1 or the promoter mutants were examined. The promoter mutants showed an excess of RFI and RFII relative to ssDNA (Fig. 6B). In both cases, the increase was particularly apparent at the later time points after infection, possibly reflecting an inhibition in the switch to accumulation of ssDNA for phage assembly. In agreement with these observations, phage titers in the rne-1 host were down 10-fold at times beyond 60 min after infection, and titers for the promoter mutants were down 20- to 30-fold. Increased synthesis of RF at the expense of ssDNA has been observed previously when pII alone or pII and pX in combination were overexpressed (5). It should be noted that the distributions of f1 DNAs seen in the two situations here and others described previously are not identical, but differences probably reflect how much pV was present to sequester ssDNA and regulate translation of genes II and X. Any differences in pV levels made from the mRNAs made in the experiments reported here may also explain some of the changes in the absolute levels of pII and pX we observed.
FIG. 6.
Southern blot hybridization of phage DNA species. (A and B) DS211 (rne+) or DS212 (rne-1) (A) and S26r1eλ− (B) left uninfected (−) or infected with the indicated phage strains (multiplicity of infection of 80). Subcultures of DS211 and DS212 grown at 30°C were shifted to 37°C just before infection. Samples (0.5 ml) were mixed with an equal volume of an ice-cold mixture of 10 mM Tris-HCl (pH 8.1), 10 mM EDTA, 10 mM NaCl, and 10 mM NaN3 and washed twice with the same buffer lacking NaN3 to remove free phage. Resuspended cells were lysed (14) at 55°C. EDTA (pH 9.3) was added to 25 mM, proteinase K was added to 150 μg/ml, and incubation was continued at 37°C for 60 min. Chromosomal DNA was sheared by passage through a 22-gauge needle. Purified DNA preparations (24) were resuspended in 20 μl of 10 mM Tris-HCl (pH 8.0)–1 mM EDTA. Standards included RFI (duplex supercoiled), RFII (nicked duplex), and ssDNA (SS) (24). RFIV DNA (relaxed) was generated by treatment of RFI DNA with Drosophila melanogaster topoisomerase II (9). Denatured DNA (Dn) was generated from RFI DNA by incubation in 1.5 N NaOH at room temperature for 15 min, followed by addition of sodium acetate (pH 4.9) to a concentration of 0.6 M. DNA samples (3 μl) were electrophoresed overnight at 40 V in 1% agarose gels (20 by 20 by 0.3 cm) in a mixture of 80 mM Tris-phosphate (pH 8.2) and 8 mM EDTA containing 0.5 μg of ethidium bromide per ml with continuous buffer recirculation. Transfer of DNA was to GeneScreen Plus after alkaline denaturation (24). The DNA probe (∼107 cpm) was generated by nick translation (24) of 0.1 μg of f1 RFI DNA in the presence of [α-32P]dCTP (3,000 Ci/mmol, DuPont NEN). DNA samples from uninfected cultures (−) and from time points in minutes after infection are indicated. In panel B, the three lanes shown for each phage strain represent samples isolated at 30, 60, and 90 min after infection.
Conclusions.
Our finding that the absence of RNase E leads to an increase in the amount of RNA B (10) and a relatively larger increase in RNA A reveals that the rne-dependent cleavage pathway does have an important role in setting the normal relative steady-state levels of these primary transcripts during phage infection. Cleavage of RNA A functions to limit its stability, the yield of pII, and hence the level of pII relative to pX. From this, it is clear that segmental differences in the stabilities of the mRNAs encoding genes II and X provide one important means of regulating their relative expression, as observed in a number of other bacterial operons (15, 20, 22, 26). Moreover, since conditions that change the stability of RNA A lead to an aberrant distribution of phage DNA species, the half-life of gene II mRNA observed in wild-type hosts appears to be an integral component of the regulatory circuit needed to maintain a balanced pattern of DNA replication. A second component is the relegation of translation to separate transcripts, so that a distinct means is available to set the level of pX. As judged from the promoter mutants, synthesis of RNA B in sufficient amounts appears to be required to achieve the normal level of pX. Finally, comparison of the relative steady-state levels of the mRNAs and proteins suggests that additional factors operate to determine how much pII is made relative to pX. There is normally about twice as much pII as pX, but the amount of mRNA template for pII is fivefold less abundant than the template for pX. This raises the possibility that differences exist at the level of translational efficiency, a possibility supported by predictions from RNA folding (32) that the 5′ end of RNA B is involved in an extensive structure positioned to interfere with ribosome binding to the gene X initiation site (11). Further experimentation is required to understand more fully how expression of genes II and X is regulated.
Acknowledgments
This research was supported by National Institutes of Health grant GM33349 to D.A.S. R.J.K. was supported in part by National Institute of General Medical Sciences Predoctoral Traineeship GM07184.
We thank S. Kushner, P. Model, and M. Russel for bacterial strains and plasmids; R. Webster for helpful discussions; and L. Arrington for assistance in preparing the manuscript.
REFERENCES
- 1.Blumer K J, Steege D A. mRNA processing in Escherichia coli: an activity encoded by the host processes bacteriophage f1 mRNAs. Nucleic Acids Res. 1984;12:1847–1861. doi: 10.1093/nar/12.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cashman J S, Webster R E, Steege D A. Transcription of bacteriophage f1: the major in vivo RNAs. J Biol Chem. 1980;255:2554–2562. [PubMed] [Google Scholar]
- 3.Cohen S N, McDowall K J. RNase E: still a wonderfully mysterious enzyme. Mol Microbiol. 1997;23:1099–1106. doi: 10.1111/j.1365-2958.1997.tb02593.x. [DOI] [PubMed] [Google Scholar]
- 4.Fulford W, Model P. Specificity of translational regulation by two DNA binding proteins. J Mol Biol. 1984;173:211–226. doi: 10.1016/0022-2836(84)90190-6. [DOI] [PubMed] [Google Scholar]
- 5.Fulford W, Model P. Regulation of bacteriophage f1 DNA replication. I. New functions for genes II and X. J Mol Biol. 1988;203:49–62. doi: 10.1016/0022-2836(88)90090-3. [DOI] [PubMed] [Google Scholar]
- 6.Goodrich, F., and D. A. Steege. Unpublished results.
- 7.Greenstein D, Horiuchi K. Interaction between the replication origin and the initiator protein of the filamentous phage f1: binding occurs in two steps. J Mol Biol. 1987;197:157–174. doi: 10.1016/0022-2836(87)90115-x. [DOI] [PubMed] [Google Scholar]
- 8.Hill D F, Petersen G B. Nucleotide sequence of bacteriophage f1 DNA. J Virol. 1982;44:32–46. doi: 10.1128/jvi.44.1.32-46.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsieh T-S. Purification and properties of type II DNA topoisomerase from embryos of Drosophila melanogaster. Methods Enzymol. 1983;100:161–170. doi: 10.1016/0076-6879(83)00052-x. [DOI] [PubMed] [Google Scholar]
- 10.Kokoska R J, Blumer K J, Steege D A. Phage f1 mRNA processing in Escherichia coli: search for the upstream products of endonuclease cleavage, requirement for the product of the altered mRNA stability (ams) locus. Biochimie. 1990;72:803–811. doi: 10.1016/0300-9084(90)90189-n. [DOI] [PubMed] [Google Scholar]
- 11.Kokoska, R. J., and D. A. Steege. Unpublished results.
- 12.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Lerner T J, Model P. The “steady state” of coliphage f1: DNA synthesis late in infection. Virology. 1981;115:282–294. doi: 10.1016/0042-6822(81)90111-2. [DOI] [PubMed] [Google Scholar]
- 15.McCarthy J E G, Gerstel B, Surin B, Wiedemann U, Ziemke P. Differential gene expression from the Escherichia coli atp operon mediated by segmental differences in mRNA stability: the roles of mRNA structure and translational efficiency. Mol Microbiol. 1991;5:2447–2458. doi: 10.1111/j.1365-2958.1991.tb02090.x. [DOI] [PubMed] [Google Scholar]
- 16.Miczak A, Kaberdin V R, Wei C-L, Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci USA. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Model P, McGill C, Mazur B, Fulford W D. The replication of bacteriophage f1: gene V protein regulates the synthesis of gene II protein. Cell. 1982;29:329–335. doi: 10.1016/0092-8674(82)90149-0. [DOI] [PubMed] [Google Scholar]
- 18.Model P, Russel M. Filamentous bacteriophage. In: Calendar R, editor. The bacteriophages. New York, N.Y: Plenum Press; 1988. pp. 375–456. [Google Scholar]
- 19.Moyle H, Waldburger C, Susskind M M. Hierarchies of base pair preferences in the P22 ant promoter. J Bacteriol. 1991;173:1944–1950. doi: 10.1128/jb.173.6.1944-1950.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nilsson P, Naureckiene S, Uhlin B E. Mutations affecting mRNA processing and fimbrial biogenesis in the Escherichia coli pap operon. J Bacteriol. 1996;178:683–690. doi: 10.1128/jb.178.3.683-690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Normark S, Bergström S, Edlund T, Jaurin B, Lindberg F P, Olsson O. Overlapping genes. Annu Rev Genet. 1983;17:499–525. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- 22.Petersen C. Control of functional mRNA stability in bacteria: multiple mechanisms of nucleolytic and non-nucleolytic inactivation. Mol Microbiol. 1992;6:277–282. doi: 10.1111/j.1365-2958.1992.tb01469.x. [DOI] [PubMed] [Google Scholar]
- 23.Py B, Higgins C F, Krisch H M, Carpousis A J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schramm H-C, Schneppe B, Birkenhager R, McCarthy J E G. The promoter-proximal, unstable IB region of the atp mRNA of Escherichia coli: an independently degraded region that can act as a destabilizing element. Biochim Biophys Acta. 1996;1307:162–170. doi: 10.1016/0167-4781(96)00034-6. [DOI] [PubMed] [Google Scholar]
- 27.Steege D A, Low B. Isolation and characterization of lambda transducing bacteriophages for the su1+ (supD−) amber suppressor of Escherichia coli. J Bacteriol. 1975;122:120–128. doi: 10.1128/jb.122.1.120-128.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stump M D, Steege D A. Functional analysis of filamentous phage f1 processing sites. RNA. 1996;2:1286–1294. [PMC free article] [PubMed] [Google Scholar]
- 29.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 30.Yen T S B, Webster R E. Bacteriophage f1 gene II and X proteins: isolation and characterization of the products of two overlapping genes. J Biol Chem. 1981;256:11259–11265. [PubMed] [Google Scholar]
- 31.Yen T S B, Webster R E. Translational control of bacteriophage f1 gene II and gene X proteins by gene V protein. Cell. 1982;29:337–345. doi: 10.1016/0092-8674(82)90150-7. [DOI] [PubMed] [Google Scholar]
- 32.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]