Abstract
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) nucleases system (CRISPR/Cas9) is a popular gene-editing technology with an expanding scope in the field of medicine. Recent studies have investigated the role of CRISPR/Cas9 system in the treatment of neurodegenerative diseases such as Alzheimer’s disease (AD) and Parkinson’s disease (PD). Since the risk of occurrence of both conditions is strongly associated with genetic mutations and variations, the use of gene-editing technologies to rectify these genetic errors becomes relevant. The CRISPR/Cas9 system has been tested in AD, which has led to a decrease in either amyloid beta deposition or tau phosphorylation in cells. Likewise, genetic mutations in cells affected by PD have been corrected with promising results in initial studies undertaken. Therefore, the use of the CRISPR/Cas9 system should be expanded among different populations to understand its efficacy and safety in depth among neurodegenerative conditions.
Keywords: Alzheimer’s disease, CRISPR/Cas9, gene editing, neurodegenerative disorders, Parkinson’s disease
Introduction
Highlights
The risk of occurrence of neurodegenerative conditions like Alzheimer’s disease and Parkinson’s disease is strongly associated with genetic mutations.
The CRISPR/Cas9 system is a gene-editing technology with a great potential to rectify the underlying genetic errors.
Initial studies on disease-affected cells treated by rectifying the identified genetic mutations have shown promising results.
Neurodegenerative diseases such as Alzheimer’s disease (AD) and Parkinson’s disease (PD) are important causes of morbidity, especially in the elderly population. Both of these are characterized by a common process of neurodegeneration, which stands for a gradual and progressive loss of neurons leading to nervous system dysfunction1,2. AD is the most common neurodegenerative disorder, causing dementia globally and affecting an estimated 24 million people worldwide3. Particularly in developed nations like Australia, AD has a prevalence of 10–30% in people over 65 years, and the incidence doubles every 10 years after 60 years4. Although the most common and strongest risk factor for AD is the genetic factor, other acquired factors such as cerebrovascular diseases, diabetes, hypertension, obesity, and dyslipidemia are associated with an increased risk of the occurrence of AD5. On the other hand, PD is the second most common neurodegenerative disease, with an annual incidence rate of 160 per 100 000 people aged 65 years and older. Use of dairy products, exposure to pesticides and methamphetamine, the presence of neoplasms like melanoma, and a history of traumatic brain injuries are some of the risk factors associated with the risk of PD6.
The mainstay of the management of AD is the use of drugs that target cholinergic or glutamatergic transmission. These drugs can relieve the symptoms and improve cognition. However, there is no curative effect whatsoever7. Likewise, PD is currently being managed by the use of drugs with different mechanisms, such as supplementation of dopamine precursors, dopamine agonism, inhibition of metabolizing enzymes, and increased anticholinergic effects. Just like AD, a definite curative treatment is not yet available8.
Newer advancements in the management of neurodegenerative disorders are focused on tackling a disease at its molecular level, usually targeting a faulty gene or protein to repair, replace, or remove it from the affected cells9. Gene editing is the intentional modification of genomic DNA by insertion, deletion, and replacement at a target site of DNA. This novel technique could be employed in the inactivation of target genes, the acquisition of new genetic traits, and the correction of gene mutations. This process of editing genes has been achieved successfully by an inexpensive and precise system known as the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) nucleases system10–12.
After the discovery of CRISPR/Cas9 technology by Ishino et al. in 1987, CRISPR/Cas9 has been identified as a breakthrough method for genome editing with great promise for treating disorders with few or no therapy choices. The CRISPR/Cas9 system is an essential component of a bacterium’s defense mechanism, providing defense against the unwanted incorporation of mobile genetic components like viruses and plasmids13. Recent comprehensive studies on CRISPR/Cas9 have greatly increased editing efficiency and reduced off-target effects while being widely employed for fundamental and translational research. The CRISPR/Cas9 system consists of two primary constituents, namely the Cas9 enzyme and the single-guide RNA (sgRNA). The sgRNA identifies the target DNA sequence, considering many criteria throughout the design phase to enhance specificity. On the other hand, the Cas9 protein functions as an endonuclease, facilitating the cleavage of DNA double strands through its molecular scissor-like activity14–16.
Genetic factors are the most essential risk factors for irreversible neurodegenerative diseases like AD and PD. Genetic mutations are responsible for around 1% of familial cases of AD. Consequently, the use of genome editing using CRISPR/Cas9 holds potential for addressing familial Alzheimer’s disease (FAD) on a broader scale while offering limited or insignificant advantages for sporadic Alzheimer’s disease (SAD)14. Likewise, PD also has two forms: sporadic Parkinson’s disease (SPD) and familial Parkinson’s disease (FPD). Both of these forms have a genetic basis for disease, apart from other environmental exposures and lifestyle factors17. A wealth of newer studies has reported inspiring results from the CRISPR/Cas9 system in the management of AD and PD. The aim of the review is to comprehensively summarize the application of the CRISPR/Cas9 system in the management of two of the most common neurodegenerative disorders: AD and PD.
Methods
A comprehensive search of electronic databases, including PubMed and Scopus, from inception to January 2023 was conducted. The search terms used included “Alzheimer’s disease”, “Parkinson’s disease”, “Parkinsonism”, “Dementia”, “Neurodegenerative disease”, “Gene therapy”, “Gene editing”, and “CRISPR-Cas9”. The Boolean operators “OR” and “AND” were placed between the search terms to create a search strategy and identify the relevant articles in each database. We also searched the reference lists of relevant articles and reviews for additional studies.
The selection process involved an assessment of the abstracts and the major findings of the studies using the following inclusion criteria: any English-language article published in a peer-reviewed journal that reported on the utility of the CRISPR/Cas9 system in the management of either AD or PD. We excluded studies that used gene-editing techniques other than the CRISPR/Cas9 system, other neurodegenerative disorders such as amyotrophic lateral sclerosis (ALS), Huntington’s disease, etc., and articles with inaccessible full-texts. Duplicate articles were removed using Mendeley Reference Manager. This narrative review study did not require either the ethical approval of the institutional review committee or the informed consent of the participants.
Discussion
Pathophysiology of Alzheimer’s disease and Parkinson’s disease
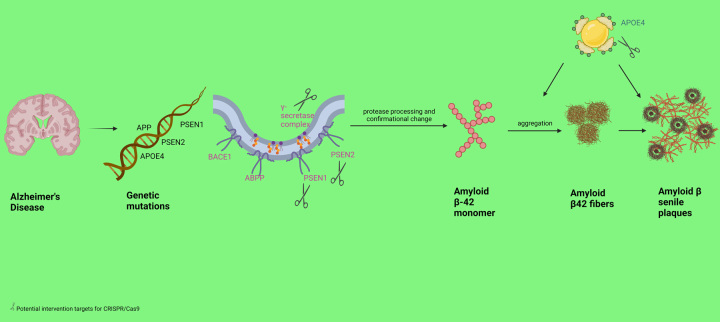
The discussion of the utility of gene-editing systems such as CRISPR/Cas9 becomes comprehendible when the underlying pathogenesis of neurodegenerative conditions is elucidated. One of the most commonly recognized hypothetical mechanisms of AD is amyloid beta (Aβ) plaque deposition in the brain. These plaques are recognized as foreign antigens by the host immune system, initiating a cascade of inflammatory responses via the activation of microglia and the release of cytokines. Continuous inflammation in different regions of the brain over a period of time leads to gradual and progressive cellular death and neurodegeneration. The Aβ plaque consists of Aβ peptides produced from amyloid precursor protein (APP) by the enzymatic breakdown of α, β, and γ secretase enzymes18. The production of plaque begins with the breakdown of APP by β-secretase enzyme, which releases C-terminal membrane amino-acid fragments. The C-terminal membrane-bound fragment gets further broken down by γ-secretase to produce Aβ1-40 and Aβ1-42 isoforms. Aβ1-42 isoform contributes to the formation of the amyloid plaque because of its propensity to easy deposition. The production of Aβ1-42 is the result of change in the splitting pattern, which is believed to be due to mutations in the APP gene, presenilin-1 (PSEN1), presenilin-2 (PSEN2), and apolipoprotein E (APoE4) gene7,19,20. The APP gene mutation results in an upregulation of β-secretase-mediated enzymatic breakdown, consequently resulting in elevated levels of Aβ protein21. Likewise, Shea et al.22 reported that PSEN1 mutations are the reason behind increased production of more Aβ42 compared to Aβ40.
Another hypothesis for the development of AD is the hyperphosphorylation of tau proteins present in neurons. Tau proteins are responsible for stabilizing the microtubule assembly that works to maintain the integrity of the cytoskeleton in neurons. Their activity is regulated by phosphorylating enzymes such as cyclin-dependent kinase-5 (CDK5). Overactivity of the CDK5 enzyme leads to hyperphosphorylation of tau proteins, which results in decreased affinity of the tau proteins for microtubules. The hyperphosphorylated tau gets deposited in the cytosol as neurofibrillary tangles (NFTs) negatively affecting synaptic transmission, axonal transport, and signal transduction7,23. Ultimately, this results in progressive neuronal degeneration of the affected cells.
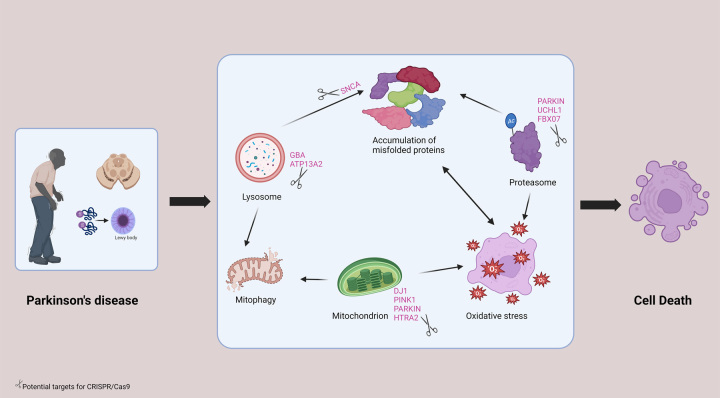
Regarding PD, the underlying ultimate pathology is the degeneration of dopaminergic neurons in the substantia nigra of the basal ganglia. A number of genetic factors that influence the risk of PD have been identified by numerous studies. Mutations in the alpha-synuclein gene (SNCA), UCH-L1, MAPT/STH, PARKIN, and PINK1 genes are widely studied in the risk of developing PD24–27. A missense mutation in SNCA gene known as Ala53Thr (A53T) was reported to have a strong association with PD, according to Spira et al.28. Both PARKIN and PINK1 genes are involved in a process called ‘mitophagy’, by which lysosomes remove dysfunctional mitochondria from the cells. Loss-of-function mutation of these genes lead to compromised mitophagy, resulting in the accumulation of dysfunctional mitochondria in neuronal cells. A continuous state of accumulation of dysfunctional mitochondria is implicated in the preferential degeneration of dopaminergic neurons17. Dominations in the DJ-1 gene, which exerts antioxidant effects through the DJ-1 protein, are also associated with an increased risk of PD29. Likewise, mutations in the LRRK2 gene are associated with autosomal dominant PD30. Another common mutation leading to an increased risk of PD is a mutation in the GBA gene. The mutated GBA gene, which has been associated with Gaucher’s disease, has an approximately four-fold increased risk of occurrence of PD31,32.
Utility of CRISPR/Cas9 in neurodegenerative diseases
Neurodegenerative disorders such as AD, PD, ALS, and Huntington’s disease are widely recognized as significant health concerns affecting a substantial number of individuals on a global scale. These diseases like AD and PD have garnered considerable attention due to their high prevalence in elderly and the considerable impact they have on affected individuals and their families1.
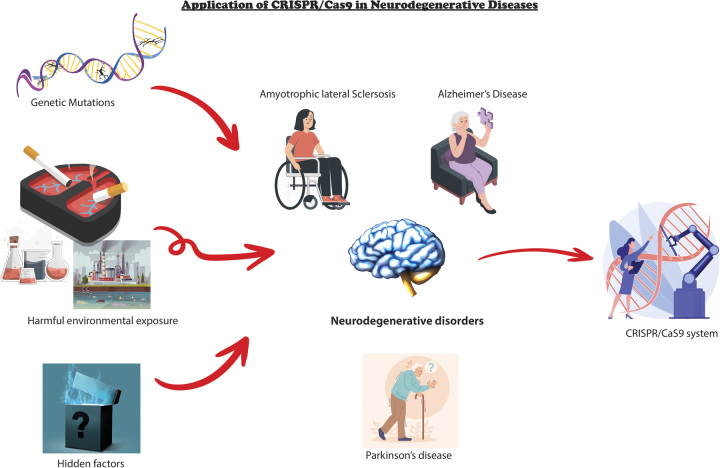
Even though we have identified numerous genetic associations and underlying pathophysiology of AD and PD in familial cases, the molecular mechanisms underlying the pathophysiology of SPD and SAD remain elusive. The majority of cases involving AD and PD manifest in a sporadic manner, as indicated by previous research6,14,22,33. The confirmation of these neurodegenerative disorders can pose challenges due to the reliance on brain autopsy, which is widely regarded as the most established and definitive diagnostic approach. Therefore, it is imperative to acquire a comprehensive understanding of the unique characteristics and presentations of the disease in question to effectively differentiate authentic AD or PD from other associated disorders. This technology is applied to manipulate genes associated with the condition through knockouts, knock-ins, or alterations. Additionally, it allows for the selective activation or repression of crucial genes, as well as the introduction of epigenetic changes. Overall, the emergence of CRISPR/Cas9 technology has presented novel opportunities for understanding and treating neurodegenerative disorders34,35. The capacity to accurately manipulate genetic material holds promise for forthcoming therapeutic interventions that have the potential to greatly enhance patient outcomes (Fig. 1).
Figure 1.
Application of CRISPR/Cas9 in neurodegenerative diseases.
Application of CRISPR/Cas9 in Alzheimer’s disease
The utilization of CRISPR/Cas9 technology has become increasingly prevalent within the realm of AD research. The efficacy of the CRISPR/Cas9 system in rectifying mutations associated with cancer has been well documented. With the same principle in mind, the scope of utility of the CRISPR/Cas9 system has been expanded from the diagnosis to the treatment of AD. Currently, there is widespread utilization of this technology in the creation of disease models, the identification of culprit genes through screening, and the implementation of targeted gene therapy36.
The CRISPR/Cas9 system has been tested to rectify the mutations in the PSEN1 gene, APP gene, and the APoE4 gene. Autosomal dominant mutations in genes such as PSEN1 can be corrected with the use of this technology, according to recent studies. In 2016, the mutations in the PSEN1 gene were rectified by using induced pluripotent stem cells derived from AD patients by two studies37,38. Furthermore, another study provided evidence that the application of the CRISPR/Cas9 system for the purpose of disrupting PSEN1 genes in N2a cells resulted in the elimination of the inherent γ-secretase background39. A recent study utilized the CRISPR/Cas9 technique to specifically interfere with the PSEN1M146L allele. This intervention may also have the ability to partially restore the imbalanced Aβ42/40 ratio40. A similar restoration in the ratio of Aβ42/40 and normal electrophysiology of affected neurons has been described when the PSEN2N141I mutation was corrected with CRISPR/Cas9 by Ortiz-Virumbrales et al.41.
APP mutations have also been selected as targets for elimination using the CRISPR/Cas9 system in a handful of studies. A study demonstrated a decrease in the expression of Aβ protein in fibroblasts derived from AD patients after eliminating Swedish APP (APPswe) mutations21. Furthermore, another study introduced a novel mutation (E674K) via a CRISPR/Cas9-mediated system to alter the APP gene. The alanine codon was modified to threonine in HEK293T cells and SH-SY5Y cells, which harbor the APP gene with deaminated cytosine 1 and cytosine 2 positions. They also demonstrated a decrease in the accumulation of Aβ peptide as a result of the successful introduction of the A673T mutation in 53% of HEK293T cells42.
One of the strongest predictors of sporadic AD development, APoE4 gene, was investigated at first using CRISPR/Cas9 technology by Lin et al. in 201843. The study revealed that the impact of APoE4 on Aβ metabolism varied across different cell types. Moreover, another study explored potential therapeutic targets associated with APoE4. The researchers employed the CRISPR/Cas9 technique to rectify the E4 allele to the E3/E3 genotype in induced pluripotent stem cells of AD patients. Following the research, it was observed that E3 neurons exhibited greater resistance to ionomycin-induced cytotoxicity and displayed a decrease in tau phosphorylation in comparison to E4 neurons44. The application of CRISPR/Cas9 in rectifying potential targets of genetic mutations of AD is illustrated in Figure 2. The details of the interventional studies that employed targeted gene therapy to achieve favorable outcomes are provided in the Table 1.
Figure 2.
Application of CRISPR/Cas9 in Alzheimer’s disease.
Table 1.
CRISPR/Cas9 interventional studies on Alzheimer’s disease
| Study | Targeted gene | Outcomes |
|---|---|---|
| Pires et al.37 | PSEN1 | Production of a gene-corrected induced pluripotent stem cell line by substituting the point mutation with the wild-type sequence |
| Poon et al.38 | PSEN1 | Production of a gene-corrected induced pluripotent stem cell line by correcting for the single base pair mutation |
| Sun et al.39 | PSEN1 | Elimination of inherent γ-secretase background by disrupting PSEN1 genes in N2a cells |
| Konstantinidis et al.40 | PSEN1 | Reduction of extracellular Aβ42/40 ratios by disrupting the PSEN1 allele in human fibroblasts |
| Ortiz-Virumbrales et al.41 | PSEN1 | Correction of the electrophysiological deficit by rectifying the PSEN2 point mutation in basal forebrain cholinergic neurons generated from induced pluripotent stem cells |
| György et al.21 | APP | Reduction of Aβ levels in fibroblasts after eliminating Swedish APP mutations |
| Guyon et al.42 | APP | Decrease in the accumulation of Aβ peptide by introduction of the A673T mutation in HEK293T cells |
| Wadhwani et al.44 | APOE4 | Production of stem-cell-derived E3 neurons that are less susceptible to ionomycin-induced cytotoxicity by correcting E4 allele to the E3/E3 genotype |
Application of CRISPR/Cas9 in Parkinson’s disease
The evidence of use of CRISPR/Cas9 gene-editing technology in the potential management of PD is limited in the literature. The details of the interventional studies that employed targeted gene therapy to achieve favorable outcomes are provided in Table 2. The studies published so far in this area have focused more on uncovering the different associated mutations and their hypothesized pathways. For instance, the CRISPR/Cas system was used to eliminate PARKIN (PRKN), DJ-1, and ATP13A2 (PARK9) genes from dopaminergic neurons in a study49. Likewise, recent studies have identified loss-of-function mutations in the DNAJC6 gene, which encodes the HSP40 auxilin protein. A study uncovered that disruptions in DNAJC6-mediated endocytosis can impede the WNT-LMX1A signal pathway in the process of midbrain dopamine (mDA) neuron development. Consequently, production of mDA neurons that are susceptible to vulnerability is created due to diminished expression of LMX1A during the developmental process50.
Table 2.
CRISPR/Cas9 interventional studies on Parkinson’s disease
| Study | Targeted gene or protein | Outcome |
|---|---|---|
| Zhou et al.45 | PINK1, PARK2 | Generation of single or double gene-targeted porcine fetal fibroblasts |
| Kantor et al.46 | SNCA | Reduction in SNCA levels of human-induced pluripotent stem cell (hiPSC)-derived dopaminergic neurons by targeted DNA methylation editing |
| Yoon et al.47 | A53T-SNCA | Reduction of the overexpression of α-synuclein, reactive microgliosis, dopaminergic neurodegeneration, and parkinsonian motor symptoms by gene deletion of A53T-SNCA significantly |
| Inoue et al.48 | p13 protein | Prevention of toxin-induced motor deficits and the loss of dopaminergic neurons in the substantia nigra of heterozygous p13 knockout mices |
As early as 2015, a study conducted an examination of the PARK2 and PINK1 genes using CRISPR/Cas9 and somatic cell nuclear transfer methodologies within a model of the domestic pig. The same study reported a success rate of ~38% in obtaining homozygous cell colonies with a double-knockout for the PARK2 and PINK1 genes45. An intriguing development of an innovative lentiviral vector that integrates CRISPR/Cas9 technology to selectively suppress the expression of SNCA mRNA and protein has been described in the literature. This targeted intervention resulted in the restoration of phenotypic abnormalities associated with PD46. A recent study demonstrated that the utilization of the CRISPR/Cas9 system for the purpose of eliminating the A53T-SNCA gene mutation resulted in notable improvements in various aspects associated with PD. These improvements included the reduction of α-synuclein overproduction, prevention of dopaminergic neurodegeneration, mitigation of reactive microgliosis, and alleviation of motor symptoms47. Furthermore, Chen et al. used human-induced pluripotent stem cells obtained from individuals with PD to study A53T and SNCA-triplication autosomal dominant mutations. The research findings presented in the study indicate that the lack of SNCA is associated with a reduced susceptibility to Lewy pathology51,52.
Inoue and colleagues provided promising evidence that the expression of a novel 13-kDa protein (p13), which is involved in inducing mitochondrial dysfunction and apoptosis in dopaminergic neurons, could be a potential target gene-editing system. Employing the CRISPR/Cas9 system, they also produced p13-deficient mice with no motor dysfunction or dopaminergic neuron destruction following treatment with neurotoxin that can lead to mitochondrial dysfunction48. This suggests that the CRISPR/Cas9 system could be explored as a potential therapeutic approach for PD. The application of CRISPR/Cas9 in rectifying potential targets of genetic mutations of PD is illustrated in Figure 3.
Figure 3.
Application of CRISPR/Cas9 in Parkinson’s disease.
Conclusion
The utilization of CRISPR/Cas9 gene-editing technology in the diagnosis and treatment of neurodegenerative disorders with a genetic pathophysiological basis is a relatively new concept. Recent studies have shown promising results in effectively targeting the known mutations of certain genes associated with AD and PD. The ability to precisely edit genes associated with these diseases offers a promising approach to understanding disease mechanisms and developing potential therapies. Future research should focus on optimizing the delivery and specificity of CRISPR/Cas9, minimizing off-target effects, and conducting rigorous preclinical and clinical trials. With these advancements, CRISPR/Cas9 gene editing could potentially revolutionize the therapeutic landscape of neurodegenerative diseases.
Ethical approval
No ethical approval was obtained for this review.
Consent
Informed consent was not obtained for this review.
Sources of funding
The authors received no funding to complete the research work.
Author contribution
Conceptualization: N.T.; methodology: N.T., M.A.F.E., N.R., and T.K.; validation: N.T. and H.S.B.; formal analysis and investigation: N.T., M.A.F.E., N.R., and T.K.; resources: N.T. and D.S.; data curation: N.T., M.A.F.E., N.R., and T.K.; writing – original draft preparation: N.T., M.A.F.E., N.R., T.K., and D.S.; writing – review and editing: N.T. and D.S.; visualization: N.T. and T.K.; supervision: T.K.; project administration: N.T. and D.S. All authors have read and agreed to the published version of the manuscript.
Conflicts of interest disclosure
The authors have no conflicts of interest to declare.
Research registration unique identifying number (UIN)
Not applicable.
Guarantor
Dr Nandita Thapar.
Data availability statement
Not applicable.
Provenance and peer review
Not commissioned, externally peer-reviewed.
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Published online 16 November 2023
Contributor Information
Nandita Thapar, Email: thapnandita92@gmail.com.
Mosab Ahmad Fathi Eid, Email: s11844942@stu.najah.edu.
Nishchita Raj, Email: drnishchita100@gmail.com.
Theodosios Kantas, Email: theodosiskantas96@gmail.com.
Harbir S. Billing, Email: forlifegooner@gmail.com.
Dhavalkumar Sadhu, Email: dhavalsadhu98@gmail.com.
References
- 1.Erkkinen MG, Kim MO, Geschwind MD. Clinical neurology and epidemiology of the major neurodegenerative diseases. Cold Spring Harb Perspect Biol 2018;10:a033118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chi H, Chang HY, Sang TK. Neuronal cell death mechanisms in major neurodegenerative diseases. Int J Mol Sci 2018;19:3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayeux R, Stern Y. Epidemiology of Alzheimer disease. Cold Spring Harb Perspect Med 2012;2:a006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masters CL, Bateman R, Blennow K, et al. Alzheimer’s disease. Nat Rev Dis Primers 2015;1:15056. [DOI] [PubMed] [Google Scholar]
- 5.Silva MVF, Loures CMG, Alves LCV, et al. Alzheimer’s disease: risk factors and potentially protective measures. J Biomed Sci 2019;26:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ascherio A, Schwarzschild MA. The epidemiology of Parkinson’s disease: risk factors and prevention. Lancet Neurol 2016;15:1257–1272. [DOI] [PubMed] [Google Scholar]
- 7.Khan S, Barve KH, Kumar MS. Recent advancements in pathogenesis, diagnostics and treatment of Alzheimer’s disease. Curr Neuropharmacol 2020;18:1106–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Church FC. Treatment options for motor and non-motor symptoms of Parkinson’s disease. Biomolecules 2021;11:612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta D, Bhattacharjee O, Mandal D, et al. CRISPR-Cas9 system: a new-fangled dawn in gene editing. Life Sci 2019;232:116636. [DOI] [PubMed] [Google Scholar]
- 10.Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science 2018;361:866–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y, Li Z. CRISPR-Cas systems: overview, innovations and applications in human disease research and gene therapy. Comput Struct Biotechnol J 2020;18:2401–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westra ER, Dowling AJ, Broniewski JM, et al. Evolution and ecology of CRISPR. Annu Rev Ecol Evol Syst 2016;47:307–331. [Google Scholar]
- 13.Ishino Y, Shinagawa H, Makino K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 1987;169:5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhardwaj S, Kesari KK, Rachamalla M, et al. CRISPR/Cas9 gene editing: new hope for Alzheimer’s disease therapeutics. J Adv Res 2022;40:207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mir A, Edraki A, Lee J, et al. Type II-C CRISPR-Cas9 biology, mechanism, and application. ACS Chem Biol 2018;13:357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li T, Yang Y, Qi H, et al. CRISPR/Cas9 therapeutics: progress and prospects. Signal Transduct Target Ther 2023;8:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon DK, Tanner CM, Brundin P. Parkinson disease epidemiology, pathology, genetics, and pathophysiology. Clin Geriatr Med 2020;36:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barage SH, Sonawane KD. Amyloid cascade hypothesis: pathogenesis and therapeutic strategies in Alzheimer’s disease. Neuropeptides 2015;52:1–18. [DOI] [PubMed] [Google Scholar]
- 19.Grimm M, Hartmann T. Recent understanding of the molecular mechanisms of Alzheimer’s disease. J Addict Res Ther 2012;5:1–27. [Google Scholar]
- 20.Raulin AC, Doss SV, Trottier ZA, et al. ApoE in Alzheimer’s disease: pathophysiology and therapeutic strategies. Mol Neurodegener 2022;17:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.György B, Lööv C, Zaborowski MP, et al. CRISPR/Cas9 mediated disruption of the Swedish APP allele as a therapeutic approach for early-onset Alzheimer’s disease. Mol Ther Nucleic Acids 2018;11:429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shea YF, Chu LW, Chan AO, et al. A systematic review of familial Alzheimer’s disease: differences in presentation of clinical features among three mutated genes and potential ethnic differences. J Formos Med Assoc 2016;115:67–75. [DOI] [PubMed] [Google Scholar]
- 23.Huang F, Wang M, Liu R, et al. CDT2-controlled cell cycle reentry regulates the pathogenesis of Alzheimer’s disease. Alzheimers Dement 2019;15:217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polymeropoulos MH, Lavedan C, Leroy E, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 1997;276:2045–2047. [DOI] [PubMed] [Google Scholar]
- 25.Kitada T, Asakawa S, Hattori N, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 1998;392:605–608. [DOI] [PubMed] [Google Scholar]
- 26.Valente EM, Abou-Sleiman PM, Caputo V, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 2004;304:1158–1160. [DOI] [PubMed] [Google Scholar]
- 27.Nalls MA, Blauwendraat C, Vallerga CL, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol 2019;18:1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spira PJ, Sharpe DM, Halliday G, et al. Clinical and pathological features of a Parkinsonian syndrome in a family with an Ala53Thr alpha-synuclein mutation. Ann Neurol 2001;49:313–319. [PubMed] [Google Scholar]
- 29.Raninga PV, Di Trapani G, Tonissen KF. The multifaceted roles of DJ-1 as an antioxidant. Adv Exp Med Biol 2017;1037:67–87. [DOI] [PubMed] [Google Scholar]
- 30.Alessi DR, Sammler E. LRRK2 kinase in Parkinson’s disease. Science 2018;360:36–37. [DOI] [PubMed] [Google Scholar]
- 31.Clark LN, Nicolai A, Afridi S, et al. Pilot association study of the beta-glucocerebrosidase N370S allele and Parkinson’s disease in subjects of Jewish ethnicity. Mov Disord 2005;20:100–103. [DOI] [PubMed] [Google Scholar]
- 32.Riboldi GM, Di Fonzo AB. GBA, Gaucher disease, and Parkinson’s disease: from genetic to clinic to new therapeutic approaches. Cells 2019;8:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Checkoway H, Lundin J, I, Kelada SN. Neurodegenerative diseases. IARC Sci Publ 2011;163:407–419. [PubMed] [Google Scholar]
- 34.Sun J, Roy S. Gene-based therapies for neurodegenerative diseases. Nat Neurosci 2021;24:297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W, Tu Z, Sun Q, et al. CRISPR/Cas9: implications for modeling and therapy of neurodegenerative diseases. Front Mol Neurosci 2016;9:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu L, Yu X, Cai Y, et al. Application of CRISPR/Cas9 in Alzheimer’s disease. Front Neurosci 2021;15:803894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pires C, Schmid B, Petræus C, et al. Generation of a gene-corrected isogenic control cell line from an Alzheimer’s disease patient iPSC line carrying a A79V mutation in PSEN1. Stem Cell Res 2016;17:285–288. [DOI] [PubMed] [Google Scholar]
- 38.Poon A, Schmid B, Pires C, et al. Generation of a gene-corrected isogenic control hiPSC line derived from a familial Alzheimer’s disease patient carrying a L150P mutation in presenilin 1. Stem Cell Res 2016;17:466–469. [DOI] [PubMed] [Google Scholar]
- 39.Sun L, Zhou R, Yang G, et al. Analysis of 138 pathogenic mutations in presenilin-1 on the in vitro production of Aβ42 and Aβ40 peptides by γ-secretase. Proc Natl Acad Sci USA 2017;114:E476–E485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konstantinidis E, Molisak A, Perrin F, et al. CRISPR-Cas9 treatment partially restores amyloid-β 42/40 in human fibroblasts with the Alzheimer’s disease PSEN 1 M146L mutation. Mol Ther Nucleic Acids 2022;28:450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortiz-Virumbrales M, Moreno CL, Kruglikov I, et al. CRISPR/Cas9-correctable mutation-related molecular and physiological phenotypes in iPSC-derived Alzheimer’s PSEN2 (N141I) neurons. Acta Neuropathol Commun 2017;5:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guyon A, Rousseau J, Bégin FG, et al. Base editing strategy for insertion of the A673T mutation in the APP gene to prevent the development of AD in vitro. Mol Ther Nucleic Acids 2021;24:253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin YT, Seo J, Gao F, et al. APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron 2018;98:1141–1154.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wadhwani AR, Affaneh A, Van Gulden S, et al. Neuronal apolipoprotein E4 increases cell death and phosphorylated tau release in alzheimer disease. Ann Neurol 2019;85:726–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou X, Xin J, Fan N, et al. Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci 2015;72:1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kantor B, Tagliafierro L, Gu J, et al. Downregulation of SNCA expression by targeted editing of DNA methylation: a potential strategy for precision therapy in PD. Mol Ther 2018;26:2638–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon HH, Ye S, Lim S, et al. CRISPR-Cas9 gene editing protects from the A53T-SNCA overexpression-induced pathology of Parkinson’s disease in vivo. CRISPR J 2022;5:95–108. [DOI] [PubMed] [Google Scholar]
- 48.Inoue N, Ogura S, Kasai A, et al. Knockdown of the mitochondria-localized protein p13 protects against experimental parkinsonism. EMBO Rep 2018;19:e44860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahfeldt T, Ordureau A, Bell C, et al. Pathogenic pathways in early-onset autosomal recessive Parkinson’s disease discovered using isogenic human dopaminergic neurons. Stem Cell Rep 2020;14:75–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wulansari N, Darsono WHW, Woo HJ, et al. Neurodevelopmental defects and neurodegenerative phenotypes in human brain organoids carrying Parkinson’s disease-linked DNAJC6 mutations. Sci Adv 2021;7:eabb1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X, Xie C, Tian W, et al. Parkinson’s disease-related Leucine-rich repeat kinase 2 modulates nuclear morphology and genomic stability in striatal projection neurons during aging. Mol Neurodegener 2020;15:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen V, Moncalvo M, Tringali D, et al. The mechanistic role of alpha-synuclein in the nucleus: impaired nuclear function caused by familial Parkinson’s disease SNCA mutations. Hum Mol Genet 2020;29:3107–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.