Abstract
The roles of the nitrogen fixation regulatory proteins NifA, FixK1, and FixK2 in the symbiotic regulation of hydrogenase structural gene expression in Bradyrhizobium japonicum have been investigated. Bacteroids from FixJ and FixK2 mutants have little or no hydrogenase activity, and extracts from these mutant bacteroids contain no hydrogenase protein. Bacteroids from a FixK1 mutant exhibit wild-type levels of hydrogenase activity. In β-galactosidase transcriptional assays with NifA and FixK2 expression plasmids, the FixK2 protein induces transcription from the hup promoter to levels similar to those induced by HoxA, the transcriptional activator of free-living hydrogenase expression. The NifA protein does not activate transcription at the hydrogenase promoter. Therefore, FixK2 is involved in the transcriptional activation of symbiotic hydrogenase expression. By using β-galactosidase transcriptional fusion constructs containing successive truncations of the hup promoter, the region of the hup promoter required for regulation by FixK2 was determined to be between 29 and 44 bp upstream of the transcription start site.
The slow-growing symbiont of the soybean plant, Bradyrhizobium japonicum, expresses a hydrogen uptake hydrogenase that oxidizes hydrogen under both free-living and symbiotic conditions. In the free-living state, the expression of the NiFe hydrogenase is regulated at the transcriptional level by hydrogen, oxygen, and nickel (21). These three signals exert their effects within a 50-bp region of DNA located between 99 and 149 bp upstream of the transcription start site of the hydrogenase structural genes (22). In addition, the hydrogenase promoter is ς54 dependent and requires integration host factor for full induction (5).
The hoxA gene (32) is located approximately 12 kb downstream of the hydrogenase structural genes, in a region of the hydrogenase gene cluster previously shown to be necessary for free-living hydrogenase activity (23). The hoxA gene encodes a protein with extensive homology to transcriptional activators of hydrogenase expression in several other organisms, including HoxA in Alcaligenes eutrophus (12), HupR1 in Rhodobacter capsulatus (30), and HydG in Escherichia coli (31), all of which are members of the NtrC-like family of response regulators (17). Subsequent studies of the role of the HoxA protein in the biosynthesis of hydrogenase by our group (11) and another (33) have confirmed that HoxA is a transcriptional activator of hydrogenase expression under free-living, microaerobic conditions. Its cognate sensor protein is presently unknown. However, bacteroids from nodules formed by B. japonicum HoxA mutants exhibit wild-type levels of hydrogenase activity and extracts from Hup+ bacteroids used in gel retardation assays do not cause a shift of a fragment of the hydrogenase promoter containing the 50-bp regulatory region (11). Because the hydrogenase promoter is ς54 dependent, there must be some other, symbiosis-specific, activator of hydrogenase expression that binds the promoter at an alternative site.
The hydrogenase enzyme in the pea symbiont, Rhizobium leguminosarum, is expressed solely in symbiotic conditions (28). Interestingly, a defective hoxA gene that has been inactivated by several frameshift and deletion mutations has been reported for this organism (6). The hydrogenase structural genes have been shown to be under the transcriptional control of the nitrogen fixation regulatory protein NifA (6). Several groups have suggested that symbiotic hydrogenase expression in B. japonicum is also linked to the regulation of nitrogen fixation (11, 33). In addition to NifA, another candidate for a symbiotic regulator is the Fnr-like, DNA binding protein FixK (2). In rhizobial species, FixK is part of an oxygen-responsive regulatory cascade controlled by the FixL-FixJ two-component system (13). B. japonicum contains two homologs of the FixK protein (13). FixK2 activates the expression of several genes, including nitrate metabolism genes, the fixNOQP operon, and fixK1, in response to low oxygen levels (13). Mutants in this gene are Nif−. The target or targets for FixK1 are unknown (2). It is possible that one or both of the FixK homologs may be involved in the regulation of other oxygen-responsive genes, including hydrogenase (11, 13).
In this work, we show that mutants in the fixLJ-fixK nitrogen fixation regulatory cascade are deficient in symbiotic hydrogenase activity. Unlike in R. leguminosarum, NifA does not seem to be involved in the symbiotic transcriptional activation of B. japonicum hydrogenase expression. Instead, the results here are consistent with FixK2 acting as a symbiotic transcriptional activator. A possible FixK2 binding site is centered at 40 bp from the transcription start site.
Bacterial strains and plasmids.
All bacterial strains and plasmids used in this work are listed in Table 1. B. japonicum JH (16) is a derivative of USDA I-110 and is considered the wild type. JHΔA (11) is derived from strain JH and contains an 886-bp in-frame genomic deletion that removes most of the hoxA gene. Strains 7360 (1), 7454 (2), and 9043 (13) are all derived from B. japonicum 110spc (29). Strains 7360 and 7454 contain an insertion of the kanamycin resistance gene that disrupts the fixJ and fixK1 genes, respectively. In strain 9043, the fixK2 gene is replaced by the spectinomycin resistance gene. Escherichia coli ET8000 (24) is a lac mutant strain that is used as a background strain in β-galactosidase transcriptional assays. Plasmid pRJ9044 (unpublished data; a gift of H. Fischer) contains the B. japonicum fixK2 gene on a 1.85-kb BamHI-SalI fragment cloned into pBluescript SK under control of the lac promoter. Plasmid pMC71A (7) contains the Klebsiella pneumoniae nifA gene cloned into the multicopy vector pACYC184 (9) under control of the promoter of the tetracycline resistance gene. Plasmid pSKA contains the B. japonicum hoxA gene on a 1.5-kb KpnI-SpeI fragment cloned into pBluescript SK in the same orientation as the vector lac promoter. Plasmid pSY7 (21) is a hup-lacZ transcriptional fusion construct derived from pGD499 (10) and contains a 2.4-kb BamHI-PstI fragment of the hydrogenase structural genes including 680 bp of the promoter region. The remaining plasmids, pGHh1, pGHp1, pGR1, pGHf1, pGBs3, and pGNSdB (21, 22), are all derived from plasmid pSY7 and contain successive deletions of the hup promoter region (listed in Table 1) fused to a promoterless lacZ gene.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strains or plasmids | Genotype or relevant features | Reference or source |
|---|---|---|
| Strains | ||
| B. japonicum | ||
| JH | Wild-type derivative of USDA I-110 | 16 |
| JHΔA | In-frame deletion of 886 bp of hoxA gene | 11 |
| 7360 | fixJ::aphII (Spcr Kanr) | 1 |
| 7454 | fixK1::aphII (Spcr Kanr) | 2 |
| 9043 | fixK2::Ω (Spcr Strr) | Unpublished data; see also reference 13 |
| E. coli ET8000 | rbs lacZ::IS1 gyrA hutCK | 24 |
| Plasmids | ||
| pRJ9044 | B. japonicum fixK2 cloned into pSK under control of the lac promoter | 13a |
| pMC71A | K. pneumoniae nifA cloned into pACYC184 under control of the Tcr gene promoter | 8 |
| pSKA | B. japonicum hoxA cloned into pSK in the same orientation as the lac promoter | This study |
| pSY7 | hup-lacZ fusion; −681 to +1649 | 21 |
| pGHh1 | hup-lacZ fusion; −171 to +39 | 21 |
| pGHp1 | hup-lacZ fusion; −149 to +171 | 21 |
| pGR1 | hup-lacZ fusion; −99 to +171 | 21 |
| pGHf1 | hup-lacZ fusion; −44 to +171 | 22 |
| pGBs3 | hup-lacZ fusion; −29 to +171 | 22 |
| pGNSdB | hup-lacZ fusion; −220 to +162 with region −64 to −29 deleted | 22 |
The hup phenotype of fixJ, fixK1, and fixK2 regulatory mutants.
Free-living B. japonicum strains were grown in modified Bergerson’s medium (3) and derepressed for hydrogenase activity by incubation for 18 to 20 h in no-carbon medium (26) under standard conditions (4, 11) of 5 μM nickel and an atmosphere of 84% nitrogen, 10% hydrogen, 5% carbon dioxide, and 1% oxygen. Whole bacteroids were prepared as previously reported (11, 20) by crushing nodules harvested from soybean plants inoculated with each B. japonicum strain and grown for 5 to 6 weeks as described previously (20, 25).
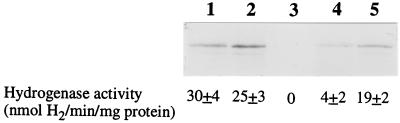
As shown in Table 2, all three of the nitrogen fixation regulatory mutants (fixJ, fixK1, and fixK2) are not affected in hydrogenase activity under free-living, microaerobic conditions. The FixK1 mutant is also Hup+ in symbiosis (Fig. 1, lane 5). However, bacteroids from the FixJ and FixK2 mutants exhibit little or no hydrogenase activity and extracts from these mutant bacteroids contain little or no hydrogenase protein (Fig. 1, lanes 3 and 4) as detected by immunoblotting with antibody to the large subunit of hydrogenase (15). NifA mutants are severely affected in the ability to form an effective symbiosis and in nodule morphology (14). Therefore, a NifA mutant could not be assayed for symbiotic hydrogenase activity. The HoxA mutant JHΔA was assayed for hydrogenase activity as a control and was Hup− in free-living conditions (Table 2) and Hup+ in symbiosis (Fig. 1, lane 2) as expected.
TABLE 2.
Hydrogenase activities of free-living B. japonicum strains
| Strain | Hydrogenase activity (nmol of H2 oxidized/108 cells/min)a |
|---|---|
| JH | 281 ± 14 |
| JHΔA | 0 |
| 9043 (fixK2) | 203 ± 55 |
| 7360 (fixJ) | 215 ± 73 |
| 7454 (fixK1) | 165 ± 71 |
Activities were measured amperometrically in samples of whole cells. Activities are averages ± standard deviations of nine separate experiments.
FIG. 1.
Immunoblotting and hydrogenase activities of wild-type and mutant bacteroid samples. The hydrogenase activity of whole bacteroids was measured amperometrically (18, 34). Activities are the averages ± standard deviations of six separate determinations. Western blots of bacteroid extracts prepared as previously described (11) were probed with antibody against the large subunit of B. japonicum hydrogenase. Lane 1, JH; lane 2, JHΔA; lane 3, 9043; lane 4, 7360; lane 5, 7454.
Since the FixJ and FixK2 mutants are defective in nitrogen fixation, the possibility exists that the Hup− phenotype observed in bacteroids from these strains is due to an indirect effect of a lack of hydrogen (a known requirement for hup transcription) produced by the nitrogenase enzyme rather than the absence of either FixJ or FixK2. To investigate this possibility, bacteroids from a B. japonicum mutant strain harboring a Tn5 insertion in the nitrogenase structural gene nifD were assayed for hydrogenase activity. Bacteroids from this nif mutant are also Hup− (data not shown). However, the data do not rule out a role for FixK2 in the symbiotic regulation of hydrogenase genes. It is possible and, in fact, probable that, as in free-living conditions, symbiotic hydrogenase expression is regulated by multiple signals. Presumably, hydrogen acts as an environmental signal in addition to oxygen and the positive signals for hup transcription are passed on by as-yet-unidentified components to the oxygen-responsive FixLJ-FixK regulatory cascade, which then acts on the hydrogenase genes.
Transcriptional control of the hup promoter by FixK2.
To demonstrate a role for the FixK2 protein in the symbiotic regulation of hydrogenase biosynthesis and to rule out the NifA protein as a symbiotic transcriptional activator, β-galactosidase transcriptional assays (27) were done in the heterologous background of the lac mutant E. coli strain ET8000 (24). Plasmid pSY7, containing the hup promoter fused to a promoterless lacZ gene, was cotransformed into ET8000 with each of the following plasmids: pRJ9044 and pSKA, which constitutively express B. japonicum FixK2 and HoxA, respectively, from the lac promoter of pBluescript SK, and pMC71A, which constitutively expresses K. pneumoniae NifA from the tetracycline resistance gene promoter of pACYC184.
Both the HoxA and FixK2 proteins induce expression from the hydrogenase promoter to levels 6- to 14-fold above the background levels represented by plasmid pSY7 and plasmid pSY7 cotransformed with pBluescript KS (Table 3). Expression of the hup promoter was not activated by the NifA protein. In addition, the B. japonicum hydrogenase promoter was provided on a multicopy plasmid in a K. pneumoniae wild-type strain and the activity of the nitrogenase enzyme was measured by acetylene reduction (19). It has been shown that multiple copies of a NifA binding sequence will reduce nitrogenase activity as measured by acetylene reduction due to the titration of NifA (6, 8). The hup promoter plasmid reduced levels of acetylene reduction to the same extent as a control vector with no insert, while a plasmid containing the B. japonicum nifH promoter region, which is known to bind NifA, eliminated acetylene reduction activity (data not shown). These data indicate that FixK2, and not NifA, has a role in the transcriptional regulation of the B. japonicum hydrogenase structural genes. This is in contrast to the situation in R. leguminosarum, in which NifA has been shown to bind and regulate the hydrogenase promoter (6).
TABLE 3.
Transcriptional activity of the hup promoter measured in a lac mutant E. coli strain (ET8000) carrying various plasmidsa
| Plasmid | β-Galactosidase activityb |
|---|---|
| pSY7 (hup-lacZ) | 11 ± 0 |
| pKS | 25 ± 1 |
| pSY7 + pKS | 26 ± 1 |
| pSY7 + pSKA (HoxA) | 364 ± 11 |
| pSY7 + pRJ9044 (FixK2) | 145 ± 3 |
| pSY7 + pMC71A (NifA) | 5 ± 0 |
The following strains were also assayed and found to have fewer than 6 Miller units of activity: ET8000 and ET8000 carrying plasmid pMC71A, pRJ9044, or pSKA.
Miller units per 108 cells. Values are averages ± standard deviations of six separate determinations.
Localization of a potential FixK2 binding site in the hydrogenase promoter region.
A visual inspection of the hydrogenase promoter region revealed the presence of two potential binding sites for FixK2 (Fig. 2A). Both sites are about 50% identical to the FixK consensus binding sequence (Fig. 2B) (13). The sites are located between 213 and 228 bp and between 32 and 47 bp upstream of the transcription start site. When compared to the placement of FixK binding sites upstream of genes known to be regulated by FixK (13), either site upstream of the hydrogenase structural genes is equally likely to be the actual binding area.
FIG. 2.
Sequence of the upstream region of the hydrogenase structural gene operon and FixK consensus binding sequence. (A) Two potential FixK binding sites extending from −213 to −228 and from −32 to −47 bp upstream of the transcription start site of the hupSL operon (marked with an asterisk) are underlined. (B) Comparison of the hydrogenase promoter region with the FixK consensus binding sequence. Identical residues in each potential binding site are in boldface type.
To determine the region of the hydrogenase promoter necessary for regulation by FixK2, plasmid pRJ9044 was cotransformed into strain ET8000 with various plasmids containing successive truncations of the hydrogenase promoter region fused to a promoterless lacZ gene (Table 1) (21, 22) and β-galactosidase activity was measured (see Table 4). Induction of the hydrogenase promoter by the FixK2 protein is unaffected when the promoter region is truncated to 99 bp upstream of the transcription start site as in plasmid pGR1 but is reduced to just twice background levels when the promoter region is truncated to 44 bp upstream of the hydrogenase structural genes as in plasmid pGHf1 (Table 4). Hydrogenase promoter activity is reduced even further, to background levels, when the upstream region is truncated to 29 bp (i.e., plasmid pGBs3). A deletion of the promoter region between 29 and 64 bp upstream of the transcription start site in plasmid pGNSdB also abolishes induction of the hup promoter by FixK2 (Table 4). In plasmids pGBs3 and pGNSdB, the integration host factor binding site shown to be necessary for full induction of the hydrogenase promoter under free-living conditions (5) is not present. However, in previous studies with pGNSdB, hup promoter activity was only reduced to 50% of the activity observed with full-length promoter constructs (5). No hup promoter activity was measured with either plasmid pGBs3 or pGNSdB in our experiment. Therefore, the reduction in hup promoter activity is due to a lack of binding of a factor (FixK2) other than integration host factor. The partial reduction of hydrogenase promoter activity with a 44-bp upstream region indicates that sequences within the putative FixK binding site closest to the transcription start site are important for activation of hup transcription by FixK2.
TABLE 4.
FixK2 induction of the hydrogenase promoter
| Fusion constructa | β-Galactosidase activityb |
|---|---|
| pSY7 (alone) | 22 ± 3 |
| pSY7 | 311 ± 52 |
| pGHh1 | 304 ± 37 |
| pGHp1 | 213 ± 55 |
| pGR1 | 409 ± 33 |
| pGHf1 | 50 ± 9 |
| pGBs3 | 30 ± 5 |
| pGNSdB | 28 ± 6 |
The exact end points of each fusion construct are described in Table 1. Except where noted, each strain contains the fusion construct indicated and the FixK2 expression plasmid pRJ9044.
Activities are the averages ± standard deviations of nine representative measurements. Units are Miller units per 108 cells.
Taken together, these data show that in contrast to R. leguminosarum, NifA is not a regulator of symbiotic hydrogenase expression in B. japonicum. Instead, the Fnr-like DNA binding protein FixK2 is involved in the symbiotic transcriptional activation of the hydrogenase genes in this organism. The possibility exists that FixK2 exerts its effect on hydrogenase expression in an indirect fashion, through some as-yet-unidentified component(s). However, the β-galactosidase transcriptional assays with the FixK2 expression plasmid provide strong evidence for the FixK2 protein being at least one point at which the regulation of symbiotic hydrogenase expression and the regulation of nitrogen fixation merge.
Acknowledgments
We thank Hans-Martin Fischer for providing several B. japonicum mutant strains and plasmid pRJ9044. We are grateful to Mike Merrick for his generous gifts of plasmid pMC71A and strain ET8000 and for his helpful suggestions. We also thank Sue Maier for her assistance with plants and Jon Olson for helpful discussions.
This work was supported by Department of Energy grant DEFG02-89-ER14011.
REFERENCES
- 1.Anthamatten D, Hennecke H. The regulatory status of the fixL- and fixJ-like genes in Bradyrhizobium japonicum may be different from that in Rhizobium meliloti. Mol Gen Genet. 1991;225:38–48. doi: 10.1007/BF00282640. [DOI] [PubMed] [Google Scholar]
- 2.Anthamatten D, Scherb B, Hennecke H. Characterization of a FixLJ-regulated Bradyrhizobium japonicum gene sharing similarity with the Escherichia coli fnr and Rhizobium meliloti fixK genes. J Bacteriol. 1992;174:2111–2120. doi: 10.1128/jb.174.7.2111-2120.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop P E, Guevarra J G, Engelke J S, Evans H J. Relation between glutamine synthetase and nitrogenase activities in the symbiotic association between Rhizobium japonicum and Glycine max. Plant Physiol. 1976;57:542–546. doi: 10.1104/pp.57.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black L K, Fu C, Maier R J. Sequences and characterization of hupU and hupV genes of Bradyrhizobium japonicum encoding a possible nickel-sensing complex involved in hydrogenase expression. J Bacteriol. 1994;176:7102–7106. doi: 10.1128/jb.176.22.7102-7106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black L K, Maier R J. IHF- and RpoN-dependent regulation of hydrogenase expression in Bradyrhizobium japonicum. Mol Microbiol. 1995;16:405–413. doi: 10.1111/j.1365-2958.1995.tb02406.x. [DOI] [PubMed] [Google Scholar]
- 6.Brito B, Martinez M, Fernandez D, Rey L, Cabrera E, Palacios J M, Imperial J, Ruiz-Argüeso T. Hydrogenase genes from Rhizobium leguminosarum bv. viciae are controlled by the nitrogen fixation regulatory protein NifA. Proc Natl Acad Sci USA. 1997;94:6019–6024. doi: 10.1073/pnas.94.12.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchanan-Wollaston V, Cannon M C, Beynon J L, Cannon F C. Role of the nifA gene product in the regulation of nif expression in Klebsiella pneumoniae. Nature. 1981;294:776–778. doi: 10.1038/294776a0. [DOI] [PubMed] [Google Scholar]
- 8.Buck M, Cannon W, Woodcock J. Mutational analysis of upstream sequences required for transcriptional activation of the Klebsiellae pneumoniae nifH promoter. Nucleic Acids Res. 1987;15:9945–9956. doi: 10.1093/nar/15.23.9945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ditta G, Schmidhauser T, Yakobson P, Liang X-W, Finlay D R, Guiney D, Helinski D R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and monitoring gene expression. Plasmid. 1985;13:149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- 11.Durmowicz M C, Maier R J. Roles of HoxX and HoxA in biosynthesis of hydrogenase in Bradyrhizobium japonicum. J Bacteriol. 1997;179:3676–3682. doi: 10.1128/jb.179.11.3676-3682.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberz G, Friedrich B. Three trans-acting regulatory functions control hydrogenase synthesis in Alcaligenes eutrophus. J Bacteriol. 1991;173:1845–1854. doi: 10.1128/jb.173.6.1845-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer H-M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–386. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Fischer, H.-M. Unpublished data.
- 14.Fischer H-M, Alvarez-Morales A, Hennecke H. The pleiotropic nature of symbiotic regulatory mutants: Bradyrhizobium japonicum nifA gene is involved in control of nif gene expression and formation of determinate symbiosis. EMBO J. 1986;5:1165–1173. doi: 10.1002/j.1460-2075.1986.tb04342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu C, Maier R J. Nickel-dependent reconstitution of hydrogenase apoprotein in Bradyrhizobium japonicum Hupc mutants and direct evidence for a nickel metabolism locus involved in nickel incorporation into the enzyme. Arch Microbiol. 1992;157:493–498. doi: 10.1007/BF00276768. [DOI] [PubMed] [Google Scholar]
- 16.Graham L A, Stults L W, Maier R J. Nitrogenase-hydrogenase relationships in Rhizobium japonicum. Arch Microbiol. 1984;140:243–246. [Google Scholar]
- 17.Gross R, Arico B, Rappouli R. Families of bacterial signal transducing proteins. Mol Microbiol. 1989;3:1661–1667. doi: 10.1111/j.1365-2958.1989.tb00152.x. [DOI] [PubMed] [Google Scholar]
- 18.Hanus F J, Carter K R, Evans H J. Techniques for measurement of hydrogen evolution by nodules. Methods Enzymol. 1980;69:731–739. [Google Scholar]
- 19.Imperial J, Ugalde R A, Shah V K, Brill W J. Role of the nifQ gene product in the incorporation of molybdenum into nitrogenase in Klebsiella pneumoniae. J Bacteriol. 1984;158:187–194. doi: 10.1128/jb.158.1.187-194.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keefe R G, Maier R J. Purification and characterization of an O2 utilizing cytochrome c oxidase complex from Bradyrhizobium japonicum bacteroid membranes. Biochim Biophys Acta. 1993;1183:91–104. doi: 10.1016/0005-2728(93)90008-4. [DOI] [PubMed] [Google Scholar]
- 21.Kim H, Maier R J. Transcriptional regulation of hydrogenase synthesis by nickel in Bradyrhizobium japonicum. J Biol Chem. 1990;265:18729–18732. [PubMed] [Google Scholar]
- 22.Kim H, Yu C, Maier R J. Common cis-acting region responsible for transcriptional regulation of Bradyrhizobium japonicum hydrogenase by nickel, oxygen, and hydrogen. J Bacteriol. 1991;173:3993–3999. doi: 10.1128/jb.173.13.3993-3999.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambert G R, Cantrell M A, Hanus F J, Russell S A, Haugland R A, Haddad K R, Evans H J. Intra- and interspecies transfer and expression of Rhizobium japonicum hydrogen uptake genes and autotrophic growth capability. Proc Natl Acad Sci USA. 1985;82:3232–3236. doi: 10.1073/pnas.82.10.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacNeil D. General method, using Mu-Mud1 dilysogens, to determine the direction of transcription of and generate deletions in the glnA region of Escherichia coli. J Bacteriol. 1981;146:260–268. doi: 10.1128/jb.146.1.260-268.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier R J, Merberg D M. Rhizobium japonicum mutants that are hypersensitive to repression of hydrogen uptake by oxygen. J Bacteriol. 1982;150:161–167. doi: 10.1128/jb.150.1.161-167.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merberg D, O’Hara E B, Maier R J. Regulation of hydrogenase in Rhizobium japonicum: analysis of mutants altered in regulation by carbon substrates and oxygen. J Bacteriol. 1983;156:1236–1242. doi: 10.1128/jb.156.3.1236-1242.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 28.Palacios J M, Murillo J, Leyva A, Ruiz-Argüeso T. Differential expression of hydrogen uptake (hup) genes in vegetative and symbiotic cells of Rhizobium leguminosarum. Mol Gen Genet. 1990;221:363–370. doi: 10.1007/BF00259401. [DOI] [PubMed] [Google Scholar]
- 29.Regensburger B, Hennecke H. RNA polymerase from Rhizobium japonicum. Arch Microbiol. 1983;135:103–109. doi: 10.1007/BF00408017. [DOI] [PubMed] [Google Scholar]
- 30.Richaud P, Colbeau A, Toussaint B, Vignais P M. Identification and sequence analysis of the hupR1 gene, which encodes a response regulator of the NtrC family required for hydrogenase expression in Rhodobacter capsulatus. J Bacteriol. 1991;173:5928–5932. doi: 10.1128/jb.173.18.5928-5932.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoker K, Reijnders W N M, Oltman L F, Stouthamer A H. Initial cloning and sequencing of hydHG, an operon homologous to ntrBC and regulating the labile hydrognase activity in Escherichia coli K-12. J Bacteriol. 1989;171:4448–4456. doi: 10.1128/jb.171.8.4448-4456.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Soom C, Verreth C, Sampaio M J, Vanderleyden J. Identification of a potential transcriptional regulator of hydrogenase activity in free-living Bradyrhizobium japonicum strains. Mol Gen Genet. 1993;239:235–240. doi: 10.1007/BF00281623. [DOI] [PubMed] [Google Scholar]
- 33.Van Soom C, de Wilde C, Vanderleyden J. HoxA is a transcriptional regulator for expression of the hup structural genes in free-living Bradyrhizobium japonicum. Mol Microbiol. 1997;23:967–977. doi: 10.1046/j.1365-2958.1997.2781648.x. [DOI] [PubMed] [Google Scholar]
- 34.Wang R. Amperometric hydrogen electrode. Methods Enzymol. 1980;69:409–412. [Google Scholar]